Abstract
The distribution of serogroups and multilocus sequence types (STs) in collections of disease-associated and carried meningococci from the period 1991 to 2000 in three European countries (the Czech Republic, Greece, and Norway) was investigated. A total of 314 patient isolates and 353 isolates from asymptomatic carriers were characterized. The frequency distributions of serogroups and clone complexes differed among countries and between disease and carrier isolate collections. Highly significant differentiation was seen at each housekeeping locus. A marked positive association of serogroup C with disease was evidenced. The ST-11 complex was strongly positively associated with disease; associations for other clone complexes were weaker. The genetic diversity of the clone complexes differed. A single ST dominated the ST-11 clone complex, while the ST-41/44 complex exhibited greater levels of diversity. These data robustly demonstrated differences in the distribution of meningococcal genotypes in disease and carrier isolates and among countries. Further, they indicated that differences in genotype diversity and pathogenicity exist between meningococcal clone complexes.
Neisseria meningitidis, which causes both epidemic and endemic disease worldwide (8), is a common human commensal that exhibits age-dependent levels of asymptomatic carriage (5, 7), ranging from 5 to 40%, in all human populations examined to date. While carried meningococci may either be acapsulate or express one of 13 serologically distinct capsules (60), only capsulate meningococci, expressing serogroups A, B, and C and to a lesser extent Y and W135, are frequently isolated from cases of invasive disease (48). The capsule is an important virulence factor in meningococcal disease that protects the bacterium from opsonophagocytosis in the bacteremic stages of meningococcal disease (58). Expressed capsular antigens of N. meningitis are readily characterized by serological means (60), and variation in serogroup frequency among disease-associated isolates over time and geographic region is a well-established feature of the epidemiology of meningococcal disease that is yet to be completely explained (48).
Typing methods based on the detection of genetic variation in housekeeping genes, initially multilocus enzyme electrophoresis (MLEE) (13) and more recently multilocus sequence typing (MLST) (41, 59), have identified extensive genetic diversity in meningococcal isolate collections. Intriguingly, despite extensive nucleotide sequence-based evidence for high levels of horizontal genetic exchange in meningococcal populations (21, 28, 30), this diversity is highly structured into groups of related genotypes that persist for decades and during global spread (8). These groups are identified in MLST data sets as groups of related sequence types (STs) known as clone complexes (41), which are thought to correspond to lineages of bacteria that have arisen from a common ancestor. Collections of meningococci recovered from asymptomatic carriers exhibit more genetic diversity than isolate collections derived from invasive disease; the majority of cases of meningococcal disease reported over the second half of the 20th century were caused by a limited number of clone complexes, the so-called hyperinvasive lineages (12). The frequency with which the various clone complexes cause disease varies with space and time, and members of the same clone complex may be associated with different serogroups, although they tend to be uniform in a given outbreak of meningococcal disease (8).
Despite the importance of asymptomatic carriage in the biology of the meningococcus and the idea that changes in the carriage prevalence of meningococcal clone complexes are responsible for changes in disease epidemiology (55), the relationships between carriage and disease are yet to be fully elucidated. Here we examined the distribution of serogroups and genotypes in a collection of 667 meningococci isolated from individuals with meningococcal disease and asymptomatic carriers in the period 1991 to 2000 in the Czech Republic, Greece, and Norway. The results confirmed that the distribution of meningococcal serogroups and clone complexes varied with geographic location and that some meningococcal genotypes were significantly overrepresented in cases of invasive disease compared with their point prevalence in meningococcal carriage. The data also indicated that clone complexes differed in their degree of diversity and in their likelihood of being associated with invasive disease.
(Part of this work was presented at the 7th Meeting of the European Monitoring Group on Meningococci, Lanzarote [Spain], September 2003.)
MATERIALS AND METHODS
Bacterial isolates.
The isolate collection (Table 1) was assembled from preexisting samples available in the Czech Republic, Greece, and Norway, representing central, southern, and northern Europe, respectively. All three countries have national reference laboratories that routinely collect meningococcal isolates obtained from cases of invasive disease and periodically carry out surveys of meningococcal carriage as part of outbreak investigation, surveillance, or research. The study was limited to a period of 10 years, 1991 to 2000 (Table 1). All bacterial isolates were characterized by Gram staining, oxidase reaction, and biochemical tests. They were serogrouped by slide agglutination with commercial antisera against meningococcal capsular polysaccharides (serogroups A, B, C, 29E, W135, X, Y, and Z; Abbot Laboratories) or monoclonal antibodies (serogroups A, B, C, W135, Y; National Institute for Biological Standards and Control, Potters Bar, United Kingdom). The isolates were also serotyped and serosubtyped with monoclonal antibodies (1, 66) (data not shown but available at http://neisseria.org/nm/emgm/eumennet/wp5).
TABLE 1.
Numbers of N. meningitidis isolates by country and year
| Yr | No. of isolates
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Czech Republic
|
Greece
|
Norway
|
Total
|
|||||
| Disease | Carriage | Disease | Carriage | Disease | Carriage | Disease | Carriage | |
| 1991 | 91 | 91 | ||||||
| 1994 | 34 | 79 | 34 | 79 | ||||
| 1995 | 1 | 1 | ||||||
| 1996 | 46 | 33 | 62 | 46 | 95 | |||
| 1999 | 49 | 88 | 69 | 118 | 88 | |||
| 2000 | 42 | 73 | 115 | |||||
| Total | 81 | 112 | 91 | 88 | 142 | 153 | 314 | 353 |
(i) Czech Republic.
A total of 112 isolates obtained from carriage studies undertaken in 1994 and 1996 were included. These comprised a random sample of 79 of 91 isolates obtained from 947 individuals (carriage rate, 10.4%) from three geographic regions during 1994 and a randomly chosen subset of 33 of 83 isolates obtained from 836 individuals (carriage rate, 9.9%) in one region in 1996. The 81 patient isolates were a random sample representing 37% of the disease isolates sent to the Czech National Reference Laboratory in 1994 and 1996.
(ii) Greece.
The 88 isolates from asymptomatic carriage were a random subset of 334 isolates obtained from a survey of 3,167 individuals (carriage rate, 10.6%) sampled mainly in 1999 in four regions of northern Greece (Ioannina, Serres, Florina, and Evros) which had experienced an increase in immigration from neighboring countries (Albania, Bulgaria, and Turkey) (34). The 91 patient isolates represented most of the isolates submitted to the Meningococcal Reference Laboratory in Athens in 1999 (49 isolates) and 2000 (42 isolates).
(iii) Norway.
The 153 carrier isolates originated in two regions: 91 isolates came from 943 randomly chosen individuals (carriage rate, 9.6%) sampled in the municipality of Lørenskog near Oslo in 1991 (11), and a further 62 isolates originated from a study of 523 individuals (carriage rate, 11.9%) in Buvika in Sør-Trøndelag County in 1996, which was initiated in response to an outbreak of ST-32 (ET-5) complex meningococci (4). The 142 patient isolates represented 85% of all cases of invasive meningococcal disease reported in Norway in the period 1999 to 2000.
Isolation of DNA and MLST.
Meningococci were grown overnight on chocolate agar plates at 37°C in a 5% CO2 atmosphere. A sample (∼10 μl) of bacterial growth was suspended in 100 μl of 1 M Tris-EDTA buffer (pH 8.0) and boiled for 10 min to disrupt the cells and inactivate DNases. Particulate matter was removed by sedimentation at 12,000 × g, and the supernatant was used in subsequent analyses. MLST was performed as described previously (28, 41). Each of the seven gene fragments was amplified, sequence extension reactions were performed with BigDye terminator cycle sequencing kits (Applied Biosystems), and products were separated with an ABI 377 or 3700 DNA analyzer in accordance with the manufacturer's instructions. Each sequence was determined at least once on each DNA strand. Members of the ST-11 complex were further sequenced at the fumC locus as described previously to see if they possessed the nucleotide sequence polymorphism characteristic of the ET-15 variant of this complex (63).
Assignment of STs and clone complexes.
Individual MLST gene fragments were assigned allele designations by querying the allelic profiles/ST Neisseria MLST database (http://pubmlst.org/neisseria). Novel alleles were submitted to the database curator for validation and assignment of allele numbers, and the allelic profiles were entered into a study-specific isolate database (http://neisseria.org/nm/emgm/eumennet/wp5). Novel STs were submitted to the Neisseria PubMLST isolate database for assignment. Potential new clone complexes were identified by analysis of any unassigned STs by BURST analysis (18), as implemented in the program START (29), to identify central STs, followed by querying these against the MLST profile database to find profiles that matched at four or more of the seven MLST loci. Matches that contained more than 20 STs, with minimal overlap with the other defined clone complexes, were forwarded to the Neisseria MLST database management committee for consideration.
Analysis of nucleotide sequence variation.
The numbers of shared polymorphisms and fixed differences among data sets were determined by using the program DNAsp (51). The degree of genetic variation between subpopulations was quantified by Wright's statistic FST, the correlation between alleles within a subpopulation relative to alleles within the total population (68, 69). If the population is thoroughly mixed, there is no correlation and FST is zero; if there is subdivision, FST is greater than zero. This parameter was analyzed by analysis of molecular variation (19) implemented in the software package ARLEQUIN, version 2.00 (53). Significance values for FST were calculated by means of a permutation test.
Statistical analyses.
Logistic regression models were used to estimate the strength of association between isolate characteristics (serogroup and clone complex) and whether the isolate originated from a healthy carrier or a clinical case. Isolates in serogroups other than the main disease-associated capsular groups (B, C, W135, and Y) were grouped as other groups (36 isolates) and as nonserogroupable or unspecified group (144 isolates). This combined other-nonserogroupable group was used as a baseline, and serogroups were compared to this and to each other in a single model. Analysis was repeated with the nonserogroupable or unspecified group excluded. The association between clone complex and disease in the data set was assessed by using a separate model for each complex, comparing isolates within the complex to all other isolates. This was undertaken for the ST-11, ST-23, ST-32, ST-35, ST-41/44, ST-162, and ST-269 complexes. All models were adjusted for country effects and year of isolation to allow for differential sampling of cases and carriers in each country and over time. The results presented are from models including a linear effect of time on prediction of source of isolation and without interaction between time and country effects. Interaction between country and organism characteristics was also assessed for serogroup and for those clone complexes showing the strongest association with disease or carriage (ST-11 and ST-23).
RESULTS
Meningococcal serogroups and association with disease.
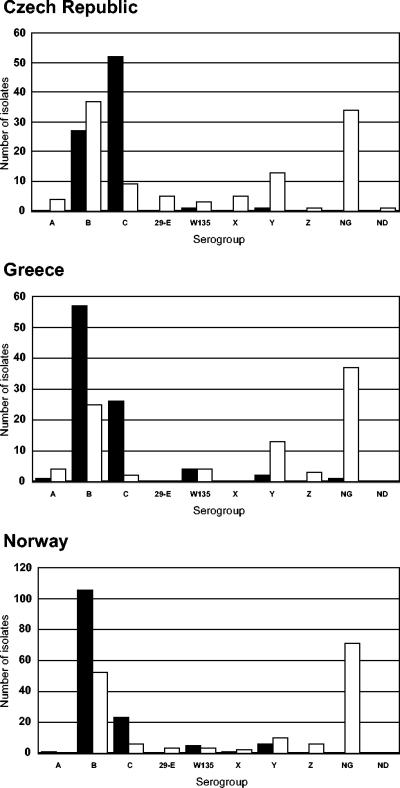
All but one of the 314 meningococci obtained from invasive disease were serogroupable, with 190 serogroup B isolates (60.5%), 101 serogroup C isolates (32.2%), 10 serogroup W135 isolates (3.2%), 9 serogroup Y isolates (2.9%), 2 serogroup A isolates, and 1 serogroup X isolate. A total of 210 of the 353 carrier isolates (59.6%) were serogroupable, with 114 serogroup B isolates (32.3%), 36 serogroup Y isolates (10.2%), 17 serogroup C isolates (4.8%), 10 serogroup W135 isolates (2.8%), 10 serogroup Z isolates (2.8%), 8 serogroup A isolates (2.3%), 8 serogroup 29E isolates (2.3%), and 7 serogroup X isolates (2%). Serogroup distributions were similar between countries in the carrier isolate collections but more variable in the disease isolate collections (Fig. 1), with, for example, serogroup B isolates predominant among disease isolates from Norway and Greece and serogroup C isolates predominant in disease isolates from the Czech Republic.
FIG. 1.
Distribution of serogroups among isolate collections. Disease isolate collections are represented by solid bars, and carriage isolate collections are represented by open bars. NG, not serogroupable; ND, not done.
A strong association between disease and expression of serogroup C capsule was shown; the odds of occurrence of serogroup C meningococci among disease isolates rather than among carrier isolates were 274 times those of serogroupable isolates expressing a serogroup other than B, C, Y, or W135 (Table 2).
TABLE 2.
Association between expressed serogroup and disease, adjusted for country and year
| Serogroup | Odds ratio (95% confidence interval)
|
|
|---|---|---|
| All isolates (n = 667) | Excluding nongroupable isolates (n = 523) | |
| Other (not B, C, Y, or W135) | 1 (baseline) | 1 (baseline) |
| B vs other | 47 (14-156) | 20 (4-98) |
| C vs other | 671 (166-2720) | 274 (48-1572) |
| Y vs other | 5 (1.0-20.2) | 1.9 (0.3-11.8) |
| W135 vs other | 35 (6-198) | 14 (2-113) |
| C vs B | 14 (6-34) | 14 (6-33) |
Genotypic characterization and geographic structuring of genetic diversity.
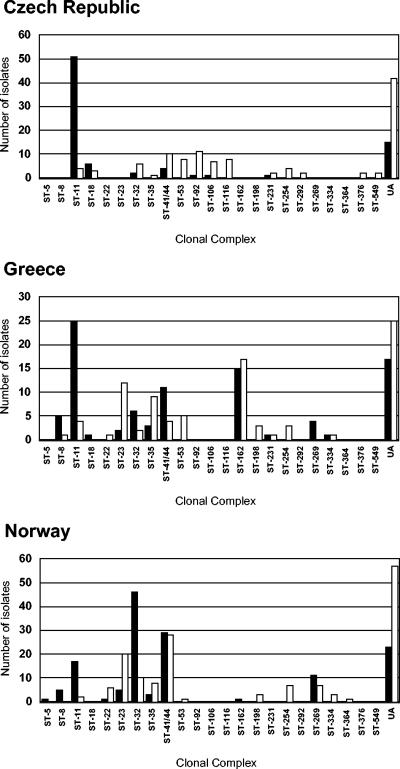
There were 308 STs among the 667 isolates, 488 (73.2%) of which were assigned to 23 clone complexes (Fig. 2). There were 139 STs among the 314 disease-associated isolates, 259 of which (82.5%) were assigned to one of 15 clone complexes. The carrier isolates were more diverse, with 197 STs among the 353 isolates, 229 of which (64.9%) were assigned to one of 22 clone complexes. Only one clone complex (ST-5 complex, one isolate) was present in the disease isolate collections but absent in the carrier isolate collections.
FIG. 2.
Distribution of clone complexes among isolate collections. Disease isolate collections are represented by solid bars, and carriage isolate collections are represented by open bars. Clone complexes are indicated by the number of their definitive ST. UA, unassigned to a clone complex.
The sequence upstream of position 776 of the fumC gene was determined for all patient isolates belonging to ST-11. The presence of A at position 640 in 47 of 49 serogroup C isolates from the Czech Republic, in 7 of the 8 serogroup C isolates from Norway, and in all ST-11 isolates from Greece confirmed that these meningococci represented the ET-15 variant of the ST-11 (ET-37) complex, as identified by multilocus enzyme electrophoresis (63). The remaining ST-11 isolates contained G at position 640.
The clone complexes were not uniformly distributed among the three countries, and the ST-162 complex, the members of which were present mostly in the isolate collections from Greece (Fig. 2), was identified in this study for the first time. The most frequent phenotype of strains of ST-162 was B:4:P1.14, but various other serotypes (including serotypes 1, 14, and 15) were found in association with the ST-162 strains. There were three other potential new clone complexes identified, with the following central genotypes: ST-103 (14 isolates); ST-60 (16 isolates), and ST-178 (5 isolates).
Comparisons of the allele sequences for all loci by analysis of molecular variation demonstrated highly significant country-to-country differentiation for both the disease and carrier isolate collections. The FST values observed ranged from 0.024 (abcZ locus) to 0.071 (aroE locus) for the carrier isolates and from 0.077 (aroE locus) to 0.198 (adk locus) for the disease isolates from all three countries. A similar analysis provided evidence for highly significant differentiation between the disease and carrier isolates within each country at all seven MLST loci (Table 3). Nevertheless, pairwise comparisons of the genetic variation among country-specific disease and carrier isolate collections identified no fixed nucleotide differences and between 385 and 482 shared polymorphic sites (data not shown).
TABLE 3.
Genetic differentiation among isolates collections
| Comparison and isolate collections | Genetic differentiation, FSTa
|
||||||
|---|---|---|---|---|---|---|---|
| abcZ | adk | aroE | fumC | gdh | pdhC | pgm | |
| Among countries, disease isolates | 0.111 | 0.198 | 0.077 | 0.177 | 0.145 | 0.085 | 0.157 |
| Among countries, carrier isolates | 0.024 | 0.042 | 0.071 | 0.027 | 0.043 | 0.041 | 0.022 |
| Between disease and carriage isolates, Czech Republic | 0.077 | 0.150 | 0.077 | 0.214 | 0.074 | 0.170 | 0.217 |
| Between disease and carriage isolates, Greece | 0.017b | 0.030c | 0.045 | 0.050 | 0.077 | 0.053 | 0.034d |
| Between disease and carriage isolates, Norway | 0.049 | 0.057 | 0.036e | 0.054 | 0.082 | 0.050 | 0.055 |
Results are significant (P < 0.0005) unless otherwise stated.
Significant (P = 0.0391).
Significant (P = 0.0029).
Significant (P = 0.0078).
Significant (P = 0.0020).
Variation within clone complexes.
There was strong evidence of the association of certain clone complexes with particular serogroups, although these associations were not absolute. For example, most of the ST-11 complex isolates were serogroup C (83 of 103; 80.1%) with a minority being serogroup B (11 of 103; 10.7%) or W135 (6 of 103; 6.8%); only three members of this clone complex (2.7%) were not serogroupable (Table 4). The ST-23 complex was predominantly serogroup Y (22 of 32; 69%), while most of the remaining clone complexes were predominantly serogroup B. The clone complexes varied in their genotypic diversity. The ST-11, ST-23, and ST-162 complexes were the most conserved, with each containing only one major ST, in each case the central ST of the clone complex. By contrast, both the ST-32 and ST-41/44 complexes were more diverse, containing multiple STs (Table 5).
TABLE 4.
Association of clone complexes with disease and serogroup, adjusted for country and year
| Clone complex | No. of isolates
|
Disease association odds ratio (95% confidence interval) | No. of isolates of serogroup:
|
|||||
|---|---|---|---|---|---|---|---|---|
| Disease | Carriage | B | C | Y | W135 | Other or not serogroupable | ||
| ST-11 | 93 | 10 | 52 (20-135) | 11 | 83 | 6 | 3 | |
| ST-23 | 7 | 32 | 0.2 (0.1-0.7) | 4 | 22 | 1 | 12 | |
| ST-32 | 54 | 18 | 0.9 (0.4-2.2) | 59 | 4 | 2 | 7 | |
| ST-35 | 6 | 18 | 0.3 (0.1-1.1) | 12 | 1 | 11a | ||
| ST-162 | 16 | 17 | 0.8 (0.4-1.18) | 27 | 2 | 4b | ||
| ST-269 | 15 | 7 | 6.1 (0.5-72.8) | 19 | 1 | 2 | ||
| ST-41/44 | 44 | 42 | 1.1 (0.5-2.3) | 67 | 2 | 2 | 15 | |
| ST-254 | 14 | NDc | 2 | 12a | ||||
| ST-53 | 14 | ND | 1 | 13a | ||||
| ST-92 | 1 | 11 | ND | 1 | 1 | 5 | 5 | |
| ST-8 | 10 | 1 | ND | 6 | 5 | |||
| ST-18 | 7 | 3 | ND | 10 | ||||
Including one serogroup A isolate.
Including two serogroup A isolates.
ND, not done.
TABLE 5.
Variation within clone complexes represented by more than 20 isolates
| Clone complex | No. of isolates | No. of STs | ST present more than twice | No. of isolates per ST
|
||
|---|---|---|---|---|---|---|
| Disease | Carriage | Total | ||||
| ST-11 | 103 | 14 | ST-11 | 81 | 8 | 89 |
| ST41/44 | 86 | 27 | ST-41 | 13 | 0 | 13 |
| ST-43 | 1 | 7 | 8 | |||
| ST-44 | 3 | 4 | 7 | |||
| ST-1127 | 7 | 0 | 7 | |||
| ST-1969 | 0 | 3 | 3 | |||
| ST-32 | 72 | 20 | ST-32 | 33 | 8 | 41 |
| ST-230 | 0 | 6 | 6 | |||
| ST-1346 | 4 | 0 | 4 | |||
| ST-1332 | 3 | 0 | 3 | |||
| ST-1357 | 3 | 0 | 3 | |||
| ST-23 | 39 | 7 | ST-23 | 5 | 28 | 33 |
| ST-162 | 33 | 8 | ST-162 | 13 | 13 | 26 |
| ST-35 | 24 | 12 | ST-35 | 3 | 8 | 11 |
| ST-160 | 1 | 1 | 2 | |||
| ST-278 | 0 | 2 | 2 | |||
| ST-269 | 22 | 14 | ST-269 | 5 | 0 | 5 |
| ST-798 | 1 | 2 | 3 | |||
| ST-96 | 0 | 2 | 2 | |||
| ST-275 | 1 | 1 | 2 | |||
Association of clone complexes with disease.
The clone complexes were not uniformly distributed among patient and carrier isolates, with some complexes occurring more frequently in the isolate collections from patients and others occurring more frequently in the carrier isolate collections (Table 4). For seven clone complexes it was possible to calculate odds ratios for association with disease. Members of the ST-11 complex were significantly more likely to be associated with disease, while, conversely, members of the ST-23 complex were significantly more likely to appear in isolate collections from asymptomatic carriers. The directions of associations were maintained following adjustment for serogroup, and in the case of the ST-11 complex, a strong and significant association remained (Table 4). Thus, notwithstanding the substantial overlap of ST-11 with serogroup C, both of these factors were independently positively associated with disease. Given the sample size and adjustments made to account for country and year effects, the confidence intervals were large and for several clone complexes crossed unity; however, the odds ratios and confidence intervals obtained for the other clone complexes were consistent with differences in the likelihood of their association with disease.
There was also evidence of differences in disease association among STs within clone complexes; in particular, ST-41 was significantly more likely to be associated with disease than the other STs in the ST-41/44 complex (Fisher's exact test, P < 0.001).
DISCUSSION
Since 1805, when meningococcal disease was first recognized (61), numerous epidemics, of various durations and intensities, have been described (54). In modern times, the annual reported incidence rates of meningococcal disease fluctuate from less than 1 case per 100,000 to greater than 500 cases per 100,000, changing with time and geographic location (50). Epidemics can last either a few weeks or many years and spread globally (10). They are normally dominated by meningococci of a particular genotype that express capsules of one of the major disease-associated serogroups (9). As N. meningitidis is transmitted primarily by asymptomatically colonized individuals, it is likely that increases in disease incidence are caused by changes in the genotypes present in the commensal flora (25). Comparisons of meningococcal isolate collections have, however, indicated major differences in the serological and genetic compositions of isolates from patients and carriers (12). Typically, carried populations are more diverse, with relatively low representation of the hyperinvasive lineages that dominate the disease-associated isolate collections (30). The data collected in the present study, which span three European countries and 10 years, confirmed these observations and permitted quantitative estimates of the invasive potential of meningococci expressing different serogroups and belonging to different clone complexes.
The disease and carrier isolate collections from all three countries were dominated by serogroup B and C meningococci. Although outbreaks of serogroup W135 and Y disease have occurred in other parts of the world in the period of our study (32, 43, 49), these serogroups were relatively uncommon among the disease isolates. Serogroup Y, however, was the third most common serogroup in carriage isolates in all three countries. Serogroup A meningococci, which have been responsible for a number of large-scale epidemics and pandemics (45) and continue to cause meningococcal disease in Africa (24), are currently rare among disease isolates from Europe and North America (49). This was reflected by their low prevalence in this study among both disease and carrier isolates.
As would be expected from the role of the capsule in the pathogenesis of meningococcal disease (31, 39, 64), all but one of the patient isolates expressed a polysaccharide capsule. The nonserogroupable carrier isolates include meningococci that expressed capsules that were not recognized by the panel of serogrouping reagents employed, along with bacteria that lacked the capsule locus, capsule-null meningococci (14, 16); neither of these would be likely to cause disease. Nonserogroupable isolates would also include meningococci in which serogroup expression has been down-regulated by one or several genetic mechanisms (26, 27, 56); at least some of these isolates would have the potential to cause disease. For this reason, the analysis of the association of disease with serogroup was performed with the whole data set and with the subset of serogroupable isolates. Both analyses showed a highly significant association of serogroup B, C, and W135 capsule expression with disease, with serogroup C estimated to be 14 times more strongly associated with disease than serogroup B. The association was less strong for serogroup Y, although other countries, most notably the United States, have reported an increased incidence of serogroup Y disease in recent years (49). As the carrier and disease isolates were not precisely matched, adjustment for country and year of isolation was made. Although this control was not complete, it is unlikely that the large associations observed were artifacts.
Meningococcal populations are highly diverse, at least partially as a consequence of high rates of horizontal genetic exchange (22, 28, 30). A number of features of our data set were consistent with these observations, including the absence of any fixed nucleotide differences and the high number of shared polymorphisms evident in the pairwise comparisons of the disease and carrier isolate collections from different geographic areas of Europe. Nevertheless, analyses of FST indicated a degree of genetic differentiation among countries, and the null model of random associations of STs with locations was rejected. Similarly, the hypothesis that STs associated with disease are a random sample of carried STs was rejected. This was also manifest in the distribution of clone complexes among the isolate collections.
Meningococci belonging to the ST-11 complex were positively associated with disease, while the ST-23 complex meningococci were negatively associated with disease. A strong relationship remained for ST-11 after adjustment for serogroup. There was some evidence for the association of the ST-269, ST-41/44, and ST-32 complexes with disease, although data sets that are slightly larger and more precisely matched in space and time would be necessary to establish these associations definitively.
Previous investigations have shown the association of particular clone complexes with certain serogroups, although the strength of this relationship varies. The ST-1, ST-4, and ST-5 complexes are strongly associated with the manosamine-based serogroup A capsule (41, 46, 65), but changes among meningococci expressing one of the four sialic acid-based capsules, i.e., serogroups B, C, Y, and W135, are more common (35, 57). These capsule changes require the horizontal genetic transfer of the siaD gene of the capsular operon and have been reported on numerous occasions (42, 57, 62). Increases in the incidence of a serogroup among cases of invasive disease can often be attributed to the spread of a particular clone within a clone complex. For example, in a 20-year period immediately preceding this study (1970 to 1992), disease in the Czech Republic was largely endemic and caused by serogroup B meningococci (37). During the early 1990s there was a rapid increase of serogroup C disease caused by the ST-11 variant characterized by a mutation in the fumC gene (also known as the ET-15 variant of the ET-37 complex) (63). After its first report in Canada (3), this variant has spread globally, causing increases in the incidence of serogroup C disease in many countries, including the Czech Republic in 1993 (36), Greece in 1993 (33), and Norway in 1994 (63). This epidemic triggered a number of major public health interventions worldwide, including mass immunization (6, 15, 52) and the accelerated introduction of the serogroup C conjugate vaccine (44). As seen in all three carrier collections, ST-11 complex meningococci normally occur at a low prevalence among carriage isolates (12, 20, 23, 47); however, during 1993, a year before the collection of most of the Czech isolates used in the present analysis, there were unusually high levels of ST-11 serogroup C carriage in the Czech Republic, possibly due to the recent spread of the variant (30). An unusually high level of carriage has also been reported for ST-11 serogroup W135 bacteria in the Gambia, possibly for the same reason (40).
The central ST dominated each of the more common clone complexes identified. These STs have been isolated on many occasions, frequently spanning decades and exhibiting a global distribution. For example, at the time of this writing, 371 ST-11 meningococci were represented in the Neisseria PubMLST database; they were isolated over a period of 39 years in 29 countries that represented all inhabited continents. The diversity of the clone complexes varied, however; the ST-11 complex was the most conserved, while the ST41/44 complex showed the most diversity. There was evidence that certain STs within the ST-32 and the ST-41/44 complexes were associated with disease, although these complexes as a whole had no positive associations with disease. The difference in apparent pathogenicity between ST-41, which is common among disease-associated meningococci (41), and ST-44, which is more common among carrier strains (30), has led to both STs being used to define this clone complex (http://www.PubMLST.org/neisseria). It is tempting to speculate that these two closely related genotypes exhibit specific, but yet unknown, genetic differences explaining their differences in disease association.
These analyses show that point prevalence in carriage is not a reliable predictor of disease incidence for hyperinvasive meningococci and that this varies among, and indeed within, clone complexes. While it is possible that this reflects differences in intrinsic pathogenicity as a consequence of the presence or absence of particular virulence determinants, it may be a consequence of differences in carriage dynamics. Highly transmissible meningococci with low duration of carriage would be acquired by new hosts more frequently than organisms with lower transmissibility and longer duration of carriage; however, the latter would be present at higher levels in point prevalence studies. As it is thought that acquisition is a risk factor for disease, with most disease onset occurring shortly after acquisition (17, 67), this would result in an overrepresentation in disease and an underrepresentation in carriage of meningococci exhibiting high transmissibility and low carriage duration. Accurate determination of the duration of carriage for clone complexes, such as the ST-11 complex, by means of appropriate longitudinal carriage studies (2) would establish whether transmission dynamics represent a likely explanation for their apparent pathogenicity.
These data confirm that, despite their high rates of horizontal genetic exchange (38), meningococcal populations are highly structured into clone complexes that persist over periods of decades and during global spread, often with a remarkable degree of genetic stability. The carriage and disease populations are genetically highly differentiated among European countries, perhaps as a result of more intense meningococcal transmission within than among countries. However, our study clearly demonstrated that not only the hyperinvasive lineages but also the prevalent carried lineages are capable of wide geographic spread.
Acknowledgments
We thank Jan Oksnes and Anne Marie Klem (Norwegian Institute of Public Health) for technical assistance.
Siamak P. Yazdankhah and Keith Jolley were supported by the EU-MenNet project of European Commission contract QLK2-CZ-2001-01436. The work in the Czech Republic was supported by research projects IGA MH CR no. NI/6882-3 and NJ/7458-3. A grant from the European Community Initiative Interreg II for Public Health in the Balkans through the Greek Ministry of Health supported the work carried out in Greece. Noel D. McCarthy is a Wellcome Trust Clinical Training Fellow, and Daniel J. Wilson is funded by a BBSRC graduate studentship. Martin C. J. Maiden is a Wellcome Trust Senior Research Fellow.
REFERENCES
- 1.Abdillahi, H., and J. T. Poolman. 1988. Typing of group-B Neisseria meningitidis with monoclonal antibodies in the whole-cell ELISA. J. Med. Microbiol. 26:177-180. [PubMed] [Google Scholar]
- 2.Andersen, J., L. Berthelsen, J. B. Bech, and I. Lind. 1998. Dynamics of the meningococcal carrier state and characteristics of the carrier strains: a longitudinal study within three cohorts of military recruits. Epidemiol. Infect. 121:85-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ashton, F. E., J. A. Ryan, A. Borczyk, D. A. Caugant, L. Mancino, and D. Huang. 1991. Emergence of a virulent clone of Neisseria meningitidis serotype 2a that is associated with meningococcal group C disease in Canada. J. Clin. Microbiol. 29:2489-2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bevanger, L., K. Bergh, G. Gisnas, D. A. Caugant, and L. O. Frøholm. 1998. Identification of nasopharyngeal carriage of an outbreak strain of Neisseria meningitidis by pulsed-field gel electrophoresis versus phenotypic methods. J. Med. Microbiol. 47:993-998. [DOI] [PubMed] [Google Scholar]
- 5.Broome, C. V. 1986. The carrier state: Neisseria meningitidis. J. Antimicrob. Chemother. 18(Suppl. A):25-34. [DOI] [PubMed] [Google Scholar]
- 6.Cardenosa, N., A. Dominguez, A. Martinez, J. Alvarez, H. Panella, P. Godoy, S. Minguell, N. Camps, and J. A. Vazquez. 2003. Meningococcal disease in Catalonia 1 year after mass vaccination campaign with meningococcal group C polysaccharide vaccine. Infection 31:392-397. [DOI] [PubMed] [Google Scholar]
- 7.Cartwright, K. A., J. M. Stuart, D. M. Jones, and N. D. Noah. 1987. The Stonehouse survey: nasopharyngeal carriage of meningococci and Neisseria lactamica. Epidemiol. Infect. 99:591-601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caugant, D. A. 1998. Population genetics and molecular epidemiology of Neisseria meningitidis. APMIS 106:505-525. [PubMed] [Google Scholar]
- 9.Caugant, D. A., P. Bol, E. A. Høiby, H. C. Zanen, and L. O. Frøholm. 1990. Clones of serogroup B Neisseria meningitidis causing systemic disease in The Netherlands, 1958-1986. J. Infect. Dis. 162:867-874. [DOI] [PubMed] [Google Scholar]
- 10.Caugant, D. A., L. O. Frøholm, K. Bøvre, E. Holten, C. E. Frasch, L. F. Mocca, W. D. Zollinger, and R. K. Selander. 1986. Intercontinental spread of a genetically distinctive complex of clones of Neisseria meningitidis causing epidemic disease. Proc. Natl. Acad. Sci. USA 83:4927-4931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Caugant, D. A., E. A. Høiby, P. Magnus, O. Scheel, T. Hoel, G. Bjune, E. Wedege, J. Eng, and L. O. Frøholm. 1994. Asymptomatic carriage of Neisseria meningitidis in a randomly sampled population. J. Clin. Microbiol. 32:323-330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Caugant, D. A., B. E. Kristiansen, L. O. Frøholm, K. Bøvre, and R. K. Selander. 1988. Clonal diversity of Neisseria meningitidis from a population of asymptomatic carriers. Infect. Immun. 56:2060-2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Caugant, D. A., L. F. Mocca, C. E. Frasch, L. O. Frøholm, W. D. Zollinger, and R. K. Selander. 1987. Genetic structure of Neisseria meningitidis populations in relation to serogroup, serotype, and outer membrane protein pattern. J. Bacteriol. 169:2781-2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Claus, H., M. C. Maiden, R. Maag, M. Frosch, and U. Vogel. 2002. Many carried meningococci lack the genes required for capsule synthesis and transport. Microbiology 148:1813-1819. [DOI] [PubMed] [Google Scholar]
- 15.De Wals, P., G. De Serres, and T. Niyonsenga. 2001. Effectiveness of a mass immunization campaign against serogroup C meningococcal disease in Quebec. JAMA 285:177-181. [DOI] [PubMed] [Google Scholar]
- 16.Dolan-Livengood, J. M., Y. K. Miller, L. E. Martin, R. Urwin, and D. S. Stephens. 2003. Genetic basis for nongroupable Neisseria meningitidis. J. Infect. Dis. 187:1616-1628. [DOI] [PubMed] [Google Scholar]
- 17.Edwards, E. A., L. F. Devine, G. H. Sengbusch, and H. W. Ward. 1977. Immunological investigations of meningococcal disease. III. Brevity of group C acquisition prior to disease occurrence. Scand. J. Infect. Dis. 9:105-110. [DOI] [PubMed] [Google Scholar]
- 18.Enright, M. C., D. A. Robinson, G. Randle, E. J. Feil, H. Grundmann, and B. G. Spratt. 2002. The evolutionary history of methicillin-resistant Staphylococcus aureus (MRSA). Proc. Natl. Acad. Sci. USA 99:7687-7692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Excoffier, L., P. E. Smouse, and J. M. Quattro. 1992. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131:479-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Feavers, I. M., S. J. Gray, R. Urwin, J. E. Russell, J. A. Bygraves, E. B. Kaczmarski, and M. C. Maiden. 1999. Multilocus sequence typing and antigen gene sequencing in the investigation of a meningococcal disease outbreak. J. Clin. Microbiol. 37:3883-3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feil, E., G. Carpenter, and B. G. Spratt. 1995. Electrophoretic variation in adenylate kinase of Neisseria meningitidis is due to inter- and intraspecies recombination. Proc. Natl. Acad. Sci. USA 92:10535-10539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feil, E. J., M. C. Maiden, M. Achtman, and B. G. Spratt. 1999. The relative contributions of recombination and mutation to the divergence of clones of Neisseria meningitidis. Mol. Biol. Evol. 16:1496-1502. [DOI] [PubMed] [Google Scholar]
- 23.Fernandez, S., L. Arreaza, I. Santiago, A. Malvar, S. Berron, J. A. Vazquez, X. Hervada, and J. J. Gestal. 1999. Carriage of a new epidemic strain of Neisseria meningitidis and its relationship with the incidence of meningococcal disease in Galicia, Spain. Epidemiol. Infect. 123:349-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Greenwood, B. 1999. Meningococcal meningitis in Africa. Trans. R. Soc. Trop. Med. Hyg. 93:341-353. [DOI] [PubMed] [Google Scholar]
- 25.Greiner, O., C. Berger, P. J. Day, G. Meier, C. M. Tang, and D. Nadal. 2002. Rates of detection of Neisseria meningitidis in tonsils differ in relation to local incidence of invasive disease. J. Clin. Microbiol. 40:3917-3921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hammerschmidt, S., R. Hilse, J. P. van Putten, R. Gerardy-Schahn, A. Unkmeir, and M. Frosch. 1996. Modulation of cell surface sialic acid expression in Neisseria meningitidis via a transposable genetic element. EMBO J. 15:192-198. [PMC free article] [PubMed] [Google Scholar]
- 27.Hammerschmidt, S., A. Muller, H. Sillmann, M. Muhlenhoff, R. Borrow, A. Fox, J. vanPutten, W. D. Zollinger, R. Gerardy-Schahn, and M. Frosch. 1996. Capsule phase variation in Neisseria meningitidis serogroup B by slipped-strand mispairing in the polysialyltransferase gene (siaD): correlation with bacterial invasion and the outbreak of meningococcal disease. Mol. Microbiol. 20:1211-1220. [DOI] [PubMed] [Google Scholar]
- 28.Holmes, E. C., R. Urwin, and M. C. Maiden. 1999. The influence of recombination on the population structure and evolution of the human pathogen Neisseria meningitidis. Mol. Biol. Evol. 16:741-749. [DOI] [PubMed] [Google Scholar]
- 29.Jolley, K. A., E. J. Feil, M. S. Chan, and M. C. Maiden. 2001. Sequence type analysis and recombinational tests (START). Bioinformatics 17:1230-1231. [DOI] [PubMed] [Google Scholar]
- 30.Jolley, K. A., J. Kalmusova, E. J. Feil, S. Gupta, M. Musilek, P. Kriz, and M. C. Maiden. 2000. Carried meningococci in the Czech Republic: a diverse recombining population. J. Clin. Microbiol. 38:4492-4498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kahler, C. M., L. E. Martin, G. C. Shih, M. M. Rahman, R. W. Carlson, and D. S. Stephens. 1998. The (alpha2→8)-linked polysialic acid capsule and lipooligosaccharide structure both contribute to the ability of serogroup B Neisseria meningitidis to resist the bactericidal activity of normal human serum. Infect. Immun. 66:5939-5947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kellerman, S. E., K. McCombs, M. Ray, W. Baughman, M. W. Reeves, T. Popovic, N. E. Rosenstein, M. M. Farley, P. Blake, and D. S. Stephens. 2002. Genotype-specific carriage of Neisseria meningitidis in Georgia counties with hyper- and hyposporadic rates of meningococcal disease. J. Infect. Dis. 186:40-48. [DOI] [PubMed] [Google Scholar]
- 33.Kremastinou, J., G. Tzanakaki, A. Kansouzidou, A. Pagalis, V. Danielides, G. Kouppari, E. Lada, P. Kriz, M. Musilek, D. M. Weir, and C. C. Blackwell. 1999. Recent emergence of serogroup C meningococcal disease in Greece. FEMS Immunol. Med. Microbiol. 23:49-55. [DOI] [PubMed] [Google Scholar]
- 34.Kremastinou, J., G. Tzanakaki, S. Levidiotou, F. Markou, E. Themeli, A. Voyiatzi, E. Psoma, M. Theodoridou, and C. C. Blackwell. 2003. Carriage of Neisseria meningitidis and Neisseria lactamica in northern Greece. FEMS Immunol. Med. Microbiol. 39:23-29. [DOI] [PubMed] [Google Scholar]
- 35.Kriz, P., D. Giorgini, M. Musilek, M. Larribe, and M. K. Taha. 1999. Microevolution through DNA exchange among strains of Neisseria meningitidis isolated during an outbreak in the Czech Republic. Res. Microbiol. 150:273-280. [DOI] [PubMed] [Google Scholar]
- 36.Krizova, P., and M. Musilek. 1995. Changing epidemiology of meningococcal invasive disease in the Czech Republic caused by new clone Neisseria meningitidis C:2a:P1.2(P1.5), ET-15/37. Cent. Eur. J. Public Health 3:189-194. [PubMed] [Google Scholar]
- 37.Krizova, P., M. Musilek, and J. Kalmusova. 1997. Development of the epidemiological situation in invasive meningococcal disease in the Czech Republic caused by emerging Neisseria meningitidis clone ET-15/37. Cent. Eur. J. Public Health 5:214-218. [PubMed] [Google Scholar]
- 38.Linz, B., M. Schenker, P. Zhu, and M. Achtman. 2000. Frequent interspecific genetic exchange between commensal neisseriae and Neisseria meningitidis. Mol. Microbiol. 36:1049-1058. [DOI] [PubMed] [Google Scholar]
- 39.Mackinnon, F. G., R. Borrow, A. R. Gorringe, A. J. Fox, D. M. Jones, and A. Robinson. 1993. Demonstration of lipooligosaccharide immunotype and capsule as virulence factors for Neisseria meningitidis using an infant mouse intranasal infection model. Microb. Pathog. 15:359-366. [DOI] [PubMed] [Google Scholar]
- 40.MacLennan, J. M., R. Urwin, S. Obaro, D. Griffiths, B. Greenwood, and M. C. Maiden. 2000. Carriage of serogroup W-135, ET-37 meningococci in The Gambia: implications for immunisation policy? Lancet 356:1078. [DOI] [PubMed] [Google Scholar]
- 41.Maiden, M. C., J. A. Bygraves, E. Feil, G. Morelli, J. E. Russell, R. Urwin, Q. Zhang, J. Zhou, K. Zurth, D. A. Caugant, I. M. Feavers, M. Achtman, and B. G. Spratt. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. USA 95:3140-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maiden, M. C., and B. G. Spratt. 1999. Meningococcal conjugate vaccines: new opportunities and new challenges. Lancet 354:615-616. [DOI] [PubMed] [Google Scholar]
- 43.Mayer, L. W., M. W. Reeves, N. Al Hamdan, C. T. Sacchi, M. K. Taha, G. W. Ajello, S. E. Schmink, C. A. Noble, M. L. Tondella, A. M. Whitney, Y. Al Mazrou, M. Al Jefri, A. Mishkhis, S. Sabban, D. A. Caugant, J. Lingappa, N. E. Rosenstein, and T. Popovic. 2002. Outbreak of W135 meningococcal disease in 2000: not emergence of a new W135 strain but clonal expansion within the electophoretic type-37 complex. J. Infect. Dis. 185:1596-1605. [DOI] [PubMed] [Google Scholar]
- 44.Miller, E., D. Salisbury, and M. Ramsay. 2001. Planning, registration, and implementation of an immunisation campaign against meningococcal serogroup C disease in the UK: a success story. Vaccine 20(Suppl. 1):S58-S67. [DOI] [PubMed] [Google Scholar]
- 45.Moore, P. S., M. W. Reeves, B. Schwartz, B. G. Gellin, and C. V. Broome. 1989. Intercontinental spread of an epidemic group A Neisseria meningitidis strain. Lancet ii:260-263. [DOI] [PubMed] [Google Scholar]
- 46.Olyhoek, T., B. A. Crowe, and M. Achtman. 1987. Clonal population structure of Neisseria meningitidis serogroup A isolated from epidemics and pandemics between 1915 and 1983. Rev. Infect. Dis. 9:665-692. [DOI] [PubMed] [Google Scholar]
- 47.Patrick, D. M., S. Champagne, S. H. Goh, G. Arsenault, E. Thomas, C. Shaw, T. Rahim, F. Taha, M. Bigham, V. Dubenko, D. Skowronski, and R. C. Brunham. 2003. Neisseria meningitidis carriage during an outbreak of serogroup C disease. Clin. Infect. Dis. 37:1183-1188. [DOI] [PubMed] [Google Scholar]
- 48.Peltola, H. 1983. Meningococcal disease: still with us. Rev. Infect. Dis. 5:71-91. [DOI] [PubMed] [Google Scholar]
- 49.Rosenstein, N. E., B. A. Perkins, D. S. Stephens, L. Lefkowitz, M. L. Cartter, R. Danila, P. Cieslak, K. A. Shutt, T. Popovic, A. Schuchat, L. H. Harrison, and A. L. Reingold. 1999. The changing epidemiology of meningococcal disease in the United States, 1992-1996. J. Infect. Dis. 180:1894-1901. [DOI] [PubMed] [Google Scholar]
- 50.Rosenstein, N. E., B. A. Perkins, D. S. Stephens, T. Popovic, and J. M. Hughes. 2001. Meningococcal disease. N. Engl. J. Med. 344:1378-1388. [DOI] [PubMed] [Google Scholar]
- 51.Rozas, J., J. C. Sanchez-DelBarrio, X. Messeguer, and R. Rozas. 2003. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19:2496-2497. [DOI] [PubMed] [Google Scholar]
- 52.Salleras, L., A. Dominguez, and N. Cardenosa. 2003. Impact of mass vaccination with polysaccharide conjugate vaccine against serogroup C meningococcal disease in Spain. Vaccine 21:725-728. [DOI] [PubMed] [Google Scholar]
- 53.Schneider, S., D. Rossli, and L. Excoffier. 2000. Arlequin version 2.00: a software for population genetic data analysis. University of Genoa, Genoa, Italy.
- 54.Schwartz, B., P. S. Moore, and C. V. Broome. 1989. Global epidemiology of meningococcal disease. Clin. Microbiol. Rev. 2:S118-S124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stephens, D. S. 1999. Uncloaking the meningococcus: dynamics of carriage and disease. Lancet 353:941-942. [DOI] [PubMed] [Google Scholar]
- 56.Swartley, J. S., J. H. Ahn, L. J. Liu, C. M. Kahler, and D. S. Stephens. 1996. Expression of sialic acid and polysialic acid in serogroup B Neisseria meningitidis: divergent transcription of biosynthesis and transport operons through a common promoter region. J. Bacteriol. 178:4052-4059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Swartley, J. S., A. A. Marfin, S. Edupuganti, L. J. Liu, P. Cieslak, B. Perkins, J. D. Wenger, and D. S. Stephens. 1997. Capsule switching of Neisseria meningitidis. Proc. Natl. Acad. Sci. USA 94:271-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tzeng, Y. L., and D. S. Stephens. 2000. Epidemiology and pathogenesis of Neisseria meningitidis. Microbes Infect. 2:687-700. [DOI] [PubMed] [Google Scholar]
- 59.Urwin, R., and M. C. Maiden. 2003. Multi-locus sequence typing: a tool for global epidemiology. Trends Microbiol. 11:479-487. [DOI] [PubMed] [Google Scholar]
- 60.Vedros, N. A. 1987. Development of meningococcal serogroups, p. 33-37. In N. A. Vedros (ed.), Evolution of meningococcal disease. CRC Press Inc., Boca, Raton Fla.
- 61.Vieusseux, G. 1805. Mémoire sur la maladie qui a régné à Genève au printemps de 1805. Journal de Medecine, Chirurgie et Pharmacie II:163-165. [Google Scholar]
- 62.Vogel, U., H. Claus, and M. Frosch. 2000. Rapid serogroup switching in Neisseria meningitidis. N. Engl. J. Med. 342:219-220. [DOI] [PubMed] [Google Scholar]
- 63.Vogel, U., H. Claus, M. Frosch, and D. A. Caugant. 2000. Molecular basis for distinction of the ET-15 clone within the ET-37 complex of Neisseria meningitidis. J. Clin. Microbiol. 38:941-942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vogel, U., and M. Frosch. 1999. Mechanisms of neisserial serum resistance. Mol. Microbiol. 32:1133-1139. [DOI] [PubMed] [Google Scholar]
- 65.Wang, J. F., D. A. Caugant, X. Li, X. Hu, J. T. Poolman, B. A. Crowe, and M. Achtman. 1992. Clonal and antigenic analysis of serogroup A Neisseria meningitidis with particular reference to epidemiological features of epidemic meningitis in the People's Republic of China. Infect. Immun. 60:5267-5282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wedege, E., E. A. Høiby, E. Rosenqvist, and L. O. Frøholm. 1990. Serotyping and subtyping of Neisseria meningitidis isolates by co-agglutination, dot-blotting and ELISA. J. Med. Microbiol. 31:195-201. [DOI] [PubMed] [Google Scholar]
- 67.Wenzel, R. P., J. A. Davies, J. R. Mitzel, and W. E. Beam, Jr. 1973. Non-usefulness of meningococcal carriage-rates. Lancet ii:205. [DOI] [PubMed] [Google Scholar]
- 68.Wright, S. 1951. The genetical structure of populations. Ann. Augenics 15:323-354. [DOI] [PubMed] [Google Scholar]
- 69.Wright, S. 1969. Evolution and the genetics of populations, vol. 2, p. 290-344. University of Chicago Press, Chicago, Ill.