Abstract
Fermented dry sausages, inoculated with Escherichia coli O157:H7 during batter preparation, were submitted to an in vitro digestion challenge to evaluate the extent to which passage through the human gastrointestinal tract could inactivate the pathogenic cells, previously stressed by the manufacturing process. The numbers of surviving E. coli O157:H7 cells remained constant after a 1-min exposure of the finely chopped sausage to synthetic saliva or during the following 120-min exposure to synthetic gastric juice at an initial pH of 2.0. However, significant (P ≤ 0.05) growth of the pathogen (1.03 to 2.16 log10 CFU/g) was observed in a subsequent 250-min exposure to a synthetic pancreatic juice at pH 8.0. In a different set of experiments, fractions from the gastric suspension were transferred into the synthetic pancreatic juice at 30-min intervals to mimic the dynamics of gastric emptying. Concurrently, the pH of the remaining gastric fluid was reduced to 3.0, 2.5, and 2.0 to simulate the gradual reacidification of the stomach contents after the initial buffering effect resulting from meal ingestion. Under these new conditions, pathogen growth during pancreatic challenge was observed for the first few fractions released from the stomach (90 min of exposure [pH 2.5]), but growth was no longer possible in the fractions submitted to the most severe gastric challenge (120 min of exposure [pH < 2.2]).
Escherichia coli O157:H7 is recognized as a foodborne pathogen of major significance. The organism colonizes the large intestine and produces one or more of the potent cytotoxins referred to as Shiga-like toxins (Stx). These toxins are responsible for severe hemorrhagic colitis in humans, with, in some cases, life-threatening complications such as hemolytic uremic syndrome or thrombotic thrombocytopenic purpura (22, 35). Outbreaks of food poisoning attributed to E. coli O157:H7 have traditionally been traced back to foods of bovine origin, in particular, improperly cooked ground beef and raw milk. However, several recent outbreaks have involved high-acid foods, previously considered safe, such as fermented dry sausages (10), mayonnaise (42), and apple cider (7, 43).
The ability of E. coli O157:H7 to withstand the acidic conditions encountered in various foods has been noted by several investigators (7, 20, 32, 34, 43). In addition, the results of studies in which medium-grown cells were introduced into synthetic gastric juice or into liquid medium acidified to pH values comparable to the pH value seen with the normal fasting stomach (pH 2) have generally suggested that passage through the stomach would be insufficient to inactivate the pathogen. This fact is consistent with the data from epidemiological investigations which indicated that as few as 10 to 100 cells of E. coli O157:H7 per g of raw ground beef were sufficient to cause illness (1, 3, 9).
In reality, the situation is considerably more complex for several reasons. Firstly, the organism always enters the digestive system within a food matrix. Waterman and Small (41) postulated that high protein content in food (such as ground beef and boiled egg white) may protect enteric bacteria against the killing effect of gastric acids. Using Fisher rats to model the effect of food on the gastric killing of bacteria, Drouault et al. (16) obtained a 7% survival rate in the stomach when a 109 CFU/ml suspension of Lactobacillus lactis was fed to the animal, compared to 100% survival when the organism was mixed with the rat food. Likewise, Charteris and coworkers (11) reported that during an in vitro upper intestinal transit tolerance study, milk proteins and porcine gastric mucin increased the gastric and small intestinal transit of some Lactobacillus and Bifidobacterium species.
Secondly, dry-sausage-borne E. coli O157:H7 cells have undergone acidic and osmotic stresses during the fermentation and drying phases of sausage manufacture, respectively. According to the hurdle theory (26), these stresses may later impair the ability of the pathogen to cope with the additional stresses encountered during human digestion, such as the extreme acidity in the stomach and the action of bile extracts (intestine) and of the enzymes lysozyme (saliva), pepsin (stomach), and pancreatin (intestine). Conversely, a general stress response, which could offer cross-protective effects leading to a higher survival rate during the subsequent gastric challenge, could also be triggered (4).
In light of the above, and with the ultimate goal of providing new data to refine risk analysis with respect to E. coli O157:H7 in fermented dry sausages, a study was undertaken to further evaluate the fate of sausageborne E. coli O157:H7 cells during human digestion under realistic conditions, i.e., using cells that had undergone the process-related stresses and were ingested as an integral part of infected sausages and exposed to a salivary, gastric, and pancreatic challenge under conditions that reflected the actual transit through the human digestive system.
MATERIALS AND METHODS
Organisms and cultures.
E. coli O157:H7 strain F-90, isolated from dry-cured salami implicated in the Washington-California outbreak of 1994, was obtained from the Food Research Institute of the University of Wisconsin. The E. coli O157:H7 strain 5-1, a salami isolate from the Vancouver outbreak of 1999, was provided by the Canadian Food Inspection Agency, Burnaby, British Columbia, Canada). The strain ATCC 43894 (previously known as CDC 932), an O157:H7 human isolate moderately resistant to acidity, was purchased from the American Type Culture Collection (ATCC) (Manassas, Va.).
The non-ATCC strains were provided as pure cultures on slant agar and kept at 4°C for a maximum of 48 h. Stock cultures were prepared by streaking individual colonies onto MacConkey sorbitol agar (MSA; Oxoid, Ontario, Canada) and incubating at 37°C for 24 h aerobically. Three presumptive E. coli O157:H7 colonies (white, non-sorbitol-fermenting) were plated as tight streaks on fresh MSA to form confluent cell monolayers. After a 24-h incubation at 37°C, the cell layers were harvested using sterile swabs and then suspended in tryptone soya broth (TSB; Oxoid) supplemented with glycerol (10% [vol/vol] final concentration) and maintained at −80°C in 0.15-ml aliquots. Strain ATCC 43894, provided as freeze-dried cultures, was first suspended in TSB and then streaked on MSA plates and incubated aerobically for 24 h at 37°C, prior to preparation of the stock cultures.
Working cultures were prepared from the second consecutive daily transfer of E. coli O157:H7 cells growing overnight at 37°C (with shaking at 150 rpm) in individual test tubes containing TSB supplemented with glucose (TSBG) (1% [wt/vol] final concentration). The cells were then harvested by centrifugation at 1,880 × g for 20 min at 4°C, washed twice, and suspended in peptone water (PW; Difco, Beckton Dickinson, Ontario, Canada) (0.1%) before experiments. Cell counts were evaluated by enumeration on duplicate MSA plates, after appropriate dilutions in PW.
Simulated environments.
Synthetic saliva was prepared as follows (39): amounts of 100 ml each of 25 mM K2HPO4, 24 mM Na2HPO, 1,570 mM KHCO3, 100 mM NaCl, and 1.5 mM MgCl2 were mixed together. To this were added 6 ml of 25 mM citric acid and 100 ml of 15 mM CaCl2. The pH was adjusted to 6.7 with 5 N NaOH or concentrated HCl (12 N), and the volume was made up to total 1 liter. The medium was sterilized by autoclaving, and when it reached the ambient temperature, α-amylase (Sigma-Aldrich Canada Ltd., Ontario, Canada) from human saliva and lysozyme (Sigma-Aldrich) from chicken egg white were added at 1 and 0.1 g/liter (final concentrations), respectively (13).
The synthetic gastric juice was prepared as described by Molly et al. (33) to contain (in grams/liter [final concentrations]) the following ingredients: glucose, 0.4; yeast extract, 3.0; Bacto Peptone, 1.0; porcine mucin, 4.0; cysteine, 0.5; NaCl, 0.08; NaHCO3, 0.4; K2HPO4, 0.04; KH2PO4, 0.04; CaCl2-2H2O, 0.008; MgSO4 · 7H2O, 0.008; xylan, 1.0; and soluble starch, 3.0; also, 2.0 USP of pectin/liter and 1 ml of Tween 80/liter were added to this mixture. The preparation was adjusted to pH 2.0 with concentrated HCl (12 N) prior to sterilizing by autoclaving, and the pH was corrected (with 12 N HCl or 5.0 N NaOH) following sterilization and equilibration to room temperature when required. Finally, pepsin (P-7000; Sigma-Aldrich) was added to achieve a final concentration of 3 g/liter (11).
Manufacture and inoculation of sausages.
Sausages were manufactured in a 65-m2 biosafety level II pilot plant for meat processing (Faculty of Veterinary Medicine of Montreal University, Saint-Hyacinthe, Quebec, Canada). Coarse ground (6-mm-diameter plate) pork boneless picnics, 90% lean beef shoulder cuts, and pork back fat were purchased locally and kept at about 4°C until batter preparation. Beef cuts and back fat were ground through a 6-mm-diameter plate shortly before use in sausage formulation. Formulation of batter was calculated using Least Cost Formulator Blending System software (Least Cost Formulation Ltd., Virginia Beach, Va.).
Raw batters were prepared in 2-kg batches which always contained 25% beef; with the remaining portion consisting of pork trimmings, pork back fat, and commercial spice and cure mixtures (Q11412; BSA Inc., Montreal, Canada), NaCl, NaNO2, and NaNO3 to obtain a target composition of 20% fat, 4% NaCl, 120 ppm of NaNO2, and 80 ppm of NaNO3. The ground meat mix was inoculated with pure cultures of E. coli O157:H7 strain F-90, 5-1, or ATCC 43894, at final concentrations of ca. 2 × 107 CFU/g (24). The batters were prepared in a 5-pound-capacity stand mixer (K5SSWH model; KitchenAid, Greenville, Ohio), with a 4-min cycle (1 min each for ground raw meats, E. coli inoculum, the spice mixture, and NaNO2). A starter culture (Rosellac A; Rosell Institute, Montreal, Quebec, Canada), composed of Lactobacillus plantarum, Micrococcus varians, and Pediococcus spp. hydrated in distilled water (15 g in 75 ml), was also added, and mixing continued for 1 additional minute. Negative controls consisting of uninoculated sausages were also prepared similarly.
The batter was then stuffed into 30-mm-diameter flat collagen casings (BSA Inc.) to form raw sausages at least twice as long as wide. Fermentation was carried out at 24 ± 0.1°C (130 fermentation-drying unit; Arcos S.a.r.l., Saint-Binigne, France) (20-kg capacity), at a relative humidity (RH) of 80 to 85% (minimum-maximum preset values), to reach a target pH value of 5.0 (actual value, 4.9 ± 0.1), which typically took 48 h. Subsequent drying proceeded at 14.0 ± 0.5°C in the drying compartment of the Acros B130 unit adjusted to initiate and interrupt the drying phase every time RH values reached 80 and 70%, respectively, to achieve a water activity value (aw) of 0.90.
Physicochemical analysis.
The moisture content of raw meat was determined in an AVC80 microwave moisture analyzer (CEM Corporation, Matthews, N.C.). Fat and protein contents were measured instrumentally, by use of TFE-200 and FP-428 analyzers (Leco Corporation, St. Joseph, Mich.), respectively. The aw was determined with a chilled-mirror dewpoint type aw meter (Aqualab CX2; Decagon Devices, Pullam, Wash.). The pH of digestive fluids was measured with an OAKTON pH/mv/°C meter (Cole Parmer Labcor Inc., Quebec, Canada), while the pH of sausages was measured using a Sentron pH meter (model 3001; Exeltec Inc., Quebec, Canada) equipped with Sentron ion-sensitive field-effect-transistor surface or penetration probes.
Microbiological analyses of the sausages.
Sausages (two sausages per sampling point), including noninoculated controls, were tested for viable E. coli O157:H7 cells by direct plating prior to stuffing, after fermentation, and after drying. At each sampling time, two 10-g portions of the batter or two 10-g composites of cross-sectional sausage slices were aseptically transferred to Whirl pack stomacher bags (VWR, Montreal, Canada) containing 90 ml of PW and homogenized for 1 min in a reciprocal blender (400c stomacher; VWR) at maximum speed, and serial dilutions of the homogenate were spread on MSA plates (100-μl aliquots) for enumeration of noninjured cells. The total counts corresponding to process-injured and uninjured cells were done using the modified overlay procedure (30). Briefly, aliquots (100 μl) from adequate dilutions were spread on tryptone soya agar plates (TSA; Oxoid), which were incubated for 2 h at 37°C to allow colony formation. This incubation time was shown to be sufficient in separate experiments, as results were not different when incubation was extended to 18 to 20 h. TSA agar was then lifted up with a sterile spatula and used to overlay corresponding sterile MSA plates. After a 2-min contact to allow colony replication, the plate was covered and inverted, and the TSA layer fell down when the plate was gently tapped against the bench. The MSA replicas were incubated aerobically at 37°C for 24 ± 2 h, while the TSA plates were discarded. White colonies were enumerated, and E. coli O157:H7 was confirmed by biotyping (API-20E; BioMerieux, Quebec, Canada) and serotyping (O157 agglutination test kit; Difco). The use of log numbers is necessary to satisfy the underlying hypotheses of the statistical analyses.
In vitro digestion tests.
Digestion tests were carried out in a closed system (37) in which the substrate (food matrix) was in contact with the enzymes, with no removal of the digestion products. Optimal pHs and temperature were used for each enzyme, or mixture of enzymes, and reactions were stopped by raising or decreasing the pH (19). Briefly, samples of dry sausage, previously inoculated during the formulation step with strain F-90, 5-1, or ATCC43894, were chopped into small pieces. To simulate mastication, which affords a mechanical breakdown of the food and adequate surface area for subsequent enzyme activity, 30-g portions were aseptically transferred to Whirl pack stomacher bags (VWR), diluted with 30 ml of artificial saliva, and homogenized for 1 min with the stomacher. To mimic the transient passage into the gastric environment, 240 ml of artificial gastric juice preadjusted to pH 2 (pH of fasting stomach) was added.
Gastric challenge was carried out in either of two modes. In the first one (static digestion experiments), the E. coli cells were challenged at 37°C in the gastric juice for 2 h, with periodic (every 15 min) mixing of the bag contents by hand massaging for about 10 sec. The pH was not adjusted to compensate for the buffering effect of the sausage. This corresponded to the minimum gastric challenge to which the sausage would be exposed prior to the reacidification of the stomach contents. In the second mode (dynamic digestion experiments), a more complex model was used in which 50-ml portions of the suspension in gastric phase were taken from the stomacher bag every 30 min (for a maximum digestion process of 2 h) and further exposed to pancreatic challenge, thus simulating normal gastric emptying (based on an exponential emptying rate of the gastric content beginning about 30 min after an individual starts to eat a solid meal) (25, 31). Concurrently, the pH of the remaining gastric fluid was adjusted to 3.0, 2.5, and 2.0 at 30, 60, and 90 min (HCl, 12 N) in an attempt to mimic the hydrochloric acid secretion (31). The enumeration of E. coli O157:H7 numbers was carried out in duplicate (on MSA and TSA for uninjured and injured cells, respectively, as described above) in the saliva, upon immersion in the gastric juice, at 60-min intervals or before and after each pH adjustment (occurring every 30 min) for the static or dynamic digestion experiments, respectively, and at the end of the gastric phases.
The pancreatic challenge was carried out in a synthetic pancreatic juice as follows: the pH of the emptied gastric contents was raised to 8.0 with 5 N NaOH, and pancreatin (P-1500) and porcine bile extract (B8631) (Sigma-Aldrich) were added at 0.1 and 0.3%, respectively. The pancreatic suspension was incubated aerobically at 37°C for 4 h with periodic homogenization, and E. coli O157:H7 cell counts were carried out immediately (ca. 1 min) after the addition of pancreatin and bile salts and at 1- and 2-h intervals for the static and dynamic digestion experiments, respectively. Negative controls, consisting of 30-g sausage samples homogenized in 270 ml of PW and incubated for 6 h at 37°C, were also prepared.
Statistical analyses.
Triplicate and duplicate in vitro digestion experiments were conducted under static and dynamic conditions, respectively, for inoculated sausage samples originating from four independent productions. Analysis of variance was carried out with the GLM procedure of the SAS statistical package (SAS Institute Inc., Cary, N.C.), with Duncan's multiple-range test used to discriminate means.
RESULTS
Sausage fermentation and drying resulted in a 1.53 ± 0.50 log10 CFU/g reduction in E. coli O157:H7 numbers from the initial concentration (ca. 2 × 107 log10 CFU/g) in the raw batter, and the numbers of cells which survived the process remained constant during subsequent refrigerated storage of the sausage for the whole experimental period (maximum of 3 weeks at 4°C). Therefore, the concentrations of the pathogen initially exposed to digestion challenge ranged from 5.11 ± 0.59 to 5.17 ± 0.79 log10 CFU/g.
Regardless of the strain used, the 1-min exposure of the contaminated sausage to saliva did not result in a decrease in E. coli O157:H7 counts (P > 0.05) compared to the results seen with the negative controls in peptone water (data not shown).
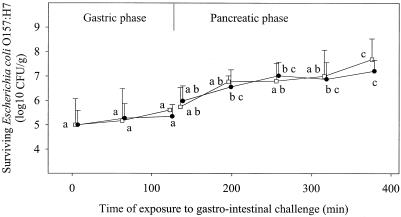
Results of the static digestion experiment are shown in Fig. 1. Mixing the sausage portion massaged in the synthetic saliva with the synthetic gastric juice (pH 2.0) resulted in an initial pH of 3.2 ± 0.1, which increased to 4.0 ± 0.2 during the 2-h gastric challenge. No inactivation of E. coli O157:H7 was observed in that period of time, regardless of the strain (P > 0.05). Significant growth (P ≤ 0.05) occurred in the pancreatic juice so that the pathogen levels at the end of the intestinal challenge were 2.69 and 2.21 log10 CFU/g higher than at the beginning of the gastric challenge for the strains 5-1 and ATCC 43894, respectively.
FIG. 1.
Survival and/or growth of E. coli O157:H7 cells experimentally inoculated in dry sausage during an in vitro digestion challenge under static conditions. Values reported represent the average results of at least two independent experiments carried out in duplicate. Bars represent standard deviations. Open squares and filled circles represent strains 5-1 and ATCC 43894, respectively. Data represent total counts for injured and uninjured cells measured on MSA after resuscitation on TSA plates. For each strain, means bearing identical letters are not significantly different (P > 0.05).
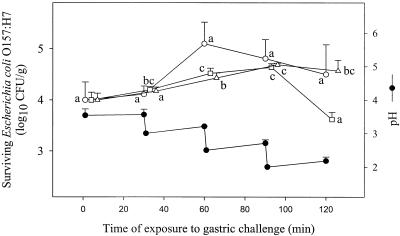
Results of the dynamic digestion experiments are presented in Fig. 2 and 3. During the early phases of the gastric challenge (Fig. 2), the pH was lowered in steps from 3.6 to 3.0 (30 min), 2.5 (60 min), and 2.0 (90 min). Limited growth of E. coli O157:H7 cells was observed for strains 5-1 and ATCC 43894 (Fig. 2), with the numbers of viable cells increasing by 0.63 and 0.69 log10 CFU/g, respectively (P < 0.05) in the first 90 min of exposure to gastric juice. A similar trend was observed for strain F-90, even though, as a result of wider distributions of values around the means, statistical analysis failed to confirm growth. Growth was no longer observed in the last phase of the gastric challenge (90 to 120 min), in which the pH was further reduced to values < 2.2, and the number of viable cells either remained stable (strains ATCC 43894 and F-90) or decreased (strain 5-1).
FIG. 2.
Survival of E. coli O157:H7 cells experimentally inoculated in dry sausage during a gastric challenge under dynamic conditions. Values reported represent the average results of at least two independent experiments carried out in duplicate. Bars represent standard deviations. Open circles, squares, and triangles represent strains F-90, 5-1, and ATCC 43894, respectively. Data represent total counts for injured and uninjured cells measured on MSA after resuscitation on TSA plates. For each strain, means bearing identical letters are not significantly different (P > 0.05).
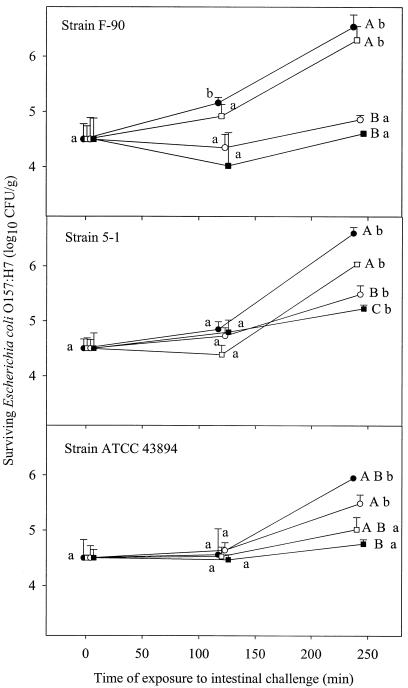
FIG. 3.
Survival or growth of E. coli O157 in pancreatic juice following 30, 60, 90, and 120 min of prior exposure to gastric challenge under dynamic conditions. Values reported represent the average results of at least two independent experiments carried out in duplicate. Filled circles, open squares, open circles, and filled squares represent cells previously challenged in the gastric juice for 30, 60, 90, and 120 min. Bars represent standard deviations. Data represent total counts for injured and uninjured cells measured on MSA after resuscitation on TSA plates. For each gastric treatment, means bearing identical lowercase letters are not significantly different (P > 0.05). For each intestinal challenge time, means bearing identical uppercase letters are not significantly different (P > 0.05).
The survival and/or growth pattern in the pancreatic juice depended on the severity of the previous gastric challenge (Fig. 3). Growth was generally not observed in the first 120 min of pancreatic challenge (P > 0.05), and the E. coli O157:H7 population remained stable. During the next 120 min in pancreatic juice, the cells that had been submitted to the milder gastric challenge (30 min; final pH of 3.56) actively grew (P < 0.05) such that their final concentrations after the 240 min of exposure to the pancreatic juice were considerably higher (by 1.44, 2.04, and 2.05 log10 CFU/g for strains ATCC 43894, F-90, and 5-1, respectively) than their initial concentrations. The capacity of the cells to grow in the pancreatic juice generally decreased with the increasing severity of the gastric challenge they had been exposed to, and growth was generally not observed with cells that had undergone the most severe gastric challenge (120 min; final pH, 2.2). Sublethal injury was not observed during the static gastric challenge or during the pancreatic challenge, since colony counts on MSA plates were not statistically different (P > 0.05), whether a prior revival phase on TSA plates had been used or not. Significant damage (P < 0.05), however, was observed as a result of the gastric challenge under dynamic conditions.
DISCUSSION
The fact that no reduction in E. coli O157:H7 numbers was seen as a result of exposure of the ground contaminated sausage to synthetic saliva (this study) is not surprising at first. It is well known that the outer membrane which shields the murein layer of gram-negative bacteria protects them from hydrolysis by lysozyme (18, 23, 29, 36). On the other hand, a recent report by Alakomi et al. (2) indicated that lactic acid (5 to 10 mM) could disrupt the outer membrane of gram-negative bacteria, including E. coli O157:H7, making it more susceptible to hydrolysis by lysozyme (10 μg/ml; 4 min). The concentration of lactic acid in sausages as a result of fermentation was not measured in our study, but it is known to be typically 45 mM in commercial fermented sausages. At these concentrations, we would expect the E. coli O157:H7 cells to be sensitized to the action of lysozyme. However, the contact time (1 min) with saliva and the concentration (0.1 μg/ml) of lysozyme have likely been insufficient to yield a noticeable inactivation of the pathogen.
Food is known to exert a buffering effect on the gastric content during human digestion, protecting enteric pathogens against the acidity of the gastric juice (41). The effect is dual and results from the interaction between the existing basic food moieties and the acids released in the stomach (31, 40), as well as from the hydrolysis of proteins by pepsin, with concurrent release of amines. This buffering effect was evidenced in the present study when gastric challenge was carried out under static conditions, with no pH adjustment, and resulted in a moderate decrease of acidity. As a result, E. coli O157:H7 cells are normally expected to be exposed to pH values between 3 and 4 in the early phases of gastric digestion, when ingested in a dry-sausage matrix, values at which the organism has been found to survive for several days in mayonnaise (42) and for at least 24 h in acidified TSB (32).
The buffering effect of food observed in early gastric challenge, which causes the postprandial rise reported in the scientific literature (31, 37), is later gradually overwhelmed by the progressive release of additional acid in the stomach, so that the gastric pH reaches its basal value of 2.0 about 2 h after the meal intake (31). In the meantime, gradual emptying of the stomach contents takes place, starting at about 30 min after ingestion, so that the E. coli O157:H7 cells contained in the contaminated dry sausages are exposed to acidic challenges of various intensity, depending on the length of time they remain in the stomach before being pushed towards the intestine.
The fact that most of E. coli O157:H7 cells can survive the severest gastric challenge (this study [dynamic digestion experiments]) is consistent with the results of various studies in which the resistance of the organism was evaluated in various synthetic media (5, 6, 32). Survival was likely ensured by the glutamate-dependent decarboxylase system, induced by the acidification (pH ∼ 5.0) resulting from glucose fermentation (8, 14, 17, 27, 28), since the cultures initially used to inoculate the sausage batter were prepared from overnight TSBG-grown E. coli O157:H7 cells. Because suitable sources of glutamate were present in the challenge medium as a result of meat hydrolysis by proteolytic enzymes, decarboxylation and subsequent proton consumption, required for maintaining a viable internal pH during exposure to extreme external acidity, were possible.
Survival of E. coli O157:H7 in the pancreatic juice (this study) was not surprising, since optimal pH conditions prevailed in this environment. Also, it is well known that the lipopolysaccharide layer of the outer membrane of gram-negative bacteria acts as a barrier for many detrimental external agents, resulting in an inherent resistance to the detergent action of bile salts and to degradation by digestive enzymes (i.e., pepsin, trypsin, and other enzymes contained in the pancreatin preparation) (2). Extrinsic factors, such as acquisition of a mucin surface coating (11) and entrapment in hydrophobic lipid moieties, such as reported for Salmonella enterica serovar Typhimurium in cheddar cheese (15), may also afford an additional physical barrier, shielding the microorganism from proteases. In the present study, the capacity of E. coli O157:H7 to further grow during the pancreatic challenge was strongly related to the duration and intensity of the previous gastric challenge. The cells that reached the intestinal compartment after a gastric exposure of no longer than 90 min (minimum pH, 2.5) were able to grow in the pancreatic juice. A lag phase (typically ca. 120 min) was, however, generally observed, during which the sublethal damages resulting from the manufacturing process and the gastric challenge were repaired. In contrast, the cells that had undergone the most severe gastric challenge (120 min; minimum pH, 2.0) could no longer grow in the pancreatic juice, even though they remain viable and could later be revived on a rich medium such as TSA.
The gastric and pancreatic juices have long been perceived as pH and enzymatic barriers to the survival of ingested microorganisms during the human digestive process (38). However, these juices offer no real challenge to E. coli O157:H7 cells ingested within contaminated dry sausages (this study). The knowledge that a large portion of these cells are transferred to the pancreatic compartment with sublethal damage that will be later repaired in the intestine to resume active growth comes in support to the previous finding that a few cells (10 to 100 cells/g) of ingested E. coli O157:H7 were sufficient to cause illness (1, 3, 4, 12, 28). From a food safety point of view, this implies that the industrial processes used to manufacture dry sausages must be designed in such a way that no viable E. coli O157:H7 cell can ever be found in an average portion of sausage for human consumption, because no additional protection will be afforded by the subsequent digestive process. This new information will be very valuable in refining our assessment of the risk associated with the manufacture of fermented dry sausages with regard to E. coli O157:H7.
Acknowledgments
We are grateful to Edward Farnworth and Isabelle Mainville for advice on the setup of the in vitro digestion experiments.
This work was made possible through a grant from the Conseil des Recherches en Pêche et en Agroalimentaire du Québec of the Government of Quebec.
REFERENCES
- 1.Abdul-Raouf, U. M., L. R. Beuchat, and M. S. Ammar. 1993. Survival and growth of Escherichia coli O157:H7 in ground beef as affected by pH, acidulant, and temperature. Appl. Environ. Microbiol. 59:2364-2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alakomi, H. L., E. Skyttä, M. Saarela, T. Mattila-Sandholm, K. Latva, and L. M. Helander. 2000. Lactic acid permeabilizes gram-negative bacteria by disrupting the outer membrane. Appl. Environ. Microbiol. 66:2001-2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anonymous. 1993. Report on the Escherichia coli O157:H7 outbreak in the western states. Food Safety and Inspection Service, U.S. Department of Agriculture, Washington, D.C.
- 4.Archer, D. L. 1996. Preservation microbiology and safety: evidence that stress enhances virulence and triggers adaptive mutations. Trends Food Sci. Technol. 7:91-95. [Google Scholar]
- 5.Arnold, K. W., and C. W. Kaspar. 1995. Starvation and stationary-phase induced acid tolerance in Escherichia coli O157:H7. Appl. Environ. Microbiol. 61:2037-2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benjamin, M. M., and A. R. Datta. 1995. Acid tolerance of enterohemorrhagic Escherichia coli. Appl. Environ. Microbiol. 61:1669-1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Besser, R. E., S. M., Lett, J. T. Weber, M. P. Doyle, T. J. Barrett, J. G. Wells, and P. M. Griffin. 1993. An outbreak of diarrhea and hemolytic uremic syndrome from Escherichia coli O157:H7 in fresh-pressed apple cider. JAMA 269:2217-2221. [PubMed] [Google Scholar]
- 8.Castanie-Cornet, M.-P., T. A. Penfound, D. Smith, J. F. Elliott, and W. Foster. 1999. Control of acid resistance in Escherichia coli. J. Bacteriol. 181:3525-3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Centers for Disease Control. 1993. Update: multistate outbreak of Escherichia coli O157:H7 infections from hamburgers—western United States, 1992-1993. Morb. Mortal. Wkly. Rep. 42:258-263. [PubMed] [Google Scholar]
- 10.Centers for Disease Control. 1995. Escherichia coli O157:H7 outbreak linked to commercially distributed dry-cured salami—Washington and California, 1994. Morb. Mortal. Wkly. Rep . 44:157-160. [PubMed] [Google Scholar]
- 11.Charteris, W. P., P. M. Kelly, L. Morelli, and J. K. Collins. 1998. Development and application of an in vitro methodology to determine the transit tolerance of potentially probiotic Lactobacillus and Bifidobacterium species in the upper human gastrotestinal tract. J. Appl. Microbiol. 84:759-768. [DOI] [PubMed] [Google Scholar]
- 12.Choi, S. H., D. J. Baumler, and C. W. Kaspar. 2000. Contribution of dps acid stress tolerance and oxydative stress tolerance in Escherichia coli O157:H7. Appl. Environ. Microbiol. 66:3911-3916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cornelius, C., C. Lentren, and A. Wink (ed.) 1981. Geigy scientific tables. Units of measurement. Body fluids, composition of the body, p. 114-122. In Nutrition, vol. 1. CIBA-GEIGY Limited, Basel, Switzerland.
- 14.Cui, S., J. Meng, and A. A. Bhagwat. 2001. Availability of glutamate and arginine during acid challenge determines cell density-dependent survival phenotype of Escherichia coli strains. Appl. Environ. Microbiol. 67:4914-4918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.D'Aoust, J. Y. 1985. Infective dose of Salmonella Typhimurium in cheddar cheese. Am. J. Epidemiol. 122:717-719. [DOI] [PubMed] [Google Scholar]
- 16.Drouault, S., G. Corthier, S. D. Ehrlish, and P. Renault. 1999. Survival, physiology and lysis of Lactococcus lactis in the digestive tract. Appl. Environ. Microbiol. 65:4881-4886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Foster, J. W. 2000. Microbial responses to acid stress, p. 99-115. In G. Storz and R. Hegge-Aronis (ed.), Bacterial stress responses. ASM Press, Washington, D.C.
- 18.García-Graells, C., B. Masschalck, and C. W. Michiels. 1999. Inactivation of Escherichia coli in milk by high hydrostatic pressure treatment in combination with antimicrobial peptides. J. Food Prot. 62:1248-1254. [DOI] [PubMed] [Google Scholar]
- 19.Gauthier, S. F., C. Vachon, and L. Savoie. 1986. Enzymatic conditions of an in vitro method to study protein digestion. J. Food Sci. 51:960-964. [Google Scholar]
- 20.Glass, K. A., J. M. Loeffelholz, J. P. Ford, and M. P. Doyle. 1992. Fate of Escherichia coli O157:H7 as affected by pH or sodium chloride and in fermented, dry sausage. Appl. Environ. Microbiol. 58:2513-2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gorden, J., and P. L. C. Small. 1992. Acid resistance in enteric bacteria. Infect. Immun. 61:364-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Griffin, P. M., and R. V. Tauxe. 1991. The epidemiology of infections caused by Escherichia coli O157:H7, other enterohemorrhagic E. coli, and the associated hemolytic uremic syndrome. Epidemiol. Rev. 13:60-97. [DOI] [PubMed] [Google Scholar]
- 23.Hauben, K. E. Wuytack, C. Soontjens, and C. W. Michiels. 1996. High pressure transient sensitization of Escherichia coli to lysozyme and nisin by disruption of outer membrane permeability. J. Food Prot. 59:350-355. [DOI] [PubMed] [Google Scholar]
- 24.Hinkens, J. C., N. G. Faith, T. D. Lorang, P. Bailey, D. Buege, C. W. Kaspar, and J. B. Luchanski. 1996. Validation of pepperoni processes for control of Escherichia coli O157:H7. J. Food Prot. 59:1260-1266. [DOI] [PubMed] [Google Scholar]
- 25.Hunt, B. J. N., and W. R. Spurrell. 1951. The pattern of emptying the human stomach. J. Physiol. 113:157-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leistner, L. 1994. Stable and safe fermented sausages world-wide, p. 160-175. In G. Campbell-Platt and P. E. Cook (ed.), Fermented meats. Blackie Academic and Professional, New York, N.Y.
- 27.Lin, J., I. S. Lee, J. Frey, J. L. Slonezewski, and J. W. Foster. 1995. Comparative analysis of extreme acid survival in Salmonella typhimurium, Shigella flexneri, and Escherichia coli. J. Bacteriol. 177:4097-4104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin, J., M. P. Smith, K. C. Chapin, H. S. Maik, G. N. Bennett, and J. W. Foster. 1996. Mechanisms of acid resistance in enterohemorrhagic Escherichia coli. Appl. Environ. Microbiol. 62:3094-3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Masschalck, B., D. Deckers, and C. Michiels. 2002. Lytic and nonlytic mechanism of inactivation of gram-positive bacteria by lysozyme under atmospheric and high hydrostatic pressure. J. Food Prot. 65:1916-1923. [DOI] [PubMed] [Google Scholar]
- 30.McCleery, D. R., and M. T. Rowe. 1995. Development of a selective plating technique for the recovery of Escherichia coli O157:H7 after heat stress. Lett. Appl. Microbiol. 21:252-256. [DOI] [PubMed] [Google Scholar]
- 31.McLauchlan, G., G. M. Fullarton, G. P. Crean, and K. E. McColl. 1989. Comparison of gastric body and antral pH: a 24 hour ambulatory study in healthy volunteers. Gut 30:573-578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miller, L. G., and C. W. Kaspar. 1994. Escherichia coli O157:H7. Acid tolerance and survival in apple cider. J. Food Prot. 57:460-464. [DOI] [PubMed] [Google Scholar]
- 33.Molly, K., M. V. Woestyne, I. De Smet, and W. Verstraete. 1994. Validation of the simulator of the human intestinal microbial ecosystem (SHIME) reactor using microorganisms-associated activities. Microb. Ecol. Health Dis. 7:191-200. [Google Scholar]
- 34.Naim, F., S. Messier, L. Saucier, and G. Piette. 2003. A model study of Escherichia coli O157: H7 survival in fermented dry sausages—influence of inoculum preparation, inoculation procedure and selected process parameters. J. Food Prot. 66:2267-2275. [DOI] [PubMed] [Google Scholar]
- 35.O'Brien, A. D., and J. B. Kaper. 1998. Shiga toxin-producing Escherichia coli: yesterday, today, and tomorrow, p. 1-11. In J. B. Kaper and A. D. O'Brien (ed.), Escherichia coli O157:H7 and other Shiga toxin-producing E. coli strains. American Society for Microbiology, Washington, D.C.
- 36.Salyers, Abigail, A., and D. D. Whitt. 1994. Bacterial pathogenesis: a molecular approach. ASM Press, Washington, D.C.
- 37.Savoie, L. 1993. Digestion and absorption of food: usefulness and limitations of in vitro models. Can J. Physiol. Pharmacol. 72:407-414. [DOI] [PubMed] [Google Scholar]
- 38.Smith, J. L. 2003. The role of gastric acid in preventing foodborne disease and how bacteria overcome acid conditions. J. Food Prot. 66:1292-1303. [DOI] [PubMed] [Google Scholar]
- 39.Söderholm, K. J. M. R. Mukherjee, and J. Longmate. 1996. Filter leachability of composites stored in distilled water or artificial saliva. J. Dent. Res. 75:1692-1699. [DOI] [PubMed] [Google Scholar]
- 40.van Herwaarden, M. A., A. Samson, and A. J. P. M Smout. 1999. 24-h recording of intragastric pH: technical aspects and clinical relevance. Scand. J. Gastroenterol. 34(Suppl. 230):9-16. [PubMed] [Google Scholar]
- 41.Waterman, S. R., and P. L. C. Small. 1998. Acid-sensitive enteric pathogens are protected from killing under extreme acidic conditions of pH 2.5 when they are inoculated onto certain solid food sources. Appl. Environ. Microbiol. 64:3882-3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhao, T., and M. P. Doyle. 1994. Fate of enterohemorrhagic Escherichia coli O157:H7 in commercial mayonnaise. J. Food Prot. 57:2526-2530. [DOI] [PubMed] [Google Scholar]
- 43.Zhao, T., M. P. Doyle, and R. E. Besser. 1993. Fate of enterohemorrhagic Escherichia coli O157:H7 in apple cider with or without preservatives. App. Environ. Microbiol. 59:2526-2530. [DOI] [PMC free article] [PubMed] [Google Scholar]