Abstract
Candida dubliniensis was first established as a novel yeast species in 1995. It is particularly associated with recurrent episodes of oral candidosis in human immunodeficiency virus (HIV)-infected patients, but it has also been detected at other anatomical sites and at a low incidence level in non-HIV-infected patients. It shares so many phenotypic characteristics with C. albicans that it is easily misidentified as such. No rapid, simple, and commercial test that allows differentiation between C. dubliniensis and C. albicans has been developed, until now. Accurate species identification requires the use of genotype-based techniques that are not routinely available in most clinical microbiology diagnostic laboratories. The present study was designed to evaluate the efficiency of a new test (the immunochromatographic membrane [ICM] albi-dubli test; SR2B, Avrillé, France) to differentiate between C. albicans and C. dubliniensis. The organisms evaluated were strains whose identities had previously been confirmed by PCR tests and freshly isolated clinical strains and included 58 C. albicans isolates, 60 C. dubliniensis isolates, and 82 isolates belonging to other species of yeast. The ICM albi-dubli test is based on the principle of immunochromatographic analysis and involves the use of two distinct monoclonal antibodies that recognize two unrelated epitopes expressed by both species or specific to only one species. The assay requires no complex instrumentation for analysis and can be recommended for routine use in clinical microbiology laboratories. Results are obtained within 2 h and 30 min and are easy to interpret. This evaluation demonstrated the good performance of this immunochromatographic test for C. albicans and C. dubliniensis isolated on Sabouraud dextrose agar, CHOROMagar Candida, and CandidaSelect, with sensitivities and specificities ranging from 93.1 to 100%. These parameters decreased, however, to 91.4% when the test was performed with yeast isolated with Candida ID.
Over the past decade there has been a significant increase in the number of reports of systemic and mucosal infections caused by Candida species, and since the recognition that Candida species differ in the expression of putative virulence factors and susceptibilities to antifungal agents, greater emphasis has been placed on the identification of isolates to the species level.
Candida dubliniensis is a recently described pathogenic Candida species which was originally identified in cases of recurrent oral candidosis in human immunodeficiency virus (HIV)-infected and AIDS patients (34). Several studies have reported an overall prevalence of 26.4 to 34% in the oral cavities of HIV-positive patients and 3.5 to 13.9% among HIV-negative patients (9, 36). Moreover, it has been suggested that C. dubliniensis can be induced to develop resistance to fluconazole in vitro, a phenomenon which may have resulted in its emergence in the HIV-infected population (23, 24, 33). More recently, it has also been identified in a small number of cases of systemic infection. In retrospective analyses of stock collections, C. dubliniensis was found to account for at least 1.2 to 2% of the yeasts initially identified as C. albicans (14, 25). C. dubliniensis shares many phenotypic characteristics with C. albicans, including the ability to form germ tubes and chlamydospores, and is phylogenetically closely related to C. albicans. It belongs to C. albicans serotype A, and chlamydospores of C. dubliniensis are produced abundantly and often in clusters or contiguous pairs (10). These similarities have caused significant problems with the identification of C. dubliniensis by the average clinical mycology laboratory. Several phenotypic methods for the identification of C. dubliniensis and discrimination from C. albicans have been reported: (i) C. dubliniensis has been shown to produce a distinctive dark green color on CHROMagar Candida (30); however, this atypical color may not persist after serial passage of the organism and may be less useful for isolate identification (32, 36); (ii) following isolation, colonies of C. dubliniensis do not exhibit fluorescence on methyl blue-Sabouraud agar under Wood's light, but this lack of fluorescence may not be reproducible with isolates subjected to storage and repeated subculture (30); (iii) unlike C. albicans, C. dubliniensis does not grow at 45°C (27), but discrimination by thermotolerance was not confirmed (16); (iv) in contrast to C. albicans, C. dubliniensis is able to reduce 2,3,5-triphenyltetrazolium chloride (37); and (v) C. dubliniensis has been found to be β-glucosidase negative, a phenotypic characteristic that may, however, also occur in C. albicans (7, 25). More recently, Staib agar was shown to allow differentiation between these two species on the basis of abundant chlamydospores and the growth of rough colonies (3, 31). Unfortunately, none of these easy-to-perform methods appear to definitively identify all C. dubliniensis isolates. To attain this goal it is necessary to perform other, time-consuming tests such as carbohydrate assimilation assays, by which C. dubliniensis strains exhibit unique assimilation profiles (26). It has recently been shown that on Pal's agar, a medium prepared with sunflower seed extract, C. dubliniensis isolates yield rough colonies surrounded by a hyphal fringe, while C. albicans colonies are smooth without any evidence of hyphae (2). Modification of this medium to make it transparent has also allowed the direct observation of chlamydospore production through the petri dish (1).
At present, the most accurate means of differentiating between C. dubliniensis and C. albicans requires the use of molecular biology-based techniques, such as DNA fingerprinting analysis with repetitive sequence-containing DNA probes, randomly amplified polymorphic DNA analysis, PCR analysis, or pulsed-field gel electrophoresis (5, 8, 11, 12, 15, 16, 21, 22, 35). Although they are very effective, these techniques are expensive, time-consuming, and not readily applicable to the identification of large numbers of isolates, nor can they be conducted routinely in most standard mycology laboratories at present.
To our knowledge, few monoclonal antibodies (MAbs), polyclonal antibodies, or single-chain variable fragments have been demonstrated to differentiate yeast cells at the species level. Marot-Leblond et al. (19, 20) isolated an MAb (MAb 3D9) that specifically allows the identification of the C. albicans mycelial phase and, more recently, a second MAb (MAb 16B1-F10) that allows the discrimination between C. albicans and C. dubliniensis hyphae by immunofluorescence assay (IFA) and enzyme-linked immunosorbent assay. Bikandi et al. (4) developed an immune serum that allows differentiation of C. dubliniensis and C. albicans blastospores; however, use of this immune serum is restricted to IFA because of cross-reactivity with a variety of intracellular antigens from other yeast species. More recently, Bliss et al. (6), using a combinatorial phage display library of human heavy and light chain variable regions, prepared two single-chain variable fragments that allowed the specific identification of C. albicans and C. dubliniensis germ tubes by IFA. However, they did not consider the possibility that the epitopes that were recognized might also be expressed within the cells of other Candida species or yeast genera (6). To date, two MAbs have led to the development of effective immunological tests for the identification of colonies of C. albicans and C. krusei (13, 18, 28, 29).
In the present study, we have investigated the potential use of an immunochromatographic assay as a basis for the identification of C. albicans and C. dubliniensis. This technique is specific, rapid, easy to perform, and applicable to large numbers of isolates and should enhance the rapid and accurate identification of C. dubliniensis strains in the future.
MATERIALS AND METHODS
Organisms and culture conditions.
Strains of C. albicans (n = 47) and C. dubliniensis (n = 43) were obtained from the Dublin Dental School and Hospital Yeast Collection, Dublin, Ireland, and from the Bilbao Facultad de Medicina Collection. All have been identified by a number of techniques, including a PCR test based on the intron sequence of the ACT1 gene or immunofluorescence with a specific MAb (19). The clinical isolates of Candida spp., and non-Candida yeast species included for comparison were obtained from the Mycological Laboratory of the Medical School Angers. These isolates were identified by using the ID 32C system (bioMérieux SA, Marcy l'Etoile, France). They consisted of 12 C. tropicalis, 14 C. krusei, 15 C. glabrata, 10 C. parapsilosis, 8 C. guilliermondii, 11 C. kefyr, 1 Rhodotorula mucilaginosa, and 11 Cryptococcus neoformans isolates. Freshly isolated clinical strains of C. albicans (n = 11) and C. dubliniensis (n = 17) were also investigated, and their identities were confirmed with modified Pal's agar (1). All yeasts were stored on cryobeads (Hardy Diagnostics, Santa Maria, Calif.) prior to use.
Cells were first subcultured twice onto Sabouraud dextrose agar (SDA; Merck, Darmstadt, Germany) at 37°C for 48 h. Colonies from the second subculture were used for testing. Colonies were transferred in parallel onto SDA, CHROMagar Candida (Becton Dickinson Microbiology Systems, Sparks, Md.), CandiSelect (Sanofi Diagnostics Pasteur, Marnes-la-Coquette, France), or Candida ID (bioMérieux SA) and were then incubated for 48 h at 37°C on these media.
ICM test.
The immunochromatographic membrane (ICM) test (the ICM albi-dubli test) is a rapid immunochromatographic test for the identification of C. albicans and C. dubliniensis that uses a sandwich assay system with two MAbs (MAb LIB 3H8 and MAb 16B1-F10), each of which binds to a repetitive epitope of two distinct antigens. MAb LIB 3H8 reacts with the protein moiety of a glycoprotein of >200 kDa specifically expressed in the C. albicans and C. dubliniensis cell wall (18). In contrast, MAb 16B1-F10 reacts with a glycoprotein component expressed uniquely on the surfaces of C. albicans hyphae (19).
The apparatus comprises a dispstick, a test tube, medium for antigen induction (minimal essential medium and calf serum bovine), and an extraction solution (glucanase) for cell component solubilization. The dipstick for the ICM test is composed of a comb-shaped device that contains a sample pad, a reagent pad, a membrane, and an absorbent pad. Each antibody is immobilized on the membrane as the capture reagent (test lines). The same antibodies, but conjugated with colloidal gold particles, are contained in the absorbent area and are used for antigen capture and detection in the sandwich-type assay.
In the presence of relevant soluble antigens, the antibody-colloidal gold conjugates bind to specific antigen in the sample. The complexes migrate upward along the membrane by capillary action and then bind to the MAbs immobilized on the solid phase in the test zone, producing red bands. Finally, the liquid continues to migrate along the membrane. In the presence of C. albicans two lines appear, and in the presence of C. dubliniensis only one line appears. For other yeast species, no line appears in the positive reaction zone (Fig. 1).
FIG. 1.
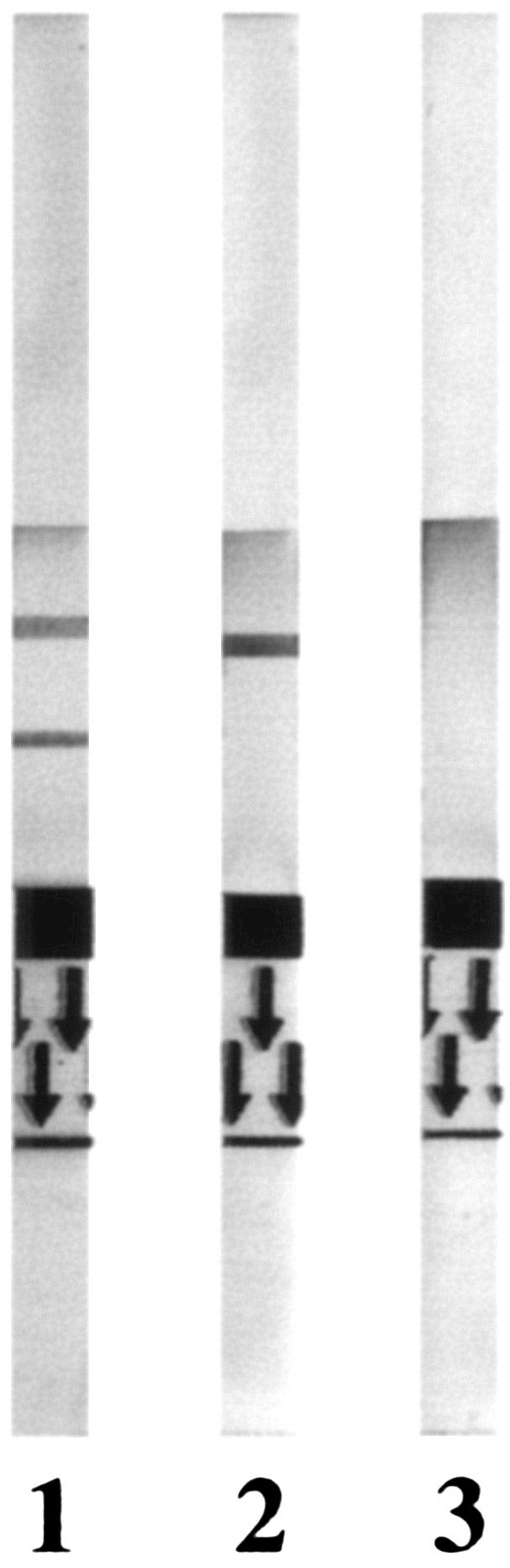
Different features of the results obtained with the ICM (albi-dubli) test kit. Lane 1, C. albicans; lane 2, C. dubliniensis; lane 3, other yeast species.
Dipstick assays. (i) Germ tube induction.
Samples were prepared according to the instructions of the manufacturer (SR2B). Briefly, two to four colonies were picked from the growth medium and transferred to the test tube included in the kit (which contained 1 ml of medium) to yield a suspension equal to a no. 3 McFarland turbidity standard. Cells were allowed to express specific antigens by incubation for 2 h at 37°C without any shaking. For some experiments, yeast suspensions with turbidity greater than a no. 3 McFarland standard were used.
(ii) Antigen extraction and dipstick assay.
Following the incubation period, the medium was discarded, and the remaining cells coated onto the plastic were scraped and resuspended in 100 μl of dissociating agent added to the tube. Antigen extraction was performed by incubation of the tube for 15 min at 37°C without shaking.
After this time, membranes were inserted into the test tube and allowed to stand at room temperature for 5 min and then removed. Assay results were read after 5 min.
All strains were tested in a blinded fashion, and results were compiled at the end of the study.
Data analysis.
The variables measured were the number of samples with true-positive results (TP), the number of samples with true-negative results (TN), the number of samples with false-positive results (FP), and the number of samples with false-negative results (FN). Sensitivity was calculated as TP/(TP + FN), and specificity was calculated as TN/(TN + FP).
RESULTS
The ICM albi-dubli test was first evaluated with yeast cells isolated on SDA. Among the 58 C. albicans strains tested, 4 were misidentified: 3 were misidentified as C. dubliniensis strains and 1 was misidentified as a non-C. albicans and non-C. dubliniensis strain. Two misidentifications were obtained among the 60 C. dubliniensis strains and isolates tested, and no false-positive results were noticed for the 82 other yeasts analyzed. These values indicated sensitivities and specificities of 93.1 and 100%, respectively, for C. albicans and 98.3 and 97.9%, respectively, for C. dubliniensis.
The influence of the growth medium on the expression of the epitopes reacting by the ICM test was also studied. Table 1 summarizes the performance characteristics of the ICM test for the identification of C. albicans and C. dubliniensis according to the growth medium used for isolation. Differences in the performance of the ICM test were observed depending on the isolation medium used, with more accurate results being obtained with SDA, CHROMagar Candida, and CandiSelect, whereas more strains were misidentified when growth was on Candida ID. Misidentifications occurred mostly with C. albicans isolates, which were wrongly identified as C. dubliniensis. For the other species tested, no difference in the performance of the ICM test according to the source of the isolates was observed, since identification as non-C. albicans and non-C. dubliniensis isolates was correct irrespective of the growth medium used. A unique C. dubliniensis strain was incorrectly identified as non-C. albicans and non-C. dubliniensis after subculture on all media.
TABLE 1.
Sensitivity and specificity of the ICM (albi-dubli) test kit
| Organisma and growth mediumb | % Sensitivity | % Specificity |
|---|---|---|
| C. albicans (58) | ||
| SDA | 93.1 | 100 |
| CH | 96.6 | 100 |
| CS | 93.1 | 100 |
| CID | 91.4 | 100 |
| C. dubliniensis (60) | ||
| SDA | 98.3 | 97.9 |
| CH | 98.3 | 99.3 |
| CS | 98.3 | 97.9 |
| CID | 98.3 | 97.2 |
| Other yeasts (82) | ||
| SDA | 100 | 98.3 |
| CH | 100 | 98.3 |
| CS | 100 | 98.3 |
| CID | 100 | 98.3 |
Clinical isolates and reference strains. Numbers in parentheses indicate the number of strains tested.
CH, CHROMagar Candida; CS, CandiSelect; CID, Candida ID.
The same C. albicans yeast strains were generally misidentified by the test regardless of the growth medium used; thus, among the 58 C. albicans strains tested, 5 yielded false-negative results. One strain was misidentified as a non-C. albicans and non-C. dubliniensis isolate, whatever medium was used, although it was clearly identified as C. albicans with the ID 32C carbohydrate assimilation system. The other four C. albicans strains were incorrectly identified as C. dubliniensis. One strain that was misidentified when it was isolated on SDA was also misidentified when it was grown on Candida ID and CandiSelect, while another strain was also misidentified following culture on these two chromogenic media. The two remaining misidentifications were observed for two independent strains grown on SDA and Candida ID, respectively. In summary, the best results were obtained when the test was performed with C. albicans strains grown on CHROMAgar Candida (sensitivity, 96.6%), while the worst results were obtained when the C. albicans strains were grown on Candida ID (sensitivity, 91.4%); however, these values are not significantly different.
The C. albicans strains with false-negative results were further investigated. Microscopic examination of cells was performed after antigen induction in the test tube and before the culture medium was discarded. From the observations it was concluded that the C. albicans isolates (n = 4) that were misidentified as C. dubliniensis could be characterized by a lack or a low rate of germination. Increasing the density of the yeast cell inoculum to greater than a no. 3 McFarland standard before germ tube induction did not significantly improve the performance of the test for these strains.
DISCUSSION
Accurate analysis of the epidemiology of infections caused by C. dubliniensis has been hampered by the lack of effective methods to identify this species in clinical samples. In addition, although C. dubliniensis and C. albicans isolates are both susceptible to azoles, fluconazole resistance has been observed in clinical isolates of C. dubliniensis from AIDS patients who have had prior exposure to fluconazole. Moreover, stable fluconazole resistance can readily be induced in C. dubliniensis isolates following direct exposure to the antifungal in vitro (23, 24). Consequently, these findings may have implications for antifungal therapy and indicate an important reason for distinction between these two species.
Identification of yeast species usually requires prior isolation on a primary isolation medium, which takes 24 to 48 h. When a nonchromogenic medium is used, recognition of C. albicans can be obtained on the basis of a single characteristic (e.g., rapid production of germ tubes in serum or chlamydosporulation). The production of germ tubes and hyphae can be induced by changes in a variety of environmental factors, including ambient pH, nutritional status, and temperature. Germ tube formation is also influenced by the inoculum size, with densities of 105 to 107 cells/ml having been found to be the most conducive to germ tube induction (17). Some other Candida spp., e.g., C. tropicalis, can also generate germ tubes or pseudohyphae which can be confused with the true germ tubes produced by C. albicans. Moreover, this strategy ignores C. dubliniensis, since this species produces germ tubes that cannot be differentiated from those of C. albicans. However, in a previous study (19) it was noted that some C. dubliniensis strains did not produce true germ tubes but were shown to exhibit elongated pseudohyphae produced by polarized cell division of the yeast cells and characterized by constrictions. Solely on the basis of the germ tube test, these strains would have been considered non-C. albicans and non-C. dubliniensis and would have been further investigated by physiologic tests, such as those used by commercial yeast identification systems. Today, many laboratories use commercial systems for Candida species identification. However, these tests are expensive and laborious, and the samples must be incubated for greater than 48 h to obtain accurate results. Comparing such systems, Pincus et al. (26) showed that, according to the kit, doubtful results or misidentifications were obtained for C. dubliniensis (30% for the ID 32C system and 19% after 48 h for the API 20C AUX system). In their study, most isolates identified as C. dubliniensis strains were concluded to be C. albicans. The results obtained with the Vitek 2 ID YST system showed a very high correlation with those obtained by molecular methods, yielding mistaken identities for only 2% of isolates. Chromogenic media allow the simultaneous isolation and identification of C. albicans and C. dubliniensis. A dark green color on CHROMAgar Candida has been described as a potential phenotypic marker for C. dubliniensis in primary cultures (30). However, not all proven C. dubliniensis isolates yield the dark green color in primary cultures (36), thus possibly leading to the potential underestimation of the prevalence of C. dubliniensis.
The reasons for the variation in the results obtained with different isolation media are not clear. However, the differences observed may be the result of differential rates of germ tube induction or antigen expression in each circumstance. According to our observations, it might be useful to examine each sample microscopically before antigen extraction and the dipstick assay are performed. In our study, if the results for C. albicans or C. dubliniensis strains that did not form germ tubes were discarded, the sensitivity of the test for C. albicans identification increased from the range of 91.4 to 93.1% to 98.2%, although no improvement was seen in the case of C. dubliniensis.
The ICM albi-dubli test is a new rapid test that allows the accurate discrimination between C. albicans and C. dubliniensis. The results obtained by this assay have a high specificity and a high sensitivity. The assay is easy to perform, and it offers early results compared to the time that the results of molecular approaches are obtained and can easily be scaled up for use with a high volume throughput.
REFERENCES
- 1.Adou-Bryn, K., C. Douchet, A. Ferrer, L. Grimaud, R. Robert, and D. Richard-Lenoble. 2003. Morphological features of Candida dubliniensis on a modified Pal's medium. Preliminary study. J. Mycol. Med. 13:99-103. [Google Scholar]
- 2.Al Mosaid, A., D. J. Sullivan, and D. C. Coleman. 2003. Differentiation of Candida dubliniensis on Pal's agar. J. Clin. Microbiol. 41:4787-4789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Al Mosaid, A., D. Sullivan, I. F. Salkin, D. Shanley, and D. C. Coleman. 2001. Differentiation of Candida dubliniensis from Candida albicans on Staib agar and caffeic acid-ferric citrate agar. J. Clin. Microbiol. 39:323-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bikandi, J., R. San Millan, M. D. Moragues, G. Cebas, M. Clarke, D. C. Coleman, D. J. Sullivan, G. Quindos, and J. Ponton. 1998. Rapid identification of Candida dubliniensis by indirect immunofluorescence based on differential localization of antigens on C. dubliniensis blastospores and Candida albicans germ tubes. J. Clin. Microbiol. 36:2428-2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Biswas, S. K., K. Yokoyama, L. Wang, K. Nishimura, and M. Miyaji. 2001. Identification of Candida dubliniensis based on the specific amplification of mitochondrial cytochrome b gene. Nippon Ishinkin Gakkai Zasshi 42:95-98. [DOI] [PubMed] [Google Scholar]
- 6.Bliss, J. M., M. A. Sullivan, J. P. Malone, and C. G. Haidaris. 2003. Differentiation of Candida albicans and Candida dubliniensis by using recombinant human antibody single-chain variable fragments specific for hyphae. J. Clin. Microbiol. 41:1152-1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boerlin, P., F. Boerlin-Petzold, and C. Durussel. 1995. Cluster of atypical Candida isolates in a group of human immunodeficiency virus-positive drug users. J. Clin. Microbiol. 33:1129-1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bors, A., B. Theelen, E. Reinders, T. Boekhout, C. Fluit, and P. H. M. Savelkoul. 2003. Use of amplified fragment length polymorphism analysis to identify medically important Candida spp., including C. dubliniensis. J. Clin. Microbiol. 41:1357-1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coleman, D. C., D. J. Sullivan, D. E. Bennett, G. P. Moran, H. J. Barry, and D. B. Shanley. 1997. Candidiasis: the emergence of a novel species, Candida dubliniensis. AIDS 11:557-567. [DOI] [PubMed] [Google Scholar]
- 10.Coleman, D. C., D. Sullivan, B. Harrington, K. Haynes, M. Henman, D. Shanley, D. Bennett, G. Moran, C. McCreary, and L. ONeill. 1997. Molecular and phenotypic analysis of Candida dubliniensis: a recently identified species linked with oral candidosis in HIV-infected and AIDS patients. Oral Dis. 3(Suppl 1):S96-S101. [DOI] [PubMed] [Google Scholar]
- 11.Donnelly, S. M., D. J. Sullivan, D. B. Shanley, and D. C. Coleman. 1999. Phylogenetic analysis and rapid identification of Candida dubliniensis based on analysis of ACT1intron and exon sequences. Microbiology 145:1871-1882. [DOI] [PubMed] [Google Scholar]
- 12.Elie, C. M., T. J. Lott, E. Reiss, and C. J. Morrison. 1998. Rapid identification of Candida species with species-specific DNA probes. J. Clin. Microbiol. 36:3260-3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Freydière, A. M., L. Buchaille, R. Guinet, and Y. Gille. 1997. Evaluation of latex reagents for rapid identification of C. albicans and C. krusei colonies. J. Clin. Microbiol. 35:877-880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jabra-Rizk, M. A., W. A. Falkler, Jr., W. G. Merz, A. A. M. A. Baqui, J. I. Kelley, and T. F. Meiller. 2000. Retrospective identification and characterization of Candida dubliniensis isolates among Candida albicans clinical laboratory isolates from human immunodeficiency virus (HIV)-infected and non-HIV-infected individuals. J. Clin. Microbiol. 38:2423-2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Joly, S., C. Pujol, M. Rysz, K. Vargas, and D. R. Soll. 1999. Development and characterization of complex DNA fingerprinting probes for the infectious yeast Candida dubliniensis. J. Clin. Microbiol. 37:1035-1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kurzai, O., W. J. Heinz, D. J. Sullivan, D. C. Coleman, M. Frosch, and F. A. Muhlschlegel. 1999. Rapid PCR test for discriminating between Candida albicans and Candida dubliniensis isolates using primers derived from the pH-regulated PHR1 and PHR2 genes of C. albicans. J. Clin. Microbiol. 37:1587-1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mackenzie, D. W. R. 1962. Serum germ tube production of Candida albicans. J. Clin. Pathol. 15:563-565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marcilla, A., C. Monteagudo, S. Mormeneo, and R. Sentandreu. 1999. Monoclonal antibody 3H8: a useful tool in the diagnosis of candidiasis. Microbiology 145:695-701. [DOI] [PubMed] [Google Scholar]
- 19.Marot-Leblond, A., L. Grimaud, S. Nail, S. Bouterige, V. Apaire-Marchais, D. J. Sullivan, and R. Robert. 2000. New monoclonal antibody specific for Candida albicans germ tube. J. Clin. Microbiol. 38:61-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marot-Leblond, A., R. Robert, J. Aubry, P. Ezcurra, and J. M. Senet. 1993. Identification and immunochemical characterization of a germ tube specific antigen of Candida albicans. FEMS Immunol. Med. Microbiol. 12:127-136. [DOI] [PubMed] [Google Scholar]
- 21.McCullough, M. J., K. V. Clemons, and D. A. Stevens. 1999. Molecular and phenotypic characterization of genotypic Candida albicans subgroups and comparison with Candida dubliniensis and Candida stellatoidea. J. Clin. Microbiol. 37:417-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McCullough, M., B. Ross, and P. Reade. 1995. Characterization of genetically distinct subgroup of Candida albicans strains isolated from oral cavities of patients infected with human immunodeficiency virus. J. Clin. Microbiol. 33:696-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moran, G. P., D. Sanglard, S. M. Donnelly, D. B. Shanley, D. J. Sullivan, and D. C. Coleman. 1998. Identification and expression of multidrug transporters responsible for fluconazole resistance in Candida dubliniensis. Antimicrob. Agents Chemother. 42:1819-1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moran, G. P., D. J. Sullivan, M. C. Henman, C. E. McCreary, B. J. Harrington, D. B. Shanley, and D. C. Coleman. 1997. Antifungal drug susceptibilities of oral Candida dubliniensis isolates from human immunodeficiency virus (HIV)-infected and non-HIV-infected subjects and generation of stable fluconazole-resistant derivatives in vitro. Antimicrob. Agents Chemother. 41:617-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Odds, F. C., L. Van Nuffel, and G. Dams. 1998. Prevalence of Candida dubliniensis isolates in a yeast stock collection. J. Clin. Microbiol. 36:2869-2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pincus, D. H., D. C. Coleman, W. R. Pruitt, A. A. Padhye, I. F. Salkin, M. Geimer, A. Bassel, D. J. Sullivan, M. Clarke, and V. Hearn. 1999. Rapid identification of Candida dubliniensis with commercial yeast identification systems. J. Clin. Microbiol. 37:3533-3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pinjon, E., D. Sullivan, I. Salkin, D. Shanley, and D. Coleman. 1998. Simple, inexpensive, reliable method for differentiation of Candida dubliniensis from Candida albicans. J. Clin. Microbiol. 36:2093-2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Quindos, G., R. San Millian, R. Robert, C. Bernard, and J. Ponton. 1997. Evaluation of Bichro-latex Albicans, a new method for rapid identification of Candida albicans. J. Clin. Microbiol. 35:1263-1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Robert, R., O. Faure, A. Carloti, B. Lebeau, C. Bernard, A. Marot-Leblond, R. Grillot, and J. M. Senet. 1998. A monoclonal antibody specific to surface antigen on Candida krusei. Clin. Diagn. Lab. Immunol. 5:121-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schoofs, A., F. C. Odds, R. Colebunders, M. Ieven, and H. Goossens. 1997. Use of specialised isolation media for recognition and identification of Candida dubliniensis isolates from HIV-infected patients. Eur. J. Clin. Microbiol. Infect. Dis. 16:296-300. [DOI] [PubMed] [Google Scholar]
- 31.Staib, P., and J. Morschhäuser. 1999. Chlamydospore formation on Staib agar as a species-specific characteristic of Candida dubliniensis. Mycoses 42:521-524. [DOI] [PubMed] [Google Scholar]
- 32.Sullivan, D., and D. Coleman. 1998. Candida dubliniensis: characteristics and identification. J. Clin. Microbiol. 36:329-334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sullivan, D., K. Haynes, J. Bille, P. Boerlin, L. Rodero, S. Lloyd, M. Henman, and D. Coleman. 1997. Widespread geographic distribution of oral Candida dubliniensis strains in human immunodeficiency virus-infected individuals. J. Clin. Microbiol. 35:960-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sullivan, D. J., G. Moran, S. Donnelly, S. Gee, E. Pinjon, B. McCartan, D. B. Shanley, and D. C. Coleman. 1999. Candida dubliniensis: an update. Rev. Iberoam. Micol. 16:72-76. [PubMed] [Google Scholar]
- 35.Sullivan, D. J., T. J. Westerneng, K. A. Haynes, D. E. Bennett, and D. C Coleman. 1995. Candida dubliniensis sp. nov.: phenotypic and molecular characterization of a novel species associated with oral candidosis in HIV-infected individuals. Microbiology 141:1507-1521. [DOI] [PubMed] [Google Scholar]
- 36.Tintelnot, K., G. Haase, M. Seibold, F. Bergmann, M. Staemmler, T. Franz, and D. Naumann. 2000. Evaluation of phenotypic markers for selection and identification of Candida dubliniensis. J. Clin. Microbiol. 38:1599-1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Velegraki, A., and M. Logotheti. 1998. Presumptive identification of an emerging yeast pathogen: Candida dubliniensis (sp. nov.) reduces 2,3,5-triphenyltetrazolium chloride. FEMS Immunol. Med. Microbiol. 20:239-241. [DOI] [PubMed] [Google Scholar]


