Abstract
Bartonella schoenbuchensis, which commonly causes bacteremia in ruminants, was isolated from the deer ked Lipoptena cervi and was shown to localize to the midgut of this blood-sucking arthropod, causing deer ked dermatitis in humans. The role of B. schoenbuchensis in the etiology of deer ked dermatitis should be further investigated.
The deer ked (Lipoptena cervi) is a common hematophagous louse fly of red deer, roe deer, elk, and sika deer in Europe, Siberia, and northern China and of white-tailed deer, elk, horses, and cattle in North America. The incidental infestation of humans with deer keds is well documented (1, 11, 12). In humans, these ectoparasites engorge on blood in 15 to 25 min. The bite is barely noticeable and initially leaves little trace. Within 3 days, the site develops into a hard, reddened welt. The accompanying itch is intense and typically lasts 14 to 20 days; occasionally, a pruritic papule may persist even for 1 year (6, 14, 15). Histological analysis of deer ked dermatitis lesions revealed C3 deposits in dermal vessels. Skin tests with deer ked whole-body extracts were positive in all patients tested, showing both immediate and delayed reactions. Moreover, 57% of the patients tested had elevated serum immunoglobulin E (IgE) levels. These findings suggest that complement, cell-mediated immune mechanisms, and IgE are involved in the pathogenesis of deer ked dermatitis (14).
While the etiological agent of this disease is unknown, all available data are consistent with the transmission of an infectious agent, either a bacterium or a parasite, through the bite of the deer ked. Deer keds have not been known to transmit any infectious agents to humans. Recent evidence has shown that several ruminant hosts of deer keds are frequently bacteremic for Bartonella spp. Bartonellae are gram-negative bacteria that have been isolated from the blood of a wide range of mammals, including humans, rodents, lagomorphs, carnivores, and ruminants (4). These hemotropic bacteria are increasingly being recognized as important human pathogens (10, 13). Until the 1990s, only Bartonella quintana (causing trench fever) and B. bacilliformis (causing Carrion's disease) were known to cause disease in humans. Since then, six additional Bartonella spp. have been associated with an increasing range of clinical manifestations, reflecting the expansion of the genus Bartonella to the currently described 20 species (7, 10, 13).
Bartonellae are transmitted by various arthropods, such as lice, fleas, and flies. Indeed, all six recently identified Bartonella spp. pathogenic for humans are anthropozoonotic agents, which are thought to be transmitted to humans by blood-sucking arthropods. Little is known about the risk of zoonosis from Bartonella spp. infecting ruminant hosts of deer keds. Recently, B. schoenbuchensis bacteremia was demonstrated in ∼80% of the roe deer analyzed in Germany (9). Given the close association of the deer ked with its ruminant hosts, including regular blood meals, and the incidental infestation of humans with this arthropod, the deer ked could serve as a vector for the transmission of B. schoenbuchensis within ruminants and to humans. The purpose of this study was to analyze whether deer keds collected from wild ruminants are colonized with B. schoenbuchensis; such colonization should represent a prerequisite for transmitting this pathogen to ruminant or human hosts.
From December 2001 to November 2002, 49 deer keds were collected from the fur of seven roe deer and eight red deer shot at three different locations in Germany (Table 1). Following surface sterilization by immersion in 70% ethanol for 5 min, 30 deer keds were individually homogenized in phosphate-buffered saline and cultured on Columbia agar containing 5% defibinated sheep blood. Following incubation at 37°C in a humidified atmosphere containing 5% CO2, 8 deer keds (27%) did not give rise to bacterial growth, while large numbers of bacterial colonies (>1,000 per deer ked) appeared on the plates for 22 deer keds (73%) after 4 to 6 days and continued to grow for several days. Except for a few instances of low-titer contamination with fast-growing bacteria (which may have resulted from incomplete surface sterilization), the uniform slow growth and colony phenotype suggested the isolation of a single bacterial species at a high titer. The growth characteristics and colony phenotype were consistent with B. schoenbuchensis, which was previously found at a similar prevalence in the blood of roe deer (80%) (3, 9). Deer keds were culture positive for seven out of eight roe deer (88%) and five out of seven red deer (71%). Interestingly, when several deer keds isolated from the same deer were analyzed, they were found to be either sterile or culture positive at a high titer (Table 1), suggesting that bacteremia of a deer results in infection of the infesting deer ked population.
TABLE 1.
Origins of L. cervi samples and Bartonella sp. isolates
| L. cervi sample | Host (deer) | Origin of host in 2002 | No. of flies collected | No. of culture-positive flies/no. tested | Bartonella sp. isolate | GenBank accession no. |
|---|---|---|---|---|---|---|
| 1 | Roe | Southern Black Forest | 2 | 2/2 | Dlf01 | AJ564632 |
| 9 | Roe | Southern Black Forest | 8 | 0/4 | ||
| 10 | Roe | Southern Black Forest | 10 | 2/2 | Dlf10 | AJ278183 |
| 11 | Roe | Southern Black Forest | 1 | 2/2 | Dlf11 | AJ278183 |
| 12 | Roe | Southern Black Forest | 5 | 2/2 | Dlf12 | AJ278183 |
| 13 | Roe | Harz | 1 | 1/1 | Dlf13 | AJ278183 |
| 14 | Roe | Harz | 1 | 1/1 | Dlf14 | AJ278183 |
| 15 | Red | Harz | 5 | 2/2 | Dlf15 | AJ564633 |
| 16 | Red | Harz | 2 | 0/2 | ||
| 17 | Red | Harz | 2 | 2/2 | Dlf17 | AJ564634 |
| 18 | Red | Harz | 2 | 0/2 | ||
| 21 | Red | Schoenbuch Nature Park | 3 | 3/3 | Dlf21 | AJ564634 |
| 22 | Red | Schoenbuch Nature Park | 5 | 3/3 | Dlf22 | AJ564635 |
| 23 | Red | Schoenbuch Nature Park | 1 | 1/1 | Dlf23 | |
| 24 | Roe | Schoenbuch Nature Park | 1 | 1/1 | Dlf24 | AJ278183 |
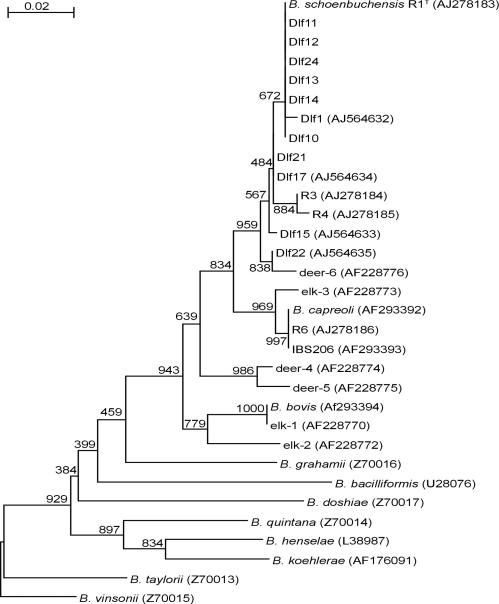
One bacterial isolate from each of the deer infested with culture-positive deer keds (except for sample 23) was subjected to DNA sequence-based typing to the species level. For this purpose, a partial fragment of the gltA gene was amplified by PCR with genus-specific primers and subjected to DNA sequence analysis as described previously (9). The partial gltA sequences of strains Dlf10, Dlf11, Dlf12, Dlf13, Dlf14, Dlf21, and Dlf24 were identical to the gltA sequence of B. schoenbuchensis type strain R1 (9), while those of strains Dlf01, Dlf15, Dlf17, and Dfl21 were found to carry several base pair substitutions (Table 1 and Fig. 1).
FIG. 1.
Neighbor-joining tree of partial gltA sequences. The tree is based on the complete aligment of 374-bp fragments (GenBank accession numbers are indicated in the tree) corresponding to bp 698 to 973 of the partial gltA sequence of B. schoenbuchensis (GenBank accession no. AJ278183). Bootstrap values resulting from 1,000 bootstrap trials are indicated for each major branch. The B. vinsonii sequence (GenBank accession no. Z70015) that was most divergent from the B. schoenbuchensis sequence was used as an outgroup.
Using CLUSTAL_X (16) and njplot software, a neighbor-joining tree was generated from the complete alignment of 374-bp gltA fragments from all deer ked isolates described here as well as from closely related Bartonella isolates and Bartonella spp. deposited in GenBank (Fig. 1). All deer ked isolates clustered within one clade together with B. schoenbuchensis strains R1T, R3, and R4 from roe deer (9) and deer isolate deer-6. The other clade belonging to the node includes B. capreoli isolates from roe deer; this species is most closely related to B. schoenbuchensis (3, 9). The next most related clades include either deer isolates (deer-4 and deer-5) or B. bovis, isolated from a bovine, together with isolates from elk (elk-1 and elk-2); all of these are ruminants known to be infested by deer keds. Together, these data suggest that the 11 deer ked isolates analyzed all belong to the species B. schoenbuchensis, which is closely related to other Bartonella spp. and to isolates obtained from ruminant hosts of deer keds.
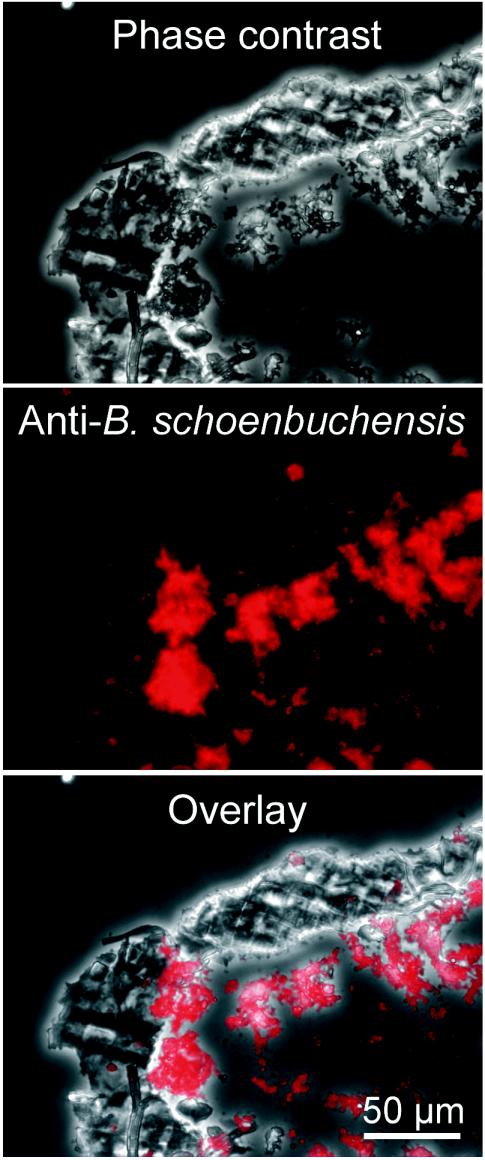
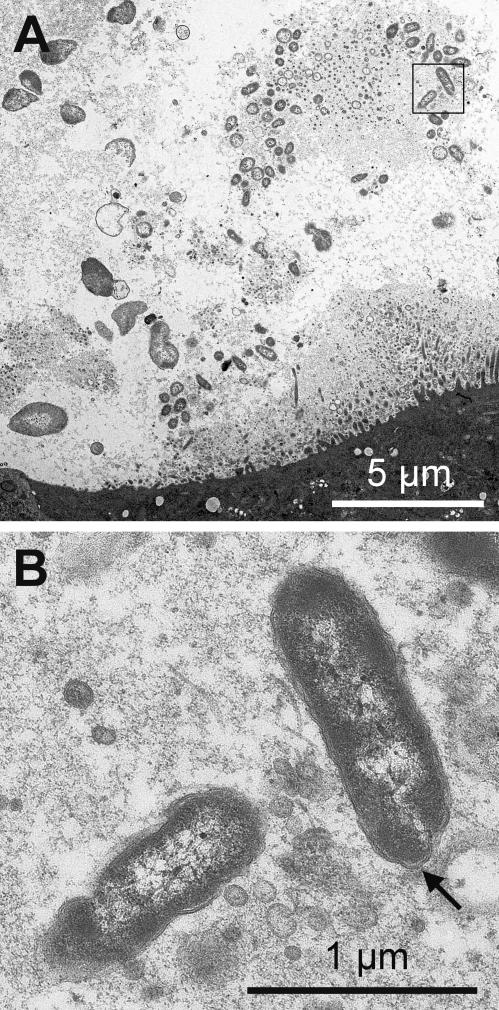
Immunohistochemical analysis allowed us to localize B. schoenbuchensis within deer keds. Cryosectioning of intact insects fixed in phosphate-buffered saline containing 3% formaldehyde and 0.5% glutaraldeyde for 30 min, followed by indirect immunofluorescence labeling with rat anti-B. schoenbuchensis serum and Cy3-labeled goat anti-rat IgG antibodies and fluorescence microscopy analysis, revealed single bacteria as well as large bacterial aggregates in the lumen of the midgut (Fig. 2). For ultrastructural analysis, the fixed midgut was postfixed with 1% OsO4 for 1 h, dehydrated, stained with 70% ethanol and 2% uranyl acetate for 1 h, and embedded in Epon. Ultrathin sections were stained with 6% uranyl acetate for 1 h and lead acetate for 2 min. Transmission electron microscopy confirmed the presence of bacterial aggregates in the midgut (Fig. 3A) and revealed typical characteristics of the gram-negative cell wall (Fig. 3B).
FIG. 2.
Immunolocalization of B. schoenbuchensis in the midgut lumen of L. cervi. Cryosections of L. cervi (sample 12) were stained with rat anti-B. schoenbuchensis serum and then with Cy3-labeled anti-rat IgG antibodies. Digital images of the specimens were recorded with phase-contrast bright-field optics (top) and with fluorescence excitation and detection with a tetramethyl rhodamine isothiocyanate fluorescence filter set (middle). (Bottom) Overlay of phase-contrast and fluorescence images.
FIG. 3.
Ultrastructural analysis of B. schoenbuchensis in the midgut lumen of L. cervi. Transmission electron micrographs of thin sections from the midgut of L. cervi (sample 12) were recorded at magnifications of ×2,000 (A) and ×16,000 (B). The image shown in panel B represents an enlargement of the inset in panel A. The electron-dense tissue in the lower part of panel A represents the midgut epithelium. The arrow in panel B indicates a typical aspect of the gram-negative cell wall.
Taken together, our findings of high-titer cultivation of B. schonbuchensis from surface-sterilized deer keds and the large bacterial aggregates observed in the midgut indicate that these arthropods represent a natural reservoir supporting the replication of this pathogen. With respect to lifestyle, the deer ked, which infests ruminants for most of its life and takes regular blood meals, appears to represent an ideal vector for efficient transmission of B. schoenbuchensis within ruminant populations. Given that deer keds incidentally take blood meals from humans, the risk for occasional transmission of B. schoenbuchensis to humans is apparent, e.g., to hunters, forestry workers, and cross-country runners. This conclusion raises the important question as to whether B. schoenbuchensis may be involved in the formation of deer ked dermatitis. Interestingly, among the known Bartonella spp., B. schoenbuchensis is most closely related to B. bacilliformis (9), an important human pathogen, which is the only other Bartonella sp. for which the vector is known to be a fly, the sandfly Lutzomyia verrucarum (5). B. schoenbuchensis strains display considerable heterogeneity, e.g., in their gltA sequences (9), a finding supported by the identification of five additional variants in this study (Table 1 and Fig. 1). Some variant strains may thus bear a larger anthropozoonotic risk than others (9).
Interestingly, the clinical scenario of deer ked dermatitis resembles a primary manifestation of cat scratch disease, caused by B. henselae. This globally distributed anthropozoonotic pathogen is transmitted from cats to humans by the bite of an infected cat flea or, alternatively, by direct contact with cats (i.e., cat scratch or bite). After a 3- to 10-day incubation period, an erythematous papule or pustule develops at the site of inoculation and regresses after 2 to 8 weeks. This primary lesion of cat scratch disease is reminiscent of the clinical scenario of deer ked dermatitis (2), while other manifestations of cat scratch disease, i.e., lymphadenopathy, are not observed in deer ked dermatitis. A positive delayed-type hypersensitivity skin test, like that characteristically observed for B. henselae antigens in cat scratch disease (2), was also reported for all cases of deer ked dermatitis when whole deer ked extracts were used for the skin test (14). Also, C3 deposits in dermal vessels like those described for deer ked dermatitis (14) are consistent with infection by vasculotropic bartonellae (8). Taken together, certain clinical and histological characteristics of deer ked dermatitis are reminiscent of human infection by bartonellae, indicating that these pathogens should be considered possible etiological agents of deer ked dermatitis.
In summary, our study has provided evidence that deer keds collected from roe deer and red deer in Germany are commonly infected by B. schoenbuchensis. Furthermore, we have shown that B. schoenbuchensis colonizes the midgut of these arthropods and that this pathogen can be cultured at high titers from surface-sterilized arthropods. Our data suggest an important risk for the transmission of B. schoenbuchensis or related bartonellae to humans by the bite of an infected deer ked and suggest that a potential role of bartonellae in the etiology of deer ked dermatitis should be investigated further.
Nucleotide sequence accession numbers.
GenBank accession numbers for the sequences determined here and sequences used for comparisons are indicated in Table 1 and Fig. 1.
Acknowledgments
We thank Charles Thompson for critical reading of the manuscript. We are grateful to Karl Ebert and Rainer Pohl (Staatliches Forstamt Tuebingen-Bebenhausen, Schoenbuch Nature Park, Baden-Wuerttemberg, Germany), Dieter Bitter-Surmann (Hunting District Herzberger Grafenforst, Herzberg im Harz, Lower Saxony, Germany), and Rainer Skierka (Hunting District Hornberg/Obergebisbach, Southern Black Forest, Baden-Wuerttemberg, Germany) for permission to collect deer keds from roe deer and red deer which were shot in their hunting districts. Christopher Snyder is acknowledged for assistance in immunization experiments.
This work was supported by a grant from the Swiss National Science Foundation (3100-061777.00/1) to C.D.
REFERENCES
- 1.Alekseev, E. A. 1985. Initial experience with individual human protection from attack by the deer louse fly Lipoptena cervi. Med. Parazitol. 1985:56-57. (In Russian.) [PubMed] [Google Scholar]
- 2.Anderson, B., C. Kelly, R. Threlkel, and K. Edwards. 1993. Detection of Rochalimaea henselae in cat-scratch disease skin test antigens. J Infect. Dis. 168:1034-1036. [DOI] [PubMed] [Google Scholar]
- 3.Bermond, D., H. J. Boulouis, R. Heller, G. Van Laere, H. Monteil, B. B. Chomel, A. Sander, C. Dehio, and Y. Piemont. 2002. Bartonella bovis Bermond et al. sp. nov. and Bartonella capreoli sp. nov., isolated from European ruminants. Int J. Syst. Evol. Microbiol. 52:383-390. [DOI] [PubMed] [Google Scholar]
- 4.Breitschwerdt, E. B., and D. L. Kordick. 2000. Bartonella infection in animals: carriership, reservoir potential, pathogenicity, and zoonotic potential for human infection. Clin. Microbiol. Rev. 13:428-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caceres, A. G. 1993. Geographic distribution of Lutzomyia verrucarum (Townsend, 1913) (Diptera, Psychodidae, Phlebotominae), vector of human bartonellosis in Peru. Rev. Inst. Med. Trop. São Paulo 35:485-490. [DOI] [PubMed] [Google Scholar]
- 6.Chitiakov, A. F. 1968. Skin lesions in people due to bites of Lipoptena cervi. Vestn. Dermatol. Venerol. 42:59-62. [PubMed] [Google Scholar]
- 7.Dehio, C. 2004. Molecular and cellular basis of Bartonella pathogenesis. Annu. Rev. Microbiol. 58:365-390. [DOI] [PubMed] [Google Scholar]
- 8.Dehio, C. 2003. Recent progress in understanding Bartonella-induced vascular proliferation. Curr. Opin. Microbiol. 6:61-65. [DOI] [PubMed] [Google Scholar]
- 9.Dehio, C., C. Lanz, R. Pohl, P. Behrens, D. Bermond, Y. Piemont, K. Pelz, and A. Sander. 2001. Bartonella schoenbuchii sp. nov., isolated from the blood of wild roe deer. Int. J. Syst. Evol. Microbiol. 51:1557-1565. [DOI] [PubMed] [Google Scholar]
- 10.Dehio, C., and A. Sander. 1999. Bartonella as emerging pathogens. Trends Microbiol. 7:226-228. [DOI] [PubMed] [Google Scholar]
- 11.Gothe, R., and H. Schol. 1994. Deer keds (Lipoptena cervi) in the accompanying equipment of the late Neolithic human mummy from the Similaun, South Tyrol. Parasitol. Res. 80:81-83. [DOI] [PubMed] [Google Scholar]
- 12.Ivanov, V. I. 1975. Anthropophilia of the deer blood sucker Lipoptena cervi L. (Diptera, Hippoboscidae). Med. Parazitol. 44:491-495. [PubMed] [Google Scholar]
- 13.Jacomo, V., P. J. Kelly, and D. Raoult. 2002. Natural history of Bartonella infections (an exception to Koch's postulate). Clin. Diagn. Lab. Immunol. 9:8-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rantanen, T., T. Reunala, P. Vuojolahti, and W. Hackman. 1982. Persistent pruritic papules from deer ked bites. Acta Dermatol. Venereol. 62:307-311. [PubMed] [Google Scholar]
- 15.Reunala, T., T. Rantanen, P. Vuojolahti, and W. Hackman. 1980. Deer ked dermatitis. Duodecim 96:897-902. [PubMed] [Google Scholar]
- 16.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]