Abstract
Small molecule inhibitors that target components of the spliceosome have great potential as tools to probe splicing mechanism and to dissect the splicing regulatory networks in cells. These compounds also hold promise as drug leads for diseases in which splicing regulation plays a critical role, including many cancers. Because the spliceosome is a complicated and dynamic macromolecular machine comprised of many RNA and protein components, a variety of compounds that interfere with different aspects of spliceosome assembly is needed to probe its function. By screening chemical libraries with high-throughput splicing assays, several labs have added to the collection of splicing inhibitors, although the mechanistic insight into splicing yielded from the initial compound hits is somewhat limited so far. In contrast, SF3B1 inhibitors stand out as a great example of what can be accomplished with small molecule tools. This group of compounds were first discovered as natural products that are cytotoxic to cancer cells, and then later shown to target the core spliceosome protein SF3B1. The inhibitors have since been used to uncover details of SF3B1 mechanism in the spliceosome and its impact on gene expression in cells. Continuing structure activity relationship analysis of the compounds is also making progress in identifying chemical features key to their function, which is critical to understanding the mechanism of SF3B1 inhibition. The knowledge is also key for the design of analogs with new and useful features for both splicing researchers and clinicians hoping to exploit splicing as pressure point to target in cancer therapy.
Keywords: Splicing, pre-mRNA, spliceosome, Inhibitor, Alternative splicing
Graphical abstract
Introduction
Pre-mRNA splicing is a key regulatory step in gene expression
During eukaryotic gene expression the genetic information in DNA that is transcribed into RNA is discontinuous due to the presence of intron sequences. Pre-messenger RNA (pre-mRNA) splicing is required to remove the introns and join the flanking exon sequences, which creates the open reading frames in mRNA that will be translated into protein. Chemically, splicing is a straightforward sequence of two phosphodiester transfer reactions (Figure 1). The 1st chemical step results in cleavage of the bond at the beginning of the intron (5' splice site) and transfer to 2' OH of an adenosine near the end of the intron (branch point). The 2nd chemical step results in cleavage of the bond at the end of the intron (3' splice site) and transfer to the 3' OH created in the first reaction, which join the flanking exons. These reactions are catalyzed by the spliceosome, a complicated assembly of five small nuclear RNAs (snRNAs) and dozens of proteins.
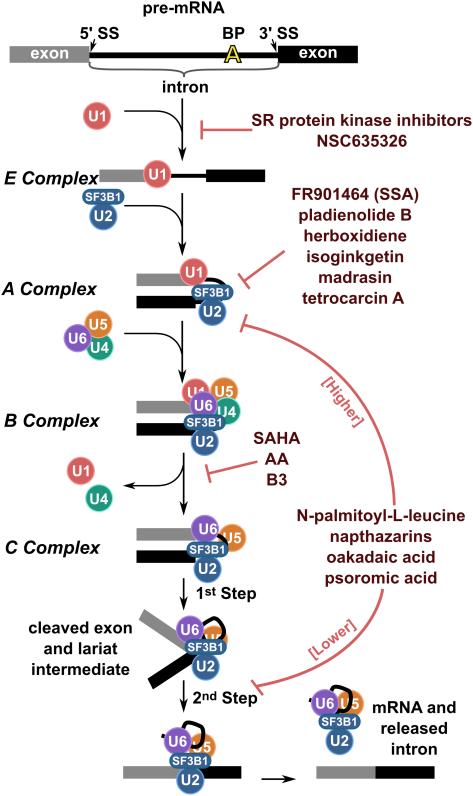
Figure 1. Spliceosome assembly pathway is blocked at different stages by splicing inhibitors.
Simplified schematic of the step-wise assembly of a spliceosome on a model pre-mRNA substrate consisting of two exons flanking an intron. Key splicing sequences are highlighted for the topmost substrate: 5' splice site (5' SS), branch point sequence (BP) and 3' splice site (3'SS). In actuality, the U snRNPs (U1, U2, U4, U5 and U6) are dynamic complexes that each contains a structured RNA and several proteins. The SF3B1 subunit of U2 snRNP, which is a target of many inhibitors, is indicated. The U snRNPs form several RNA-RNA interactions with each other and the pre-mRNA to first recognize the splice sites in E and A complex, before rearranging in B complex to create the catalytically active C complex. After the 1st and 2nd steps of splicing chemistry take place, the spliceosome disassociates into released mRNA product and intron lariat complex. Not illustrated are the dozens of additional protein components of the spliceosome. The assembly transitions affected by select splicing inhibitors are also highlighted.
In addition to the challenge of orienting the phosphodiester bonds at the 5' and 3' splice sites for catalysis and coordinating the two reactions, the spliceosome is responsible for the equally important task of selecting the right pair of splice sites, which are situated 100's to 10,000's of nucleotides apart. A choice of the wrong phosphodiester bond, even by a single nucleotide, will disrupt the final transcript message by creating a frameshift in the open reading frame. Notably, many human genetic diseases are linked to aberrant splicing 1.
In many organisms, the output of splicing is often further elaborated through alternative splicing, which tweaks the protein coding capacity and the stability of the resulting mRNAs by the selective inclusion or skipping of exons, retention of introns, and choice of alternate 5' or 3' splice sites. Alternative splicing adds an important layer of gene regulation that is exploited in many biological pathways including developmental programs, neuron identity and metabolic potential, to name just a few. Regulation of alternative splicing is often described in terms of splicing factors that guide or block spliceosome assembly at particular splice sites. Additionally, competition between splice sites that is influenced by the strength of splice site sequences, the order of their emergence from RNA polymerase II, and the availability of the spliceosome, among other factors, are other themes of regulation.
Spliceosome assembly / function is complicated
Likely reflecting the complexity involved in recognizing a wide variety of intron substrates and alternative splicing regulation, the spliceosome is a complicated macromolecule. It assembles on each intron to be spliced through the addition and rearrangement of the U1, U2, U4, U5 and U6 small nuclear ribonucleoproteins (snRNPs), which along with their snRNAs associate with specific and shared proteins. Dozens of additional proteins also participate in the assembly. The spliceosome is not a static entity and can be modeled as evolving through series of stereotypic intermediate splicing complexes (Figure 1). In a very simplistic view, spliceosome assembly starts when the U1 snRNP recognizes the 5' splice site to form E complex, followed by U2 snRNP addition at the branch point sequence to form A complex. The U4/U6.U5 tri-snRNP is then recruited to form B complex. Next, structural rearrangements that include ejection of U1 and U4 snRNPs create the catalytic C complex, in which splicing chemistry takes place. For the purpose of this review, we will refer to this simplified model of the spliceosome, but note that characterization of more RNA/RNA and RNA/protein rearrangements have led to additional assembly intermediates to the splicing process (e.g. Bact, B*, P complex) 2-4. These new intermediates point to our increasing understanding of the intricate interactions that take place during splicing, but they likely still represent only a subset of the conformations that the snRNAs and myriad of spliceosome proteins take on in the process.
The dynamics of the spliceosome pose a challenge to detailed mechanistic studies. Furthermore, while the list of spliceosome components is long, the list of known functions for these components is small, in large part because the means to manipulate them is lacking. Small molecules that target individual spliceosome components provide a flexible way to probe their function and uncover new details of spliceosome assembly. In this review, we summarize the identification and characterization of compounds that have been shown to interfere with spliceosome function, which we term splicing inhibitors. We also consider the potential for inhibitors to study the numerous cellular pathways that are impacted by splicing, and as drug leads for diseases in which splicing regulation plays a critical role. Table 1 highlights some of the most pertinent information gathered from a wide literature for representative groups of splicing inhibitors. With a few exceptions, we will not discuss molecules that have been identified to change alternative splicing choices of single genes but do not appear to generally inhibit splicing at the level of the spliceosome. Such compounds presumably target factors that regulate select splicing events upstream of the spliceosome.
Table 1.
Splicing inhibitors
|
Inhibitor class:
Prototype and select members |
Discovery as splicing
inhibitor |
~ IC50 for in vitro
splicing / first ssome block |
Features of prototype inhibitor | Other activities |
|---|---|---|---|---|
|
FR901464
FR901463/5 spliceostatins A-G thailanstatins A-C meamycins |
Spliceosome protein SF3B1 identified as anti-cancer compound target23 |
50 nM – 200 μM | Targets SF3B1; natural product isolated from FERM BP-3421 (Burkholderia sp.); affects cellular splicing in low nM range; SAR data summarized in Fig. 2A; synthetic pathway available |
Causes mega-speckles phenotype; cytotoxic at low nM range in multiple cell lines; arrests cell cycle at G1 and G2/M; anti-tumor activity in mouse xenograft model23, 84 |
| A complex | ||||
|
Pladienolide B
pladienolides A-G E7107 FD-895 |
Spliceosome protein SF3B identified as anti- cancer compound target22 |
0.9 – >200 μM | Targets SF3B1; natural product isolated from Streptomyces platensis Mer-11107; 12- memberedmacrolide; affects cellular splicing in low nM range; SAR data summarized in Fig. 2B; synthetic pathway available |
Same as listed for FR90164; derivative E7107 was tested in preliminary clinical trials 85 |
| A complex | ||||
|
Herboxidiene
(GEX1A) GEX1Q1-5 RQN-18690A (18- deoxyherboxidiene) |
Spliceosome protein SF3B1 identified as anti-cancer compound target17 |
230 – 800 nM | Targets SF3B1; natural product isolated from Streptomyces sp.; polyketide; affects cellular splicing in 1 μM range; SAR data available; synthetic pathway available |
Same as listed for FR90164; herbicidal activity108, inhibits angiogenesis 109, 110 |
| A complex | ||||
|
Sudemycins
C1, D1, F1, E, D6 |
Design based on pharmacophore model between FR901464 and pladienolide B65 |
N.D. | Targets SF3B1; affects cellular splicing in low nM range of some cell lines; multiple rounds of SAR simplified structure and synthesis for potential drug use/clinical trials; synthetic pathway available. |
Cytotoxic at low nM range in multiple cell lines; anti- tumor activity in mouse xenograft model; synergistic effect with some known anticancer drugs 65, 81 |
| N.D. | ||||
| Isoginkgetin | HTS with cell-based splicing reporter assay10 |
30 μM | Natural product found in a variety of plants including Ginkgo biloba; biflavonoid; affects cellular splicing in low nM range; inhibits multiple pre-mRNA substrates in vitro and major and minor spliceosome in cells |
Inhibits tumor cell invasion111; cyto-protective effects against Ab42-induced toxicity in Alzheimer’s disease112 |
| A complex | ||||
|
Madrasin
(DDD00107587) |
HTS with in vitro
splicing assay with enzyme-linked antibody to DDX417 |
<62.5-150 μM | Derived by SAR analysis of hit compound from screen; affects cellular splicing at 30 μM; inhibits multiple substrates in vitro and in cells (HeLa and HEK293), affects alternative splicing |
Cytotoxic; arrests cell cycle; reduces DNA synthesis, disrupts Cajal bodies7 |
| A complex | ||||
|
Tetrocarcin A
(NSC333856) |
HTS using in vitro
splicing assay with RT- qPCR readout6 |
25 μM | Natural product isolated from the culture broth of Micromonospora chalcea KY11091; also inhibits splicing in yeast extracts |
Anti-tumor activity 113, 114; Cytotoxic in several cell lines; antibiotic activity115; promotes apoptosis by blocking BCL2, activating caspase-9 or inhibiting PI3K kinase116-118 |
| A complex | ||||
|
N-palmitoyl-L-
leucine |
HTS using in vitro
splicing assay with RT- qPCR readout5 |
35 μM | Natural product isolated from Streptomyces sp. RL10- 300-HVF-B (marine sediment sample, Dolan Canyon, CA); inhibits multiple pre-mRNA substrates in vitro; tail length most important for activity with head group and absolute orientation also contributing; synthesizable |
Impacts HeLa cytological features in 35 μM range5 |
| B/C complex | ||||
| Psoromic acid | HTS using in vitro
splicing assay with enzyme-linked antibody to Abstrakt8 |
56 μM | Cell-membrane permeable depsidone produced as secondary metabolite by lichens; SAR suggest the functional core is a depsidone containing 3-aldehyde and 4-hydroxy groups |
Induces apoptosis in rat hepatocytes, activates caspase 3119, inhibits Rab geranylgeranyl- transferase120 |
| B/C complex | ||||
| Clotrimazole | HTS using cell-based splicing reporter assay11 |
N.D. | Imidazole derivative; inhibits the major and minor spliceosome in cells but is not a general splicing inhibitor, changes 874 alternative splicing events |
IC50 45 μM for cell-based splicing-reporter assay; cytotoxic for several cell lines; antifungal medication; inhibits cytochrome P450; interferes with cellular calcium homeostasis 11 |
| N.D. | ||||
| NSC635326 | HTS using in vitro
splicing assay with RT- qPCR readout6 |
50 μM | Indole derivative from an NCI library; SAR shows that presence and orientation of nitrophenyl ring is important |
|
| H/E complex | ||||
|
Napthazarin:
NSC659999 BN82865 NSC95397 |
HTS using in vitro
splicing assay with RT- qPCR readout6 |
10 – 20 μM | Naphthazarin derivative from an NCI library; inhibition of 2nd step is more sensitive than inhibition of 1st step; inhibits yeast splicing; mechanism involves modification of redox-sensitive cysteine(s) by reactive oxygen species; SAR suggests nahthazarin scaffold is important; similar compounds include naphthoquinone derivatives |
Suppresses tumor growth; NSC95397 inhibits CDC25 dual-specificity phosphatase 121, 122 |
| B/C complex | ||||
|
Translation
Inhibitors: Erythromycin Cl-tetracycline streptomycin |
Candidate-approach screen using in vitro splicing assay 36, 123 |
160 – 230 μM | Natural product produced by Saccharopolyspora
erythraea; macrolide; likely inhibits indirectly by non- specific binding to pre-mRNA; commercially available |
Antibiotic; translation inhibitor; binds ribosome peptide exit tunnel 38, 39 |
| B/C complex | ||||
|
HDAC inhibitors:
SAHA Splitomicin DHC |
Candidate-approach screen using in vitro splicing assay 24 |
1.5 – 2 mM | Synthetic hydroxamic acid suberoylanilide derivative; inhibits major and minor spliceosome in vitro; stalled complexes are functional intermediates; commercially available |
Inhibitor of Zn2+-dependent classes I and II of HDACs; antineoplastic activity; used for treatment of cutaneous T cell lymphoma 124, 125 |
| B complex | ||||
|
HAT inhibitors:
Garcinol AA BA3 |
Candidate-approach screen using in vitro splicing assay 24 |
25 – 500 μM | Polyisoprenylated benzophenone derivative isolated from Garcinia indica; inhibits major and minor spliceosome in vitro; stalled complexes are functional intermediates; commercially available |
Inhibitor for the p300/CBP and Gcn5/PCAF families of HATs126, 127 |
| A complex B complex B complex | ||||
|
PP1/PP2A
inhibitors: Okadaic acid Tautomycin microcystin-LR |
Testing importance of Ser/Thr protein phosphatases in splicing 34 |
<1 μM | Ser/Thr phosphatase inhibitor; polyketal fatty acids produced by several dinoflagellates; addition of purified catalytic PP1 or PP2A subunits restores splicing; commercially available |
|
| B/C complex | ||||
|
SR Protein Kinase
inhibitors: NB-506 diospyrin Chlorhexidine TG003 |
Testing importance of DNA topoisomerase I phosphorylation of SR proteins in splicing32 |
50 – <100 μM | DNA topoisomerase I kinase function inhibitor; 13-/V- glucopyranosyl and 6-JV-formylamino derivative of BE- 13793C; isolated from Streptoverticillium sp.; affects cellular splicing at 1 μM; inhibits SF2/ASF phosphorylation; synthesizable |
Anti-tumor activity in mouse xenograft model 128; Chlorhexidine and TG003 are Clk family of SR protein kinase inhibitors 11, 129 |
| H/E complex | ||||
| Ubistatin A | Testing importance of ubiquitination in splicing25 |
<15 mM | Small molecule from a NCI library; decreases U4/U6 unwinding time ~10-fold |
Inhibits proteasome-dependent degradation of ubiquitinated proteins130 |
| N.D. | ||||
| G5 | Candidate-approach using in vitro splicing assay in yeast extract123 |
0.8 mM (in yeast extract) |
Oxaspiro compound; SAR shows requirement of spirolactone ring and contribution of spirolactone structure with larger R1 groups improve splicing inhibition |
Oxaspiro compounds have antibacterial, insecticidal, tumoricidal, and anti-inflammatory activities123, 131 |
| B/C complex |
Searching for splicing inhibitors
High throughput screens with in vitro splicing assays
One approach to finding splicing inhibitors has been through cell-free in vitro splicing assays, which can be adapted for high-throughput screening of small molecule libraries. By bypassing other steps of gene expression, use of in vitro systems increase the chances of finding direct spliceosome inhibitors. They are also not limited to identifying molecules that can penetrate cell membranes. On the down side, compounds that affect splicing regulation and coordination with other processes, such as transcription, are likely to be missed.
Several labs have identified splicing inhibitors from range of compound libraries by assaying in vitro splicing in HeLa nuclear extract, although different strategies were used to measure splicing. Our group used RT-qPCR to detect production of spliced mRNA in the presence of known bioactive compounds and natural products from marine bacteria and characterized four new splicing inhibitors 5, 6. Three other groups employed enzyme-linked-antibodies to detect different proteins associated with the formation of catalytic spliceosomes on an immobilized pre-mRNA splicing substrate and together screened small molecule libraries ranging from ~2000-70,000 compounds 7-9. So far, all the compounds identified from the different screens display relatively low potency, with IC50 for in vitro splicing in the micromolar range. It is possible that the activity of some of these compounds could be improved by structure activity relationship (SAR) approaches, but those efforts have been limited 5. There is still impetus to continue high throughput screening for splicing inhibitors. With over 100 components participating in the dynamic assembly pathway, many activities in the spliceosome remain available as small molecules targets, such as the enzymatic function of distinct RNA-dependent ATPases that facilitate structural rearrangements. Furthermore, a huge swath of chemical space that likely includes new spliceosome-targeting compounds still waits to be explored.
High throughput screens with cell-based assays
Cell-based assays have also been used to identify splicing inhibitors in a more physiological setting. Compounds with activity will necessarily have an ability to penetrate cell membranes and make their way to the nucleus to affect splicing. These characteristics are especially important if the compounds will be used in vivo or as drug leads. Two groups have developed mini-gene splicing reporters expressed in cells to screen for inhibitors. The Moore lab engineered a HEK293 cell line that expresses luciferase when the intron in the reporter is not removed, with which they identified the natural product isoginkgetin in a library of about 8000 compounds 10. They showed that isoginkgetin interferes with spliceosome assembly in vitro, suggesting a direct effect on splicing. Taking an opposite tact, the Dreyfuss lab created a HeLa cell line that expressed luciferase only when intron in the reporter is removed and found three inhibitors in a screen of more than 23,000 small molecules 11. However, the compounds did not inhibit splicing of a model pre-mRNA in vitro, suggesting that they target a splicing regulator that is specific to the reporter intron. In line with that idea, one compound, chlorhexidine, was shown to inhibit Clks, i.e. the kinases that regulate SR proteins. SR proteins are a family of RNA binding proteins that are controlled by phosphorylation of a domain consisting of extended repeats of Arg/Ser. Several SR proteins are characterized as enhancers of spliceosome assembly at exons and often modulate alternative splicing.
In some cases, searching for molecules that alter splicing of a specific gene leads to compounds that nonetheless target spliceosome components, and in some cases enhance, rather than inhibit splicing. Two groups used luciferase mini-gene reporter based on the SMN2 gene to screen for compounds that selectively promote inclusion of exon 7 12, 13. This splicing event has been shown as a potential avenue for treating the disease spinal muscular atrophy (SMA). Both groups identified separate sets of related compounds that enhanced exon 7 inclusion and, notably, also relieved SMA disease symptoms in a mouse model. At the concentrations tested, the compounds are each very selective for a small of set splicing substrates, which suggests that their targets are gene specific splicing factors or regulatory sequences. Remarkably, however, Palacino et al showed that their compounds stabilize U1 snRNP interactions with the 5' splice site of SMN2 exon 7 to affect splicing 13.
Inhibitor Effects on the Spliceosome Are Varied
Interfering with spliceosome assembly
Splicing inhibitors, by definition, block splicing. Their effect can be quantified by measuring the amounts of 1st step and 2nd chemical step splicing products from in vitro splicing relative to inhibitor concentration. However, a compound can interfere with any number of spliceosome assembly steps prior to catalysis to confer a loss of splicing chemistry. For example, they could inhibit enzymatic activity of RNA-dependent ATPases that promote specific rearrangements, block interactions between spliceosome components, hyperstabilize interactions between spliceosome components and or freeze components in non-productive conformations, any of which could stall assembly. Spliceosome assembly can be visualized in native gels by the discrete shifts in mobility of a model pre-mRNA as intermediate complexes form in vitro in nuclear extract (E -> A -> B -> C complex) 14, 15. When an intermediate splicing complex accumulate in the presence of a splicing inhibitor, the list of potential components/steps that the compound may be targeting is limited to some extent (Figure 1). The following discussion of compounds that interfere with early to late assembly stages provides several examples.
A number of splicing inhibitors, including NSC635326 and several related pyrido-carbazoles, benzo-pyrido-indoles, and pyrido-pyrrolo-isoquinolines (termed "indole derivatives") interrupt the earliest stage of spliceosome assembly such that no splicing complex formation is observed 6, 16. With such an early block, it is not easy to rule out the possibility that the compounds are affecting the HeLa nuclear extract nonspecifically, instead of targeting a specific spliceosome component to elicit splicing inhibition. In the case of some indole derivatives, however, the compounds interact with the SR protein SRSF1 (ASF/SF2). SRSF1 is known to regulate initiation of spliceosome assembly by recognizing enhancer elements in many different exons, which is consistent with the effect of the inhibitor 16.
Many splicing inhibitors identified to date cause spliceosome assembly to stall at A complex, the stage where U2 snRNP joins the branch point sequence. With the exception of the compound madrasin, the inhibitors are natural products with distinct structures. They include isoginkgetin, tetrocarcin A and members of the FR901464, pladienolide B and herboxidiene families 6, 7, 10, 17-21. Notably, compounds from these three different families have been shown to target the same spliceosome protein, SF3B1, which is a component of the U2 snRNP 17, 22, 23. We will cover details of SF3B1 inhibitor mechanism in a following section. Whether the function of the natural products in the organisms that produce them has anything to do with splicing is not known, but if so, it suggests that the A complex assembly stage and/or SF3B1 may be a particularly responsive target.
Several splicing inhibitors interfere with late assembly stages. However, the effect is often concentration dependent because increasing drug begins to elicit earlier assembly defects 5, 6, 8, 9. For example, naphthazarine compounds and psoromic acid at 100 μM both cause assembly to stall at a B/C-like complex, but at 250 μM produce an A-like complex accumulation 6, 8. Correspondingly, at lower concentrations, the compounds predominantly inhibit 2nd step chemistry, but as concentrations increase, 1st step chemistry is also affected. It is important to note that many of the late spliceosome assembly inhibitors are quinones that are capable of generating reactive oxygen species (ROS), and in some cases their effect can be rescued by adding the reducing agent dithiothreitol (DTT) 6. Therefore, the inhibitors may not act by simply binding spliceosome components, but instead trigger oxidation of a labile cysteine required for the activity of a spliceosome protein.
Targeting known enzymatic activities
Some compounds that inhibit splicing are also known to target certain classes of enzymes, which points to a potential mechanism of action. For example, in a candidate approach to identifying splicing inhibitors, three histone deacetylase (HDAC) inhibitors, suberoylanili dehydroxamic acid (SAHA), splitomicin, and dihydrocoumarin (DHC), were shown to inhibit splicing in vitro and stall assembly at a B-like complex 24. Three histone acetyltransferase (HAT) inhibitors, garcinol, AA, and BA3, also inhibit in vitro splicing, but do not block complex formation at the same stages 24. Garcinol and AA cause accumulation of an A-like complex, while BA3 stalls at a B-like complex. It is possible that these compounds disrupt the acetylation state of a spliceosome protein that is important for splicing, but a specific target has yet to be identified. In the same vein, ubistatin A, which interferes with ubiquitin protein/protein interactions, inhibits in vitro splicing in yeast extracts 25. A specific ubiquitinated target in the spliceosome is not known, but ubistatin A's effect was linked to the destabilization of the U5.U4/U6 tri-snRNP. With all these compounds, however, relatively high IC50 values for splicing (μM to mM range) raises concerns of specificity, and limits their use to in vitro model systems.
Some splicing inhibitors are known to target kinases and phosphatases, and may be related to phosphorylation events already linked to multiple stages of spliceosome assembly. For example, changes in phosphorylation of the core spliceosome factors SF3B1 and DDX23 (hPrp28) and the activity of the kinase PRP4K correlate with spliceosome rearrangements 26-30. As previously noted, SR protein activity is also regulated by phosphorylation 31. Inhibitors of the kinase activities of topoisomerase I, MNB-506 32, 33 and the Cdc2-like kinases (Clks) 11 interfere with splicing in vitro of particular substrates known to be regulated by SR protein phosphorylation. The compounds, however, do not interfere with splicing of all substrates, which may reflect the function of SR proteins as alternative splicing factors and not necessarily as core spliceosome components.
The phosphatases okadaic acid, tautomycin and microcystin-LR, which are known to inhibit PP1 and/or PP2A, inhibit splicing in vitro but do not disrupt spliceosome assembly up to B/C complex formation 34, 35. Interestingly, their effects on splicing chemistry vary with concentration 34. At lower concentrations, 1st step chemistry takes place while 2nd step chemistry is lost. As the amounts of inhibitors increase, 1st step chemistry is also lost. These results are consistent with at least two dephosphorylation events regulating both steps of splicing catalysis. Again, if/how in vitro splicing inhibition is linked to loss of phosphatase action on specific spliceosome proteins is not known.
Antibiotics as splicing inhibitors
Three compounds described as splicing inhibitors, Cl-tetracycline, streptomycin and erythromycin, are also antibiotics 36. These drugs are known to bind specific RNA structure in the ribosome and inhibit translation 37. The spliceosome is also an RNA-dependent enzyme, which could imply a similar mechanism of inhibition. However, the IC50 values of the drugs for in vitro splicing inhibition are in the high micromolar range, supporting a less specific mode of action. Indeed, tetracycline and streptomycin form complexes with pre-mRNA at high concentration, which interferes with the earliest stages of spliceosome assembly likely through steric effects 36. On the other hand, erythromycin does not impede spliceosome assembly and effects 2nd step chemistry most strongly 36. In the ribosome, erythromycin blocks the exit tunnel of the growing peptide 38, 39, so there may be a similar structure in the spliceosome. Perhaps there is a relationship with the tunnel-like structure that the Moore lab suggested plays a role in 3' splice site selection during 2nd step chemistry 40.
SF3B1 Inhibitors: Powerful Tools to Study Splicing and Beyond
Inhibitors Suggest Multiple Roles for SF3B1 in the Spliceosome
As noted previously, the spliceosome protein SF3B1 was identified as the target of three compounds, pladienolide B, spliceostatin A and herboxidiene, which all strongly inhibit splicing 17, 22, 23. SF3B1 and six other proteins comprise the SF3B complex, which is necessary for U2 snRNP function in the initiation of spliceosome assembly 41. Genetic interactions between SF3B and U2 snRNA in yeast suggest that the complex has a role in U2 snRNA structural dynamics involved in early intron recognition at the branch point region 42. Individually, SF3B1 has been linked to the recruitment and/or stabilization of U2 snRNP at the branch point sequence during A complex formation 43-46. SF3B1 is a large protein predicted to form an extended set of alpha helical bundles and to have an intrinsically disordered N-terminal domain 47. SF3B1 interacts with the early spliceosome assembly factor U2AF65 43, 44 and with another SF3B component called p14, which directly contacts the branch point adenosine 48. SF3B1 also directly cross-links to the intron upstream and downstream of the branch point sequence coincident with the ATP-dependent stabilization of A complex 45, 46
As research tools, SF3B1 inhibitors exemplify how small molecule can be used to explore spliceosome mechanism. For example, the Reed lab reported that a pladienolide B derivative (E7107) effects U2 snRNP interactions with a branch point sequence, and concluded that SF3B1 plays a role in mediating a conformational change in U2 snRNP 20. The Valcarcel lab showed that the fidelity of U2 snRNA interactions with the branch point decreases in the presence of spliceostatin A, suggesting that SF3B1 participates in branch point discrimination 18. Our lab recently showed that after bypassing the first effects of the drugs, the SF3B1 inhibitors pladienolide B, spliceostatin A and herboxidiene interfere with spliceosome progression after A complex stabilization 49. We also probed the catalytic stage of splicing by taking the spliceosome through the 1st step chemistry before adding drug, and found that SF3B1 inhibitors block the 2nd step chemistry, i.e. exon ligation 49. These results suggest that there are additional roles for SF3B1 throughout the splicing process. That function may be linked to changes in the affinity of the SF3B complex for U2 snRNP during spliceosome activation and with exon ligation 2, 50, 51.
SF3B1 Inhibitor Links to Cancer
Discoveries from two different lines of cancer research have raised the potential of the splicing machinery as a target for chemotherapeutics. First, common mutations in a number of splicing factors have been identified by whole-exome sequencing of cancer cells 52. Notably, specific SF3B1 mutations are frequent in malignant cells from patients with myelodysplastic syndrome (MDS), chronic lymphocytic leukemia (CLL), breast cancer, and pancreatic cancer 53-56. The presence of the mutations implies that modulation of SF3B1 function perturbs gene expression programs that contribute to cancer, and recent data links SF3B1 mutations to alterations in branch point selection that leads to aberrant 3' splice site choice 12, 57, 58. Second, as note previously, several of the splicing inhibitors that target SF3B1 were first identified in screens for cell growth inhibitors. With most cancer cells in culture, SF3B1 inhibitors arrest growth at low nanomolar concentrations 17, 22, 23, 59-79. Not all cell lines are equally sensitive, and members of the sudemycin family, in particular, have been shown to inhibit growth of only a subset of cancer cell lines 65, 70, 72, 80. Cells derived from normal tissues show more resistance to SF3B1 inhibitors relative to cancer cells 70, 77, 81, 82, and tumor cells in mouse xenografts die at concentrations of these compounds that appear to have no effect on the host animal 70, 76, 81, 83, 84. Unfortunately, an early clinical trial with the pladienolide B derivative E7107 in human patients was cut short because of side effects related to toxicity 85. Still, there is therapeutic promise that the compounds can be either directly targeted to tumors or more selectively used to treat specific cancers that show higher sensitivity to the drugs, such a CLL 81-83, 86, 87.
Cellular splicing changes with inhibitors
Because of the essential role that splicing plays in gene regulation, it is not surprising that splicing inhibitors significantly impact cellular function. With many inhibitors, however, a direct link between the inhibitor effects and cellular splicing has not been demonstrated. Without that determination, the off-target effects of low potency compounds are a substantial concern. Again, the SF3B1 inhibitors stand out as highly potent and selective compounds. Where examined, the impact of SF3B1 inhibitors on cells closely parallel those of SF3B1 knockdown, which is consistent with inhibitor effects being indeed mediated by SF3B1 18, 23, 88, 89. Furthermore, identification of a point mutation in the protein (R1074H) that conferred resistance to the cytotoxicity of pladienolide B strongly links SF3B1 as the target 90. We also observed that the activity of SF3B1 inhibitor derivatives for splicing in vitro strictly correlates with the phenotypic changes that they produce in cells 19.
Looking specifically at cellular splicing, the influence of SF3B1 inhibitors was first examined on a gene-by-gene basis. Instances of intron retention, aberrant splicing, and alternative splicing changes were all noted in a small collection of gene transcripts 22, 23. Later, Corrionero et al. used splicing specific microarrays to examine 1800 alternative splicing events in HeLa cells treated with spliceostatin A and found that less than 10% of the monitored splicing events were affected 18. The authors noted that there were splicing changes in genes associated with cell-cycle regulation and apoptosis, which are pathways both regulated and connected at the level of alternative splicing 91. That result is consistent with SF3B1 inhibitors ability to inhibit growth of cancer cells in culture 76, 77, 79, 92. More recently, a number of groups have shown that SF3B1 inhibitor effects can be linked to splicing changes in the apoptosis genes MCL1 81, 82, 86, 88, 93 and BCL-X 86. Both of these genes are alternatively spliced such that one isoform encodes a pro-apoptotic protein, while the other encodes an anti-apoptotic protein. 94, 95. Kashyap et al. reported that pladienolide B and its analog FD-985 promote pro-apoptotic splicing of MCL1 and BCL-X, but that the cellular context is important: CLL cells showed the splicing isoform shifts with treatment, but normal blood cells did not 86. Larrayoz et al. also showed MCL1 splicing changes in CLL cells with SSA treatment, and further found that overexpression of the anti-apoptotic MCL1-L isoform in a human Burkitt's lymphoma cell line rescued drug-induced apoptosis 82. Consistent with these findings, SF3B1 inhibitors show synergism with pro-apoptotic compounds in culture 81, 82, 88.
One question raised by the cellular effects of SF3B1 inhibitors is "How does targeting a core splicing factor only affect splicing of select genes and result in cell-type specific phenotypes?" On its face, it seems that blocking splicing in cells should be equally catastrophic for the expression of every intron-containing gene. Splicing inhibitors would thereby appear to be very blunt tools with limited selectivity for cancer cells versus healthy cells. Hints to a resolution of this paradox come from a recent study of the meiotic splicing program in yeast, which pointed to competition between introns as a means to regulate gene expression programs 96. With that framework in mind, two related circumstances are important to consider for splicing inhibitor effects in cells. 1) The spliceosome does not have a unique substrate, but a large number of competing substrate variants, and 2) the cellular effects of splicing inhibitors are typically evaluated on the basis of cell growth.
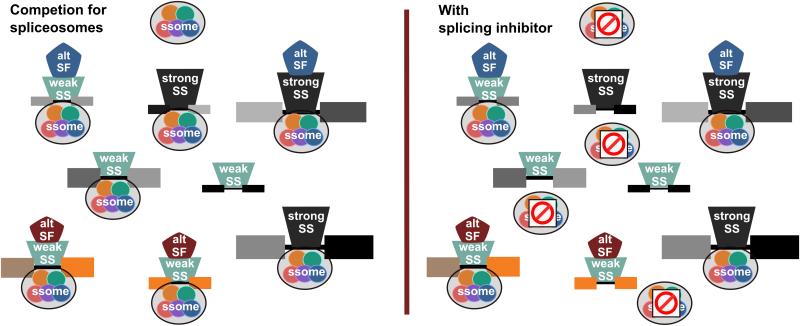
Splice sites vary in a number of ways including splicing sequence strength, the existence of alternative splicing choices and the influence of factors that regulate those choices. All of these parameters influence the rate of spliceosome recognition and assembly at different splice sites. Furthermore, the levels a pre-mRNA transcript will influence the likelihood that the splicing machinery will engage particular splice sites. In this context, splice sites that are overall weaker competitors for the spliceosome will be affected first as even low concentrations of a splicing inhibitor begins to limit the amount of active enzyme (Figure 2). With alternative splice sites, which typically have weak splicing consensus sequences, a change in competition would most likely manifest as a switch of splice isoforms, like those seen with MCL1 and BCL-X. In low abundance transcripts, even constitutive splice sites with strong splice sites may no longer compete well, resulting in intron retention. Importantly, a decrease in efficiency of only one splicing event in a transcript could have a big impact on the cells, depending on the function of the resulting mRNA. In the context of cell growth, splicing of genes controlling proliferation or apoptosis will therefore act as the meter by which splicing inhibitors activity is evaluated.
Figure 2. Splicing inhibitors may impact competition between splicing substrates in cells.
Different splice sites in pre-mRNA substrates (indicated by differently shaded exons flanking introns) compete for the spliceosome (ssome). Competitiveness is determined by a combination of splice site (SS) sequence strength, the presence of positive alternative splicing factors (alt SF) and total amount of pre-mRNA transcript (indicated by pre-mRNA size). Under normal cellular conditions (left panel) the weakest competitors "lose" for splicing (not engaged by ssome), typically resulting in alternative splicing changes. With splicing inhibitors (red and white "no" sign) additional splice sites will lose for splicing (right panel), resulting in alternative splicing changes and intron retention. Splicing events required for a cells growth (golden pre-mRNAs) will drive the response to the splicing inhibitor. Importantly, the relative competitiveness of an intron will differ in different cell types and under different conditions because the presence of alternative splicing factors differ along with pre-mRNA transcript levels.
The sensitivity of specific splicing events likely explains the difference in concentration of SF3B1 inhibitors typically required to affect cell growth (low nanomlar) vs. inhibit splicing in vitro (high nanomolar). It also can point to differences between cancer vs. normal cells in the relative abundance of different transcripts and their competitiveness for the splicing machinery as a potential foothold for therapeutics. A recent study looking at cancers driven by the Myc oncogene illustrates this point 83. A decrease in the levels of core spliceosome proteins, including SF3B1, resulted in death of cancer cells expressing activated Myc oncogene relative to cells not driven by Myc. Notably, the presence of activated Myc in several cancer cell lines also correlated with higher sensitivity to sudemycin D6, a SF3B1 inhibitor structurally related to spliceostatin A 83. Myc activity increases the overall transcription load in cells97. More transcripts means more splice sites competing for the splicing machinery, which in combination with decreased spliceosome function, may result in a lethal splicing change for one or more important genes. Further systematic analysis of the splicing changes induced by SF3B1 inhibitors will be needed to fully determine which introns drive the cellular response to the drugs. In addition to informing therapeutic strategies for splicing inhibitors, the data may also help untangle the intricate web of splicing regulation.
Structure Activity Relationships of SF3B1 Inhibitors
Relationships between SF3B1 inhibitor families
The SF3B1 inhibitors can be classified into three general structural families. The FR901464 79 family includes spliceostatin A 23, meayamycin71 and thailanstatins 64. The architecture of these compounds is based on two highly functionalized tetrahydropyran rings connected by diene chain and differs the most in structure relative to the other two compounds. Members of the pladienolide B family, which includes E7107 and FD-895 22, 67, are polyketides with a large macrolide ring. The GEX1 family of compounds 77, including herboxidiene, has a similar structure, but with a smaller tetrahydropyran ring. Despite these structural differences, the compounds exhibit nearly identical effects on spliceosome assembly and in cells, presumably by interfering with SF3B1 function. Still, specifically how and where the compounds interact with SF3B1 to interfere with splicing is not currently understood. We recently reported that inactive analogs of pladienolide B, spliceostatin A and herboxidiene, in terms of their ability to inhibit splicing, nevertheless are able to somehow out-compete their active counterparts 49. Our data are consistent with both active and inactive versions being able to interact with SF3B1, although only the active version impacts SF3B1 activity. Remarkably, we also found that the inactive version of each compound also competes with the active versions of the other two compounds (i.e. inactive pladienolide competes with both active spliceostatin A and herboxidiene). This result suggests that all three compounds have the same interaction with SF3B1, despite their different structures, and lends credence to the idea of a common pharmacophore. Still, we likely will need structures of the different inhibitor scaffolds interacting with SF3B1 to know whether a shared pharmacophore exists.
SF3B1 Inhibitor analogs
Structure activity relationship (SAR) studies identify important chemical features by altering individual moieties of the inhibitor structure and testing the resulting derivative for biological activity. If activity is lost, the feature is required, but if the derivative has the same activity as the original compound, the feature is dispensable. Multiple rounds of SAR analysis will hopefully lead to improvements in inhibitor potency, stability, membrane permeability, and simplicity of chemical synthesis. Furthermore, the information can be used to guide addition of functional groups to the inhibitor and generate biological probes. For example, SAR studies guided attachment of biotin and cross-linker moieties that were critical to first identifying SF3B1 as the target of SSA, pladienolide B and herboxidiene 17, 22, 23. Biotinylated SSA was also used to show that the drug is present in stalled spliceosomes 21. Addition of a fluorescent label showed that pladienolide B molecule localizes to the nucleus of cells in a SF3B dependent manner 22.
One critical constraint for SAR studies is the availability of a wide range of related compounds to test. In some cases, nature provides related molecules. For example, several naturally occurring analogs of SF3B1 inhibitors show differential activity in cell growth assays 64, 74, 76, 79, 84, 92 and, in some case, in vitro splicing 20, 64. Unfortunately, the access to the compounds is limited, which impedes medicinal chemistry efforts with natural products. In some cases, semi-synthetic approaches in which natural products are chemically modified have also been informative 22, 60. Still, detailed pharmacological studies of these molecules have not been reported.
Complete chemical synthesis provides the most flexibility in systematically creating derivatives to test specific features of a molecule. However, the complex chemical structures of SF3B1 inhibitors, in particular, make total synthesis very challenging. Nonetheless, several groups successfully generated SF3B1 inhibitors and derivatives for the three main structural families: FR901464 23, 68, 71, 73, 75, 78, 98-100, pladienolides (including FD-985) 19, 62, 67, 69, 101, 102, and herboxidiene 59, 103, 104. Taking SAR analysis another step further, the Webb lab compared the structures of FR901464 and pladienolide B to predict common pharmacophore consisting of epoxy and carbonyloxy groups joined and relatively positioned by a diene linker 72. Based on this scaffold, they synthesized a series of compounds termed "sudemycins" 65, 70, 80, 105. An optimized compound, sudemycin D6, is in the preclinical development stage and has fewer stereocenters and increased stability relative to the natural products 65.
Assaying SF3B1 inhibitor activity
To build a picture of key and potentially shared features that confer splicing inhibition by SF3B1 inhibitor, we ideally want to directly compare the activity of all the derivatives. The situation is complicated, however, by the different assays used to test compound activity reported in the literature. The majority of studies use growth inhibition or cytotoxicity of cells in culture, which are straightforward and biologically relevant assays 17, 22, 23, 59-79. However, results for the same compounds vary between different cell lines and assay conditions (e.g. 60, 68-70, 79) and cytotoxicity is, at best, an indirect readout for splicing inhibition. As discussed above, effects of SF3B1 inhibitor in cells hinges on the dependency on specific splicing events, which also vary between cells types. Other cell-based assays used to test SF3B1 inhibitors measured splicing of select endogenous gene transcripts, the morphology of nuclear speckles, which is linked to splicing machinery activity, or a constellation of cellular features to create a cytological profile 18, 19, 22, 23, 59, 65, 66, 69, 71, 80, 106. With cell-based assays, it is not easy to determine whether differences in activity are due to the effect of the analog on splicing or to properties like solubility, stability and membrane permeability. For the few cases where these characteristics have been reported, there is generally a correlation between activity and stability 64, 68, 75 or permeability 60. In vitro assays bypass some of these issues by more directly measuring splicing activity. Several FR901464, pladienolide B, and herboxidiene derivatives have been tested for effects on in vitro splicing in HeLa cell extract 18-21, 23, 49, 62, 71, 99, 104, 107. Generally, in vitro data correlates well with cell-based assay results. One exception is the FR901464 analog spliceostatin E, which is cytotoxic 60 but does not inhibit in vitro splicing or cause nuclear speckles to aggregate, as seen with other SF3B1 inhibitors 99. The discrepancy may be base on residual activity that cannot be detected with a model splicing substrate, but nonetheless is sufficient to alter splicing of an important gene transcript in cells.
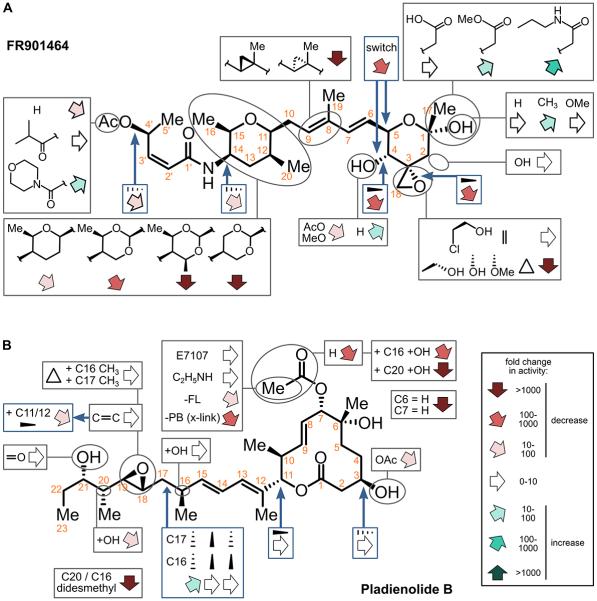
With these issues in mind, in Figure 3 we graphically summarized SAR analysis for FR901464 and pladienolide B inhibitor families based on the relative activity of natural analogs and synthetic derivatives that have been tested. Consistent with a critical function, most modifications of FR901464 removing the C3 epoxy group results in a loss of measurable activity 68, 78. However, substitution at that position with chlorine reduces cell growth inhibition by only ~2-4 fold, depending on the cell line, which is still in the low nanomolar range 68. The same substitution also drops in vitro splicing inhibition by 10-fold 68. Changes in the C11-15 tetrahydropyran ring can also reduce potency by several orders of magnitude 68. An acetyl group at the C4' position (the presumable carbonyloxy of the Sudemycin scaffold) contributes to potency, but it loss still produces low nanomolar cell growth inhibition 68, 78. The relative conformations of other FR901464 chemical features throughout the molecule are also important, as changes in stereochemistry negatively affects potency by over 100-fold 78, 107. In contrast, changes in side groups at C1 and C4 in the other tetrahydropyran ring have been shown to improved potency, evidently by improving compound stability in aqueous solution 60, 68, 73. Remarkably, Koide and co-workers achieved low picomolar potency for cell growth inhibition with the derivative meaymycin B 68.
Figure 3. Graphical summary of structure activity relationship studies for two SF3B1 inhibitors.
A. FR901464. Functional group changes (in black outlined boxes) are linked by lines to the corresponding positions in the FR901464 structure (circled). Blue arrows point to the position of bond orientations changes (in blue outlined boxes). The effect and relative magnitude of specific changes on the compound's activity is indicated by the open arrow direction and color as indicated in the inset box legend. B. Pladienolide B. Same as panel A.
The epoxide group in pladienolide B at C18-19 also contributes to, but is not required for, activity, because its absence causes up to a five-fold decrease in potency 19, 63. The presence of the carbonyloxyl group off the C7 position is critical, but several functional groups can be appended to it with out a significant loss of activity 22. Derivatives of pladienolide B-related compounds indicate that constituents at the C6 position in the macrolide ring also contribute to 63, as well as at the C16/C20 positions in the extended "arm" 19. Several stereocenters of pladienolide B, such as C11-C12, are not important 19. Modifying the stereochemistry C16/C17 is the only change identified thus far that improve activity 67.
Conclusion
Small molecules that target different components of the spliceosome can be powerful tools that allow researchers to study such a complex and dynamic macromolecular machine. They also can serve as promising drug leads for treating human disease. SF3B1 inhibitors, in particular, demonstrate that potential through the progress they have fuelled in the combined arenas of medicinal chemistry, cancer biology and splicing molecular biology. The inhibitors are revealing the role of SF3B1 in the spliceosome and showing that affecting splicing in cells is complicated. Rather than targeting specific splicing events, the compounds likely affect the competition between introns for the spliceosome. Fortunately, the introns that lose out when SF3B1 is inhibited are involved in apoptosis and cell cycle control, leading to the death of selective cancer types. Continued SAR analysis will hopefully further improve the SF3B1 inhibitors as research tools and chemotherapeutics. Finally, several other splicing inhibitors have been found by screening and candidate approaches, but they have yet to be fully characterized in terms of their targets and mechanisms of inhibition. Additional SAR and mechanistic studies of these molecules and expanded screens for new inhibitor compounds hold many opportunities for splicing researchers in the future.
Acknowledgements
We wish thank Dr. Arun Ghosh for valuable input, and acknowledge support from the Santa Cruz Cancer Benefit Society, the National Institutes of Health (Grant R01GM72649) and the Paul and Anne Irwin Graduate Fellowship in Cancer Research.
Footnotes
Competing interests: The authors have declared that no competing interests exist.
References
- 1.Singh RK, Cooper TA. Pre-mRNA splicing in disease and therapeutics. Trends Mol Med. 2012;18:472–482. doi: 10.1016/j.molmed.2012.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ilagan JO, Chalkley RJ, Burlingame AL, Jurica MS. Rearrangements within human spliceosomes captured after exon ligation. RNA. 2013;19:400–412. doi: 10.1261/rna.034223.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bessonov S, Anokhina M, Will CL, Urlaub H, Luhrmann R. Isolation of an active step I spliceosome and composition of its RNP core. Nature. 2008;452:846–850. doi: 10.1038/nature06842. [DOI] [PubMed] [Google Scholar]
- 4.Makarov EM, Makarova OV, Urlaub H, Gentzel M, Will CL, Wilm M, Luhrmann R. Small nuclear ribonucleoprotein remodeling during catalytic activation of the spliceosome. Science. 2002;298:2205–2208. doi: 10.1126/science.1077783. [DOI] [PubMed] [Google Scholar]
- 5.Effenberger KA, James RC, Urabe VK, Dickey BJ, Linington RG, Jurica MS. The Natural Product N-Palmitoyl-l-leucine Selectively Inhibits Late Assembly of Human Spliceosomes. J Biol Chem. 2015;290:27524–27531. doi: 10.1074/jbc.M115.673210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Effenberger KA, Perriman RJ, Bray WM, Lokey RS, Ares M, Jr., Jurica MS. A high-hhroughput splicing assay identifies new classes of inhibitors of human and yeast spliceosomes. J Biomol Screen. 2013;18:1110–1120. doi: 10.1177/1087057113493117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pawellek A, McElroy S, Samatov T, Mitchell L, Woodland A, Ryder U, Gray D, Luhrmann R, Lamond AI. Identification of small molecule inhibitors of pre-mRNA splicing. J Biol Chem. 2014;289:34683–34698. doi: 10.1074/jbc.M114.590976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Samatov TR, Wolf A, Odenwalder P, Bessonov S, Deraeve C, Bon RS, Waldmann H, Luhrmann R. Psoromic acid derivatives: a new family of small-molecule pre-mRNA splicing inhibitors discovered by a stage-specific high-throughput in vitro splicing assay. Chembiochem. 2012;13:640–644. doi: 10.1002/cbic.201100790. [DOI] [PubMed] [Google Scholar]
- 9.Berg MG, Wan L, Younis I, Diem MD, Soo M, Wang C, Dreyfuss G. A quantitative high-throughput in vitro splicing assay identifies inhibitors of spliceosome catalysis. Mol Cell Biol. 2012;32:1271–1283. doi: 10.1128/MCB.05788-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.O'Brien K, Matlin AJ, Lowell AM, Moore MJ. The biflavonoid isoginkgetin is a general inhibitor of Pre-mRNA splicing. J Biol Chem. 2008;283:33147–33154. doi: 10.1074/jbc.M805556200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Younis I, Berg M, Kaida D, Dittmar K, Wang C, Dreyfuss G. Rapid-response splicing reporter screens identify differential regulators of constitutive and alternative splicing. Mol Cell Biol. 2010;30:1718–1728. doi: 10.1128/MCB.01301-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Darman RB, Seiler M, Agrawal AA, Lim KH, Peng S, Aird D, Bailey SL, Bhavsar EB, Chan B, Colla S, et al. Cancer-Associated SF3B1 Hotspot Mutations Induce Cryptic 3? Splice Site Selection through Use of a Different Branch Point. Cell Rep. 2015;13:1033–1045. doi: 10.1016/j.celrep.2015.09.053. [DOI] [PubMed] [Google Scholar]
- 13.Palacino J, Swalley SE, Song C, Cheung AK, Shu L, Zhang X, Van Hoosear M, Shin Y, Chin DN, Keller CG, et al. SMN2 splice modulators enhance U1-pre-mRNA association and rescue SMA mice. Nat Chem Biol. 2015;11:511–517. doi: 10.1038/nchembio.1837. [DOI] [PubMed] [Google Scholar]
- 14.Padgett RA, Hardy SF, Sharp PA. Splicing of adenovirus RNA in a cell-free transcription system. Proc Natl Acad Sci U S A. 1983;80:5230–5234. doi: 10.1073/pnas.80.17.5230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Konarska MM, Sharp PA. Electrophoretic separation of complexes involved in the splicing of precursors to mRNAs. Cell. 1986;46:845–855. doi: 10.1016/0092-8674(86)90066-8. [DOI] [PubMed] [Google Scholar]
- 16.Soret J, Bakkour N, Maire S, Durand S, Zekri L, Gabut M, Fic W, Divita G, Rivalle C, Dauzonne D, et al. Selective modification of alternative splicing by indole derivatives that target serine-arginine-rich protein splicing factors. Proc Natl Acad Sci U S A. 2005;102:8764–8769. doi: 10.1073/pnas.0409829102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hasegawa M, Miura T, Kuzuya K, Inoue A, Won Ki S, Horinouchi S, Yoshida T, Kunoh T, Koseki K, Mino K, et al. Identification of SAP155 as the target of GEX1A (Herboxidiene), an antitumor natural product. ACS Chem Biol. 2011;6:229–233. doi: 10.1021/cb100248e. [DOI] [PubMed] [Google Scholar]
- 18.Corrionero A, Minana B, Valcarcel J. Reduced fidelity of branch point recognition and alternative splicing induced by the anti-tumor drug spliceostatin A. Genes Dev. 2011;25:445–459. doi: 10.1101/gad.2014311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Effenberger KA, Anderson DD, Bray WM, Prichard BE, Ma N, Adams MS, Ghosh AK, Jurica MS. Coherence between cellular responses and in vitro splicing inhibition for the anti-tumor drug pladienolide B and its analogs. J Biol Chem. 2014;289:1938–1947. doi: 10.1074/jbc.M113.515536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Folco EG, Coil KE, Reed R. The anti-tumor drug E7107 reveals an essential role for SF3b in remodeling U2 snRNP to expose the branch point-binding region. Genes Dev. 2011;25:440–444. doi: 10.1101/gad.2009411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roybal GA, Jurica MS. Spliceostatin A inhibits spliceosome assembly subsequent to prespliceosome formation. Nucleic Acids Res. 2010;38:6664–6672. doi: 10.1093/nar/gkq494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kotake Y, Sagane K, Owa T, Mimori-Kiyosue Y, Shimizu H, Uesugi M, Ishihama Y, Iwata M, Mizui Y. Splicing factor SF3b as a target of the antitumor natural product pladienolide. Nat Chem Biol. 2007;3:570–575. doi: 10.1038/nchembio.2007.16. [DOI] [PubMed] [Google Scholar]
- 23.Kaida D, Motoyoshi H, Tashiro E, Nojima T, Hagiwara M, Ishigami K, Watanabe H, Kitahara T, Yoshida T, Nakajima H, et al. Spliceostatin A targets SF3b and inhibits both splicing and nuclear retention of pre-mRNA. Nat Chem Biol. 2007;3:576–583. doi: 10.1038/nchembio.2007.18. [DOI] [PubMed] [Google Scholar]
- 24.Kuhn AN, van Santen MA, Schwienhorst A, Urlaub H, Luhrmann R. Stalling of spliceosome assembly at distinct stages by small-molecule inhibitors of protein acetylation and deacetylation. Rna. 2009;15:153–175. doi: 10.1261/rna.1332609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bellare P, Small EC, Huang X, Wohlschlegel JA, Staley JP, Sontheimer EJ. A role for ubiquitin in the spliceosome assembly pathway. Nat Struct Mol Biol. 2008;15:444–451. doi: 10.1038/nsmb.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang C, Chua K, Seghezzi W, Lees E, Gozani O, Reed R. Phosphorylation of spliceosomal protein SAP 155 coupled with splicing catalysis. Genes Dev. 1998;12:1409–1414. doi: 10.1101/gad.12.10.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schwelnus W, Richert K, Opitz F, Gross T, Habara Y, Tani T, Kaufer NF. Fission yeast Prp4p kinase regulates pre-mRNA splicing by phosphorylating a non-SR-splicing factor. EMBO Rep. 2001;2:35–41. doi: 10.1093/embo-reports/kve009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bottner CA, Schmidt H, Vogel S, Michele M, Kaufer NF. Multiple genetic and biochemical interactions of Brr2, Prp8, Prp31, Prp1 and Prp4 kinase suggest a function in the control of the activation of spliceosomes in Schizosaccharomyces pombe. Curr Genet. 2005;48:151–161. doi: 10.1007/s00294-005-0013-6. [DOI] [PubMed] [Google Scholar]
- 29.Mathew R, Hartmuth K, Mohlmann S, Urlaub H, Ficner R, Luhrmann R. Phosphorylation of human PRP28 by SRPK2 is required for integration of the U4/U6-U5 tri-snRNP into the spliceosome. Nat Struct Mol Biol. 2008;15:435–443. doi: 10.1038/nsmb.1415. [DOI] [PubMed] [Google Scholar]
- 30.Schneider M, Hsiao HH, Will CL, Giet R, Urlaub H, Luhrmann R. Human PRP4 kinase is required for stable tri-snRNP association during spliceosomal B complex formation. Nat Struct Mol Biol. 2010;17:216–221. doi: 10.1038/nsmb.1718. [DOI] [PubMed] [Google Scholar]
- 31.Zhou Z, Fu XD. Regulation of splicing by SR proteins and SR protein-specific kinases. Chromosoma. 2013;122:191–207. doi: 10.1007/s00412-013-0407-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pilch B, Allemand E, Facompre M, Bailly C, Riou JF, Soret J, Tazi J. Specific inhibition of serine- and arginine-rich splicing factors phosphorylation, spliceosome assembly, and splicing by the antitumor drug NB-506. Cancer Res. 2001;61:6876–6884. [PubMed] [Google Scholar]
- 33.Tazi J, Bakkour N, Soret J, Zekri L, Hazra B, Laine W, Baldeyrou B, Lansiaux A, Bailly C. Selective inhibition of topoisomerase I and various steps of spliceosome assembly by diospyrin derivatives. Mol Pharmacol. 2005;67:1186–1194. doi: 10.1124/mol.104.007633. [DOI] [PubMed] [Google Scholar]
- 34.Mermoud JE, Cohen P, Lamond AI. Ser/Thr-specific protein phosphatases are required for both catalytic steps of pre-mRNA splicing. Nucleic Acids Res. 1992;20:5263–5269. doi: 10.1093/nar/20.20.5263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tazi J, Daugeron MC, Cathala G, Brunel C, Jeanteur P. Adenosine phosphorothioates (ATP alpha S and ATP tau S) differentially affect the two steps of mammalian pre-mRNA splicing. J Biol Chem. 1992;267:4322–4326. [PubMed] [Google Scholar]
- 36.Hertweck M, Hiller R, Mueller MW. Inhibition of nuclear pre-mRNA splicing by antibiotics in vitro. Eur J Biochem. 2002;269:175–183. doi: 10.1046/j.0014-2956.2001.02636.x. [DOI] [PubMed] [Google Scholar]
- 37.Wilson DN. The A-Z of bacterial translation inhibitors. Crit Rev Biochem Mol Biol. 2009;44:393–433. doi: 10.3109/10409230903307311. [DOI] [PubMed] [Google Scholar]
- 38.Schlunzen F, Zarivach R, Harms J, Bashan A, Tocilj A, Albrecht R, Yonath A, Franceschi F. Structural basis for the interaction of antibiotics with the peptidyl transferase centre in eubacteria. Nature. 2001;413:814–821. doi: 10.1038/35101544. [DOI] [PubMed] [Google Scholar]
- 39.Gabashvili IS, Gregory ST, Valle M, Grassucci R, Worbs M, Wahl MC, Dahlberg AE, Frank J. The polypeptide tunnel system in the ribosome and its gating in erythromycin resistance mutants of L4 and L22. Mol Cell. 2001;8:181–188. doi: 10.1016/s1097-2765(01)00293-3. [DOI] [PubMed] [Google Scholar]
- 40.Chen S, Anderson K, Moore MJ. Evidence for a linear search in bimolecular 3' splice site AG selection. Proc Natl Acad Sci U S A. 2000;97:593–598. doi: 10.1073/pnas.97.2.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brosi R, Hauri HP, Kramer A. Separation of splicing factor SF3 into two components and purification of SF3a activity. J Biol Chem. 1993;268:17640–17646. [PubMed] [Google Scholar]
- 42.Yan D, Ares M., Jr Invariant U2 RNA sequences bordering the branchpoint recognition region are essential for interaction with yeast SF3a and SF3b subunits. Mol Cell Biol. 1996;16:818–828. doi: 10.1128/mcb.16.3.818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cass DM, Berglund JA. The SF3b155 N-terminal domain is a scaffold important for splicing. Biochemistry. 2006;45:10092–10101. doi: 10.1021/bi060429o. [DOI] [PubMed] [Google Scholar]
- 44.Thickman KR, Swenson MC, Kabogo JM, Gryczynski Z, Kielkopf CL. Multiple U2AF65 binding sites within SF3b155: thermodynamic and spectroscopic characterization of protein-protein interactions among pre-mRNA splicing factors. J Mol Biol. 2006;356:664–683. doi: 10.1016/j.jmb.2005.11.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gozani O, Feld R, Reed R. Evidence that sequence-independent binding of highly conserved U2 snRNP proteins upstream of the branch site is required for assembly of spliceosomal complex A. Genes Dev. 1996;10:233–243. doi: 10.1101/gad.10.2.233. [DOI] [PubMed] [Google Scholar]
- 46.Gozani O, Potashkin J, Reed R. A potential role for U2AF-SAP 155 interactions in recruiting U2 snRNP to the branch site. Mol Cell Biol. 1998;18:4752–4760. doi: 10.1128/mcb.18.8.4752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Korneta I, Magnus M, Bujnicki JM. Structural bioinformatics of the human spliceosomal proteome. Nucleic Acids Res. 2012;40:7046–7065. doi: 10.1093/nar/gks347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Will CL, Schneider C, MacMillan AM, Katopodis NF, Neubauer G, Wilm M, Luhrmann R, Query CC. A novel U2 and U11/U12 snRNP protein that associates with the pre-mRNA branch site. EMBO J. 2001;20:4536–4546. doi: 10.1093/emboj/20.16.4536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Effenberger KA, Urabe VK, Prichard BE, Ghosh AK, Jurica MS. Interchangeable SF3B1 inhibitors interfere with pre-mRNA splicing at multiple stages. RNA. 2016;22:350–359. doi: 10.1261/rna.053108.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lardelli RM, Thompson JX, Yates JR, 3rd, Stevens SW. Release of SF3 from the intron branchpoint activates the first step of pre-mRNA splicing. RNA. 2010;16:516–528. doi: 10.1261/rna.2030510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Coltri P, Effenberger K, Chalkley RJ, Burlingame AL, Jurica MS. Breaking up the C complex spliceosome shows stable association of proteins with the lariat intron intermediate. PLoS One. 2011;6:e19061. doi: 10.1371/journal.pone.0019061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chabot B, Shkreta L. Defective control of pre-messenger RNA splicing in human disease. J Cell Biol. 2016;212:13–27. doi: 10.1083/jcb.201510032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Biankin AV, Waddell N, Kassahn KS, Gingras MC, Muthuswamy LB, Johns AL, Miller DK, Wilson PJ, Patch AM, Wu J, et al. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature. 2012;491:399–405. doi: 10.1038/nature11547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ellis MJ, Ding L, Shen D, Luo J, Suman VJ, Wallis JW, Van Tine BA, Hoog J, Goiffon RJ, Goldstein TC, et al. Whole-genome analysis informs breast cancer response to aromatase inhibition. Nature. 2012;486:353–360. doi: 10.1038/nature11143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Landau DA, Carter SL, Stojanov P, McKenna A, Stevenson K, Lawrence MS, Sougnez C, Stewart C, Sivachenko A, Wang L, et al. Evolution and impact of subclonal mutations in chronic lymphocytic leukemia. Cell. 2013;152:714–726. doi: 10.1016/j.cell.2013.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yoshida K, Sanada M, Shiraishi Y, Nowak D, Nagata Y, Yamamoto R, Sato Y, Sato-Otsubo A, Kon A, Nagasaki M, et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature. 2011;478:64–69. doi: 10.1038/nature10496. [DOI] [PubMed] [Google Scholar]
- 57.DeBoever C, Ghia EM, Shepard PJ, Rassenti L, Barrett CL, Jepsen K, Jamieson CH, Carson D, Kipps TJ, Frazer KA. Transcriptome sequencing reveals potential mechanism of cryptic 3' splice site selection in SF3B1-mutated cancers. PLoS Comput Biol. 2015;11:e1004105. doi: 10.1371/journal.pcbi.1004105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Alsafadi S, Houy A, Battistella A, Popova T, Wassef M, Henry E, Tirode F, Constantinou A, Piperno-Neumann S, Roman-Roman S, et al. Cancer-associated SF3B1 mutations affect alternative splicing by promoting alternative branchpoint usage. Nat Commun. 2016;7:10615. doi: 10.1038/ncomms10615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lagisetti C, Yermolina MV, Sharma LK, Palacios G, Prigaro BJ, Webb TR. Pre-mRNA splicing-modulatory pharmacophores: the total synthesis of herboxidiene, a pladienolide-herboxidiene hybrid analog and related derivatives. ACS Chem Biol. 2014;9:643–648. doi: 10.1021/cb400695j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.He H, Ratnayake AS, Janso JE, He M, Yang HY, Loganzo F, Shor B, O'Donnell CJ, Koehn FE. Cytotoxic Spliceostatins from Burkholderia sp. and Their Semisynthetic Analogues. J Nat Prod. 2014;77:1864–1870. doi: 10.1021/np500342m. [DOI] [PubMed] [Google Scholar]
- 61.Convertini P, Shen M, Potter PM, Palacios G, Lagisetti C, de la Grange P, Horbinski C, Fondufe-Mittendorf YN, Webb TR, Stamm S. Sudemycin E influences alternative splicing and changes chromatin modifications. Nucleic Acids Res. 2014;42:4947–4961. doi: 10.1093/nar/gku151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Arai K, Buonamici S, Chan B, Corson L, Endo A, Gerard B, Hao MH, Karr C, Kira K, Lee L, et al. Total Synthesis of 6-Deoxypladienolide D and Assessment of Splicing Inhibitory Activity in a Mutant SF3B1 Cancer Cell Line. Org Lett. 2014;16:5560–5563. doi: 10.1021/ol502556c. [DOI] [PubMed] [Google Scholar]
- 63.Villa R, Kashyap MK, Kumar D, Kipps TJ, Castro JE, La Clair JJ, Burkart MD. Stabilized Cyclopropane Analogs of the Splicing Inhibitor FD-895. J Med Chem. 2013 doi: 10.1021/jm400861t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liu X, Biswas S, Berg MG, Antapli CM, Xie F, Wang Q, Tang MC, Tang GL, Zhang L, Dreyfuss G, et al. Genomics-guided discovery of thailanstatins A, B, and C As pre-mRNA splicing inhibitors and antiproliferative agents from Burkholderia thailandensis MSMB43. J Nat Prod. 2013;76:685–693. doi: 10.1021/np300913h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lagisetti C, Palacios G, Goronga T, Freeman B, Caufield W, Webb TR. Optimization of antitumor modulators of pre-mRNA splicing. J Med Chem. 2013;56:10033–10044. doi: 10.1021/jm401370h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gao Y, Vogt A, Forsyth CJ, Koide K. Comparison of splicing factor 3b inhibitors in human cells. Chembiochem. 2013;14:49–52. doi: 10.1002/cbic.201200558. [DOI] [PubMed] [Google Scholar]
- 67.Villa R, Mandel AL, Jones BD, La Clair JJ, Burkart MD. Structure of FD-895 revealed through total synthesis. Org Lett. 2012;14:5396–5399. doi: 10.1021/ol3023006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Osman S, Albert BJ, Wang Y, Li M, Czaicki NL, Koide K. Structural requirements for the antiproliferative activity of pre-mRNA splicing inhibitor FR901464. Chemistry. 2011;17:895–904. doi: 10.1002/chem.201002402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gundluru MK, Pourpak A, Cui X, Morris SW, Webb TR. Design, synthesis and initial biological evaluation of a novel pladienolide analog scaffold. Medchemcomm. 2011;2:904–908. doi: 10.1039/C1MD00040C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lagisetti C, Pourpak A, Goronga T, Jiang Q, Cui X, Hyle J, Lahti JM, Morris SW, Webb TR. Synthetic mRNA splicing modulator compounds with in vivo antitumor activity. J Med Chem. 2009;52:6979–6990. doi: 10.1021/jm901215m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Albert BJ, Mcpherson PA, O’brien K, Czaicki NL, Destefino V, Osman S, Li M, Day BW, Grabowski PJ, Moore MJ, et al. Meayamycin inhibits pre-messenger RNA splicing and exhibits picomolar activity against multidrug-resistant cells. Mol Cancer Therapeutics. 2009;8:2308–2318. doi: 10.1158/1535-7163.MCT-09-0051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lagisetti C, Pourpak A, Jiang Q, Cui X, Goronga T, Morris SW, Webb TR. Antitumor compounds based on a natural product consensus pharmacophore. J Med Chem. 2008;51:6220–6224. doi: 10.1021/jm8006195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Albert BJ, Sivaramakrishnan A, Naka T, Czaicki NL, Koide K. Total syntheses, fragmentation studies, and antitumor/antiproliferative activities of FR901464 and its low picomolar analogue. J Am Chem Soc. 2007;129:2648–2659. doi: 10.1021/ja067870m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sakai T, Sameshima T, Matsufuji M, Kawamura N, Dobashi K, Mizui Y. Pladienolides, new substances from culture of Streptomyces platensis Mer-11107. I. Taxonomy, fermentation, isolation and screening. J Antibiot (Tokyo) 2004;57:173–179. doi: 10.7164/antibiotics.57.173. [DOI] [PubMed] [Google Scholar]
- 75.Motoyoshi H, Horigome M, Ishigami K, Yoshida T, Horinouchi S, Yoshida M, Watanabe H, Kitahara T. Structure-activity relationship for FR901464: a versatile method for the conversion and preparation of biologically active biotinylated probes. Biosci Biotechnol Biochem. 2004;68:2178–2182. doi: 10.1271/bbb.68.2178. [DOI] [PubMed] [Google Scholar]
- 76.Mizui Y, Sakai T, Iwata M, Uenaka T, Okamoto K, Shimizu H, Yamori T, Yoshimatsu K, Asada M. Pladienolides, new substances from culture of Streptomyces platensis Mer-11107. III. In vitro and in vivo antitumor activities. J Antibiot (Tokyo) 2004;57:188–196. doi: 10.7164/antibiotics.57.188. [DOI] [PubMed] [Google Scholar]
- 77.Sakai Y, Tsujita T, Akiyama T, Yoshida T, Mizukami T, Akinaga S, Horinouchi S, Yoshida M, Yoshida T. GEX1 compounds, novel antitumor antibiotics related to herboxidiene, produced by Streptomyces sp. II. The effects on cell cycle progression and gene expression. J Antibiot (Tokyo) 2002;55:863–872. doi: 10.7164/antibiotics.55.863. [DOI] [PubMed] [Google Scholar]
- 78.Thompson CF, Jamison TF, Jacobsen EN. FR901464: total synthesis, proof of structure, and evaluation of synthetic analogues. J Am Chem Soc. 2001;123:9974–9983. doi: 10.1021/ja016615t. [DOI] [PubMed] [Google Scholar]
- 79.Nakajima H, Sato B, Fujita T, Takase S, Terano H, Okuhara M. New antitumor substances, FR901463, FR901464 and FR901465. I. Taxonomy, fermentation, isolation, physico-chemical properties and biological activities. J Antibiot (Tokyo) 1996;49:1196–1203. doi: 10.7164/antibiotics.49.1196. [DOI] [PubMed] [Google Scholar]
- 80.Fan L, Lagisetti C, Edwards CC, Webb TR, Potter PM. Sudemycins, novel small molecule analogues of FR901464, induce alternative gene splicing. ACS Chem Biol. 2011;6:582–589. doi: 10.1021/cb100356k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Xargay-Torrent S, Lopez-Guerra M, Rosich L, Montraveta A, Roldan J, Rodriguez V, Villamor N, Aymerich M, Lagisetti C, Webb TR, et al. The splicing modulator sudemycin induces a specific antitumor response and cooperates with ibrutinib in chronic lymphocytic leukemia. Oncotarget. 2015;6:22734–22749. doi: 10.18632/oncotarget.4212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Larrayoz M, Blakemore SJ, Dobson RC, Blunt MD, Rose-Zerilli MJ, Walewska R, Duncombe A, Oscier D, Koide K, Forconi F, et al. The SF3B1 inhibitor spliceostatin A (SSA) elicits apoptosis in chronic lymphocytic leukaemia cells through downregulation of Mcl-1. Leukemia. 2015 doi: 10.1038/leu.2015.286. [DOI] [PubMed] [Google Scholar]
- 83.Hsu TY, Simon LM, Neill NJ, Marcotte R, Sayad A, Bland CS, Echeverria GV, Sun T, Kurley SJ, Tyagi S, et al. The spliceosome is a therapeutic vulnerability in MYC-driven cancer. Nature. 2015;525:384–388. doi: 10.1038/nature14985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Nakajima H, Hori Y, Terano H, Okuhara M, Manda T, Matsumoto S, Shimomura K. New antitumor substances, FR901463, FR901464 and FR901465. II. Activities against experimental tumors in mice and mechanism of action. J Antibiot (Tokyo) 1996;49:1204–1211. doi: 10.7164/antibiotics.49.1204. [DOI] [PubMed] [Google Scholar]
- 85.Eskens FA, Ramos FJ, Burger H, O'Brien JP, Piera A, de Jonge MJ, Mizui Y, Wiemer EA, Carreras MJ, Baselga J, et al. Phase I, Pharmacokinetic and Pharmacodynamic Study of the First-in-Class Spliceosome Inhibitor E7107 in Patients with Advanced Solid Tumors. Clin Cancer Res. 2013 doi: 10.1158/1078-0432.CCR-13-0485. [DOI] [PubMed] [Google Scholar]
- 86.Kashyap MK, Kumar D, Villa R, La Clair JJ, Benner C, Sasik R, Jones H, Ghia EM, Rassenti LZ, Kipps TJ, et al. Targeting the spliceosome in chronic lymphocytic leukemia with the macrolides FD-895 and pladienolide-B. Haematologica. 2015;100:945–954. doi: 10.3324/haematol.2014.122069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lee SC, Dvinge H, Kim E, Cho H, Micol JB, Chung YR, Durham BH, Yoshimi A, Kim YJ, Thomas M, et al. Modulation of splicing catalysis for therapeutic targeting of leukemia with mutations in genes encoding spliceosomal proteins. Nat Med. 2016 doi: 10.1038/nm.4097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Laetsch TW, Liu X, Vu A, Sliozberg M, Vido M, Elci OU, Goldsmith KC, Hogarty MD. Multiple components of the spliceosome regulate Mcl1 activity in neuroblastoma. Cell Death Dis. 2014;5:e1072. doi: 10.1038/cddis.2014.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Papasaikas P, Tejedor JR, Vigevani L, Valcarcel J. Functional splicing network reveals extensive regulatory potential of the core spliceosomal machinery. Mol Cell. 2015;57:7–22. doi: 10.1016/j.molcel.2014.10.030. [DOI] [PubMed] [Google Scholar]
- 90.Yokoi A, Kotake Y, Takahashi K, Kadowaki T, Matsumoto Y, Minoshima Y, Sugi NH, Sagane K, Hamaguchi M, Iwata M, et al. Biological validation that SF3b is a target of the antitumor macrolide pladienolide. FEBS J. 2011;278:4870–4880. doi: 10.1111/j.1742-4658.2011.08387.x. [DOI] [PubMed] [Google Scholar]
- 91.Moore MJ, Wang Q, Kennedy CJ, Silver PA. An alternative splicing network links cell-cycle control to apoptosis. Cell. 2010;142:625–636. doi: 10.1016/j.cell.2010.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Seki-Asano M, Okazaki T, Yamagishi M, Sakai N, Takayama Y, Hanada K, Morimoto S, Takatsuki A, Mizoue K. Isolation and characterization of a new 12-membered macrolide FD-895. J Antibiot (Tokyo) 1994;47:1395–1401. doi: 10.7164/antibiotics.47.1395. [DOI] [PubMed] [Google Scholar]
- 93.Gao Y, Koide K. Chemical perturbation of Mcl-1 pre-mRNA splicing to induce apoptosis in cancer cells. ACS Chem Biol. 2013;8:895–900. doi: 10.1021/cb300602j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bae J, Leo CP, Hsu SY, Hsueh AJ. MCL-1S, a splicing variant of the antiapoptotic BCL-2 family member MCL-1, encodes a proapoptotic protein possessing only the BH3 domain. J Biol Chem. 2000;275:25255–25261. doi: 10.1074/jbc.M909826199. [DOI] [PubMed] [Google Scholar]
- 95.Boise LH, Gonzalez-Garcia M, Postema CE, Ding L, Lindsten T, Turka LA, Mao X, Nunez G, Thompson CB. bcl-x, a bcl-2-related gene that functions as a dominant regulator of apoptotic cell death. Cell. 1993;74:597–608. doi: 10.1016/0092-8674(93)90508-n. [DOI] [PubMed] [Google Scholar]
- 96.Munding EM, Shiue L, Katzman S, Donohue JP, Ares M., Jr. Competition between pre-mRNAs for the splicing machinery drives global regulation of splicing. Mol Cell. 2013;51:338–348. doi: 10.1016/j.molcel.2013.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lin CY, Loven J, Rahl PB, Paranal RM, Burge CB, Bradner JE, Lee TI, Young RA. Transcriptional amplification in tumor cells with elevated c-Myc. Cell. 2012;151:56–67. doi: 10.1016/j.cell.2012.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ghosh AK, Chen ZH. Enantioselective Syntheses of FR901464 and Spliceostatin A: Potent Inhibitors of Spliceosome. Org Lett. 2013;15:5088–5091. doi: 10.1021/ol4024634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ghosh AK, Veitschegger AM, Sheri VR, Effenberger KA, Prichard BE, Jurica MS. Enantioselective synthesis of spliceostatin E and evaluation of biological activity. Org Lett. 2014;16:6200–6203. doi: 10.1021/ol503127r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Albert BJ, Sivaramakrishnan A, Naka T, Koide K. Total synthesis of FR901464, an antitumor agent that regulates the transcription of oncogenes and tumor suppressor genes. J Am Chem Soc. 2006;128:2792–2793. doi: 10.1021/ja058216u. [DOI] [PubMed] [Google Scholar]
- 101.Ghosh AK, Anderson DD. Enantioselective total synthesis of pladienolide B: a potent spliceosome inhibitor. Org Lett. 2012;14:4730–4733. doi: 10.1021/ol301886g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kanada RM, Itoh D, Nagai M, Niijima J, Asai N, Mizui Y, Abe S, Kotake Y. Total synthesis of the potent antitumor macrolides pladienolide B and D. Angew Chem Int Ed Engl. 2007;46:4350–4355. doi: 10.1002/anie.200604997. [DOI] [PubMed] [Google Scholar]
- 103.Ghosh AK, Li J. A stereoselective synthesis of (+)-herboxidiene/GEX1A. Org Lett. 2011;13:66–69. doi: 10.1021/ol102549a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ghosh AK, Ma N, Effenberger KA, Jurica MS. Total Synthesis of GEX1Q1, Assignment of C-5 Stereoconfiguration and Evaluation of Spliceosome Inhibitory Activity. Org Lett. 2014;16:3154–3157. doi: 10.1021/ol501345d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Goronga T, Boyd VA, Lagisetti C, Jeffries C, Webb TR. Radiosynthesis of antitumor spliceosome modulators. Appl Radiat Isot. 2011;69:1231–1234. doi: 10.1016/j.apradiso.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Furumai R, Uchida K, Komi Y, Yoneyama M, Ishigami K, Watanabe H, Kojima S, Yoshida M. Spliceostatin A blocks angiogenesis by inhibiting global gene expression including VEGF. Cancer Science. 2010;101:2483–2489. doi: 10.1111/j.1349-7006.2010.01686.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ghosh AK, Chen ZH, Effenberger KA, Jurica MS. Enantioselective Total Syntheses of FR901464 and Spliceostatin A and Evaluation of Splicing Activity of Key Derivatives. J Org Chem. 2014;79:5697–5709. doi: 10.1021/jo500800k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Miller-Wideman M, Makkar N, Tran M, Isaac B, Biest N, Stonard R. Herboxidiene, a new herbicidal substance from Streptomyces chromofuscus A7847. Taxonomy, fermentation, isolation, physico-chemical and biological properties. J Antibiot (Tokyo) 1992;45:914–921. doi: 10.7164/antibiotics.45.914. [DOI] [PubMed] [Google Scholar]
- 109.Jung HJ, Kim Y, Shin JY, Sohng JK, Kwon HJ. Antiangiogenic activity of herboxidiene via downregulation of vascular endothelial growth factor receptor-2 and hypoxia-inducible factor-1alpha. Arch Pharm Res. 2015;38:1728–1735. doi: 10.1007/s12272-015-0625-4. [DOI] [PubMed] [Google Scholar]
- 110.Kakeya H, Kaida D, Sekiya H, Nagai K, Yoshida M, Osada H. RQN-18690A (18-deoxyherboxidiene) targets SF3b, a spliceosome component, and inhibits angiogenesis. J Antibiot (Tokyo) 2015 doi: 10.1038/ja.2015.94. [DOI] [PubMed] [Google Scholar]
- 111.Sasaki H, Kitoh Y, Tsukada M, Miki K, Koyama K, Juliawaty LD, Hakim EH, Takahashi K, Kinoshita K. Inhibitory activities of biflavonoids against amyloid-beta peptide 42 cytotoxicity in PC-12 cells. Bioorg Med Chem Lett. 2015;25:2831–2833. doi: 10.1016/j.bmcl.2015.04.106. [DOI] [PubMed] [Google Scholar]
- 112.Yoon SO, Shin S, Lee HJ, Chun HK, Chung AS. Isoginkgetin inhibits tumor cell invasion by regulating phosphatidylinositol 3-kinase/Akt-dependent matrix metalloproteinase-9 expression. Mol Cancer Ther. 2006;5:2666–2675. doi: 10.1158/1535-7163.MCT-06-0321. [DOI] [PubMed] [Google Scholar]
- 113.Morimoto M, Fukui M, Ohkubo S, Tamaoki T, Tomita F. Tetrocarcins, new antitumor antibiotics. 3. Antitumor activity of tetrocarcin A. J Antibiot (Tokyo) 1982;35:1033–1037. doi: 10.7164/antibiotics.35.1033. [DOI] [PubMed] [Google Scholar]
- 114.Tamaoki T, Kasai M, Shirahata K, Ohkubo S, Morimoto M, Mineura K, Ishii S, Tomita F. Tetrocarcins, novel antitumor antibiotics. II. Isolation, characterization and antitumor activity. J Antibiot (Tokyo) 1980;33:946–950. doi: 10.7164/antibiotics.33.946. [DOI] [PubMed] [Google Scholar]
- 115.Tomita F, Tamaoki T. Tetrocarcins, novel antitumor antibiotics. I. Producing organism, fermentation and antimicrobial activity. J Antibiot (Tokyo) 1980;33:940–945. doi: 10.7164/antibiotics.33.940. [DOI] [PubMed] [Google Scholar]
- 116.Nakajima H, Sakaguchi K, Fujiwara I, Mizuta M, Tsuruga M, Magae J, Mizuta N. Apoptosis and inactivation of the PI3-kinase pathway by tetrocarcin A in breast cancers. Biochem Biophys Res Commun. 2007;356:260–265. doi: 10.1016/j.bbrc.2007.02.136. [DOI] [PubMed] [Google Scholar]
- 117.Anether G, Tinhofer I, Senfter M, Greil R. Tetrocarcin-A--induced ER stress mediates apoptosis in B-CLL cells via a Bcl-2--independent pathway. Blood. 2003;101:4561–4568. doi: 10.1182/blood-2002-08-2501. [DOI] [PubMed] [Google Scholar]
- 118.Nakashima T, Miura M, Hara M. Tetrocarcin A inhibits mitochondrial functions of Bcl-2 and suppresses its anti-apoptotic activity. Cancer Res. 2000;60:1229–1235. [PubMed] [Google Scholar]
- 119.Correche ER, Enriz RD, Piovano M, Garbarino J, Gomez-Lechon MJ. Cytotoxic and apoptotic effects on hepatocytes of secondary metabolites obtained from lichens. Altern Lab Anim. 2004;32:605–615. doi: 10.1177/026119290403200611. [DOI] [PubMed] [Google Scholar]
- 120.Deraeve C, Guo Z, Bon RS, Blankenfeldt W, DiLucrezia R, Wolf A, Menninger S, Stigter EA, Wetzel S, Choidas A, et al. Psoromic acid is a selective and covalent Rab-prenylation inhibitor targeting autoinhibited RabGGTase. J Am Chem Soc. 2012;134:7384–7391. doi: 10.1021/ja211305j. [DOI] [PubMed] [Google Scholar]
- 121.Shoemaker RH. The NCI60 human tumour cell line anticancer drug screen. Nat Rev Cancer. 2006;6:813–823. doi: 10.1038/nrc1951. [DOI] [PubMed] [Google Scholar]
- 122.Lazo JS, Nemoto K, Pestell KE, Cooley K, Southwick EC, Mitchell DA, Furey W, Gussio R, Zaharevitz DW, Joo B, et al. Identification of a potent and selective pharmacophore for Cdc25 dual specificity phosphatase inhibitors. Mol Pharmacol. 2002;61:720–728. doi: 10.1124/mol.61.4.720. [DOI] [PubMed] [Google Scholar]
- 123.Aukema KG, Chohan KK, Plourde GL, Reimer KB, Rader SD. Small molecule inhibitors of yeast pre-mRNA splicing. ACS Chem Biol. 2009;4:759–768. doi: 10.1021/cb900090z. [DOI] [PubMed] [Google Scholar]
- 124.Marks PA. Discovery and development of SAHA as an anticancer agent. Oncogene. 2007;26:1351–1356. doi: 10.1038/sj.onc.1210204. [DOI] [PubMed] [Google Scholar]
- 125.Grant S, Easley C, Kirkpatrick P. Vorinostat. Nat Rev Drug Discov. 2007;6:21–22. doi: 10.1038/nrd2227. [DOI] [PubMed] [Google Scholar]
- 126.Sarli V, Giannis A. Selective inhibition of CBP/p300 HAT. Chem Biol. 2007;14:605–606. doi: 10.1016/j.chembiol.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 127.Balasubramanyam K, Altaf M, Varier RA, Swaminathan V, Ravindran A, Sadhale PP, Kundu TK. Polyisoprenylated benzophenone, garcinol, a natural histone acetyltransferase inhibitor, represses chromatin transcription and alters global gene expression. J Biol Chem. 2004;279:33716–33726. doi: 10.1074/jbc.M402839200. [DOI] [PubMed] [Google Scholar]
- 128.Arakawa H, Iguchi T, Morita M, Yoshinari T, Kojiri K, Suda H, Okura A, Nishimura S. Novel indolocarbazole compound 6-N-formylamino-12,13-dihydro-1,11-dihydroxy- 13-(beta-D-glucopyranosyl)-5H-indolo[2,3-a]pyrrolo-[3,4-c]carbazole- 5,7(6H)-dione (NB-506): its potent antitumor activities in mice. Cancer Res. 1995;55:1316–1320. [PubMed] [Google Scholar]
- 129.Muraki M, Ohkawara B, Hosoya T, Onogi H, Koizumi J, Koizumi T, Sumi K, Yomoda J, Murray MV, Kimura H, et al. Manipulation of alternative splicing by a newly developed inhibitor of Clks. J Biol Chem. 2004;279:24246–24254. doi: 10.1074/jbc.M314298200. [DOI] [PubMed] [Google Scholar]
- 130.Verma R, Peters NR, D'Onofrio M, Tochtrop GP, Sakamoto KM, Varadan R, Zhang M, Coffino P, Fushman D, Deshaies RJ, et al. Ubistatins inhibit proteasome-dependent degradation by binding the ubiquitin chain. Science. 2004;306:117–120. doi: 10.1126/science.1100946. [DOI] [PubMed] [Google Scholar]
- 131.Sattler I, Thiericke R, Zeeck A. The manumycin-group metabolites. Nat Prod Rep. 1998;15:221–240. doi: 10.1039/a815221y. [DOI] [PubMed] [Google Scholar]