Abstract
Humans are exposed to Campylobacter spp. in a range of sources via both food and environmental pathways. For this study, we explored the frequency and distribution of thermophilic Campylobacter spp. in a 10- by 10-km square rural area of Cheshire, United Kingdom. The area contains approximately 70, mainly dairy, farms and is used extensively for outdoor recreational activities. Campylobacter spp. were isolated from a range of environmental samples by use of a systematic sampling grid. Livestock (mainly cattle) and wildlife feces and environmental water and soil samples were cultured, and isolates were presumptively identified by standard techniques. These isolates were further characterized by PCR. Campylobacter jejuni was the most prevalent species in all animal samples, ranging from 11% in samples from nonavian wildlife to 36% in cattle feces, and was isolated from 15% of water samples. Campylobacter coli was commonly found in water (17%) and sheep (21%) samples, but rarely in other samples. Campylobacter lari was recovered from all sample types, with the exception of sheep feces, and was found in moderate numbers in birds (7%) and water (5%). Campylobacter hyointestinalis was only recovered from cattle (7%) and birds (1%). The spatial distribution and determinants of C. jejuni in cattle feces were examined by the use of model-based spatial statistics. The distribution was consistent with very localized within-farm or within-field transmission and showed little evidence of any larger-scale spatial dependence. We concluded that there is a potentially high risk of human exposure to Campylobacter spp., particularly C. jejuni, in the environment of our study area. The prevalence and likely risk posed by C. jejuni-positive cattle feces in the environment diminished as the fecal material aged. After we took into account the age of the fecal material, the absence or presence of rain, and the presence of bird feces, there was evidence of significant variation in the prevalence of C. jejuni-positive cattle feces between grazing fields but no evidence of spatial clustering beyond this resolution. The spatial pattern of C. jejuni is therefore consistent with that for an organism that is ubiquitous in areas contaminated with cattle feces, with a short-scale variation in infection intensity that cannot be explained solely by variations in the age of the fecal material. The observed pattern is not consistent with large-scale transmission attributable to watercourses, wildlife territories, or other geographical features that transcend field and farm boundaries.
Campylobacter infections resulting in gastrointestinal disease are recognized as an emerging problem worldwide. In the United Kingdom, the number of reported cases per annum rose from 32,636 to 56,420 between 1991 and 2001 (35). Recent estimates indicate that, after allowing for underreporting, there are about 500,000 cases every year in England and Wales (14, 41). The majority of cases (≈90%) are attributed to infections with Campylobacter jejuni (19), although Campylobacter coli is increasingly recognized as an important pathogen and is estimated to account for 25,000 cases of infectious intestinal disease, at a cost of four million pounds, by the UK National Health Service (41). Although most infections are self-limiting, some are associated with chronic, debilitating sequelae such as Guillain-Barré syndrome and Miller Fischer syndrome (30). Most infections are believed to result from the ingestion of contaminated food, although the role of other, nonfood exposures in the epidemiology of sporadic campylobacteriosis is still unknown. C. jejuni has been isolated from a range of food sources, including poultry (33), red meat, and milk (12, 18). Point source outbreaks are thought to be relatively uncommon compared to those by other major enteric pathogens, although there is increasing evidence for localized transmission (6). The primary source of contamination is believed to be animal feces. This is consistent with high carriage rates in poultry, pigs, and cattle (23) and with molecular evidence showing similar genotypes in farm animals and humans (9, 13). Contamination of the environment by domestic and wild animal feces (and that of humans) presents an alternative exposure pathway for human infection, for example, via drinking (10, 16, 37) and recreational water use (1). Humans may also be exposed to voided animal feces in the environment through outdoor activities such as camping, walking, and picnicking.
The work described here forms part of a larger study of the epidemiology and genetic diversity of thermophilic Campylobacter spp. isolated by structured sampling of an area of the United Kingdom that is intensely farmed and also widely used for recreation. This paper describes the sampling protocol and the distribution of campylobacters in the environment to the level of the species, with particular emphasis on C. jejuni isolated from cattle feces. We examined the spatial distribution of C. jejuni, C. coli, Campylobacter lari, and Campylobacter hyointestinalis in cattle feces in the environment and tested the null hypothesis that Campylobacter spp. are distributed randomly, with no evidence of spatial clustering. We used model-based spatial statistical methods to describe the nature and extent of spatial and nonspatial clustering and to explore the determinants of the observed pattern of C. jejuni in cattle feces in the environment. In particular, we looked for evidence of spatial effects beyond the within-field effect that could be attributed to larger-scale spatial processes. Further species typing based on restriction fragment length polymorphism analysis (flaA and pulsed-field gel electrophoresis) and multilocus sequence typing will be reported elsewhere (25).
MATERIALS AND METHODS
Study area.
The study area is a 10- by 10-km square region of farmland in Cheshire, United Kingdom. The area has a varying topography (ranging from 9 to 222 m above sea level), various soil types, major roads, two small towns, a canal, a large river, and active and disused railway lines. It was selected because it has several specific characteristics, as follows: it contains approximately 70, mainly dairy, farms of various sizes; a detailed wildlife survey of the entire area was conducted between 1998 and 1999; the site is located within 30 min of the Veterinary Field Station and testing laboratories; and the area is used for recreational activities such as canal barging, angling, walking, and camping. The wildlife survey provided details of field usage and habitat and gained excellent compliance from farmers (only two refused access to farmland).
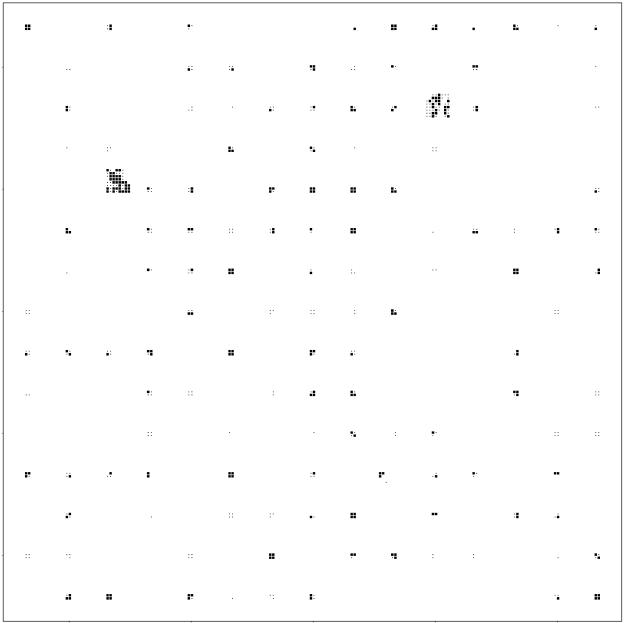
Figure 1a shows, for a subregion of the study area, the locations from which fecal material and other samples were collected. For the most part, the sampling locations were situated in clusters of four points forming the corners of squares with 50-m-long sides. These squares are referred to as “primary squares,” the centers of which form a 15- by 15-km grid and are spaced 667 m apart. Each of the secondary points in the primary squares was in turn surrounded by four tertiary points (not shown) that were each 25 m apart. Soil samples were taken from the tertiary points, and fecal pats were counted and sampled in circles with a 5-m radius centered on the tertiary points. In three areas, a primary square was surrounded by 15 more primary squares, thus creating “fill-in” areas of 400 by 400 m. A small number of secondary points lay in areas which were not suitable for sampling (e.g., areas covered by buildings or roads) or on land for which sampling permission had not been given.
FIG. 1.
Locations of environmental samples for isolation of Campylobacter spp. in a subregion of the study area. (a) Secondary points where feces were sought. (b) Secondary points where cattle feces were present.
The sampling scheme was designed as a compromise between two desirable characteristics, namely, coverage of the whole study region and coverage of a range of spatial scales, within a fixed constraint on the total cost of field sampling and subsequent laboratory analysis. As is usually the case, choosing an appropriate balance between these two considerations was hampered by the lack of advance knowledge of which spatial scales would contribute most variability to the process.
The sampling points were overlaid onto a 1:2,500-scale digitized map of the study area and were identified in the field by use of a combination of geographical features (e.g., field boundaries and buildings), a compass, and a tape measure. All sampling was carried out between 22 May and 26 July 2000. Figure 1b shows the secondary points in the subregion where cattle feces were present. Notice that no feces were collected from many primary squares and that other primary squares contained fewer than four secondary points where cattle feces were present. Only two of the fill-in squares in the study area contained cattle feces.
Sample collection.
Up to two cattle fecal samples were taken from the vicinity of each tertiary point: the nearest sample, defined as the sample nearest to the sampling point, and the freshest sample, defined as the sample which, on visual inspection, was considered to be the most recently voided. The nearest samples provided an indication of a typical pat in the sample region, whereas the freshest samples were expected to be the most reliable indicator of the presence of Campylobacter spp. in the area. When there was only one fecal pat, it was only sampled once and was considered the nearest sample, with the freshest pat being considered missing. Samples from the tertiary points were merged into two pooled samples for each secondary point, with one pool of the nearest samples and another of the freshest samples. Pooling was performed because the number of pats from tertiary points was too large for each one to be tested.
Soil samples were taken from the surface down to a depth of approximately 2 cm. If the sampling point was covered with feces, soil was taken from an uncontaminated location adjacent to the sampling point. Wildlife and sheep feces were identified by scanning the entire area of the primary square and were pooled in the field at the level of the primary square for wildlife samples and the secondary point for sheep samples. Wild mammal feces (mainly from rabbits and badgers) were easily identified by their size, shape, and in the case of badger samples, location in well-marked pits (latrines). Wild bird fecal samples were identified by their color, consistency, and location. Water samples were identified when we scanned the area of the primary square, but the samples were not pooled.
Isolation and species identification.
One milliliter of homogenized pooled fecal sample was added to 9 ml of campylobacter enrichment broth (Lab M, Bury, United Kingdom). The broth was incubated at 42°C for 24 h under microaerophilic conditions in a variable atmosphere incubator, inoculated onto campylobacter blood-free agar (Lab M) containing a CA antibiotic supplement (X112), and incubated as described above for a further 48 h. Three suspect colonies, representing the variability in colony morphology, were subcultured onto Columbia agar (Lab M) supplemented with 5% defibrinated horse blood (Southern Group Labs, Corby, United Kingdom) and incubated as described above for 24 to 48 h. Each isolate was presumptively identified by standard methods (no growth in O, presence of catalase and oxidase, Gram stain, cell size, and morphology) and then frozen in Microbank tubes (Pro-Lab Diagnostics, Neston, United Kingdom) at −70°C.
Presumptive positive isolates were identified as being either C. jejuni, C. coli, C. lari, C. hyointestinalis, or genus-specific Campylobacter by a series of single-reaction PCRs. Isolates were inoculated onto Columbia blood agar and grown microaerophilically for 48 h at 37°C. One colony was arbitrarily selected and suspended in 500 μl of sterile distilled water. The DNA was extracted by boiling the suspension for 20 min. One microliter of the boiled cell suspension was used in each 25-μl reaction volume.
The primers for C. jejuni were JEJ1 (5′ CCTGCTACGGTGAAAGTTTTGC 3′) and JEJ2 (5′ GATCTTTTTGTTTTGTGCTGC 3′), resulting in an amplicon of 793 bp, and the primers for C. coli were COL1 (5′ ATGAAAAAATATTTAGTTTTTGCA 3′) and COL2 (5′ ATTTTATTATTTGTAGCAGCG 3′), resulting in an amplicon of 894 bp (20), and the target for both was the putative virulence marker ceuE. The reaction mixtures for C. jejuni and C. coli contained the following: 0.2 mM (each) dATP, dCTP, dGTP, and dTTP, a 1 μM concentration of each primer, 20 mM (NH4)2SO4, 75 mM Tris-HCl (pH 8.8), 0.01% (wt/vol) Tween 20, 3.5 mM MgCl2, and 0.5 U of Taq DNA polymerase (Abgene). The reaction mixtures were initially held at 94°C for 5 min. The amplification cycle was performed 30 times and consisted of denaturing at 94°C for 30 s, annealing at 57°C for 30 s, and extension at 72°C for 1 min. At the end of cycling, the samples were held at 72°C for 5 min. The primers for C. lari were CL594F (5′ CAAGTCTCTTGTGAAATCCAAC 3′) and CL1155R (5′ATTTAGAGTGCTCACCCGAAG 3′), resulting in an amplicon of 561 bp (26). The primers for C. hyointestinalis were CFCH57F (5′ GCAAGTCGAACGGAGTATTA 3′) and CH1344R (5′ GCGATTCCGGCTTCATGCTC 3′), resulting in an amplicon of 1,287 bp (26). The primers for genus-specific Campylobacter PCRs were C412F (5′ GGATGACACTTTTCGGAGC 3′) and C1288R (5′ CATTGTAGCACGTGTGTC 3′), resulting in an amplicon of 816 bp (26). The reaction conditions for C. lari, C. hyointestinalis, and genus-specific Campylobacter were the same as those for C. jejuni and C. coli except that 2.5 mM MgCl2 was used. The target for genus-specific Campylobacter, C. lari, and C. hyointestinalis PCRs was the 16S ribosomal DNA region of the genome. Samples were initially held at 94°C for 4 min. The amplification consisted of 25 cycles of denaturation at 94°C for 1 min, annealing for 1 min at 64°C for C. lari, 65°C for C. hyointestinalis, and 55°C for genus-specific Campylobacter, and extension at 72°C for 1 min. At the end of cycling, the samples were held at 72°C for 5 min. The amplified DNAs were analyzed by electrophoresis in 1.5% agarose gels run at 120 V for 75 min, and the gels were stained with ethidium bromide.
Data recorded.
The age of each pat was scored by use of a four-point scale as follows: 1, freshly voided with no signs of aging (animal may be observed defecating, no crust); 2, recently voided with some early signs of aging (thin skin-like crust on surface); 3, clear signs of aging but still retaining integrity (thicker rigid crust, mold on surface, obvious insect activity); 4, aged (degenerated, fibrous, and dehydrated, with grass growing into fecal pat). The scoring system was designed and piloted with all observers to ensure consistency of scoring. The degree of consistency was continuously monitored throughout the period of study. Although the scoring is categorical and not continuous, intermediate scores, such as 2.5, were also permitted. A small number of missing values for pat age corresponded to pats for which the age was not recorded.
The weather conditions at the time of sampling were observed as a means to adjust for the potential effect of UV light on the isolation of Campylobacter spp. and the effect of rain on age scoring of cattle feces. Weather conditions were given simple scores (scores for rain, none, light, heavy; scores for sun, cloudy, overcast, sunny). The principal habitat at each secondary point was also known. The number of pats within 5 m of each tertiary point was recorded, and the average of these counts within a secondary point was considered to be a potential influence on the presence of C. jejuni in a pooled sample.
Statistical methods.
As discussed in Results, the spatial structure of each species was examined by the production of semivariograms (7). To construct a variogram, we calculated the spatial separations (Uij = ‖Si − Sj‖) and the squared differences [Vij = (Yi − Yj)2/2] for each pair of observations, Yi and Yj, located at points Si and Sj, respectively. Here Si refers to the location of the secondary point from around which the fecal pats comprising the pooled sample were taken. The collection of squared differences (Vij) and distances (Uij) is the variogram cloud, consisting of a large number of points which must be smoothed or averaged before they yield useful information. Typically, the spatial separations (Uij) are grouped into n equally spaced bins, with the bin width being termed the step size and with the variogram consisting of the bin centers (Uk, where k = 1…n) and the averages (V̂k) of all variogram ordinates (Vij) whose spatial separation (Uij) is in bin k.
Because of the regular distribution of sampling points in this study, the bins were given various sizes, which allowed for the fact that there were many points separated by distances of multiples of 666 m (the distance between primary squares) and very few with a separation of, say, 300 m. The first bin contained all pairs with a spatial separation of zero, i.e., the freshest and nearest samples from each secondary point. The second and third bins corresponded to distances within a primary square, namely, a 50-m separation for horizontal and vertical distances and a 71-m separation for diagonal distances. The seven subsequent bins corresponded to distances within fill-in squares, which were larger than the 50 m within primary squares but smaller than the 666 m between primary squares. The final bins related to pairs in neighboring primary squares and pairs whose primary squares were second neighbors. The variograms computed were omnidirectional variograms, meaning that only the separation distance and not the angle of pairs was taken into account.
In order to obtain a measure of uncertainty for variograms computed from the data, we compared them to variograms obtained from permuted data, for which the spatial locations were randomly permuted 99 times. Permuted data should not exhibit spatial dependence, and a species can be judged to have spatial dependence if its variogram shows more spatial structure than any of the permuted data sets. The analysis was done with the R package geoR (36).
A statistical model for the prevalence of C. jejuni was used to judge the contribution of various factors to the presence of C. jejuni. The relatively small numbers of positive isolates for other Campylobacter species and sample types precluded a similar analysis of these data. In this model, the subscripts i, j, and k are used to denote the ith primary square, the jth secondary point, and the kth tertiary point. We use ℓ to refer to the type of sample, either the freshest or nearest pooled sample. Yijℓ refers to the presence or absence of C. jejuni in the pooled sample made from the pats near the tertiary points ijk of type ℓ surrounding secondary point ij, taking the values 1 for the presence and 0 for the absence of C. jejuni.
In the model, each pooled sample Yijℓ is assumed to test positive for C. jejuni with the probability pijℓ, according to the formula Yijℓ ∼ Bernoulli(pijℓ).
The probability pijℓ varied from site to site and depended on the covariate Xijℓ. The environmental variables listed in Table 3 were considered possible covariates. Two weather-related covariates were the presence or absence of rain and a categorical variable denoting the amount of sun on the day of sampling. Other covariates were the predominant habitat within the square, the number of cattle pats in the sampling area, the presence or absence of bird or rabbit feces in the square, and whether the bird and rabbit feces present were positive for C. jejuni.
TABLE 3.
Percentages of C. jejuni-positive cattle fecal samples stratified by various environment-related categories
| Category | No. of samples | % of samples testing positive |
|---|---|---|
| Intensity of fecal contamination (Avg no. of cattle fecal pats per 5-m-radius circle) | ||
| 1-5 | 245 | 28 |
| 6-10 | 280 | 32 |
| 11-20 | 152 | 35 |
| >20 | 22 | 21 |
| Rain | ||
| None | 796 | 33 |
| Light | 172 | 21 |
| Heavy | 12 | 0 |
| Habitat type | ||
| Unimproved grassland | 16 | 19 |
| Semi-improved grassland | 286 | 34 |
| Improved grassland | 668 | 31 |
| Short-term grass leys | 24 | 33 |
| Sun score | ||
| Overcast | 433 | 27 |
| Some clouds | 395 | 36 |
| Clear | 152 | 24 |
| Wildlife feces | ||
| Bird feces absent from square | 388 | 27 |
| Bird feces present and negative for C. jejuni | 393 | 31 |
| Bird feces present and positive for C. jejuni | 233 | 39 |
| Rabbit feces absent from square | 421 | 32 |
| Rabbit feces present and negative for C. jejuni | 499 | 28 |
| Rabbit feces present and positive for C. jejuni | 94 | 43 |
The age of pat ijkℓ was denoted Aijkℓ, and the age score used to describe each pooled sample was Ãijℓ = mink(Aijkℓ), or the age of the youngest pat of type ℓ at point ij. The values assigned to the age categories were arbitrary, and there was no reason to expect that a pat of age 4 was twice as old as a pat of age 2. The average age score of the pats in the pooled sample was therefore unlikely to be meaningful. Furthermore, fitting the minimum age as a categorical variable, in which each age category has its own parameter, is likely to fit the data better than fitting age as a continuous variable, with the age effect being Ãijℓβ.
Since pijℓ must be between 0 and 1, we modeled the inverse logit transform of the probabilities by using the following equation:
 |
(1) |
This model is a generalized linear model (28) and is fitted by the method of maximum likelihood, using a Fisher scoring algorithm as implemented in the function glm in the R software (8). Models were compared by using Akaike's information criteria (AIC).
A random effects model was used to ascertain the extent of spatial dependence in the data. The question of interest was as follows: after the effects of pat age and other covariates have been taken into account, are cattle fecal pats that are closer to each other more likely to have similar degrees of C. jejuni prevalence than pats which are further apart? Since the primary squares formed a regular lattice, a convenient model for these data was a Markov random field model (7).
In this spatial model, we allowed pijℓ to depend on the fixed effects, a latent spatial process (Ai), and a spatially uncorrelated random effect (Bi). Thus, all of the pats within a primary square had the same probability of being positive for C. jejuni (subject to the covariate Xijℓ values being equal), and neighboring primary squares were potentially more similar than primary squares that were further apart. The full model is Sijℓ = Xijℓβ + Ai + Bi. Ai follows a Markov random field with the following formula:
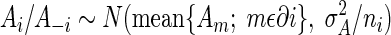 |
(2) |
Bi values are mutually independent random variables according to the equation Bi ∼ N(0,σB2). Here A−i = {Aj; j ≠ i}, ∂i refers to all of the primary squares which are neighbors of i, with the neighborhood being the four squares to the immediate north, south, east, and west, and ni is the number of points for this neighborhood in the study region. The fill-in areas were deemed to correspond to one primary square located at the center of the group to preserve the lattice structure.
The nature of the spatial transmission of C. jejuni can be inferred by examining the marginal variances of Ai and Bi in equation 2. Small values for the variances of both Ai and Bi would indicate no spatial dependence in the data. A large variance of Ai would be expected if there was a spatial correlation at distances of 600 m or more. If there was a spatial structure at short scales but not at 600 m or more, σB2 would be large, but var(Ai) would be negligible.
Markov random field models of this type are usually implemented within a Bayesian framework and fitted by use of the Gibbs sampler (17). For this study, we used BayesX (24) software, which has by default gamma(1, 0.005) distributions as priors on variance parameters, as recommended by Besag et al. (3), and uniform priors on the coefficients of the fixed effects. These priors are used throughout this paper. The chain was thinned by taking only every 300th sample and was run until 6,000 samples were obtained after a burn-in. Autocorrelation plots and time traces were used to monitor chain convergence.
Field-level variation.
Anticipating the results shown below, we explored random effects at the field level rather than the level of primary squares. One possible reason for the strong square effect shown below could be that the spatial correlation decays to zero somewhere between 50 and 600 m. Another possibility is that pats from the same primary square were likely to be located in the same field and therefore to come from the same herd or even from the same animal. For this study, detailed information on the herds and animals that had grazed each pasture was not available, although the field that each sample belonged to could be inferred from the spatial locations and habitat types recorded.
Recall that a sample tested for C. jejuni was derived from between one and four pats located at tertiary points. A sample was deemed to belong to a given field if all of the pats comprising the sample were located in that field. Samples consisting of pats from more than one field were not included in the analysis.
Using Yij to refer to pooled sample j in field i, we used the following model to incorporate field effects:
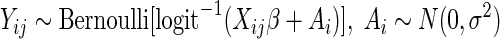 |
(3) |
where Xij is a vector of covariates and Ai values are mutually independent random effects.
RESULTS
Data summary.
Table 1 shows the number of samples of each type tested and the proportion of samples that were positive for each species. C. jejuni was the most commonly isolated species for all animal samples, ranging from 11% for samples from nonavian wildlife to 36% for cattle feces. A total of 518 secondary points contained cattle feces, with 36% of the freshest and 27% of the nearest pooled cattle fecal samples testing positive for C. jejuni. C. coli was commonly isolated from water (17%) and sheep (21%) samples but was rarely isolated from other samples. C. lari was isolated from all sample types, with the exception of sheep feces, and was found in moderate numbers in birds (7%) and water (5%). C. hyointestinalis was only isolated from cattle (7%) and birds (1%).
TABLE 1.
Percentages of samples that tested positive for Campylobacter spp., listed by species and sourcea
| Sample type | No. of samples | % of samples testing positive for indicated species
|
||||
|---|---|---|---|---|---|---|
| C. jejuni | C. coli | C. lari | C. hyointestinalis | Other | ||
| Cattle feces | ||||||
| Freshest | 496 | 36 | 1 | 3 | 7 | 18 |
| Nearest | 518 | 27 | 1 | 2 | 7 | 12 |
| Total | 1,014 | 31 | 1 | 2 | 7 | 15 |
| Water | 137 | 15 | 17 | 5 | 0 | 8 |
| Bird feces | 180 | 26 | 1 | 7 | 1 | 5 |
| Sheep feces | 24 | 25 | 21 | 0 | 0 | 8 |
| Other wildlife feces | 271 | 11 | 0.4 | 3 | 0 | 2 |
| Soil | 1,015 | 0.2 | 0.1 | 0.2 | 0 | 0.4 |
Samples were identified as being positive by standard culture methods, and isolates were confirmed by PCR.
Cattle feces in the environment.
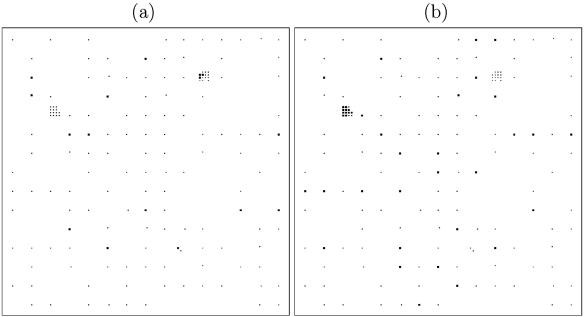
Figure 2 shows the locations of cattle fecal samples from which C. jejuni was isolated. Each point in Fig. 2 corresponds to a secondary sampling point, from which up to two samples were collected, and is marked as positive if C. jejuni was isolated from at least one of the samples. Figure 3 shows the locations of C. hyointestinalis- and C. lari-positive samples, aggregated at the primary square level. Up to eight samples were tested from each primary square, and a primary square is marked as positive in Fig. 4 if Campylobacter spp. were isolated from at least one of these samples.
FIG. 2.
Locations of secondary sampling points that were negative (·) and positive (▪) for C. jejuni in cattle feces. Samples were identified as being positive by standard culture methods, and isolates were confirmed by PCR.
FIG. 3.
Centers of primary squares from which at least one sample of cattle feces tested positive (▪) and from which no samples tested positive (·) for C. lari and C. hyointestinalis. (a) C. lari. (b) C. hyointestinalis.
FIG. 4.
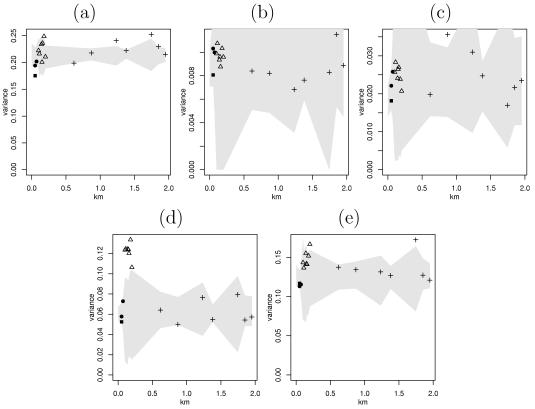
Variograms with observations at the same secondary point (▪) and at distances within primary squares (•), within fill-in squares (▵), and between primary squares (+), along with permutation envelopes. (a) C. jejuni. (b) C. coli. (c) C. lari. (d) C. hyointestinalis. (e) Other Campylobacter spp.
Table 2 shows the number of pooled samples and proportion of Campylobacter sp.-positive pooled samples according to the minimum age score of the pats comprising each sample.
TABLE 2.
Percentages of positive samples for each Campylobacter spp. according to the age of the youngest pat comprising the pooled samplea
| Minimum age score | No. of samples | % of samples testing positive for indicated species
|
||||
|---|---|---|---|---|---|---|
| C. jejuni | C. hyointestinalis | C. lari | C. coli | Other | ||
| 1 | 23 | 39 | 9 | 0 | 0 | 9 |
| 1.5 | 61 | 52 | 7 | 0 | 3 | 21 |
| 2 | 285 | 42 | 8 | 4 | 1 | 20 |
| 2.5 | 240 | 35 | 5 | 4 | 2 | 18 |
| 3 | 243 | 23 | 6 | 0 | 0 | 10 |
| 3.5 | 97 | 11 | 9 | 1 | 0 | 6 |
| 4 | 65 | 3 | 5 | 0 | 0 | 0 |
Age scores are as follows: 1, freshly voided with no signs of aging; 2, recently voided with some early signs of aging; 3, clear signs of aging but still retaining integrity; 4, aged. Samples were identified as being positive by standard culture methods, and isolates were confirmed by PCR.
Table 3 lists the proportions of C. jejuni-positive samples for several environment-related categories. The prevalence increased if bird feces were present and were negative for C. jejuni and further increased if the bird feces tested positive for C. jejuni. There was an apparent negative relationship with regards to rain. These environmental measurements were formally incorporated into a statistical model of C. jejuni prevalence, as discussed below.
Spatial exploratory analysis.
Figure 4 shows variograms for the Campylobacter sp. data along with permutation envelopes. If the dissemination of a Campylobacter sp. were spatial, we would expect an upward trend on the variogram, with variogram ordinates to the left of the plot lying below the gray confidence region. If spatial effects were not important, the variogram ordinates should be flat and lie within the confidence region.
The various widths of the confidence region were due to the different numbers of points for different spatial separations. A primary square with two samples at each of the four secondary points yielded four pairs at coincident points and eight pairs each at separations of 50 and 71 m, causing the confidence regions to all be slightly wider at the first point than at the subsequent two points. The permutation envelopes were widest at distances between 75 and 600 m due to the fact that there were only small numbers of pairs at these separations, which are in the fill-in squares.
For the most part, the variograms lay inside the permutation envelopes, meaning that there is little evidence of spatial dependence. C. jejuni seems to have less variance within primary squares than do the permutations, which suggests that the data exhibit dependence at short ranges.
Modeling the presence of C. jejuni in cattle feces.
A statistical model was used to test the influence of several possible covariates, specifically the pat age, the presence of rain and sun, the presence of bird or rabbit feces and whether they were contaminated with C. jejuni, the number of pats in the area, and the habitat type of the sampling region. The first covariate considered was the ages of the fecal pats in order to test the hypothesis that older pats were less likely to test positive than fresher fecal material. As expected, the minimum age score proved a better predictor than the mean age score, and treating age scores as categorical was an improvement over fitting the age scores as continuous. The estimates of the age effect for young pats were broadly similar and had large standard errors. Combining age categories 1, 1.5, and 2 into a single category had a negligible effect on the deviance and reduced the number of parameters in the model. The presence of C. jejuni in bird feces and the presence of rain were significantly related to the isolation of C. jejuni, whereas the other variables were not statistically significant.
Table 4 shows parameter estimates and standard errors for the model incorporating the significant covariates. P values refer to the test that the true parameter value is zero, which along with the standard errors were based on a normal approximation of the model.
TABLE 4.
Maximum likelihood estimates and standard errors for factors related to the isolation of C. jejuni from cattle feces
| Parametera | Estimate | SE | P value |
|---|---|---|---|
| Intercept | −0.21 | 0.12 | 0.07 |
| Minimum age of fecal pat = 2.5 | −0.39 | 0.18 | 0.03 |
| Minimum age of fecal pat = 3 | −0.97 | 0.19 | <0.001 |
| Minimum age of fecal pat = 3.5 | −1.7 | 0.34 | <0.001 |
| Minimum age of fecal pat = 4 | −3.0 | 0.71 | <0.001 |
| Presence of C. jejuni in bird feces | 0.77 | 0.28 | 0.005 |
| Rain | −0.77 | 0.21 | <0.001 |
Age scores are as follows: 2,recently voided with some early signs of aging; 3, clear signs of aging but still retaining integrity; 4, aged.
The higher the age of the fecal pats (over a score of 2), the smaller the chance of isolating C. jejuni. The presence of C. jejuni-positive bird feces in the area of the sample was associated with a higher probability of isolating C. jejuni from cattle feces, while samples taken in the rain were associated with a lower likelihood of testing positive.
Modeling of spatial effects.
The nature of the spatial variation of C. jejuni could be inferred by examining the estimated variances of the independent square effect Bi and the spatially correlated square effect Ai. The posterior mean of the independent effect σB2 was 1.29, with a posterior standard deviation of 0.38. The marginal variance of the spatially correlated term Ai, calculated as the sample variance of the simulated Ai at each iteration, was an order of magnitude smaller, with a posterior mean of 0.040 and a posterior standard deviation of 0.084. The posterior means of the coefficients on the fixed effects were all within half of a standard error of those obtained above, and the posterior standard deviations were similar to the standard errors for the data described above.
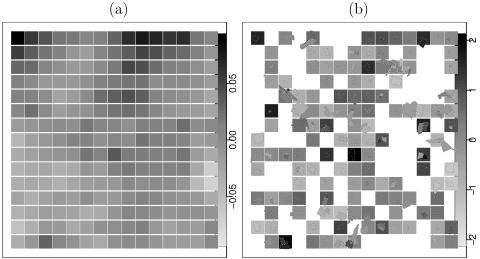
The estimated variance of the spatial process suggests that, although feces sampled within the same primary square tended to have a similar likelihood of being positive for C. jejuni (as confirmed by the large σB2), neighboring primary squares were mostly independent of one another. Figure 5 shows the posterior means of Ai and Bi, with the shading of the squares indicating the values of the primary square effects. The irregularly shaped regions in Fig. 5 are explained below. Notice the different scales for the two figure panels, with the Bi values spanning a larger range than the Ai values. This larger variability in Bi is more evidence that the independent random effect term Bi has more influence on C. jejuni prevalence than the spatially correlated term Ai.
FIG. 5.
Posterior means of the spatial random effects, square random effects, and field random effects. White areas correspond to locations without observations. (a) Spatial effect Ai. (b) Independent random effect Bi.
Estimates of Ai were available for every square on the grid. For primary squares from which no cattle fecal samples were collected, the value of Ai could be inferred from adjacent squares. In Fig. 5b, primary squares without observations are shown in white, as there was no dependence structure which would permit inference at those locations.
The relative importance of the two effects can be affected by the priors used for the variance parameters. As a diagnostic tool, we fitted the model by using only the spatial Ai process, in effect forcing the algorithm to find a spatial pattern. The result of fitting this model was that the estimated spatial effects were similar to the Bi values fitted before. Thus, the conditionally independent square effect Bi proves to be the more important effect, persisting even when the model forces the latent process to be spatially correlated.
The two fill-in squares provided information about how the isolation of C. jejuni from cattle feces varies over short distances. Since the secondary points within the two squares formed a lattice, a Markov random field model could be fitted for these regions, with the spatial effect Aj and the independent effect Bj now representing the risk of C. jejuni presence at a secondary point j in a given fill-in square. The posterior means of the Aj values, with the mean corrected, were all <0.02 in magnitude, and the 90% credible intervals all contained zero. While the two fill-in squares had different risks for C. jejuni, there was no evidence that within a fill-in square neighboring points were more similar than squares that were further apart.
Modeling of field effects.
Table 5 shows the parameter estimates and credible intervals for the coefficient β and the vari-ance of the field effect Ai in equation 3. These parameters are similar to those found above, as for the most part the fields and primary square groups were coincident.
TABLE 5.
Parameter estimates for a model in which C. jejuni prevalence depends on the covariates listed and a random effect at the field level
| Variablea | Mean | SD | Quantile
|
||
|---|---|---|---|---|---|
| 2.5% | 50% | 97.5% | |||
| Intercept | −0.44 | 0.19 | −0.82 | −0.43 | −0.086 |
| Minimum age of fecal pat = 2.5 | −0.36 | 0.22 | −0.80 | −0.36 | 0.063 |
| Minimum age of fecal pat = 3 | −1.04 | 0.25 | −1.53 | −1.03 | −0.56 |
| Minimum age of fecal pat = 3.5 | −1.76 | 0.42 | −2.65 | −1.76 | −0.92 |
| Minimum age of fecal pat = 4 | −3.62 | 0.94 | −5.96 | −3.54 | −2.05 |
| Presence of C. jejuni in bird feces | 0.94 | 0.40 | 0.18 | 0.94 | 1.77 |
| Rain | −0.98 | 0.33 | −1.66 | −0.97 | −0.34 |
| Field effect variance (σ2) | 1.30 | 0.43 | 0.64 | 1.28 | 2.46 |
Age scores are as follows: 2, recently voided with some early signs of aging; 3, clear signs of aging but still retaining integrity; 4, aged.
The irregularly shaped regions in Fig. 5b show the posterior means of the field effects, with the remaining spaces in the graph being fields for which no data were available. For squares which only contained one field, the square effect and the field effect were mostly identical, whereas when a square contained more than one field, the square effect was close to the average of the field effects. Discrepancies between the field model and the squares model could occur because the variance of the field effect was higher than that of the square effect and because there were points that were discarded from the field model due to fecal samples obtained from more than one field being pooled together.
Comparing the fit of the field model to the square model was difficult because the models are not nested. The variance of the field effect was only slightly higher than the square effect variance. The models could be fitted by using penalized pseudo-likelihood and then could be compared by examining their approximate AICs, although this showed an AIC that was only marginally in favor of the field model. We conclude here that the field effect is more appropriate for these data because of the more satisfactory interpretation of boundaries based on fields rather than primary squares.
A further question of interest was whether fields that are close together tend to have risk factors which are more similar than fields which are further apart. Figure 6 shows variograms of the posterior means of the field effects Ai, in which the location of each field is defined as the average of the x-axis and y-axis coordinates of the boundary vertices. The gray region is a 95% permutation-based confidence region, which is wide at short distances due to the small number of fields at a spatial separation of <600 m. The variogram ordinates at short ranges are close to the edge of the confidence region, and although the variogram does not indicate a statistically significant amount of spatial correlation, the issue warrants further investigation.
FIG. 6.
Variogram of the posterior means of the field risk factors for C. jejuni (○) and of the permutation-based confidence region (shaded area).
Do Campylobacter spp. tend to occur together?
The analysis described above suggests that pats within a field have similar probabilities of testing positive for C. jejuni. In this section, we examine whether fields at high risk for the isolation of C. jejuni are more likely to contain C. hyointestinalis or other species than fields with low levels of C. jejuni.
The model described above was applied to the data for C. hyointestinalis, C. lari, and C. coli. Positive results for C. lari and C. coli were very sparse, which led to numerical problems when fitting the model both in the Bayesian framework and with pseudo-likelihood. The prevalence of these two species was therefore too low to permit the modeling of field effects, and we restricted our attention to C. hyointestinalis and C. jejuni.
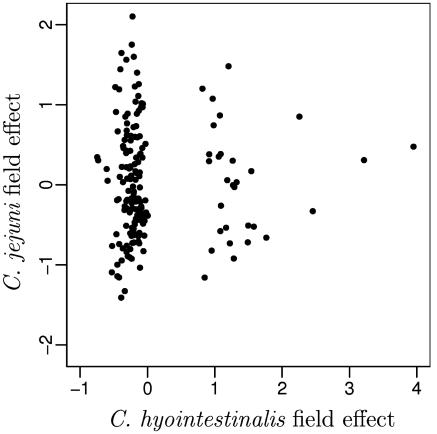
Due the sparsity of C. hyointestinalis-positive results, the parameter estimates for C. hyointestinalis are less precise than those for C. jejuni. Figure 7 shows the field effects for C. jejuni plotted against the field effects for C. hyointestinalis. If the two species were likely to occur together, we would expect fields at a high risk of C. hyointestinalis to also be at a high risk for C. jejuni. Figure 7 shows no such relationship, suggesting that the species are independent of one another.
FIG. 7.
Posterior means of field risk factors for C. jejuni plotted against the field risk factors for C. hyointestinalis.
The posterior means of the field effects for C. hyointestinalis are not normally distributed; there is a collection of points about zero and another of points scattered above one. These two groups of points correspond to fields that were free from C. hyointestinalis (the field effects of zero) and those where C. hyointestinalis was present. Using a model in which the presence of C. hyointestinalis in a field was added as a fixed effect, we examined whether the fields that were free of C. hyointestinalis were less susceptible to the presence of C. jejuni than those where C. hyointestinalis was present. The estimate for this effect was very close to zero, adding further evidence that C. hyointestinalis and C. jejuni are independent. Similar models were fitted to allow for elevated risk when C. coli or C. lari was present in a field, with neither proving to be significant. It does not appear that the likelihood of isolating one species in a field is associated with the likelihood of isolating another.
DISCUSSION
This study formed part of a larger investigation into the distribution and diversity of human enteric pathogens in a well-characterized study area. We were particularly interested in the prevalence and distribution of pathogens on a small spatial scale, over a short time period, in an area that presents multiple exposure pathways to the human population. Campylobacter spp., particularly C. jejuni, were highly prevalent and widespread. Furthermore, as described elsewhere (15, 25), many of the C. jejuni genotypes isolated from cattle, wildlife, and water were indistinguishable from those recovered from human clinical cases by flaA and multilocus sequence typing.
For this study, thermophilic Campylobacter spp. were commonly isolated from a wide range of samples. In addition to producing food for both local and national consumption, the intensively farmed area contains two small towns (with a population of approximately 2,000 in each) and is visited by a larger population for outdoor leisure activities. The presence of C. jejuni in environmental samples of domestic and wild animal feces and water found in this study suggests that humans are likely to be exposed to this microorganism through recreational and occupational activities in addition to food pathways. We have demonstrated a moderately high prevalence of C. jejuni in environmental cattle feces throughout the study area. The spatial pattern is consistent with that of a ubiquitous organism, with a short-scale variation in infection intensity that cannot be explained by variations in the ages of the fecal material.
The high prevalence of C. jejuni in fresh cattle feces is consistent with findings from other studies (21, 39). The role of ruminants as reservoirs of Campylobacter spp. and their potential importance as sources of human infection were reviewed by Stanley and Jones (38). In addition to direct contact through occupational and recreational exposure, C. jejuni-positive cattle feces may contaminate food products (milk and meat), public and private water supplies (10, 37), and recreational waters (1). There are more than 50 dairy farms in the study area that produce milk products for local and national consumption, including an “open” farm on which visitors may have direct contact with livestock. Milk and other dairy products have been associated with Campylobacter outbreaks (2, 16, 18), and there is evidence of an association between particular genotypes of C. jejuni and cattle (27).
In contrast to food pathways, the role of environmental contamination with cattle feces in human disease is less clear. It is possible that direct and indirect exposures to cattle feces through these multiple, nonfood pathways account for many of the apparently sporadic cases of human Campylobacter infection. There was a large amount of C. jejuni-positive fecal material in the environment of our study area. The recovery of C. jejuni from cattle feces was highest for freshly voided feces and declined with the apparent age of the fecal pat, but C. jejuni was still isolated from highly degraded fecal material. The C. jejuni prevalence was lower in samples collected during rainy periods, although this was most likely due to the effect of moisture on the age scoring of fecal material, but there may also have been a dilution effect due to the higher water content of samples collected during periods of wet weather. The decline in recovery with age was likely due to the death of the organism over time but may also have been due to the organism entering a viable but nonculturable state (29). Thermophilic Campylobacter spp. have been shown to survive well in dairy cattle slurry (40). Surface runoff and slurry spreading are therefore likely to disseminate Campylobacter spp., thereby increasing the potential for human exposure.
C. lari was isolated mainly from water and from cattle and wild bird feces in the study area. This species was identified as an opportunistic pathogen of an immunocompromised patient in 1984 (31) and as a human enteric pathogen in 1985 (42), although the origin of human infections is unclear. C. lari occurs at a much lower prevalence than C. jejuni and C. coli, being responsible for 0.1% of reported cases in the United Kingdom in 2000 (5). C. lari has been isolated from a range of environmental sources in other studies and has been associated with birds (43), shellfish (11), and water environments (22).
After we adjusted the data for the age of the fecal material, cattle feces were more likely to be positive for C. jejuni if they were in an area in which wild bird feces were present and were positive for C. jejuni. There are several possible explanations for this finding. Firstly, there may be a direct transmission between cattle and wild birds. Wild birds may be a source of transmission to cattle, with C. jejuni-positive wild bird feces presenting a fresh pool of infected fecal material for direct transmission, or alternatively, wild birds may be infected by the ingestion of infected cattle feces. This explanation was supported by a preliminary analysis of the genotypes of C. jejuni found in wild birds and cattle (flaA, pulsed-field gel electrophoresis, and multilocus sequence typing) that showed that many indistinguishable genotypes were present in cattle and wild bird feces in the study area (15). However, this finding may be attributable to factors unrelated to direct transmission that are associated with the isolation of C. jejuni-positive cattle and wild bird feces (e.g., another common source of infection or other common risk factors for carriage and shedding). Other studies have shown that wild birds are carriers of C. jejuni (34, 43), including strains that have been isolated from human infections (4).
Our analysis found no evidence of spatial dependence for C. jejuni in cattle feces at distances of >600 m. This observed pattern was therefore not consistent with a large-scale transmission attributable to watercourses, wildlife territories, or other geographical features that transcended field and farm boundaries. Similarly, although the data were sparse, there was no evidence for spatial dependence for any of the other Campylobacter spp. isolated, and the presence of one species of Campylobacter was not associated with the presence of another. After we allowed for the ages of the feces in the environment, samples taken from the same field proved to be correlated, most likely because these pats would have come from the same groups of animals within the same herd. In this study, we focused entirely on feces found in the environment, with the aim of systematically sampling the entire area with minimal disruption to the farmers and landowners. Therefore, the type and identity of the animals that had grazed the fields were not always available. Groups of cattle grazing the same field were likely to belong to the same age and management group, and these factors have been shown to be associated with the prevalence and level of fecal carriage (32, 39). However, since the spatial resolution was not sufficiently fine and since we could not obtain reliable information on the identities of farms using each field, we were unable to examine whether different fields within the same farm tended to be similar and whether neighboring fields from different farms were correlated. A further study of short-range spatial correlation is therefore warranted, with a focus on the effects of fields and herds. A Markov random field model could then be used to assess the transmission of C. jejuni between neighboring fields and herds.
Assessing the dependence of different species was difficult because of the scarcity of positive results for species other than C. jejuni. This may be due to the dominant species masking the presence of other, less common species. This phenomenon would weaken the dependence between the species, as fields with a high prevalence of C. jejuni would hide the C. hyointestinalis even if C. hyointestinalis was also present in large numbers. Since the intensities of the other species were low, many more samples per field would have to be tested and more colonies would need to be examined for inferences about field-level effects to be possible.
Acknowledgments
We thank the farmers and landowners for their cooperation with this study.
This work was funded by DEFRA UK.
REFERENCES
- 1.Adak, G. K., J. M. Cowden, S. Nicholas, and H. S. Evan. 1995. The public-health laboratory service national case-control study of primary indigenous sporadic cases of campylobacter infection. Epidemiol. Infect. 115:15-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barrett, N. J. 1986. Communicable disease associated with milk and dairy-products in England and Wales—1983-1984. J. Infect. 12:265-272. [DOI] [PubMed] [Google Scholar]
- 3.Besag, J., P. Green, D. Higdon, and K. Mengersen. 1995. Bayesian computation and stochastic systems. Statist. Sci. 10:3-66. [Google Scholar]
- 4.Broman, T., H. Palmgren, S. Bergstrom, M. Sellin, J. Waldenstrom, M. L. Danielsson-Tham, and B. Olsen. 2002. Campylobacter jejuni in black-headed gulls (Larus ridibundus): prevalence, genotypes, and influence on C. jejuni epidemiology. J. Clin. Microbiol. 40:4594-4602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.CDSC. 2000. Sentinel surveillance of Campylobacter in England and Wales. Commun. Dis. Rep. Wkly. 10:169, 172. [PubMed] [Google Scholar]
- 6.Charlett, A., J. M. Cowden, J. A. Frost, I. A. Gillespie, J. Millward, K. R. Neal, S. J. O'Brien, M. J. Painter, Q. Syed, D. S. Tompkins, G. K. Adak, C. C. Tam, and F. J. Bolton. 2003. Point source outbreaks of Campylobacter jejuni infection—are they more common than we think and what might cause them? Epidemiol. Infect. 130:367-375. [PMC free article] [PubMed] [Google Scholar]
- 7.Cressie, N. A. C. 1993. Statistics for spatial data. John Wiley and Sons, Inc., New York, N.Y.
- 8.Dalgaard, P. 2002. Introductory statistics with R. Springer-Verlag, New York, N.Y.
- 9.Dingle, K., F. Colles, D. Wareing, R. Ure, A. Fox, F. Bloton, H. Bootsma, R. Willems, R. Urwin, and M. Maiden. 2001. Multilocus sequence typing system for Campylobacter jejuni. J. Clin. Microbiol. 39:14-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duke, L. A., A. S. Breathnach, D. R. Jenkins, B. A. Harkis, and A. W. Codd. 1996. A mixed outbreak of Cryptosporidium and Campylobacter infection associated with a private water supply. Epidemiol. Infect. 116:303-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Endtz, H. P., J. S. Vliegenthart, P. Vandamme, H. W. Weverinkm, N. P. van den Braak, H. A. Verburgh, and A. van Belkum. 1997. Genotypic diversity of Campylobacter lari isolated from mussels and oysters in The Netherlands. Int. J. Food Microbiol. 34:79-88. [DOI] [PubMed] [Google Scholar]
- 12.Fahey, T., D. Morgan, C. Gunneburg, G. K. Adak, F. Majid, and E. Kaczmarski. 1995. An outbreak of Campylobacter jejuni enteritis associated with failed milk pasteurization. J. Infect. 31:137-143. [DOI] [PubMed] [Google Scholar]
- 13.Fitzgerald, C., K. Stanley, S. Andrew, and K. Jones. 2001. Use of pulsed-field gel electrophoresis and flagellin gene typing in identifying clonal groups of Campylobacter jejuni and Campylobacter coli in farm and clinical environments. Appl. Environ. Microbiol. 67:1429-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Food Standards Agency. 2000. Report of the study of infectious intestinal disease in England. Technical report. The Stationery Office, London, United Kingdom.
- 15.French, N. P., M. Barrigas, A. Fox, H. Leatherbarrow, P. E. Brown, and N. Williams. 2004. Multi-locus sequence typing of Campylobacter jejuni isolated from cattle faeces: implications for public health. Res. Vet. Sci. 76(Suppl. A):15. [Google Scholar]
- 16.Frost, J. A., I. A. Gillespie, and S. J. O'Brien. 2002. Public health implications of campylobacter outbreaks in England and Wales, 1995-9: epidemiological and microbiological investigations. Epidemiol. Infect. 128:111-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gilks, W. R., S. Richardson, and D. J. Spiegelhalter (ed.). 1996. Markov chain Monte Carlo in practice. Chapman & Hall, London, United Kingdom.
- 18.Gillespie, I. A., G. K. Adak, S. J. O'Brien, and F. J. Bolton. 2003. Milkborne general outbreaks of infectious intestinal disease, England and Wales, 1992-2000. Epidemiol. Infect. 130:461-468. [PMC free article] [PubMed] [Google Scholar]
- 19.Gillespie, I. A., S. J. O'Brien, J. A. Frost, G. K. Adak, P. Horby, A. V. Swan, M. J. Painter, and K. R. Neal. 2002. A case-case comparison of Campylobacter coli and Campylobacter jejuni infection: a tool for generating hypotheses. Emerg. Infect. Dis. 8:937-942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gonzalez, I., K. A. Grant, P. T. Richardson, S. F. Park, and M. D. Collins. 1997. Specific identification of the enteropathogens Campylobacter jejuni and Campylobacter coli by using a PCR test based on the ceuE gene encoding a putative virulence determinant. J. Clin. Microbiol. 35:759-763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grau, F. H. 1988. Campylobacter jejuni and Campylobacter hyointestinalis in the intestinal-tract and on the carcasses of calves and cattle. J. Food Prot. 51:857-861. [DOI] [PubMed] [Google Scholar]
- 22.Jones, K. 2001. Campylobacters in water, sewage and the environment. J. Appl. Microbiol. 90:68S-79S. [DOI] [PubMed] [Google Scholar]
- 23.Kramer, J. M., J. A. Frost, F. J. Bolton, and D. R. A. Wareing. 2000. Campylobacter contamination of raw meat and poultry at retail sale: identification of multiple types and comparison with isolates from human infection. J. Food Prot. 63:1654-1659. [DOI] [PubMed] [Google Scholar]
- 24.Lang, S., and A. Brezger. 2002. BayesX: software for Bayesian inference based on Markov chain Monte Carlo simulation techniques. University of Munich, Munich, Germany. [Online.] www.stat.uni-muenchen.de/∼lang/bayesx/bayesx.html.
- 25.Leatherbarrow, A. J. H., C. A. Hart, R. Kemp, N. J. Williams, A. Ridley, M. Sharma, P. J. Diggle, E. J. Wright, J. Sutherst, and N. P. French. 2004. Genotypic and antibiotic susceptibility characteristics of a population of Campylobacter coli isolated from dairy farmland in the United Kingdom. Appl. Environ. Microbiol. 70:822-830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Linton, D., R. J. Owen, and J. Stanley. 1996. Rapid identification by PCR of the genus Campylobacter and of five Campylobacter species enteropathogenic for man and animals. Res. Microbiol. 147:707-718. [DOI] [PubMed] [Google Scholar]
- 27.Manning, G., C. G. Dowson, C. Bagnall, I. H. Ahmed, M. West, and D. G. Newell. 2003. Multilocus sequence typing for comparison of veterinary and human isolates of Campylobacter jejuni. Appl. Environ. Microbiol. 69:6370-6379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McCullagh, P., and J. A. Nelder. 1983. Generalized linear models. Monographs on statistics and applied probability. Chapman & Hall, London, United Kingdom.
- 29.Moore, J. E. 2001. Bacterial dormancy in campylobacter: abstract theory or cause for concern? Int. J. Food Sci. Technol. 36:593-600. [Google Scholar]
- 30.Nachamkin, I., B. M. Allos, and T. Ho. 1998. Campylobacter species and Guillain-Barré syndrome. Clin. Microbiol. Rev. 11:555-567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nachamkin, I., C. Stowell, D. Skalma, A. M. Jones, R. M. Hoop, and R. M. Smilbert. 1984. Campylobacter laridis causing bacteremia in an immunosuppressed patient. Ann. Intern. Med. 101:55-57. [DOI] [PubMed] [Google Scholar]
- 32.Nielsen, E. M. 2002. Occurrence and strain diversity of thermophilic campylobacters in cattle of different age groups in dairy herds. Lett. Appl. Microbiol. 35:85-89. [DOI] [PubMed] [Google Scholar]
- 33.Pearson, A. D., M. H. Greenwood, J. Donaldson, T. D. Healing, D. M. Jones, M. Shahamat, R. K. A. Feltham, and R. R. Colwell. 2000. Continuous source outbreak of campylobacteriosis traced to chicken. J. Food Prot. 63:309-314. [DOI] [PubMed] [Google Scholar]
- 34.Petersen, L., E. M. Nielsen, J. Engberg, S. L. W. On, and H. H. Dietz. 2001. Comparison of genotypes and serotypes of Campylobacter jejuni isolated from Danish wild mammals and birds and from broiler flocks and humans. Appl. Environ. Microbiol. 67:3115-3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Public Health Laboratory Service. 1986-2001. Campylobacter sp. laboratory reports, England and Wales, faecal isolates. Technical report. [Online.] http://www.phls.co.uk.
- 36.Ribeiro, P. J., Jr., and P. Diggle. 2001. geoR: a package for geostatistical analysis. R-NEWS 1:15-18. [Online.] http://cran.R-project.org/doc/Rnews. [Google Scholar]
- 37.Said, B., F. Wright, G. L. Nichols, M. Reacher, and M. Rutter. 2003. Outbreaks of infectious disease associated with private drinking water supplies in England and Wales 1970-2000. Epidemiol. Infect. 130:469-479. [PMC free article] [PubMed] [Google Scholar]
- 38.Stanley, K., and K. Jones. 2003. Cattle and sheep farms as reservoirs of campylobacter. J. Appl. Microbiol. 94:104S-113S. [DOI] [PubMed] [Google Scholar]
- 39.Stanley, K. N., J. S. Wallace, J. E. Currie, P. J. Diggle, and K. Jones. 1998. The seasonal variation of thermophilic campylobacters in beef cattle, dairy cattle and calves. J. Appl. Microbiol. 85:472-480. [DOI] [PubMed] [Google Scholar]
- 40.Stanley, K. N., J. S. Wallace, and K. Jones. 1998. Thermophilic campylobacters in dairy slurries on Lancashire farms: seasonal effects of storage and land application. J. Appl. Microbiol. 85:405-409. [Google Scholar]
- 41.Tam, C. C., S. J. O'Brien, G. K. Adak, S. M. Meakins, and J. A. Frost. 2003. Campylobacter coli—an important foodborne pathogen. J. Infect. 47:28-32. [DOI] [PubMed] [Google Scholar]
- 42.Tauxe, R. V., C. M. Patton, P. Edmonds, T. J. Barret, D. J. Brenner, and P. A. Blake. 1985. Illness associated with Campylobacter laridis, a new recognized Campylobacter species. J. Clin. Microbiol. 21:222-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Waldenstrom, J., T. Broman, I. Carlsson, D. Hasselquist, R. P. Achterberg, J. A. Wagenaar, and B. Olsen. 2002. Prevalence of Campylobacter jejuni, Campylobacter lari, and Campylobacter coli in different ecological guilds and taxa of migrating birds. Appl. Environ. Microbiol. 68:5911-5917. [DOI] [PMC free article] [PubMed] [Google Scholar]