Abstract
This study involves a first evaluation of 25 novel spacer oligonucleotides in addition to the 43 routine spacers for molecular characterization of a panel of 65 isolates of tubercle bacilli from different geographic origins that were initially classified as Mycobacterium africanum based on phenotypic characters. The 68-spacer format defined four additional patterns, and three groups were identified. The relatively homogeneous groups A1 and A2 included strains from West Africa, and A3-1 included strains from East Africa. The presence of deletion region RD9 confirmed the reclassification of the M. africanum subtype II spoligopattern within group A3-1 as Mycobacterium tuberculosis. These isolates may represent a diverging branch of M. tuberculosis in Africa. The use of new spacers also suggested an undergoing evolution of M. africanum subtype I in West Africa. Our results showed that the strain differentiation within the M. tuberculosis complex is improved by using novel spacers, and extensive studies using new-generation spoligotyping may be helpful to better understand the evolution of M. africanum.
Many recent studies have used spoligotyping, a commonly used PCR-based method, for Mycobacterium tuberculosis complex identification and genotyping (13, 19). Recently, Viana-Niero et al. showed that a collection of isolates tentatively identified as Mycobacterium africanum could be successfully subdivided into three groups, A1 to A3, by spoligotyping, IS6110-restriction fragment length polymorphism analysis, and analysis by variable number of tandem DNA repeats and that a specific spoligotyping signature of M. africanum was the absence of spacers 8, 9, and 39 (25). This was followed by a study on isolates from Uganda, which showed that M. africanum subtype II differed from the classical M. africanum subtype I isolates by the absence of spacers 33 to 36 and 40 (16, 21).
Although spoligotyping is easier to perform than IS6110-restriction fragment length polymorphism analysis, its discriminatory power is not optimal when it is used in a routine 43-spacer format (14). Recently, a panel of novel spacers was used in spoligotyping to improve M. tuberculosis complex differentiation (23, 24). As these studies did not investigate any M. africanum strains, we thought it desirable to investigate the variability of a set of novel sequences in addition to the routine spoligotyping spacers with a panel of isolates of tubercle bacilli from different geographic origins that were initially classified as M. africanum based on phenotypic characters. The present study involves a first evaluation of the 25 novel spoligotyping spacers for M. africanum characterization.
MATERIALS AND METHODS
Mycobacterial strains.
This study used a collection of 65 clinical isolates from Benin, Cameroon, France, Guinea-Bissau, Nigeria, Senegal, Sierra-Leone, and Uganda and from African patients residing in the United States. Twenty-nine clinical isolates were from the National Reference Laboratory for Mycobacteria, Institut Pasteur, Paris, France, and 36 were from the Clinical Mycobacteriology Laboratory, Wadsworth Center, Albany, New York. Among the latter, two were M. africanum American Type Culture Collection strains (ATCC 25420T and ATCC 35711), and six were isolates that were recently reclassified as variants of M. tuberculosis by genomic deletion analysis (3, 17).
Genotyping.
DNA was extracted by a simple thermolysis method. Spoligotyping using 43 spacers was performed as previously described by Kamerbeek et al. (13). The 68-spacer format with 25 novel spacers was used according to the scheme of van Embden et al., which numbers the spacers according to their position in the direct repeat locus (24). It should be emphasized that this representation of spacers, although genomically true, is completely different from the initial arbitrary designation of Kamerbeek et al. (13); e.g., spacer 12 in the scheme of van Embden et al. corresponds to spacer 4 in that of Kamerbeek et al. The hybridization results (positive or negative) were documented in the form of a binary code for each spacer and entered in an Excel spreadsheet file. The results were further analyzed by using the Bionumerics software (Applied Maths, St. Maarten Latem, Belgium), following the instruction manual for similarity analysis with the Jaccard index. Dendrograms were built by the unweighted pair group method of arithmetic averages method. The relative discriminatory powers of the 43- and 68-spacer spoligotyping methods were determined by using the Hunter-Gaston discriminatory index (11). A more exhaustive analysis, using a full set of deletion regions RD1 to RD11 and TbD1, was also performed on some isolates for each of the group defined by 68-spacer spoligotyping (3, 17).
RESULTS AND DISCUSSION
Characterization of M. africanum by 43-spacer spoligotyping.
Classical spoligotyping showed a total of 41 different patterns for 65 isolates studied (results not shown). The results obtained corroborated the typical absence of spacers 8, 9, and 39 in 57 subtype I isolates (25). The six subtype II pattern strains (strains 960096, 960104, 960106, 960107, 960108, and 960109 in Table 1) showed a positive hybridization with these three spacers, which corroborated their classification as M. tuberculosis (3, 17). Two other isolates were also atypical, since one showed positive hybridization signals with spacers 8 and 9 but not 39 (isolate 960091), whereas the other (isolate 220476) was positive for spacer 8 but not for spacers 9 and 39.
TABLE 1.
Summary of spoligotypes, geographic origins, groups, and subgroups of the 65 strains studied
| Strain | Origina | Type | Group defined by using:
|
|
|---|---|---|---|---|
| 43 spacers | 68 spacers | |||
| 960090 | SLE | Orphan 1 | A2-2 | A2-2 |
| 960091 | SLE | 1338 | A3 | A3-2 |
| 960092 | SLE | 438 | A2-2 | A2-2 |
| 960093 | SLE | Orphan 2 | A1 | A1 |
| 960094 | SLE | Orphan 3 | A1 | A1 |
| 960095 | NGA | 438 | A2-2 | A2-2 |
| 960096 | UGA | 52 | A3 | A3-1 |
| 960097 | GIN | Orphan 4 | A1 | A1 |
| 960098 | NGA | 331cb | A2-1 | A2-1 |
| 960099 | NGA | 331ab | A2-1 | A2-1 |
| 960100 | NGA | Orphan 5 | A2-1 | A2-1 |
| 960101 | NGA | 438 | A2-2 | A2-2 |
| 960102 | GIN | 181ab | A1 | A1 |
| 960103 | GIN | 326 | A1 | A1 |
| 960104 | GIN | 50 | A3 | A3-1 |
| 960105 | GIN | 181bb | A1 | A1 |
| 960106 | UGA | 135 | A3 | A3-1 |
| 960107 | UGA | 52 | A3 | A3-1 |
| 960108 | UGA | 52 | A3 | A3-1 |
| 960109 | UGA | 420 | A3 | A3-1 |
| 970697 | Xc | 181ab | A1 | A1 |
| 973402 | X | 181ab | A1 | A1 |
| 990984 | X | 101 | A2-1 | A2-1 |
| 991541 | X | Orphan 6 | A1 | A1 |
| 993083 | X | 181ab | A1 | A1 |
| 220064 | X | 181ab | A1 | A1 |
| 221703 | X | 187ab | A1 | A1 |
| 990315 | X | 187bb | A1 | A1 |
| 210271 | X | Orphan 7 | A2-2 | A2-2 |
| 210390 | X | 187bb | A1 | A1 |
| 211535 | X | 331bb | A2-1 | A2-1 |
| 220097 | X | 331bb | A2-1 | A2-1 |
| 220476 | X | 1153 | A3 | A3-2 |
| 221372 | X | Orphan 8 | A1 | A1 |
| 200100d | SEN | 181ab | A1 | A1 |
| 200101e | X | 181ab | A1 | A1 |
| 940960 | FXX | 325 | A1 | A1 |
| 940961 | FXX | 181ab | A1 | A1 |
| 950468 | FXX | 181ab | A1 | A1 |
| 951936 | FXX | 326 | A1 | A1 |
| 961262 | FXX | 181ab | A1 | A1 |
| 961389 | FXX | Orphan 9 | A2-1 | A2-1 |
| 970133 | FXX | 181ab | A1 | A1 |
| 970866 | X | Orphan 10 | A3 | A3-2 |
| 970869 | CIV | 319 | A2-1 | A2-1 |
| 970872 | CIV | 331ab | A2-1 | A2-1 |
| 970874 | CIV | Orphan 11 | A2-1 | A2-1 |
| 970877 | CIV | 318 | A1 | A1 |
| 980338 | CMR | 856 | A2-3 | A2-3 |
| 980698 | CMR | 328 | A2-1 | A2-1 |
| 980699 | CMR | Orphan 12 | A2-3 | A2-3 |
| 980701 | CMR | 328 | A2-1 | A2-1 |
| 980702 | CMR | Orphan 13 | A2-3 | A2-3 |
| 981444 | CMR | 320 | A2-1 | A2-1 |
| 981445 | CMR | 861 | A2-1 | A2-1 |
| 981446 | CMR | 858 | A2-3 | A2-3 |
| 981448 | CMR | 328 | A2-1 | A2-1 |
| 981449 | CMR | 329 | A2-1 | A2-1 |
| 981538 | BEN | Orphan 14 | A2-2 | A2-2 |
| 14003001 | SEN | 799 | A1 | A1 |
| 14003009 | SEN | Orphan 15 | A2-1 | A2-1 |
| 14003011 | SEN | 323 | A1 | A1 |
| 14003028 | CMR | Orphan 16 | A2-3 | A2-3 |
| 14003041 | CMR | Orphan 17 | A3 | A3-2 |
| 14003048 | SEN | Orphan 18 | A3 | A3-2 |
BEN, Benin; CIV, Ivory Coast; CMR, Cameroon; FXX, metropolitan France; GIN, Guinea-Bissau; NGA, Nigeria; SEN, Senegal; SLE, Sierra Leone; UGA, Uganda.
Strain for which a different pattern was observed when spoligotyping with 68 spacers was used.
X, strain isolated from African patients residing in the United States.
Strain 200100 was M. africanum type strain ATCC 25420T.
Strain 200101 was M. africanum type strain ATCC 35711.
The spoligotyping patterns were compared to those found in the international spoligotyping database at the Institut Pasteur de Guadeloupe (7). Its recent version (SpolDB4), which is under development, contained patterns from 23,200 clinical isolates, split into 1,340 shared types (STs) and 2,000 unique types, at the time of the comparison. Eighteen unique patterns and 23 STs (two or more isolates with the same pattern in the SpolDB4 database) were observed. Among the 23 STs, seven clusters containing 2 to 12 isolates were defined: 1 cluster of 12 strains (ST 181), 1 of 5 strains (ST 331), 4 of 3 strains (STs 52, 187, 328, and 438) and 1 of 2 strains (ST 326). Detailed analysis of the patterns (p), and not the strains, led to the identification of three main groups (Table 1), designated A1 (p-12), A2 (p- 20), and A3 (p-9). It should be emphasized that the classification of strains within various groups in the present study does not follow the scheme of Viana-Niero et al. (25).
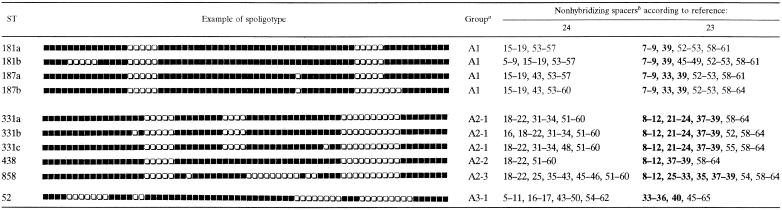
Groups A1 and A2 were relatively homogeneous, whereas A3 was more heterogeneous and included isolates that could not be grouped as either A1 or A2. The group A1 signature appeared to be the absence of spacers 7 to 9 and 39 (Table 2).Group A2 could be divided into three subgroups: A2-1 (p-11), whose characteristic was the absence of spacers 8 to 12, 21 to 24, and 37 to 39; subgroup A2-2 (p-4), with the absence of spacers 8 to 12 and 37 to 39; and subgroup A2-3 (p-5), which showed no hybridization with spacers 8 to 12, 25 to 33, 35, and 37 to 39 (Table 2). A3 arbitrarily contained all of the isolates that were not grouped as either A1 or A2. A tentative profile for some of the A3 strains was the absence of spacers 33 to 36 with or without spacer 40 (subtype II [16]). The absence of spacer 40 in strains from Africa has been reported (20).
TABLE 2.
Rules describing specific spoligotyping signatures for strains within each of the groups or subgroups defined by 68-spacer spoligotyping
A3-2 did not show a specific signature, and hence no deletion pattern could be defined.
Numbers in boldface indicate the spoligotyping signatures based on the classical 43-spacer format.
Characterization of M. africanum by new-generation spoligotyping.
As summarized in Tables 1 and 2 and Fig. 1, a total of 45 different patterns were obtained for the 65 isolates by 68-spacer spoligotyping. Group A1 (p 14) was characterized by the absence of spacers 15 to 19 and 53 to 57 (Table 2). Group A2 (p 22) was divided into three different subgroups (Table 2): A2-1 (p 13), characterized by the absence of spacers 18 to 22, 31 to 34, and 51 to 60; A2-2 (p 4), characterized by the absence of spacers 18 to 22 and 51 to 60; and A2-3 (p 5), characterized by the absence of spacers 18 to 22, 25, 35 to 43, and 51 to 60. This method also highlighted a new subgroup (p 4) with a potential signature characterized by the absence of spacers 5 to 11, 16 to 17, 43 to 50, and 54 to 62 within the previously defined group A3. It contained the six subtype II pattern strains that did not show a typical M. africanum signature in the 43-spacer format (missing spacers 8, 9, and 39) and was designated A3-1. Nonetheless, five other patterns did not fit any of the above-described groups and were arbitrarily designated A3-2 (Table 1; Fig. 1).
FIG. 1.
Dendrogram and schematic representation of spoligotype patterns obtained by using the new 68-spacer format. The degree of similarity of the spoligotypes was calculated with the 1-Jaccard index, and the relationship between patterns was assessed by the unweighted pair group method of arithmetic averages. For descriptions of the various groups and subgroups and the STs listed, see the text.
Regarding the geographic variability (Table 1), groups A1 and A2 included strains from West Africa (Benin, Cameroon, African patients in France, Guinea-Bissau, Ivory Coast, Nigeria, Senegal, and Sierra Leone), whereas A3-1 included strains that were mostly from East Africa (five out of six strains).
The addition of novel spacers enhanced the discriminatory power of the spoligotyping. It increased the number of patterns identified from 41 to 45, with a Hunter-Gaston discriminatory index of 0.97, instead of 0.95 for the 43-spacer format. Indeed, the 20 clinical isolates that clustered in three STs numbered 181, 187, and 331 in the classical 43-bit representation, were further subdivided by using the 68-bit representation system. The seven subtypes thus obtained are shown in Table 2. Interestingly, ST 181, which defines the majority of M. africanum isolates described so far (25), was characterized by an increased deletion step (absence of a block of five spacers). The same phenomenon is also apparent for shared types 187 and 331 (Table 2). These results corroborate and further extend those of van der Zanden et al. showing that the strain differentiation of M. bovis, M. microti, and M. canettii within the M. tuberculosis complex was improved by using novel spacers (23), and they suggest that extensive studies using new-generation spoligotyping may be helpful to better understand the evolution of M. africanum.
PCR-deletion region analysis.
Out of 20 variable regions described for the M. tuberculosis complex (2, 3, 8), RD9 and RD10 are particularly useful to differentiate M. africanum, since it lacks RD9 (and sometimes RD10), regions that are present in M. tuberculosis (3, 10, 17). Our results showed that RD9 was present only among the 6 subtype II strains (A3-1 subgroup) and not in the remaining 59 strains. In contrast, the TbD1 region was systematically present in all M. africanum groups studied with the exception of six isolates in subgroup A3-1, suggesting that these isolates are “modern” strains of M. tuberculosis (3). This finding shows that the latter isolates are clearly different from other M. africanum strains seen in this and previous studies (12, 25) and is consistent with their reclassification as M. tuberculosis (17). Further analysis showed that RD11 was absent in all strains belonging to group A1 but was present in strains in groups A2 and A3, whereas RD10 was variable. The distribution of the RDs in conjunction with the current knowledge of M. tuberculosis phylogeny suggests a possible way in which M. tuberculosis evolution may have proceeded (3, 10, 15). It seems likely that M. bovis is the final member of a separate lineage represented by M. africanum, M. microti, M. caprae, and M. bovis and that M. africanum may be an offspring of M. tuberculosis which has lost RD9. This evolutionary hypothesis regarding M. africanum applies to M. africanum subtype I but not to M. africanum subtype II, which may represent a diverging branch of the M. tuberculosis lineage.
Phylogeographical considerations.
The M. tuberculosis complex currently encompasses M. tuberculosis, M. africanum, M. bovis, M. bovis BCG, M. bovis subsp. caprae, M. canettii, M. microti, and a recently described species, M. pinnipedii (4), with characteristic animal and/or human epidemiologies (4, 18). M. tuberculosis and M. bovis are most commonly isolated in clinical laboratories and may be easily distinguished by biochemical tests, phenotypic characters, and several genetic markers (5). However, the taxonomical status of M. africanum isolates, which were shown to introduce substantial phenotypic heterogeneity and genetic diversity within M. bovis and M. tuberculosis clusters, remained controversial (6). A distinction between West African strains (from Senegal, Mauritania, and Cameroon), which are more likely to have M. bovis-like phenotypic characteristics (M. africanum type I), and East African strains (from Burundi and Rwanda), which are more likely to have M. tuberculosis-like characteristics (M. africanum type II), was previously made by David et al. (6). Our results with the 68-spacer spoligotyping format showed that the patterns for these strains lacking spacers 33 to 36 and 40, did resemble those previously labeled as M. tuberculosis (24). Nonetheless, spoligotyping alone may not always allow us to gain sufficient information to establish a likely evolutionary history of M. tuberculosis; e.g., generation of identical spoligotype patterns among different strain families may also arise due to IS6110-mediated deletion polymorphism (26). For a precise phylogeographical attribution of the various M. africanum clades, larger studies using 68-spacer format spoligotyping in conjunction with other markers, such as mycobacterial interspersed repetitive units (22), RDs, and single-nucleotide polymorphisms (1, 9), may be rewarding.
Acknowledgments
This work was supported through grants from the Réseau International des Instituts Pasteur et Instituts Associés, Institut Pasteur, Paris, France, and EU Project QLK2-CT-2000-630, entitled “New generation genetic markers and techniques for the epidemiology and control of tuberculosis.” K.B. was cofinanced by Institut Pasteur and European Social Funds provided through the Regional Council of Guadeloupe.
L.M.P. is grateful to Gisela Bretzel for access to isolates of M. africanum from African patients.
REFERENCES
- 1.Alland, D., T. S. Whittam, M. B. Murray, M. D. Cave, M. H. Hazbon, K. Dix, M. Kokoris, A. Duesterhoeft, J. A. Eisen, C. M. Fraser, and R. D. Fleischmann. 2003. Modeling bacterial evolution with comparative-genome-based marker systems: application to Mycobacterium tuberculosis evolution and pathogenesis. J. Bacteriol. 185:3392-3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Behr, M. A., M. A. Wilson, W. P. Gill, H. Salamon, G. K. Schoolnik, S. Rane, and P. M. Small. 1999. Comparative genomics of BCG vaccines by whole-genome DNA microarray. Science 284:1520-1523. [DOI] [PubMed] [Google Scholar]
- 3.Brosch, R., S. V. Gordon, M. Marmiesse, P. Brodin, C. Buchrieser, K. Eiglmeier, T. Garnier, C. Gutierrez, G. Hewinson, K. Kremer, L. M. Parsons, A. S. Pym, S. Samper, D. van Soolingen, and S. T. Cole. 2002. A new evolutionary scenario for the Mycobacterium tuberculosis complex. Proc. Natl. Acad. Sci. USA 99:3684-3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cousins, D. V., R. Bastida, A. Cataldi, V. Quse, S. Redrobe, S. Dow, P. Duignan, A. Murray, C. Dupont, N. Ahmed, D. M. Collins, W. R. Butler, D. Dawson, D. Rodriguez, J. Loureiro, M. I. Romano, A. Alito, M. Zumarraga, and A. Bernardelli. 2003. Tuberculosis in seals caused by a novel member of the Mycobacterium tuberculosis complex: Mycobacterium pinnipedii sp. nov. Int. J. Syst. Evol. Microbiol. 53:1305-1314. [DOI] [PubMed] [Google Scholar]
- 5.David, H., V. Levy-Frebault, and M. F. Thorel. 1989. Méthodes de laboratoire pour Mycobactériologie clinique, p. 1-87. Institut Pasteur, Paris, France.
- 6.David, H. L., M. T. Jahan, A. Jumin, J. Grandry, and E. H. Lehmann. 1978. Numerical taxonomy of Mycobacterium africanum. Int. J. Syst. Bacteriol. 28:467-472. [Google Scholar]
- 7.Filliol, I., J. R. Driscoll, D. van Soolingen, B. N. Kreiswirth, K. Kremer, G. Valetudie, D. A. Dang, R. Barlow, D. Banerjee, P. J. Bifani, K. Brudey, A. Cataldi, R. C. Cooksey, D. V. Cousins, J. W. Dale, O. A. Dellagostin, F. Drobniewski, G. Engelmann, S. Ferdinand, D. Gascoyne-Binzi, M. Gordon, M. C. Gutierrez, W. H. Haas, H. Heersma, E. Kassa-Kelembho, M. L. Ho, A. Makristathis, C. Mammina, G. Martin, P. Mostrom, I. Mokrousov, V. Narbonne, O. Narvskaya, A. Nastasi, S. N. Niobe-Eyangoh, J. W. Pape, V. Rasolofo-Razanamparany, M. Ridell, M. L. Rossetti, F. Stauffer, P. N. Suffys, H. Takiff, J. Texier-Maugein, V. Vincent, J. H. de Waard, C. Sola, and N. Rastogi. 2003. Snapshot of moving and expanding clones of Mycobacterium tuberculosis and their global distribution assessed by spoligotyping in an international study. J. Clin. Microbiol. 41:1963-1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gordon, S. V., R. Brosch, A. Billault, T. Garnier, K. Eiglmeier, and S. T. Cole. 1999. Identification of variable regions in the genomes of tubercle bacilli using bacterial artificial chromosome arrays. Mol. Microbiol. 32:643-655. [DOI] [PubMed] [Google Scholar]
- 9.Gutacker, M. M., J. C. Smoot, C. A. Migliaccio, S. M. Ricklefs, S. Hua, D. V. Cousins, E. A. Graviss, E. Shashkina, B. N. Kreiswirth, and J. M. Musser. 2002. Genome-wide analysis of synonymous single nucleotide polymorphisms in Mycobacterium tuberculosis complex organisms. Resolution of genetic relationships among closely related microbial strains. Genetics 162:1533-1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huard, R. C., L. C. de Oliveira Lazzarini, W. R. Butler, D. van Soolingen, and J. L. Ho. 2003. PCR-based method to differentiate the subspecies of the Mycobacterium tuberculosis complex on the basis of genomic deletions. J. Clin. Microbiol. 41:1637-1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hunter, P. R., and M. A. Gaston. 1988. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J. Clin. Microbiol. 26:2465-2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Källenius, G., T. Koivula, S. Ghebremichael, S. E. Hoffner, R. Norberg, E. Svensson, F. Dias, B. Marklund, and S. B. Svenson. 1999. Evolution and clonal traits of Mycobacterium tuberculosis in Guinea-Bissau. J. Clin. Microbiol. 37:3872-3878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kamerbeek, J., L. Schouls, A. Kolk, M. van Agterveld, D. Van Soolingen, S. Kuijper, A. Bunschoten, H. Molhuizen, R. Shaw, M. Goyal, and J. D. A. van Embden. 1997. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J. Clin. Microbiol. 35:907-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kremer, K., D. van Soolingen, R. Frothingham, W. H. Haas, P. W. M. Hermans, C. Martin, P. Palittapongarnpim, B. B. Plikaytis, L. W. Riley, M. A. Yakrus, J. M. Musser, and J. D. A. van Embden. 1999. Comparison of methods based on different molecular epidemiologial markers for typing of Mycobacterium tuberculosis strains: interlaboratory study of discriminatory power and reproducibility. J. Clin. Microbiol. 37:2607-2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mostowy, S., and M. A. Behr. 2002. Comparative genomics in the fight against tuberculosis: diagnostics, epidemiology, and BCG vaccination. Am. J. Pharmacogenomics 2:189-196. [DOI] [PubMed] [Google Scholar]
- 16.Niemann, S., S. Rusch-Gerdes, M. L. Joloba, C. C. Whalen, D. Guwatudde, J. J. Ellner, K. Eisenach, N. Fumokong, J. L. Johnson, T. Aisu, R. D. Mugerwa, A. Okwera, and S. K. Schwander. 2002. Mycobacterium africanum subtype II is associated with two distinct genotypes and is a major cause of human tuberculosis in Kampala, Uganda. J. Clin. Microbiol. 40:3398-3405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parsons, L. M., R. Brosch, S. T. Cole, A. Somoskovi, A. Loder, G. Bretzel, D. Van Soolingen, Y. M. Hale, and M. Salfinger. 2002. Rapid and simple approach for identification of Mycobacterium tuberculosis complex isolates by PCR-based genomic deletion analysis. J. Clin. Microbiol. 40:2339-2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rastogi, N., E. Legrand, and C. Sola. 2001. The mycobacteria: an introduction to nomenclature and pathogenesis. Rev. Sci. Tech. Off. Int. Epiz. 20:21-46. [DOI] [PubMed] [Google Scholar]
- 19.Soini, H., X. Pan, A. Amin, E. A. Graviss, A. Siddiqui, and J. M. Musser. 2000. Characterization of Mycobacterium tuberculosis isolates from patients in Houston, Texas, by spoligotyping J. Clin. Microbiol. 38:669-676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sola, C., I. Filliol, E. Legrand, I. Mokrousov, and N. Rastogi. 2001. Mycobacterium tuberculosis phylogeny reconstruction based on combined numerical analysis with IS1081, IS6110, VNTR and DR-based spoligotyping suggests the existence of two new phylogeographical clades. J. Mol. Evol. 53:680-689. [DOI] [PubMed] [Google Scholar]
- 21.Sola, C., N. Rastogi, M. C. Gutierrez, V. Vincent, R. Brosch, L. M. Parsons, S. Niemann, S. Rüsch-Gerdes, and S. K. Schwander. 2003. Is Mycobacterium africanum subtype II (Uganda I and Uganda II) a genetically well-defined subspecies of the Mycobacterium tuberculosis complex? J. Clin. Microbiol. 41:1345-1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Supply, P., E. Mazars, S. Lesjean, V. Vincent, B. Gicquel, and C. Locht. 2000. Variable human minisatellite-like regions in the Mycobacterium tuberculosis genome. Mol. Microbiol. 36:762-771. [DOI] [PubMed] [Google Scholar]
- 23.van der Zanden, A. G., K. Kremer, L. M. Schouls, K. Caimi, A. Cataldi, A. Hulleman, N. J. Nagelkerke, and D. van Soolingen. 2002. Improvement of differentiation and interpretability of spoligotyping for Mycobacterium tuberculosis complex isolates by introduction of new spacer oligonucleotides. J. Clin. Microbiol. 40:4628-4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Embden, J. D. A., T. van Gorkom, K. Kremer, R. Jansen, B. A. M. van der Zeijst, and L. M. Schouls. 2000. Genetic variation and evolutionary origin of the direct repeat locus of Mycobacterium tuberculosis complex bacteria. J. Bacteriol. 182:2393-2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Viana-Niero, C., C. Gutierrez, C. Sola, I. Filliol, F. Boulahbal, V. Vincent, and N. Rastogi. 2001. Genetic diversity of Mycobacterium africanum clinical isolates based on IS6110-restriction fragment length polymorphism analysis, spoligotyping, and variable number of tandem DNA repeats. J. Clin. Microbiol. 39:57-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Warren, R. M., E. M. Streicher, S. L. Sampson, G. D. Van Der Spuy, M. Richardson, D. Nguyen, M. A. Behr, T. C. Victor, and P. D. Van Helden. 2002. Microevolution of the direct repeat region of Mycobacterium tuberculosis: implications for interpretation of spoligotyping data. J. Clin. Microbiol. 40:4457-4465. [DOI] [PMC free article] [PubMed] [Google Scholar]