Abstract
Genetic characterization of a human cerebrospinal fluid West Nile virus isolate from Beaumont, Texas, revealed several nucleotide changes and amino acid substitutions that differentiated it from all other North American strains isolated to date, suggesting that isolates from the Texas Gulf Coast may form a unique genetic group among North American strains.
West Nile virus (WNV) was first described in a febrile patient in the West Nile district of Uganda in 1937 (10) and arrived in the western hemisphere in New York in 1999 (7). Subsequently, WNV has spread across the United States, including to Texas in 2002 (2). WNV has been transmitted to humans by transplacental, transfusion, or mosquito routes (9). If symptomatic, patients may present with various signs and symptoms, including fever, headache, nuchal rigidity, weakness, and myalgia (1, 3, 8). More recently, presentations have included acute flaccid paralysis, extrapyramidal symptoms, and rhabdomyolysis (3, 7).
The genome of WNV is approximately 11,000 nucleotides in length, depending on the strain, with three structural genes (capsid [C], membrane [prM-M], and envelope [E]) and seven nonstructural genes (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5) (6). Phylogenetically, WNV is divided into two major lineages. Lineage I predominates in North America, Asia, Europe, the Middle East, and northern Africa. Lineage II is found primarily in Africa (7).
An isolate of WNV was obtained from the cerebrospinal fluid (CSF) of a University of Texas Medical Branch patient from Beaumont, Texas (Jefferson County), admitted in August 2002, who exhibited fever, rigors, mild memory deficits, and progressive difficulty maintaining his airway at the time of lumbar puncture (11). This isolate was passaged once in Vero cells. A sample from a mosquito pool from Orange County, Texas, adjacent to Jefferson County and also near the Gulf of Mexico, was also obtained, (4) and both were sequenced. Reverse transcription-PCRs were performed with a TITAN One Tube RT-PCR kit (Roche) or by using two steps involving Moloney murine leukemia virus or avian myeloblastosis virus reverse transcriptase to generate double-stranded DNA, which was subsequently amplified by PCR using Taq polymerase (Roche). Primers used to generate PCR fragments were designed by using the prototypical New York 1999 strain (7). Samples of cDNA were separated by 1.2% agarose gel electrophoresis, excised, purified by a QIAquick gel extraction kit (QIAGEN), and directly sequenced with an Applied Biosystems 377A automated sequencer at the University of Texas Medical Branch Protein Core Facility. The direct sequencing utilized the same primers used to generate PCR fragments as well as using sequencing primers, when needed, to sequence long PCR fragments. Whole-genome sequences were aligned using Vector NTI Suite software (version 8.0; Informax, Inc.), which utilizes the ClustalW algorithm. Phylogenetic trees were constructed using maximum parsimony and neighbor-joining methods with PAUP version 4.04a (Sinauer Associates, Sutherland, Mass.).
The genome of WNV strain Beaumont was 11,029 nucleotides in length. There were 30 nucleotide differences (0.27%) between the Beaumont strain and the prototypical New York 1999 strain 382-99 (NY99) (7). In contrast, there were 44 nucleotide differences (0.40%) between the Beaumont strain and the 2001 human isolate from New York (AF533540) (5). Although most changes were noncoding mutations, there were five coding mutations (18.5% of total mutations) compared to the sequence of NY 382-99 and seven coding mutations (16.7% of total mutations) compared to the sequence of the human New York isolate from 2001 (5). Comparison of the Beaumont strain and the New York isolates, however, showed that nucleotide differences were clustered in NS2A, NS2B, and E protein genes as well as in the 3′ noncoding region (3′NCR) (data not shown). Significantly, several of these nucleotide differences were unique to the Beaumont strain among all published full-length West Nile genome sequences, including Kunjin and the prototypical lineage II strain Uganda 1937. The overall nucleotide identity was >99.6% among published North American isolates.
Comparison of the polyproteins of Jefferson County and other North American strains revealed that the Beaumont strain had five amino acid substitutions that were unique among all published lineage I virus isolates (E-76, NS1-94, NS2A-138, NS4B-173, and NS5-526) (Table 1). Four out of those five (all except NS1-94) were unique among all available WNV full-length virus sequences (Table 1). Of note, the same amino acid substitutions were found in an isolate (the Orange County strain) from a mosquito pool from Orange County, Texas, which is near Beaumont. As with the nucleotide differences, the amino acid substitutions were distributed throughout the genome, with one each located in the E, NS1, NS2A, NS4B, and NS5 proteins. Additionally, there were two unique nucleotide changes among all full-length sequences of WNV in the 3′NCR of the Beaumont strain (nucleotides 3′NCR 10494 and 10768) (Table 1). We do not believe that the unique nucleotide and amino acid differences of the human and mosquito isolates are due to mixed populations, as it is very unlikely that the human CSF isolate is a mixed population and there was no evidence that the mosquito isolate was a mixed population based on plaque morphology or analysis of the nucleotide sequencing chromatograms.
TABLE 1.
Comparison of amino acid substitutions and unique nucleotide changes of the Beaumont isolate to various published full-length amino acid sequencesa
| Amino acid substitution or nucleotide change | Virus strain or geographic location
|
||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Beau- mont, Tex. | Orange, Tex. | NY 2001 | NJ 2000 MQ | MD 2000 Crow | NY 2000 Crow | NY 2000 Grouse | CN 1999 Mosquito | NY 382-99 | NY 1999 Equine | NY 1999 Human | Israel 1998 Stork | RO 97-50 Mosquito | Italy 1998 Equine | Astra- khan 1999 | Kenya 1998 | Volgo- grad 1999 | Volgo- grad 1999, brain | Volgo- grad 2000 | Eg101 1951 | Kunjin | Caucasus 1998 | Uganda 1937 | |
| E-76 | A | A | T | T | T | T | T | T | T | T | T | T | T | T | T | T | T | T | T | T | T | T | T |
| NS1-94 | G | G | E | E | E | E | E | E | E | E | E | E | E | E | E | E | E | E | E | E | E | S | N |
| NS2A-90 | M | M | V | M | M | M | M | M | M | M | M | M | M | M | M | M | M | M | M | M | M | M | L |
| NS2A-138 | I | I | V | V | V | V | V | V | V | V | V | V | V | V | V | V | V | V | V | V | V | V | V |
| NS3-188 | Q | Q | K | Q | Q | Q | Q | Q | Q | Q | Q | Q | Q | Q | Q | Q | Q | Q | Q | Q | Q | Q | Q |
| NS4B-173 | I | I | V | V | V | V | V | V | V | V | V | V | V | V | V | V | V | V | V | V | V | V | V |
| NS5-526 | I | I | T | T | T | T | T | T | T | T | T | T | T | T | T | T | T | T | T | T | T | T | T |
| 3′NCR-10494 | C | C | T | T | T | T | T | T | T | T | T | T | T | T | T | T | T | T | T | T | T | T | ND |
| 3′NCR-10768 | A | A | T | T | T | T | T | T | T | T | T | T | T | T | T | T | T | T | T | T | ND | T | T |
The Beaumont human isolate and nearby Orange County mosquito isolate share five amino acid substitutions and two noncoding nucleotide changes that are unique among published full-length amino acid sequences. See Fig. 1 for sources of genomic sequences. ND, not determined.
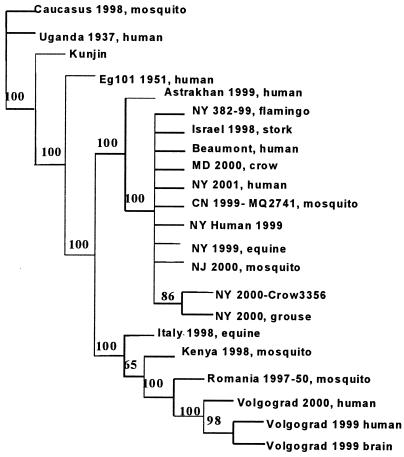
Complete genomic nucleotide sequences of WNV from recent outbreaks in Italy, Romania, Israel, and Russia, as well as all published North American sequences and Kunjin and Ugandan strains were used to generate phylogenetic trees by using PAUP version 4.04b with maximum parsimony and neighbor-joining methods. Although bootstrap values significantly differentiated the Italian, Romanian, Israeli, and Russian strains from the North American strains, bootstrap values did not show significant divergence of the Beaumont strain from the other North American strains (Fig. 1). Nonetheless, the presence of unique nucleotide changes and amino acid substitutions suggest that the Texas Gulf Coast isolates described here are distinct from the other North American isolates. In the phylogram generated by the neighbor-joining method (data not shown), the Beaumont isolate groups with Israel 1998, an antecedent to the New York isolates of 1999.
FIG. 1.
Phylogenetic tree of nucleotide sequence alignment of WNVs, including the human isolate Beaumont strain. Nucleotide sequences are full-length genomes, approximately 11,000 nucleotides. The phylogram was constructed by using maximum parsimony and heuristic methods, with bootstrap values shown at nodes. The following full-length genome sequences were obtained from GenBank for comparison, with accession numbers in parentheses: Egypt 1951 human (Eg 101) (AF260968), Italy 1998-equine (AF404757), Volgograd 1999 human brain isolate (AF317203), Volgograd 1999 human brain isolate (AY277252), Volgograd 2000 human blood isolate (AY278442), Caucasus 1998 mosquito isolate (AY277251), Astrakhan 1999 human isolate (AY278441), Israel 1998-Stork (AF481864), Uganda 1937 human isolate (M12294), Kunjin (D00246), CN 1999-MQ 2741 (AF206518), RO97-50 mosquito isolate from Romania (AF260969), New York 382-99 Flamingo isolate (AF196835), New York human isolate 2001 (AF533540), Maryland 2000-Crow 265 (AF404753), New York 2000-Crow 3356 (AF404756), New York 2000-Grouse 3282 (AF404755), New York 1999-equine (AF260967), New York human isolate 1999 (AF202541), New Jersey 2000-MQ5488 mosquito isolate (AF404754), and Kenya 1998 mosquito isolate (AY262283).
Overall, genetic analysis based on complete genome sequences of WNV isolated from the CSF of a patient from Beaumont, in Jefferson County, Texas, and a mosquito isolate from Orange County, Texas, revealed that these strains were the most divergent of all the North American strains examined to date, with up to 0.40% nucleotide divergence from the sequence of the recently reported 2001 human isolate from New York (5). The unique amino acid substitutions shared by the human and mosquito isolates, from the southeastern coast of the Texas Gulf of Mexico, suggest that there are probably no human-specific molecular determinants of WNV infection and that strains associated with encephalitis in humans to date are probably not unique genetic variants. There are many possible explanations for the unique mutations in the Texas Gulf Coast strains, but these are too speculative without additional data beyond the genetic characterization reported here. Clearly, additional studies are required to understand the evolution and virulence of WNV in North America.
Nucleotide sequence accession number. The genome of WNV strain Beaumont has been deposited in GenBank under accession no. AY289214.
Acknowledgments
This study was supported by the National Institutes of Health (T32 AI 007536 and T32 AI 007526) and the State of Texas Advanced Research Program.
We thank Robert Tesh for providing the Jefferson County virus isolate and Peter Mason and Melissa Whiteman for helpful discussions. We also thank the Protein Chemistry Laboratory at the University of Texas Medical Branch in Galveston for nucleotide sequencing.
REFERENCES
- 1.Asnis, D. S., R. Conetta, A. A. Teixeira, G. Waldman, and B. A. Sampson. 2000. The West Nile Virus outbreak of 1999 in New York: the Flushing Hospital experience. Clin. Infect. Dis. 30:413-418. [DOI] [PubMed] [Google Scholar]
- 2.Campbell, G. L., A. A. Marfin, R. S. Lanciotti, and D. J. Gubler. 2002. West Nile virus. Lancet Infect. Dis. 2:519-529. [DOI] [PubMed] [Google Scholar]
- 3.Centers for Disease Control and Prevention. 2002. Acute flaccid paralysis syndrome associated with West Nile virus infection—Mississippi and Louisiana. Morb. Mortal. Wkly. Rep. 51:825-828. [PubMed] [Google Scholar]
- 4.Davis, C. T., D. W. C. Beasley, H. Guzman, P. Raj, M. D'Anton, R. J. Novak, T. R. Unnasch, R. B. Tesh, and A. D. T. Barrett. 2003. Genetic variation among temporally and geographically distinct West Nile virus isolates, United States, 2001, 2002. Emerg. Infect. Dis. 9:1423-1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang, C., B. Slater, R. Rudd, N. Parchuri, R. Hull, M. Dupuis, and A. Hindenberf. 2002. First isolation of West Nile virus from a patient with encephalitis in the United States. Emerg. Infect. Dis. 8:1367-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lanciotti, R. S., G. D. Ebel, V. Deubel, A. J. Kerst, S. Murri, R. Meyer, M. Bowen, N. McKinney, W. E. Morrill, M. B. Crabtree, L. D. Kramer, and J. T. Roehrig. 2002. Complete genome sequences and phylogenetic analysis of West Nile Virus strains isolated from the United States, Europe, and the Middle East. Virology 299:96-105. [DOI] [PubMed] [Google Scholar]
- 7.Lanciotti, R. S., J. T. Roehrig, V. Deubel, J. Smith, M. Parker, K. Steele, B. Crise, K. E. Volpe, M. B. Crabtree, J. H. Scherret, R. A. Hall, J. S. MacKenzie, C. B. Cropp, B. Panigrahy, E. Ostlund, B. Schmitt, M. Malkinson, C. Banet, J. Weissman, N. Komar, H. M. Savage, W. Stone, T. McNamara, and D. J. Gubler. 1999. Origin of the West Nile Virus responsible for an outbreak of encephalitis in the northeastern United States. Science 286:2333-2337. [DOI] [PubMed] [Google Scholar]
- 8.Marfin, A. A., and D. J. Gubler. 2001. West Nile encephalitis: an emerging disease in the United States. Clin. Infect. Dis. 33:1713-1719. [DOI] [PubMed] [Google Scholar]
- 9.Mostashari, F., M. L. Bunning, P. T. Kitsutani, D. A. Singer, D. Nash, M. J. Cooper, N. Katz, K. A. Liljebjelke, B. J. Biggerstaff, A. D. Fine, M. C. Layton, S. M. Mullin, A. J. Johnson, D. A. Martin, E. B. Hayes, and G. L. Campbell. 2001. Epidemic West Nile encephalitis, New York, 1999: results of a household-based seroepidemiological survey. Lancet 358:261-264. [DOI] [PubMed] [Google Scholar]
- 10.Smithburn, K. C., T. P. Hughes, A. W. Burke, and J. H. Paul. 1940. A neurotropic virus isolated from the blood of a native of Uganda. Am. J. Trop. Med. 20:471-492. [Google Scholar]
- 11.Solomon, T. S., A. F. Fisher, D. W. Beasley, P. Mandava, B. P. Granwehr, H. Langsjoen, A. P. Travassos Da Rosa, A. D. T. Barrett, and R. B. Tesh. 2003. Natural and nosocomial infection in a patient with West Nile encephalitis and extrapyramidal movement disorders. Clin. Infect. Dis. 36:140-145. [DOI] [PubMed] [Google Scholar]