Abstract
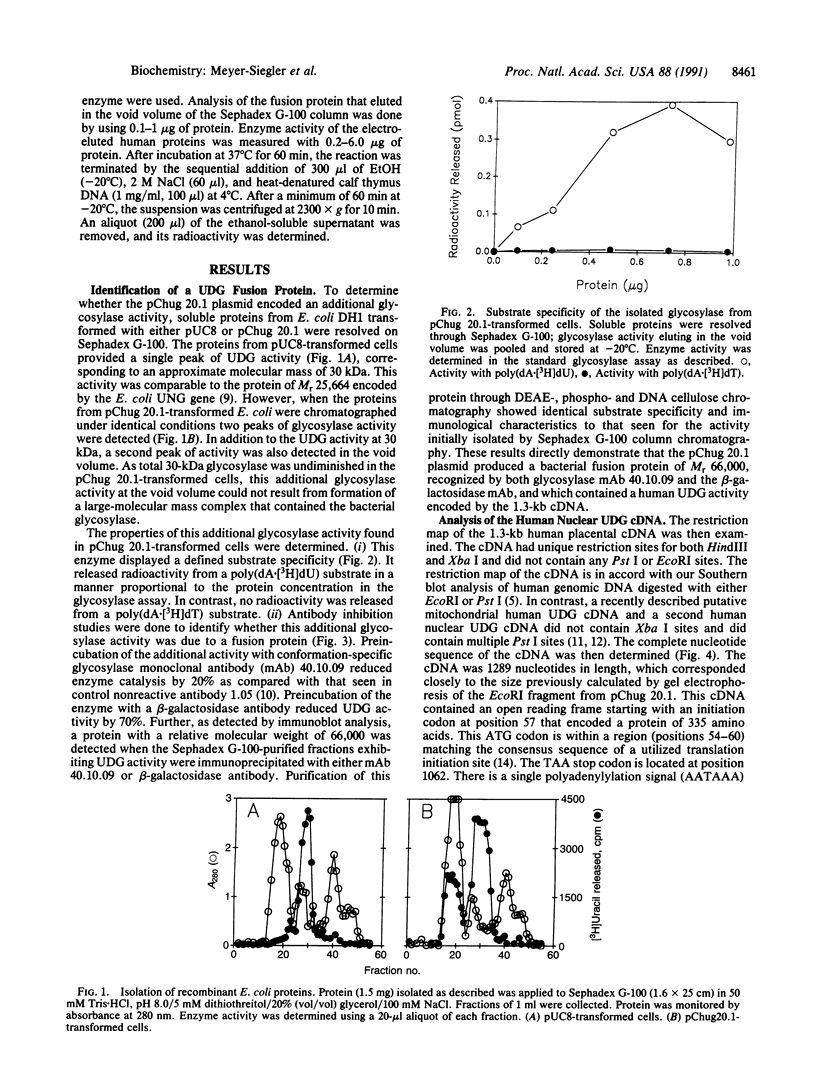
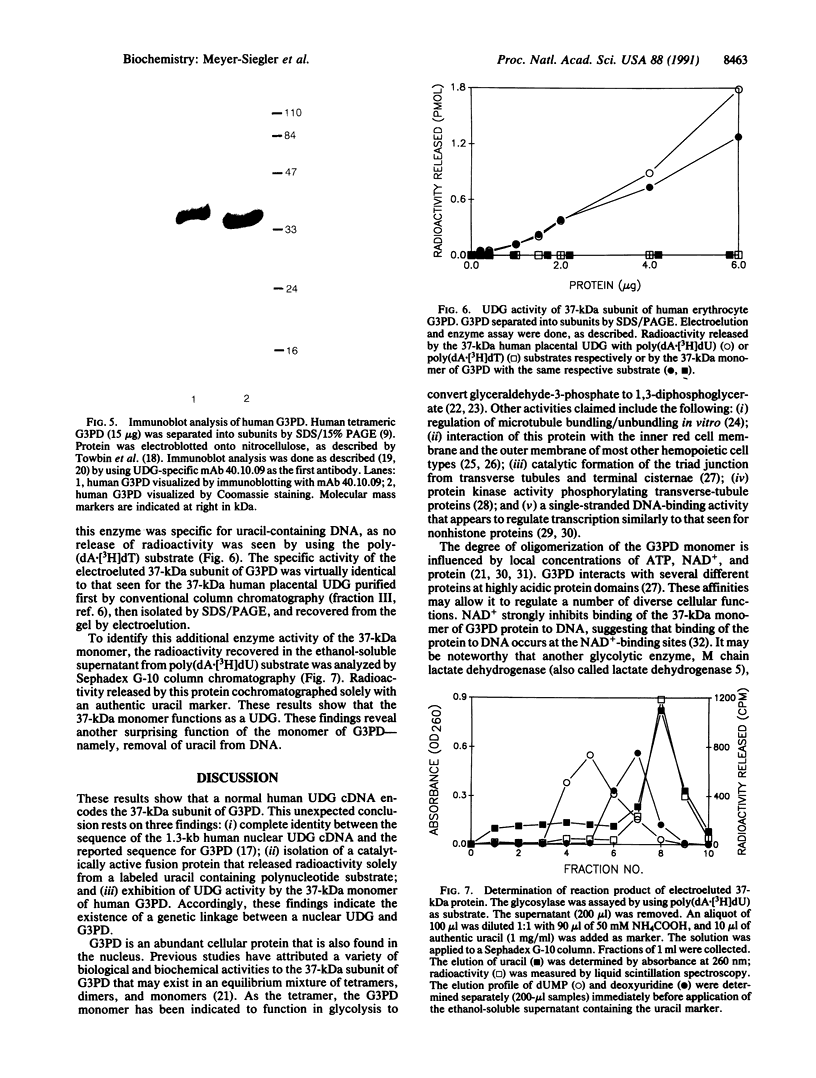
We have isolated and characterized a plasmid (pChug 20.1) that contains the cDNA of a nuclear uracil DNA glycosylase (UDG) gene isolated from normal human placenta. This cDNA directed the synthesis of a fusion protein (Mr 66,000) that exhibited UDG activity. The enzymatic activity was specific for a uracil-containing polynucleotide substrate and was inhibited by a glycosylase antibody or a beta-galactosidase antibody. Sequence analysis demonstrated an open reading frame that encoded a protein of 335 amino acids of calculated Mr 36,050 and pI 8.7, corresponding to the Mr 37,000 and pI 8.1 of purified human placental UDG. No homology was seen between this cDNA and the UDG of herpes simplex virus, Escherichia coli, and yeast; nor was there homology with the putative human mitochondrial UDG cDNA or with a second human nuclear UDG cDNA. Surprisingly, a search of the GenBank data base revealed that the cDNA of UDG was completely homologous with the 37-kDa subunit of human glyceraldehyde-3-phosphate dehydrogenase. Human erythrocyte glyceraldehyde-3-phosphate dehydrogenase was obtained commercially in its tetrameric form. A 37-kDa subunit was isolated from it and shown to possess UDG activity equivalent to that seen for the purified human placental UDG. The multiple functions of this 37-kDa protein as here and previously reported indicate that it possesses a series of activities, depending on its oligomeric state. Accordingly, mutation(s) in the gene of this multifunctional protein may conceivably result in the diverse cellular phenotypes of Bloom syndrome.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allen R. W., Trach K. A., Hoch J. A. Identification of the 37-kDa protein displaying a variable interaction with the erythroid cell membrane as glyceraldehyde-3-phosphate dehydrogenase. J Biol Chem. 1987 Jan 15;262(2):649–653. [PubMed] [Google Scholar]
- Arcari P., Martinelli R., Salvatore F. The complete sequence of a full length cDNA for human liver glyceraldehyde-3-phosphate dehydrogenase: evidence for multiple mRNA species. Nucleic Acids Res. 1984 Dec 11;12(23):9179–9189. doi: 10.1093/nar/12.23.9179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arenaz P., Sirover M. A. Isolation and characterization of monoclonal antibodies directed against the DNA repair enzyme uracil DNA glycosylase from human placenta. Proc Natl Acad Sci U S A. 1983 Oct;80(19):5822–5826. doi: 10.1073/pnas.80.19.5822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessman M. J., Lehman I. R., Adler J., Zimmerman S. B., Simms E. S., Kornberg A. ENZYMATIC SYNTHESIS OF DEOXYRIBONUCLEIC ACID. III. THE INCORPORATION OF PYRIMIDINE AND PURINE ANALOGUES INTO DEOXYRIBONUCLEIC ACID. Proc Natl Acad Sci U S A. 1958 Jul 15;44(7):633–640. doi: 10.1073/pnas.44.7.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caswell A. H., Corbett A. M. Interaction of glyceraldehyde-3-phosphate dehydrogenase with isolated microsomal subfractions of skeletal muscle. J Biol Chem. 1985 Jun 10;260(11):6892–6898. [PubMed] [Google Scholar]
- Chan J. Y., Becker F. F., German J., Ray J. H. Altered DNA ligase I activity in Bloom's syndrome cells. Nature. 1987 Jan 22;325(6102):357–359. doi: 10.1038/325357a0. [DOI] [PubMed] [Google Scholar]
- Constantinides S. M., Deal W. C., Jr Reversible dissociation of tetrameric rabbit muscle glyceraldehyde 3-phosphate dehydrogenase into dimers or monomers by adenosine triphosphate. J Biol Chem. 1969 Oct 25;244(20):5695–5702. [PubMed] [Google Scholar]
- Dehazya P., Sirover M. A. Regulation of hypoxanthine DNA glycosylase in normal human and Bloom's syndrome fibroblasts. Cancer Res. 1986 Aug;46(8):3756–3761. [PubMed] [Google Scholar]
- Duester G., Jörnvall H., Hatfield G. W. Intron-dependent evolution of the nucleotide-binding domains within alcohol dehydrogenase and related enzymes. Nucleic Acids Res. 1986 Mar 11;14(5):1931–1941. doi: 10.1093/nar/14.5.1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FOX J. B., Jr, DANDLIKER W. B. A study of some of the physical properties of glyceraldehyde-3-phosphate dehydrogenase. J Biol Chem. 1956 Jan;218(1):53–57. [PubMed] [Google Scholar]
- Gupta P. K., Sirover M. A. Altered temporal expression of DNA repair in hypermutable Bloom's syndrome cells. Proc Natl Acad Sci U S A. 1984 Feb;81(3):757–761. doi: 10.1073/pnas.81.3.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayatsu H. Co-operative mutagenic actions of bisulfite and nitrogen nucleophiles. J Mol Biol. 1977 Sep;115(1):19–31. doi: 10.1016/0022-2836(77)90243-1. [DOI] [PubMed] [Google Scholar]
- Huitorel P., Pantaloni D. Bundling of microtubules by glyceraldehyde-3-phosphate dehydrogenase and its modulation by ATP. Eur J Biochem. 1985 Jul 15;150(2):265–269. doi: 10.1111/j.1432-1033.1985.tb09016.x. [DOI] [PubMed] [Google Scholar]
- Kawamoto R. M., Caswell A. H. Autophosphorylation of glyceraldehydephosphate dehydrogenase and phosphorylation of protein from skeletal muscle microsomes. Biochemistry. 1986 Feb 11;25(3):657–661. doi: 10.1021/bi00351a022. [DOI] [PubMed] [Google Scholar]
- Kenne K., Ljungquist S. Expression of a DNA-ligase-stimulatory factor in Bloom's syndrome cell line GM1492. Eur J Biochem. 1988 Jun 15;174(3):465–470. doi: 10.1111/j.1432-1033.1988.tb14121.x. [DOI] [PubMed] [Google Scholar]
- Kim S., Vollberg T. M., Ro J. Y., Kim M., Sirover M. A. O6-methylguanine methyltransferase increases before S phase in normal human cells but does not increase in hypermutable Bloom's syndrome cells. Mutat Res. 1986 Feb;173(2):141–145. doi: 10.1016/0165-7992(86)90092-8. [DOI] [PubMed] [Google Scholar]
- Kozak M. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell. 1986 Jan 31;44(2):283–292. doi: 10.1016/0092-8674(86)90762-2. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lin T., Allen R. W. Isolation and characterization of a 37,000-dalton protein associated with the erythrocyte membrane. J Biol Chem. 1986 Apr 5;261(10):4594–4599. [PubMed] [Google Scholar]
- Mezzina M., Nardelli J., Nocentini S., Remault G., Sarasin A. DNA ligase activity in human cell lines from normal donors and Bloom's syndrome patients. Nucleic Acids Res. 1989 Apr 25;17(8):3091–3106. doi: 10.1093/nar/17.8.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgenegg G., Winkler G. C., Hübscher U., Heizmann C. W., Mous J., Kuenzle C. C. Glyceraldehyde-3-phosphate dehydrogenase is a nonhistone protein and a possible activator of transcription in neurons. J Neurochem. 1986 Jul;47(1):54–62. doi: 10.1111/j.1471-4159.1986.tb02830.x. [DOI] [PubMed] [Google Scholar]
- Muller S. J., Caradonna S. Isolation and characterization of a human cDNA encoding uracil-DNA glycosylase. Biochim Biophys Acta. 1991 Feb 16;1088(2):197–207. doi: 10.1016/0167-4781(91)90055-q. [DOI] [PubMed] [Google Scholar]
- Olsen L. C., Aasland R., Wittwer C. U., Krokan H. E., Helland D. E. Molecular cloning of human uracil-DNA glycosylase, a highly conserved DNA repair enzyme. EMBO J. 1989 Oct;8(10):3121–3125. doi: 10.1002/j.1460-2075.1989.tb08464.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovãdi J., Telegdi M., Batke J., Keleti T. Functional non-identity of subunits and isolation of active dimers of D-glyceraldehyde-3-phosphate dehydrogenase. Eur J Biochem. 1971 Oct 14;22(3):430–438. doi: 10.1111/j.1432-1033.1971.tb01561.x. [DOI] [PubMed] [Google Scholar]
- Percival K. J., Klein M. B., Burgers P. M. Molecular cloning and primary structure of the uracil-DNA-glycosylase gene from Saccharomyces cerevisiae. J Biol Chem. 1989 Feb 15;264(5):2593–2598. [PubMed] [Google Scholar]
- Perucho M., Salas J., Salas M. L. Identification of the mammalian DNA-binding protein P8 as glyceraldehyde-3-phosphate dehydrogenase. Eur J Biochem. 1977 Dec;81(3):557–562. doi: 10.1111/j.1432-1033.1977.tb11982.x. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seal G., Arenaz P., Sirover M. A. Purification and properties of the human placental uracil DNA glycosylase. Biochim Biophys Acta. 1987 Aug 13;925(2):226–233. doi: 10.1016/0304-4165(87)90113-9. [DOI] [PubMed] [Google Scholar]
- Seal G., Brech K., Karp S. J., Cool B. L., Sirover M. A. Immunological lesions in human uracil DNA glycosylase: association with Bloom syndrome. Proc Natl Acad Sci U S A. 1988 Apr;85(7):2339–2343. doi: 10.1073/pnas.85.7.2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro R., Klein R. S. The deamination of cytidine and cytosine by acidic buffer solutions. Mutagenic implications. Biochemistry. 1966 Jul;5(7):2358–2362. doi: 10.1021/bi00871a026. [DOI] [PubMed] [Google Scholar]
- Sirover M. A. Induction of the DNA repair enzyme uracil-DNA glycosylase in stimulated human lymphocytes. Cancer Res. 1979 Jun;39(6 Pt 1):2090–2095. [PubMed] [Google Scholar]
- Stancel G. M., Deal W. C., Jr Reversible dissociation of yeast glyceraldehyde 3-phosphate dehydrogenase by adenosine triphosphate. Biochemistry. 1969 Oct;8(10):4005–4011. doi: 10.1021/bi00838a017. [DOI] [PubMed] [Google Scholar]
- Towbin H., Staehelin T., Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshney U., Hutcheon T., van de Sande J. H. Sequence analysis, expression, and conservation of Escherichia coli uracil DNA glycosylase and its gene (ung). J Biol Chem. 1988 Jun 5;263(16):7776–7784. [PubMed] [Google Scholar]
- Vollberg T. M., Seal G., Sirover M. A. Monoclonal antibodies detect conformational abnormality of uracil DNA glycosylase in Bloom's syndrome cells. Carcinogenesis. 1987 Nov;8(11):1725–1729. doi: 10.1093/carcin/8.11.1725. [DOI] [PubMed] [Google Scholar]
- Vollberg T. M., Siegler K. M., Cool B. L., Sirover M. A. Isolation and characterization of the human uracil DNA glycosylase gene. Proc Natl Acad Sci U S A. 1989 Nov;86(22):8693–8697. doi: 10.1073/pnas.86.22.8693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weksberg R., Smith C., Anson-Cartwright L., Maloney K. Bloom syndrome: a single complementation group defines patients of diverse ethnic origin. Am J Hum Genet. 1988 Jun;42(6):816–824. [PMC free article] [PubMed] [Google Scholar]
- Williams K. R., Reddigari S., Patel G. L. Identification of a nucleic acid helix-destabilizing protein from rat liver as lactate dehydrogenase-5. Proc Natl Acad Sci U S A. 1985 Aug;82(16):5260–5264. doi: 10.1073/pnas.82.16.5260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis A. E., Lindahl T. DNA ligase I deficiency in Bloom's syndrome. Nature. 1987 Jan 22;325(6102):355–357. doi: 10.1038/325355a0. [DOI] [PubMed] [Google Scholar]
- Worrad D. M., Caradonna S. Identification of the coding sequence for herpes simplex virus uracil-DNA glycosylase. J Virol. 1988 Dec;62(12):4774–4777. doi: 10.1128/jvi.62.12.4774-4777.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]




