Abstract
Acute respiratory distress syndrome (ARDS) is a devastating clinical syndrome with a considerable case fatality rate (~30-40%). Health disparities exist with African descent subjects (ADs) exhibiting greater mortality than European descent individuals (EDs). Myosin light chain kinase (MLCK) is encoded by MYLK whose genetic variants are implicated in ARDS pathogenesis and may influence ARDS mortality. As baseline population-specific epigenetic changes, i.e. cytosine modifications, have been observed between AD and ED individuals, epigenetic variations in MYLK may provide insights into ARDS disparities. We compared methylation levels of MYLK CpGs between ARDS patients and ICU controls overall and by ethnicity in a nested case control study of 39 ARDS cases and 75 non-ARDS intensive care unit controls. Two MYLK CpG sites (cg03892735, cg23344121) were differentially modified between ARDS subjects and controls (p<0.05; q<0.25) in a logistic regression model, where no effect modification from ethnicity or age was found. One CpG site was associated with ARDS in patients less than 58 years old, cg19611163 (intron 19,20). Two CpG sites were associated with ARDS in EDs only, gene body CpG (cg01894985, intron 2,3) and CpG (cg16212219, intron 31,32), with higher modification levels exhibited in ARDS subjects than controls. Cis-acting mQTL (modified cytosine quantitative trait loci) were identified using linear regression between local genetic variants and modification levels for two ARDS-associated CpGs (cg23344121, cg16212219). In summary, these ARDS-associated MYLK CpGs with effect modification by ethnicity and local mQTL, suggest that MYLK epigenetic variation and local genetic background may contribute to health disparities observed in ARDS.
Introduction
Acute respiratory distress syndrome (ARDS) is a devastating clinical syndrome defined by increased vascular leakiness and pulmonary edema resulting from underlying inflammatory disease conditions. Despite improvements in supportive therapy in the management of ARDS, ARDS mortality remains at ~30-40% (1-8). Previous US epidemiologic studies identified a health disparity between persons of African descent (ADs) and European descent (EDs) in mortality from ARDS, with ADs displaying a higher mortality rate than EDs (9, 10). A study in the National Institutes of Health ARDS Network found higher mortality in persons of ADs than EDs before controlling for severity of illness at presentation (11). Additionally, a multicenter observational study found that ADs were less likely to develop the syndrome than EDs (12). These studies of differences in ARDS susceptibility and mortality between these and other ethnic groups (10-14), highlight the need for elucidating the underlying mechanisms of ARDS health disparities (15).
The search for implicated genes involved in ARDS initially focused on studying gene expression in various pre-clinical ARDS models and human case-control studies (16, 17). These studies, combined with pathway analysis of vascular barrier-regulatory genes and genome-wide association studies (GWAS) led to the selection of MYLK (encoding myosin light chain kinase; also known as MLCK) as an attractive ARDS candidate gene (18). Genetic studies have demonstrated that MYLK haplotypes confer ethnic-specific susceptibility to sepsis and sepsis-/trauma-induced ARDS (19-21). Functionally, MLCK regulates endothelial cell permeability via contractile pathways in response to inflammation (22-24). Genetic haplotypes may affect MLCK expression and/or functionality, thus providing a mechanism for ARDS susceptibility (25).
Specifically, the non-muscle isoform of MLCK (nmMLCK) has been implicated in increased lung injury in response to endotoxin administration both in vitro and in vivo (26-29). Furthermore, MYLK genetic variants in the form of single nucleotide polymorphisms (SNPs) were found to contribute to nmMLCK expression and/or protein function as expression quantitative trait loci (eQTL) (25, 30, 31). In addition to eQTL, epigenetic systems can also play a critical regulatory role in MYLK expression. However, the only MYLK epigenetic mechanism in ARDS explored to date involves microRNA-mediated regulation (32-34). Another form of epigenetic regulation, i.e. cytosine modifications (primarily DNA methylation of cytosines at CpG dinucleotides) in MYLK, have not yet been evaluated for its potential role in the pathogenesis of ARDS. Interestingly, differentially modified cytosines in MYLK were recently identified in samples from healthy AD and ED individuals, thereby indicating significant baseline variation in methylation between these two populations (35, 36). Given the existence of natural genetic and epigenetic variations between individuals of ED and AD ancestry, understanding ethnicity-specific MYLK epigenetic regulation will enhance our knowledge of the molecular mechanisms underlying the observed ARDS disparities (37). In this study, we sought to first identify ARDS-associated epigenetic modification of MYLK by comparing cytosine methylation levels in ARDS patients with ICU controls; secondly find ethnicity-associated epigenetic variants among ARDS patients; and thirdly, to determine if common genetic variants contribute to the discovered epigenetic variations.
Materials and Methods
Human Subjects
A nested case control design was used to select whole blood DNA samples from two Chicago-based ARDS cohorts: Consortium for Investigating Intensive Care Unit Genetics (CIICUG, 2006 – 2009, University of Chicago) and Genomic Association Studies (GAS, 2010 – 2013, University of Illinois at Chicago). ARDS cases were defined by the 1994 American-European Consensus Conference (AECC) definition (38). Exclusion criteria in all patients included Hispanic heritage, diagnosis of cancer, history of organ transplant, or concurrent drug overdose, to maintain a clear AD/ED comparison and to exclude known confounders of cytosine modification (39). All samples were identified using a DNA identification number that was linked to demographic and clinical data, but not to personal identifiable data. Final distribution of cases and controls included 39 ARDS cases (18 EDs, 21 ADs) and 75 ICU controls (39 EDs, 36 ADs) for a total of 114 samples (Table 1).
Table 1.
General characteristics, clinical indices and comorbidities of the study population
| African Descent |
European Descent |
|||||
|---|---|---|---|---|---|---|
| ARDS Cases (n = 21) |
ICU Controls (n = 36) |
p | ARDS Cases (n = 18) |
ICU Controls (n = 39) |
p | |
| Age, mean yrs (+/−SD) | 49 (+/−17.4) | 55 (+/−20.6) | 0.26* | 56 (+/−17.6) | 61 (+/−15.7) | 0.27* |
| Male, n (%) | 12 (57%) | 18 (50%) | 0.60† | 11 (61%) | 24 (62%) | 0.98† |
| APACHE II, (25-75%) (n available) |
22 (18-28) (n = 14) |
25 (21-29) (n = 27) |
0.45‡ | 21 (19-36) (n = 7) |
22 (15-26) (n = 25) |
0.39‡ |
| SIRS, n (%) | 17 (81%) | 34 (94%) | 0.18§ | 12 (71%) | 33 (85%) | 0.28§ |
| Sepsis, n (%) | 17 (81%) | 33 (94%) | 018§ | 13 (76%) | 25 (64%) | 0.36† |
| Septic shock, n (%) | 12 (60%) | 21 (60%) | 1.00† | 9 (53%) | 17 (44%) | 0.52† |
| Died ICU/Hosp, n (%) | 7 (35%) | 5 (16%) | 018§ | 8 (47%) | 6 (20%) | 0.05† |
| Obese, n (%) | 5 (29%) | 16 (62%) | 0.04 † | 5 (63%) | 16 (64%) | 1.00§ |
| Lung disease, n (%) | 21 (100%) | 28 (78%) | 0.02 § | 18 (100%) | 20 (51%) | <.001 § |
| Pneumonia | 12 (57%) | 9 (25%) | 0.02 † | 11 (65%) | 5 (13%) | <.001 § |
| Endocrine, n (%) | 4 (19%) | 18 (50%) | 0.02 † | 5 (29%) | 23 (59%) | 0.04 † |
| Diabetes mellitus | 4 (19%) | 12 (33%) | 0.25† | 4 (24%) | 21 (54%) | 0.04 † |
| Hypertension, n (%) | 10 (48%) | 20 (56%) | 0.56† | 9 (53%) | 19 (49%) | 0.77† |
| Cardiac disease, n (%) | 10 (48%) | 19 (53%) | 0.71† | 7 (41%) | 24 (52%) | 0.16† |
| Renal disease, n (%) | 11 (52%) | 26 (72%) | 0.12† | 9 (53%) | 21 (54%) | 0.95† |
| Rheumatologic, n (%) | 5 (24%) | 10 (28%) | 0.74† | 2 (12%) | 8 (21%) | 0.43† |
| Smoker, n (%) | 10 (63%) | 15 (65%) | 0.86† | 8 (80%) | 15 (60%) | 0.43§ |
One-way ANOVA,
Chi-square,
Wilcoxon-Mann-Whitney median (25-75%),
Fisher’s exact are the associated statistical tests for each analysis between ARDS cases and ICU controls within AD and ED individuals as indicated on each row. There were no significant differences found between AD and ED ARDS cases. Percentages represent the percent subjects with the condition out of the total number of subjects with available data within each group.
Available data for all patients included: age, sex, and ethnicity. Clinical data available included presence of sepsis, septic shock, systemic inflammatory response syndrome (SIRS), and ICD-9 Codes (International Statistical Classification of Diseases and Related Health Problems). Data including weight and height, smoking status, whether the patient died in the ICU or in-hospital and APACHE II score (Acute Physiology and Chronic Health Evaluation II) were present for some but not all patients.
The study was carried out according to The Code of Ethics of the World Medical Association (Declaration of Helsinki) and all sample handling and analyses included in this study was determined to not involve “human subjects” as defined in 45 CFR 46.102(f) by the Office for the Protection of Research Subjects at the University of Illinois at Chicago, where sample handling and analyses were based. The original Chicago-based cohort sample collections were approved by the Institutional Review Board at each participating institution, were also carried out according to The Declaration of Helsinki, and informed consent was obtained from all patients before blood draw.
DNA Sample Preparation and DNA Methylation Quantification
For the CIICUG cohort, DNA was isolated from individual patient’s whole blood using the FlexiGene DNA Kit (QIAGEN) according to company protocol and subsequently maintained in −20 to −80°C freezers. For the GAS cohort, DNA was isolated from individual patient’s whole blood using the iPrep PureLink gDNA Blood Kit (Invitrogen) according to company protocol and subsequently maintained in −20 to −80°C freezers. For each selected sample, 800ng DNA was submitted for bisulfite conversion (EZ-96 DNA Methylation Kit, Zymo Research). The efficacy of the bisulfite conversion was checked by PCR amplification and DNA sequencing of selected genomic regions. Approximately 150 ng of bisulfite-converted DNA from each sample was used for profiling DNA methylation levels using the Illumina Infinium Human Methylation 450 BeadChip (450K array) (40), according to the manufacturers’ protocols at the University of Chicago Genomics Core.
DNA Methylation Data Processing
The 450K array data were processed according to a previous a publication (35). Briefly, the M-values (log2 ratio of the intensities of modified probe versus unmodified probe) (41) of CpG probes that passed quality control based on detection p-value (p<0.01) across at least 95% of samples were summarized and batch corrected using COMBAT (42). We also detected 15 CpG probes in MYLK that are mapped to multiple loci in the human genome (43) or contain known common genetic variants based on the dbSNP v135 (44). The final test set was comprised of 37 CpG sites located within the MYLK gene. The genomic location groups (e.g., promoter, gene body) were assigned according to the annotations provided by Illumina. The raw and processed 450K array data have been deposited into NCBI Gene Expression Omnibus database (accession number: GSE67530).
Statistical Analysis
Data analysis was performed with SAS 9.4 (Cary, NC) with a dichotomous dependent variable ARDS case or ICU control. Characteristics of the patient and control groups were compared using one-way ANOVA, Wilcoxon-Mann-Whitney, Chi-square or Fisher’s exact tests, as appropriate (Table 1). Normally distributed M-values (logit transformation of the β-value, i.e., the ratio of methylated probe intensity and overall intensity) were used (41). Logistic regression modeling with ARDS status as the dependent variable was used to assess the association of M-values of individual CpGs and potential effect modification by age (continuous), sex and ethnicity by including interaction terms. For CpGs with significant (p<0.10) ethnicity*M-value or age*M-value interaction terms, analysis was stratified by ethnicity or age (<58 years of age and ≥58 years of age categories) as appropriate. Sex, age and ethnicity were retained as covariates if the model was not already stratified by one of these variables. Including these covariates improved the fit of the model, as analyzed by the log likelihood when adding or removing terms and the Hosmer-Lemeshow Goodness-of-Fit Test, and it is consistent clinically to control for these factors (45-47). Effect modification was determined to be present if the interaction term was statistically significant in a logistic regression model and if the directionality or magnitude of association differed substantially. A nominal p-value of < 0.05 was utilized to identify differential CpGs, and the false discovery rate (FDR) was controlled by q-value for the crude unadjusted associations (48). Some crude unadjusted associations did not meet the nominal p or FDR, however stratified associations met the nominal p-value of < 0.05 requirement (Table 2).
Table 2.
M-values: Differentially modified MYLK CpGs, with M-value as the main effect, in logistic regression with ICU controls as the reference for the dependent variable, adjusting for: age, sex and ethnicity (cg03892735, cg23344121); age and sex (cg01894985, cg16212219); sex and ethnicity (cg19611163). Adj. = adjusted odds ratio, Cr. = crude unadjusted odds ratio, AD and ED or <58 yrs and ≥58 yrs = stratified odds ratios, indicating effect modification by ethnicity or age. β = regression coefficient, SE = standard error.
| CpG Location Base Position |
Odds Ratio |
95% CI | β | SE | p | |
|---|---|---|---|---|---|---|
| cg01894985 | Cr. | 3.01 | 0.57, 15.76 | 1.10 | 0.84 | 0.192 |
| Intron (2,3) | Adj. | 3.98 | 0.74, 21.52 | 1.38 | 0.86 | 0.109 |
| 123589179 | AD | 0.81 | 0.10, 6.48 | −0.21 | 1.06 | 0.845 |
| ED | *205 | 4.50, >999 | 5.32 | 1.95 | 0.006 | |
|
| ||||||
| cg03892735 | Cr. | *0.09 | 0.01, 0.55 | −2.44 | 0.94 | 0.009 |
| Intron (4,5) | Adj. | *0.09 | 0.01, 0.56 | −2.45 | 0.95 | 0.010 |
| 123506296 | ||||||
|
| ||||||
| cg23344121 | Cr. | *0.07 | 0.008, 0.59 | −2.67 | 1.09 | 0.015 |
| Exon 18 | Adj. | *0.06 | 0.006, 0.63 | −2.79 | 1.19 | 0.018 |
| 123419622 | ||||||
|
| ||||||
| cg19611163 | Cr. | *0.10 | 0.01, 0.78 | −2.32 | 1.06 | 0.028 |
| Intron (19,20) | Adj. | *0.09 | 0.01, 0.75 | −2.40 | 1.08 | 0.026 |
| 123411211 | <58 yrs | *0.03 | 0.001, 0.58 | −3.53 | 1.53 | 0.021 |
| ≥58 yrs | 0.19 | 0.007, 4.96 | −1.65 | 1.66 | 0.320 | |
|
| ||||||
| cg16212219 | Cr. | 1.94 | 0.92, 4.07 | 0.66 | 0.38 | 0.082 |
| Intron (31,32) | Adj. | 1.91 | 0.92, 3.99 | 0.65 | 0.37 | 0.083 |
| 123339918 | AD | 1.07 | 0.44, 2.57 | 0.06 | 0.45 | 0.889 |
| ED | *6.14 | 1.37, 27.58 | 1.81 | 0.77 | 0.018 | |
odds ratio with a nominal p value <0.05.
Mapping of mQTL
Genotyping was performed on some of the DNA samples from our on-going GWAS (unpublished data) using the Affymetrix Axiom Genome-Wide PanAFR (Pan-African array) and Affymetrix Genome-Wide Human SNP Array 6.0 according to the manufacturer’s recommendation at the University of Illinois DNA Services Facility. SNPs 1Mb up-/down-stream from the MYLK gene were imputed to the 1000 Genomes Project (49) Phase I integrated data using IMPUTE2 (50). Both directly genotyped and imputed SNPs with MAF (minor allele frequency) > 0.01 and missing rate < 25% (314 local SNPs, in total) were retained for mQTL mapping. Given that mQTL are predominantly local to target CpGs (39), we included SNPs within 1Mb of target CpGs in MYLK to map cis-acting (local) mQTL associated with the CpG modification levels using a linear regression model with sex as a covariate. FDR for mQTL mapping was controlled using the Benjamini-Hochberg (BH) approach (51).
Results
Comparisons of General Characteristics, Clinical Characteristics and Comorbidities
ARDS cases and ICU controls had similar distributions of age and sex (Table 1). Clinical indices, including APACHE II scores and other variables associated with severity, were not statistically different between ARDS cases and ICU controls, except for occurrence of death during the current ICU or hospital stay (Died ICU/Hosp.) where ARDS cases where the odds of dying was higher during the current stay compared to ICU controls (OR = 3.1, 95% CI 1.22-7.82, p = 0.014). Presence of comorbidities was also not statistically different between cases and controls except for pneumonia, being more common among ARDS cases; endocrine disease, being more common among ICU controls; and obesity, being less common among AD ARDS cases (Table 1).
Identification of CpGs in MYLK Associated with ARDS
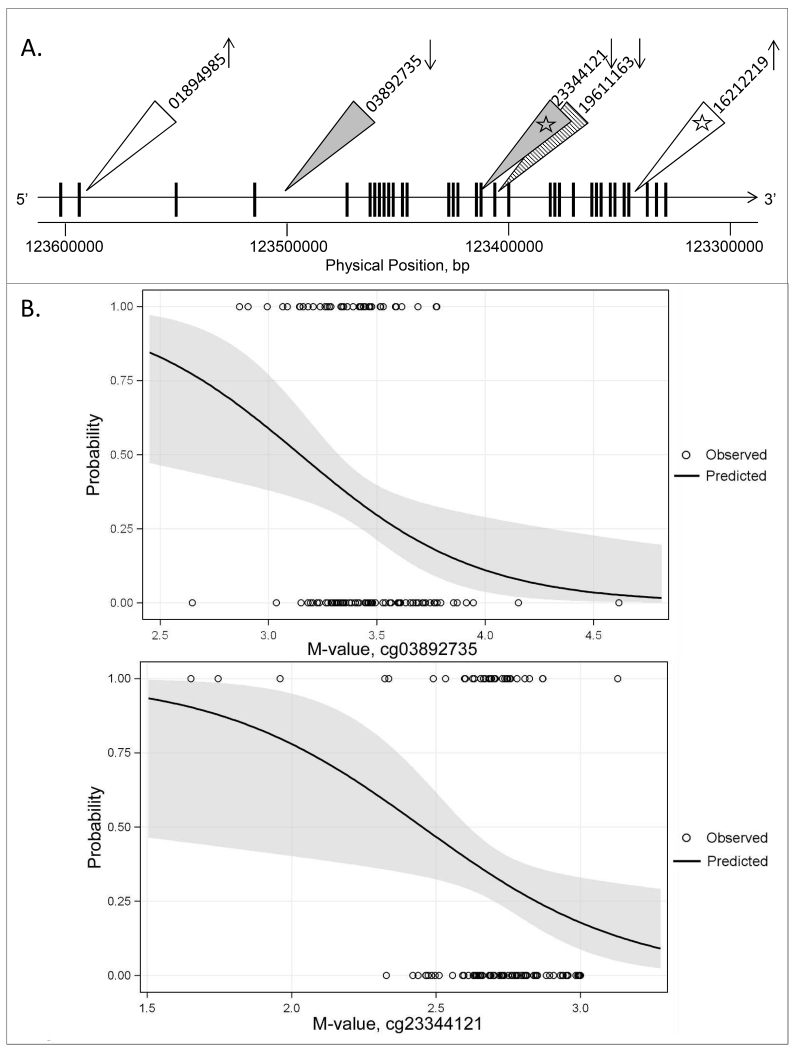
In logistic regression models, of the 33 unambiguous, high quality CpG probes located within MYLK for which ethnicity or age were found not to be an effect modifier for methylation level, two CpG probes, cg03892735 (intron 4,5) and cg23344121 (exon 18) were found to be significantly associated with ARDS status (p<0.05; q<0.25, Table 2, Figure 1B). One CpG, cg19611163 (intron 19,20), for which age was found to be an effect modifier was analyzed in stratified logistic regression models and was found to be differentially methylated between ARDS patients and ICU controls among those less than 58 years of age, but not those 58 years or older, where patients less than 58 years of age were less methylated in ARDS cases compared to ICU controls (Table 2, Figure 1D). Two CpGs for which ethnicity was found to be an effect modifier were evaluated in stratified logistic regression models and were found to be differentially methylated between ARDS subjects and ICU controls among EDs, but not ADs (Table 2, Figure 1C). No modifications unique to AD ARDS cases were found. Of the MYLK cytosines modified in ED ARDS cases, cg01894985 (intron 2,3) and cg16212219 (intron 31,32) were more methylated in ARDS cases compared to ICU controls (Figure 1A, 1C).
Figure 1. ARDS-associated and population-modified CpGs in MYLK.
(A). Locations of significant CpGs after controlling for age and sex and stratifying by ethnicity (cg01894985, cg16212219), for age and ethnicity and stratifying by age (cg19611163), and for age, ethnicity and age (cg03892735, cg23344121), in logistic regression are indicated based on hg19. Vertical bars are exons; white triangles indicate significantly differentially modified CpG sites for ED ARDS cases vs. ED ICU controls; striped triangle indicates a significant CpG for less than 58 year old ARDS vs. less than 58 year old ICU controls; light gray triangles indicate significant CpG sites for ARDS cases vs. ICU controls with no effect modification by age or ethnicity; arrows indicate higher modification (up arrow) or lower modification (down arrow) for ARDS cases vs. ICU controls; numbers correlate to CpG probe ID numbers in the 450k array; white stars indicate significant mQTL detected for two target CpG sites.
(B). Significantly modified CpG sites for ARDS cases vs. ICU controls controlling for age, sex and ethnicity for which no significant M-value interaction terms were identified: cg03892735, cg23344121, located in intron (4,5) and exon 18, respectively. Effect plots of the probability that a case has ARDS (y-axis) according to M-value (x-axis). Open circles indicate observed case status (1 = ARDS, 0 = ICU control), line the predicted probability across M-value, shaded area the 95% confidence limits for the predicted probabilities.
(C). Significantly modified CpG sites for ARDS cases vs. ICU controls among ED, controlling for age and sex: cg01894985, cg16212219, located in intron (2,3) and intron (31,32) respectively. Effect plots of the probability that a case has ARDS (y-axis) according to M-value (x-axis). Plus signs indicate observed ED case status and open circles observed AD case status (1 = ARDS, 0 = ICU control), predicted probability across M-value is indicated by a dashed line for ED patients and a solid line for AD patients, and the shaded areas (light gray for ED, dark gray for AD) indicate the 95% confidence limits for the predicted probabilities.
(D). Significantly modified CpG site for ARDS cases vs. ICU controls among those less than 58 years of age, controlling for sex and ethnicity: cg19611163, located in intron (19,20). Effect plot of the probability that a case has ARDS (y-axis) according to M-value (x-axis). Plus signs indicate observed <58 year old patient case status, open circles indicate observed ≥58 year old patient case status (1 = ARDS, 0 = ICU control), predicted probability across M-value is indicated by a dashed line for <58 year old patients and a solid line for ≥58 year old patients, and the shaded areas (dark gray for <58 year, light gray for ≥ 58 year) indicate the 95% confidence limits for the predicted probabilities.
Mapping of mQTL for ARDS-associated CpGs in MYLK
To assess the potential effect of genetic variation on cytosine modification, mQTL were mapped for the ARDS-associated and ethnicity-interacting CpGs in MYLK (Figure 1A). At the FDR corrected p < 0.05, there were six local SNPs (rs7650050, rs7630208, rs7652791, rs6780229, rs61281424, and rs59204624) (Table 3) significantly associated with cg23344121 (p = 6.84e-5, r2 = 0.23). All six SNPs were located in a linkage disequilibrium (LD) block (r2=1). Eight local mQTL were detected for cg16212219 with a BH-adjusted p-value of <0.05 (Figure 2, Table 3). For other tested CpGs: cg01894985, cg03892735 and cg19611163 no local mQTL was detected at a BH-adjusted p < 0.10.
Table 3.
Beta-values: Six and eight mQTL detected for cg23344121 and cg16212219, respectively. β = regression coefficient, SE= standard error, r2 = correlation coefficient, p = unadjusted p-value, BH-p = Benjamini-Hochberg-adjusted p-value.
| SNP | β | SE | r2 | p | BH-p |
|---|---|---|---|---|---|
| cg23344121 | |||||
| rs112200373 | −0.0191 | 0.0044 | 0.2318 | 5.6E-05 | 0.0031 |
| rs59204624 | −0.0191 | 0.0045 | 0.2305 | 6.8E-05 | 0.0031 |
| rs61281424 | −0.0191 | 0.0045 | 0.2305 | 6.8E-05 | 0.0031 |
| rs6780229 | −0.0191 | 0.0045 | 0.2305 | 6.8E-05 | 0.0031 |
| rs7652791 | −0.0191 | 0.0045 | 0.2305 | 6.8E-05 | 0.0031 |
| rs7630208 | −0.0191 | 0.0045 | 0.2305 | 6.8E-05 | 0.0031 |
| SNP | β | SE | r2 | p | BH-p |
| cg16212219 | |||||
| rs820350 | −0.0225 | 0.0063 | 0.1676 | 0.0006 | 0.0037 |
| rs820334 | −0.0163 | 0.0048 | 0.1287 | 0.0012 | 0.0037 |
| rs1902348 | −0.0173 | 0.0052 | 0.1298 | 0.0014 | 0.0037 |
| rs151080191 | −0.0231 | 0.0079 | 0.1095 | 0.0045 | 0.0080 |
| rs150265167 | −0.0234 | 0.0081 | 0.1133 | 0.0054 | 0.0080 |
| rs114536425 | −0.0234 | 0.0083 | 0.1129 | 0.0062 | 0.0080 |
| rs1947510 | −0.0135 | 0.0048 | 0.1169 | 0.0070 | 0.0080 |
| rs820359 | −0.0130 | 0.0048 | 0.0936 | 0.0080 | 0.0080 |
Figure 2. mQTL mapping for an ARDS-associated CpG in MYLK.
(A). Box and whisker plots show the beta-values of cg16212219 and genotypes of one of its local mQTL, rs820359. The first and third quartiles are indicated at box ends, median by the horizontal line, the minimum and maximum by the end of the whiskers, and outliers by the dots.
(B). The physical positions (hg19) of cg16212219 and its eight detected local mQTL on chromosome 3 are shown.
Discussion
This study identified two epigenetic variants, cg03892735 (intron 4,5), cg23344121 (exon 18), associated with ARDS in both ethnicities, one CpG site associated with ARDS only among those less than 58 years of age, and two CpG sites associated with ARDS only among EDs, when controlling for sex, age and/or ethnicity as appropriate. The two MYLK CpGs associated with ARDS in EDs were more methylated (cg01894985 and cg16212219, located in the gene body) in ARDS cases compared with ICU controls. Of these two CpG sites, gene body cg16212219 was found to have eight associated local SNPs, indicating possible contribution of local genetic variation to this epigenetic variant. Although we hypothesize that the observed variations in CpG modification may result in altered MYLK expression, future studies are necessary to test this hypothesis.
ARDS is a devastating clinical syndrome with a substantial mortality, annually affecting an estimated 190,600 people and attributing to 3.6 million hospital days in the United States (1). Although AD patients display a higher mortality rate than ED patients even after adjusting for demographic and clinical variables (9-11), in the current study, significant differences were not identified for in-hospital mortality between AD and ED ARDS patients. These results highlight several limitations of the study including the limited sample size due to the number of available samples and the applied exclusion criteria. Socioeconomic data and cause of death were not available. Height and weight and APACHE II score were also not available for all patients; therefore, it was not feasible to enter BMI or APACHE II scores into the final regression analyses as it would dramatically reduce the number of samples analyzed. DNA was obtained from peripheral whole blood with multiple cell types, and complete blood cell counts were not available. As multiple physicians phenotyped the ARDS cases, there is a chance for misclassification of cases (52), although the 1994 AECC definition was used in chart evaluation. Type I error is possible due to the false discovery rate of 25% and alpha threshold of 0.05 over 37 CpG sites evaluated. This analysis was not performed in a replicate manner, and it would be of benefit to reproduce these data in another cohort.
While the functionality of the differentially modified CpGs identified in this study needs to be further studied, it is evident that there are novel epigenetic differences in the MYLK gene, associated with genetic variation and modified by ethnicity, between ARDS cases and ICU controls. Understanding these differences and further elucidating the effect of the specific SNPs and CpG modification levels on gene expression will be key next steps in mechanistically understanding the genetic-epigenetic effects on health disparities in ARDS.
Brief Commentary.
Background
Acute respiratory distress syndrome (ARDS) is accompanied with substantial mortality, especially in African descent patients. Despite recent advances, underlying mechanisms influencing ethnic health disparities in ARDS are not fully understood.
Translational Significance
Our nested case-control study found epigenetic modification variation of the well-described ARDS candidate gene, MYLK encoding myosin light chain kinase to be associated with ARDS patients and European ancestry. Epigenetic differences were partially associated with local genetic variation as well. Understanding the mechanisms of these ethnicity-specific and ARDS-associated modifications will be the next step in illuminating the genetic-epigenetic influences on health disparities in ARDS.
Acknowledgements
This work was supported by USDA/NIFA grant AG 2010-34283-21243 (K.L.S., W.Z), NIH grants HG006367 (W.Z), HL 91889 (J.G.N.G.), HL 125615 (J.G.N.G.); P01 HL126609 (J.G.N.G.), and OD010914 (J.D.F.). The authors would like to thank Dr. Rick Kittles for valuable discussions and Michael Wade for his work in sample preparation.
Abbreviations
- ARDS
(acute respiratory distress syndrome)
- AD
(African descent)
- ED
(European descent)
- MLCK
(myosin light chain kinase protein)
- MYLK
(myosin light chain kinase gene)
- CpG
(cytosine-guanine dinucleotide)
- ICU
(intensive care unit)
- mQTL
(modified cytosine quantitative trait loci)
- BMI
(body mass index)
- APACHE II
(Acute Physiology and Chronic Health Evaluation)
- OR
(odds ratio)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
All authors have read the journal’s policy on disclosure of potential conflicts of interest and have no potential conflicts of interest to declare. There were no sources of editorial support for the preparation of this manuscript. All authors have read the journal’s authorship agreement and the manuscript has been reviewed and approved by all named authors.
References
- 1.Rubenfeld GD, Caldwell E, Peabody E, Weaver J, Martin DP, Neff M, et al. Incidence and outcomes of acute lung injury. N Engl J Med. 2005;353(16):1685–93. doi: 10.1056/NEJMoa050333. doi: 10.1056/NEJMoa050333. PubMed PMID: 16236739. [DOI] [PubMed] [Google Scholar]
- 2.Phua J, Badia JR, Adhikari NK, Friedrich JO, Fowler RA, Singh JM, et al. Has mortality from acute respiratory distress syndrome decreased over time?: A systematic review. Am J Respir Crit Care Med. 2009;179(3):220–7. doi: 10.1164/rccm.200805-722OC. doi: 10.1164/rccm.200805-722OC. PubMed PMID: 19011152. [DOI] [PubMed] [Google Scholar]
- 3.Blank R, Napolitano LM. Epidemiology of ARDS and ALI. Crit Care Clin. 2011;27(3):439–58. doi: 10.1016/j.ccc.2011.05.005. doi: 10.1016/j.ccc.2011.05.005. PubMed PMID: 21742210. [DOI] [PubMed] [Google Scholar]
- 4.Villar J, Blanco J, Anon JM, Santos-Bouza A, Blanch L, Ambros A, et al. The ALIEN study: incidence and outcome of acute respiratory distress syndrome in the era of lung protective ventilation. Intensive Care Med. 2011;37(12):1932–41. doi: 10.1007/s00134-011-2380-4. Epub 2011 Oct 14. doi: 10.1007/s00134-011-2380-4. PubMed PMID: 21997128. [DOI] [PubMed] [Google Scholar]
- 5.Sigurdsson MI, Sigvaldason K, Gunnarsson TS, Moller A, Sigurdsson GH. Acute respiratory distress syndrome: nationwide changes in incidence, treatment and mortality over 23 years. Acta Anaesthesiol Scand. 2013;57(1):37–45. doi: 10.1111/aas.12001. doi: 10.1111/aas.12001. PubMed PMID: 23216361. [DOI] [PubMed] [Google Scholar]
- 6.Wang CY, Calfee CS, Paul DW, Janz DR, May AK, Zhuo HJ, et al. One-year mortality and predictors of death among hospital survivors of acute respiratory distress syndrome. Intens Care Med. 2014;40(3):388–96. doi: 10.1007/s00134-013-3186-3. doi: 10.1007/s00134-013-3186-3. PubMed PMID: WOS:000332457400009; PubMed Central PMCID: PMCPMC3943651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Villar J, Blanco J, Kacmarek RM. Current incidence and outcome of the acute respiratory distress syndrome. Curr Opin Crit Care. 2016;22(1):1–6. doi: 10.1097/MCC.0000000000000266. doi: 10.1097/MCC.0000000000000266. PubMed PMID: 26645551; PubMed Central PMCID: PMC26645551. [DOI] [PubMed] [Google Scholar]
- 8.Bellani G, Laffey JG, Pham T, Fan E, Brochard L, Esteban A, et al. Epidemiology, Patterns of Care, and Mortality for Patients With Acute Respiratory Distress Syndrome in Intensive Care Units in 50 Countries. JAMA. 2016;315(8):788–800. doi: 10.1001/jama.2016.0291. doi: 10.1001/jama.2016.0291. PubMed PMID: 26903337; PubMed Central PMCID: PMC26903337. [DOI] [PubMed] [Google Scholar]
- 9.Moss M, Mannino DM. Race and gender differences in acute respiratory distress syndrome deaths in the United States: an analysis of multiple-cause mortality data (1979-1996) Crit Care Med. 2002;30(8):1679–85. doi: 10.1097/00003246-200208000-00001. PubMed PMID: 12163776. [DOI] [PubMed] [Google Scholar]
- 10.Cochi SE, Kempker JA, Annangi S, Kramer MR, Martin GS. Mortality Trends of Acute Respiratory Distress Syndrome in the United States from 1999-2013. Ann Am Thorac Soc. 2016 doi: 10.1513/AnnalsATS.201512-841OC. doi: 10.1513/AnnalsATS.201512-841OC. PubMed PMID: 27403914; PubMed Central PMCID: PMC27403914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Erickson SE, Shlipak MG, Martin GS, Wheeler AP, Ancukiewicz M, Matthay MA, et al. Racial and ethnic disparities in mortality from acute lung injury. Crit Care Med. 2009;37(1):1–6. doi: 10.1097/CCM.0b013e31819292ea. doi: 10.1097/CCM.0b013e31819292ea. PubMed PMID: 19050621; PubMed Central PMCID: PMC2696263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lemos-Filho LB, Mikkelsen ME, Martin GS, Dabbagh O, Adesanya A, Gentile N, et al. Sex, race, and the development of acute lung injury. Chest. 2013;143(4):901–9. doi: 10.1378/chest.12-1118. doi: 10.1378/chest.12-1118. PubMed PMID: 23117155; PubMed Central PMCID: PMCPMC3747719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Khan NA, Palepu A, Norena M, Ayas N, Wong H, Chittock D, et al. Differences in hospital mortality among critically ill patients of Asian, Native Indian, and European descent. Chest. 2008;134(6):1217–22. doi: 10.1378/chest.08-1016. Epub 2008 Aug 8. doi: 10.1378/chest.08-1016. PubMed PMID: 18689577. [DOI] [PubMed] [Google Scholar]
- 14.Ryb GE, Cooper C. Race/ethnicity and acute respiratory distress syndrome: a National Trauma Data Bank study. J Natl Med Assoc. 2010;102(10):865–9. doi: 10.1016/s0027-9684(15)30700-8. PubMed PMID: 21053700. [DOI] [PubMed] [Google Scholar]
- 15.Garcia JG, Sznajder JI. Healthcare disparities in patients with acute respiratory distress syndrome. Toward equity. Am J Respir Crit Care Med. 2013;188(6):631–2. doi: 10.1164/rccm.201307-1394ED. doi: 10.1164/rccm.201307-1394ED. PubMed PMID: 24032377; PubMed Central PMCID: PMC24032377. [DOI] [PubMed] [Google Scholar]
- 16.Grigoryev DN, Finigan JH, Hassoun P, Garcia JG. Science review: searching for gene candidates in acute lung injury. Crit Care. 2004;8(6):440–7. doi: 10.1186/cc2901. doi: 10.1186/cc2901. PubMed PMID: 15566614; PubMed Central PMCID: PMCPMC1065043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Christie JD. Genetic epidemiology of acute lung injury: choosing the right candidate genes is the first step. Crit Care. 2004;8(6):411–3. doi: 10.1186/cc2931. doi: 10.1186/cc2931. PubMed PMID: 15566603; PubMed Central PMCID: PMCPMC1065050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kamp R, Sun X, Garcia JG. Making genomics functional: deciphering the genetics of acute lung injury. Proc Am Thorac Soc. 2008;5(3):348–53. doi: 10.1513/pats.200709-152DR. doi: 10.1513/pats.200709-152DR. PubMed PMID: 18403332; PubMed Central PMCID: PMC2645247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gao L, Grant A, Halder I, Brower R, Sevransky J, Maloney JP, et al. Novel polymorphisms in the myosin light chain kinase gene confer risk for acute lung injury. Am J Respir Cell Mol Biol. 2006;34(4):487–95. doi: 10.1165/rcmb.2005-0404OC. doi: 10.1165/rcmb.2005-0404OC. PubMed PMID: 16399953; PubMed Central PMCID: PMCPMC2644210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garcia JG, Moreno Vinasco L. Genomic insights into acute inflammatory lung injury. Am J Physiol Lung Cell Mol Physiol. 2006;291(6):L1113–7. doi: 10.1152/ajplung.00266.2006. Epub 2006 Jul 28. doi: 10.1152/ajplung.00266.2006. PubMed PMID: 16877634; PubMed Central PMCID: PMC16877634. [DOI] [PubMed] [Google Scholar]
- 21.Christie JD, Ma SF, Aplenc R, Li M, Lanken PN, Shah CV, et al. Variation in the myosin light chain kinase gene is associated with development of acute lung injury after major trauma. Crit Care Med. 2008;36(10):2794–800. doi: 10.1097/ccm.0b013e318186b843. PubMed PMID: 18828194. [DOI] [PubMed] [Google Scholar]
- 22.Garcia JG, Davis HW, Patterson CE. Regulation of endothelial cell gap formation and barrier dysfunction: role of myosin light chain phosphorylation. J Cell Physiol. 1995;163(3):510–22. doi: 10.1002/jcp.1041630311. doi: 10.1002/jcp.1041630311. PubMed PMID: 7775594. [DOI] [PubMed] [Google Scholar]
- 23.Garcia JG, Lazar V, Gilbert-McClain LI, Gallagher PJ, Verin AD. Myosin light chain kinase in endothelium: molecular cloning and regulation. Am J Respir Cell Mol Biol. 1997;16(5):489–94. doi: 10.1165/ajrcmb.16.5.9160829. doi: 10.1165/ajrcmb.16.5.9160829. PubMed PMID: 9160829. [DOI] [PubMed] [Google Scholar]
- 24.Garcia JG, Verin AD, Herenyiova M, English D. Adherent neutrophils activate endothelial myosin light chain kinase: role in transendothelial migration. J Appl Physiol. 1998;84(5):1817–21. doi: 10.1152/jappl.1998.84.5.1817. PubMed PMID: 9572834. [DOI] [PubMed] [Google Scholar]
- 25.Shen K, Ramirez B, Mapes B, Shen GR, Gokhale V, Brown ME, et al. Structure-Function Analysis of the Non-Muscle Myosin Light Chain Kinase (nmMLCK) Isoform by NMR Spectroscopy and Molecular Modeling: Influence of MYLK Variants. PLoS One. 2015;10(6):e0130515. doi: 10.1371/journal.pone.0130515. doi: 10.1371/journal.pone.0130515. PubMed PMID: 26111161; PubMed Central PMCID: PMCPMC4482139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wainwright MS, Rossi J, Schavocky J, Crawford S, Steinhorn D, Velentza AV, et al. Protein kinase involved in lung injury susceptibility: evidence from enzyme isoform genetic knockout and in vivo inhibitor treatment. Proc Natl Acad Sci U S A. 2003;100(10):6233–8. doi: 10.1073/pnas.1031595100. doi: 10.1073/pnas.1031595100. PubMed PMID: 12730364; PubMed Central PMCID: PMC156355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu J, Gao XP, Ramchandran R, Zhao YY, Vogel SM, Malik AB. Nonmuscle myosin light-chain kinase mediates neutrophil transmigration in sepsis-induced lung inflammation by activating beta2 integrins. Nat Immunol. 2008;9(8):880–6. doi: 10.1038/ni.1628. doi: 10.1038/ni.1628. PubMed PMID: 18587400; PubMed Central PMCID: PMC2553242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mirzapoiazova T, Moitra J, Moreno-Vinasco L, Sammani S, Turner JR, Chiang ET, et al. Non-muscle myosin light chain kinase isoform is a viable molecular target in acute inflammatory lung injury. Am J Respir Cell Mol Biol. 2011;44(1):40–52. doi: 10.1165/rcmb.2009-0197OC. doi: 10.1165/rcmb.2009-0197OC. PubMed PMID: 20139351; PubMed Central PMCID: PMC3028257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shimizu Y, Camp SM, Sun X, Zhou T, Wang T, Garcia JG. Sp1-mediated nonmuscle myosin light chain kinase expression and enhanced activity in vascular endothelial growth factor-induced vascular permeability. Pulm Circ. 2015;5(4):707–15. doi: 10.1086/684124. doi: 10.1086/684124. PubMed PMID: 26697178; PubMed Central PMCID: PMCPMC4671745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Han YJ, Ma SF, Wade MS, Flores C, Garcia JG. An intronic MYLK variant associated with inflammatory lung disease regulates promoter activity of the smooth muscle myosin light chain kinase isoform. J Mol Med (Berl) 2012;90(3):299–308. doi: 10.1007/s00109-011-0820-9. doi: 10.1007/s00109-011-0820-9. PubMed PMID: 22015949. [DOI] [PubMed] [Google Scholar]
- 31.Wang T, Zhou T, Saadat L, Garcia JG. A MYLK variant regulates asthmatic inflammation via alterations in mRNA secondary structure. Eur J Hum Genet. 2015;23(6):874–6. doi: 10.1038/ejhg.2014.201. Epub 2014 Oct 1. doi: 10.1038/ejhg.2014.201. PubMed PMID: 25271083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou T, Garcia JG, Zhang W. Integrating microRNAs into a system biology approach to acute lung injury. Transl Res. 2011;157(4):180–90. doi: 10.1016/j.trsl.2011.01.010. Epub 2011 Feb 4. doi: 10.1016/j.trsl.2011.01.010. PubMed PMID: 21420028; PubMed Central PMCID: PMCPMC3073780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Adyshev DM, Moldobaeva N, Mapes B, Elangovan V, Garcia JG. MicroRNA regulation of nonmuscle myosin light chain kinase expression in human lung endothelium. Am J Respir Cell Mol Biol. 2013;49(1):58–66. doi: 10.1165/rcmb.2012-0397OC. doi: 10.1165/rcmb.2012-0397OC. PubMed PMID: 23492194; PubMed Central PMCID: PMCPMC3727884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Adyshev DM, Elangovan VR, Moldobaeva N, Mapes B, Sun X, Garcia JG. Mechanical stress induces pre-B-cell colony-enhancing factor/NAMPT expression via epigenetic regulation by miR-374a and miR-568 in human lung endothelium. Am J Respir Cell Mol Biol. 2014;50(2):409–18. doi: 10.1165/rcmb.2013-0292OC. doi: 10.1165/rcmb.2013-0292OC. PubMed PMID: 24053186; PubMed Central PMCID: PMCPMC3930953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moen EL, Zhang X, Mu W, Delaney SM, Wing C, McQuade J, et al. Genome-wide variation of cytosine modifications between European and African populations and the implications for complex traits. Genetics. 2013;194(4):987–96. doi: 10.1534/genetics.113.151381. doi: 10.1534/genetics.113.151381. PubMed PMID: 23792949; PubMed Central PMCID: PMCPMC3730924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang W, Gamazon ER, Zhang X, Konkashbaev A, Liu C, Szilagyi KL, et al. SCAN database: facilitating integrative analyses of cytosine modification and expression QTL. Database (Oxford) 2015:2015. doi: 10.1093/database/bav025. doi: 10.1093/database/bav025. PubMed PMID: 25818895; PubMed Central PMCID: PMCPMC4375357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Szilagyi KL, Garcia JG, Zhang W. Exploring DNA Methylation of MYLK as a Contributor to Acute Respiratory Distress Syndrome Disparities. Journal of Pulmonary & Respiratory Medicine. 2013;03(04):e127. doi: 10.4172/2161-105X.1000e127. [Google Scholar]
- 38.Bernard GR, Artigas A, Brigham KL, Carlet J, Falke K, Hudson L, et al. The American-European Consensus Conference on ARDS. Definitions, mechanisms, relevant outcomes, and clinical trial coordination. Am J Respir Crit Care Med. 1994;149(3 Pt 1):818–24. doi: 10.1164/ajrccm.149.3.7509706. doi: 10.1164/ajrccm.149.3.7509706. PubMed PMID: 7509706. [DOI] [PubMed] [Google Scholar]
- 39.Zhang X, Moen EL, Liu C, Mu W, Gamazon ER, Delaney SM, et al. Linking the genetic architecture of cytosine modifications with human complex traits. Hum Mol Genet. 2014;23(22):5893–905. doi: 10.1093/hmg/ddu313. doi: 10.1093/hmg/ddu313. PubMed PMID: 24943591; PubMed Central PMCID: PMCPMC4204771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bibikova M, Barnes B, Tsan C, Ho V, Klotzle B, Le JM, et al. High density DNA methylation array with single CpG site resolution. Genomics. 2011;98(4):288–95. doi: 10.1016/j.ygeno.2011.07.007. doi: 10.1016/j.ygeno.2011.07.007. PubMed PMID: 21839163. [DOI] [PubMed] [Google Scholar]
- 41.Du P, Zhang X, Huang CC, Jafari N, Kibbe WA, Hou L, et al. Comparison of Beta-value and M-value methods for quantifying methylation levels by microarray analysis. BMC Bioinformatics. 2010;11(11):587. doi: 10.1186/1471-2105-11-587. doi: 10.1186/1471-2105-11-587. PubMed PMID: 21118553; PubMed Central PMCID: PMCPMC3012676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8(1):118–27. doi: 10.1093/biostatistics/kxj037. doi: 10.1093/biostatistics/kxj037. PubMed PMID: 16632515. [DOI] [PubMed] [Google Scholar]
- 43.Zhang X, Mu W, Zhang W. On the analysis of the illumina 450k array data: probes ambiguously mapped to the human genome. Front Genet. 2012;3(3):73. doi: 10.3389/fgene.2012.00073. doi: 10.3389/fgene.2012.00073. PubMed PMID: 22586432; PubMed Central PMCID: PMCPMC3343275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29(1):308–11. doi: 10.1093/nar/29.1.308. PubMed PMID: 11125122; PubMed Central PMCID: PMCPMC29783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weidner CI, Wagner W. The epigenetic tracks of aging. Biol Chem. 2014;395(11):1307–14. doi: 10.1515/hsz-2014-0180. doi: 10.1515/hsz-2014-0180. PubMed PMID: 25205717; PubMed Central PMCID: PMC25205717. [DOI] [PubMed] [Google Scholar]
- 46.Singmann P, Shem-Tov D, Wahl S, Grallert H, Fiorito G, Shin SY, et al. Characterization of whole-genome autosomal differences of DNA methylation between men and women. Epigenetics Chromatin. 2015;8(43):43. doi: 10.1186/s13072-015-0035-3. doi: 10.1186/s13072-015-0035-3. PubMed PMID: 26500701; PubMed Central PMCID: PMCPMC4615866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Toperoff G, Kark JD, Aran D, Nassar H, Ahmad WA, Sinnreich R, et al. Premature aging of leukocyte DNA methylation is associated with type 2 diabetes prevalence. Clin Epigenetics. 2015;7(1):35. doi: 10.1186/s13148-015-0069-1. doi: 10.1186/s13148-015-0069-1. PubMed PMID: 25829970; PubMed Central PMCID: PMCPMC4379765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 2003;100(16):9440–5. doi: 10.1073/pnas.1530509100. doi: 10.1073/pnas.1530509100. PubMed PMID: 12883005; PubMed Central PMCID: PMCPMC170937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Genomes Project C. Abecasis GR, Altshuler D, Auton A, Brooks LD, Durbin RM, et al. A map of human genome variation from population-scale sequencing. Nature. 2010;467(7319):1061–73. doi: 10.1038/nature09534. doi: 10.1038/nature09534. PubMed PMID: 20981092; PubMed Central PMCID: PMCPMC3042601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5(6):e1000529. doi: 10.1371/journal.pgen.1000529. doi: 10.1371/journal.pgen.1000529. PubMed PMID: 19543373; PubMed Central PMCID: PMCPMC2689936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society Series B (Methodological) 1995;57(1):289–300. [Google Scholar]
- 52.Rubenfeld GD, Caldwell E, Granton J, Hudson LD, Matthay MA. Interobserver variability in applying a radiographic definition for ARDS. Chest. 1999;116(5):1347–53. doi: 10.1378/chest.116.5.1347. PubMed PMID: 10559098; PubMed Central PMCID: PMC10559098. [DOI] [PubMed] [Google Scholar]