Abstract
IMPORTANCE
Recurrent and/or metastatic head and neck cancer is usually incurable. Implementation of precision oncology for these patients has been limited by incomplete understanding of the molecular alterations underlying advanced disease. At the same time, the molecular profiles of many rare head and neck cancer types are unknown. These significant gaps in knowledge need to be addressed to rationally devise new therapies.
OBJECTIVE
To illuminate the distinct biology of recurrent and metastatic head and neck cancers and review implementation of precision oncology for patients with advanced disease.
DESIGN, SETTING, AND PARTICIPANTS
After exclusions, 151 patients with advanced, treatment-resistant head and neck tumors, including squamous cell carcinoma (HNSCC), adenoid cystic carcinoma (ACC), and other salivary and cutaneous cancers, whose tumors were sequenced between January 2014 and July 2015 at Memorial Sloan Kettering were recruited. Next-generation sequencing of tumors as part of clinical care included high-depth (median 600×) exonic coverage of 410 cancer genes and whole-genome copy number analysis.
INTERVENTIONS
Next-generation sequencing of tumors and matched normal DNA.
MAIN OUTCOMES AND MEASURES
Feasibility, the frequency of actionable molecular alterations, the effect on decision making, and identification of alterations associated with recurrent and metastatic disease.
RESULTS
Overall, 151 patients (95 men and 56 women; mean [range] age, 61.8 [17-100] years) were included in the study. Next-generation sequencing ultimately guided therapy in 21 of 151 patients (14%) (13 of 53 [25%] of patients with HNSCC) by refining diagnoses and matching patients to specific therapies, in some cases with dramatic responses on basket studies. Molecular alterations were potentially actionable in 28 of 135 patients (21%). The genetic profiles of recurrent and metastatic tumors were often distinct from primary tumors. Compared to primary human papillomavirus (HPV)-positive tumors, many recurrent and metastatic HPV-positive tumors exhibited a molecular profile more similar to HPV-negative tumors, including enriched frequencies of TP53 mutation (3 of 20 tumors [15%]), whole genome duplication (5 of 20 tumors [25%]), and 3p deletion (11 of 20 tumors [55%]). There were high rates of TERT promoter mutation in recurrent and metastatic HPV-negative HNSCC (13 of 30 tumors [43%]), cutaneous SCC (11 of 21 tumors [52%]), basal cell carcinoma (3 of 4 tumors [75%]), and ACC (5 of 36 tumors [14%]). Activating NOTCH1 mutations were enriched in metastatic ACCs (8 of 36 tumors [22%]).
CONCLUSIONS AND RELEVANCE
These findings reveal the molecular landscape of advanced disease and rare cancer subtypes, both predominant challenges in head and neck oncology. To understand the repertoire of targetable alterations in advanced cancers, it is necessary to sequence recurrent and metastatic tumors. These data are important first steps toward implementation of precision head and neck oncology.
Precision oncology is a clinical approach that seeks to match every patient’s tumor with the best drug, based on molecular alterations. This approach also expands our knowledge of cancer biology, by revealing the molecular landscape of tumor types in which existing knowledge is limited: recurrent, metastatic, and rare cancers.
Head and neck cancers are the sixth most common cancer worldwide, and arise from the upper aerodigestive mucosa, salivary glands, and skin.1 Despite differences in histology and biology across cancer types, most treatment protocols are extrapolated from head and neck squamous cell carcinoma (HNSCC)—the most common histology. Recurrent and metastatic disease is usually incurable. There are significant gaps in knowledge. Unlike other cancer types, there are no predictive biomarkers, and no new targeted therapies have been approved for HNSCC other than cetuximab in 2006. Currently, our understanding of the molecular landscape of head and neck cancer is drawn from exome studies of primary tumors representing the more common histologic subtypes.2-10
Genetic profiles of the most clinically challenging tumors—incurable, recurrent and metastatic disease—remain poorly defined. Less common cancer types have not heretofore been molecularly profiled. Here, we describe our multidisciplinary head and neck cancer team’s implementation of a precision oncology approach, review therapeutic successes and limitations for patients, and analyze sequencing data to glean new insights into the alterations that define recurrent and metastatic head and neck cancer.
Methods
The MSK-IMPACT (Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets) assay is a targeted next-generation sequencing (NGS) assay approved through Clinical Laboratory Improvement Amendments.11 The assay is optimized for DNA from formalin-fixed, paraffin-embedded samples, and targets single nucleotide variants, indels, and structural variants in 410 genes functionally relevant to cancer or clinically actionable targets (eTable 1 in Supplement 2), and genome-wide copy number.12,13
After receiving institutional review board approval from the Memorial Sloan Kettering Cancer Center, we reviewed patients treated by our multidisciplinary head and neck oncology team whose tumors were sequenced after informed written consent to an institutional review board–approved protocol (NCT01775072). Of 224 patients enrolled, 151 with minimum 6-month follow-up, whose tumors were sequenced between January 2014 and July 2015, were included. Treatment(s) initiated after NGS were considered guided by molecular data if results led to, or justified continuation with, specific treatments. Molecular alterations were classified by evidence indicating standard or investigational therapies (eTable 2 in Supplement 2).
Copy number analyses with FACETS14 were performed to identify whole-genome duplication, 3p deletion (eFigures 1 and 2 in Supplement 1), and subclonal population structure. Mutational signatures (APOBEC, tobacco, ultraviolet light carcinogenesis) were analyzed using definitions previously described.15,16
Somatic alterations in HNSCC were examined using a support vector machine algorithm to classify human papillomavirus (HPV) status, and using enrichment analysis for comparison to primary tumors in the The Cancer Genome Atlas (TCGA) cohort.3 Germline sequencing from matched normal blood was analyzed using a previously described pipeline.13 Additional technical details of the assay, variant calling, support vector machine algorithm, bioinformatic and biostatistical analyses are in the eAppendix in Supplement 1.
Results
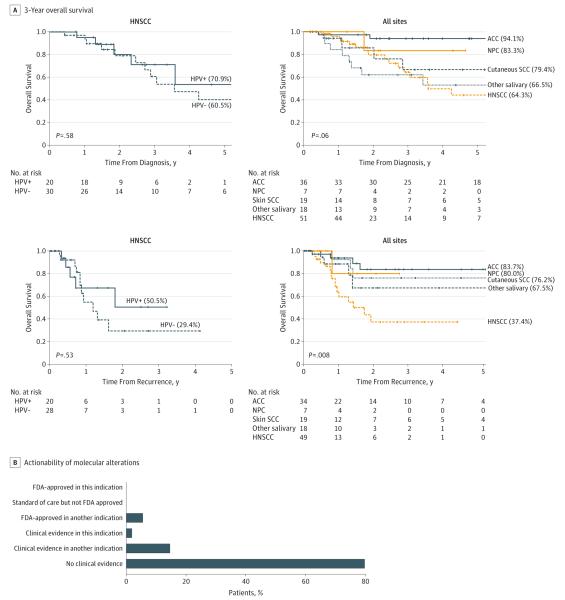
MSK-IMPACT was used to sequence 151 head and neck tumors, including 53 HNSCC, 56 salivary carcinomas, 27 cutaneous carcinomas, 8 nasopharyngeal carcinomas, 3 neuroendocrine carcinomas, 2 odontogenic carcinomas, and 2 olfactory neuroblastomas (eFigure 11 in Supplement 1), in 95 men and 56 women; 66 patients had locoregional recurrence, 106 had distant metastases, and 6 had locally advanced tumors that were surgically unresectable. Clinical data are summarized in eTable 3 in Supplement 2. Median (range) depth of sequencing was 600 × (82 × −1165 ×). Survival and genomic data are summarized in Figure 1 and eFigures 12 and 13 in Supplement 1.
Figure 1. Overall Survival for Patients Undergoing Clinical Tumor Sequencing.
A, Overall survival from time of index treatment and time of disease recurrence and/or metastasis, for selected tumor types. Survival probabilities were calculated with the Kaplan-Meier method, and tumor types were compared using the log-rank test. B, Distribution of molecular alterations, classified by levels of evidence for actionability. HNSCC, head and neck squamous cell carcinoma. ACC, adenoid cystic carcinoma; HPV, human papillomavirus; NPC, nasopharyngeal carcinoma; Other salivary, other salivary cancer types; SCC, squamous cell carcinoma.
Actionable Alterations
There are no standard-of-care biomarkers in head and neck cancer, but 28 of 135 patients (21%) with tumors harboring alterations had findings with potential investigational implications—7 patients (5%) with a US Food and Drug Administration (FDA)-approved biomarker and drug in another indication, and 21 patients (16%) with clinical evidence linking the alteration to drug response (Figure 1) (eTable 2 in Supplement 2). Ultimately, next-generation sequencing data guided therapy in 21 of 151 patients (14%).
Landscape of HNSCC
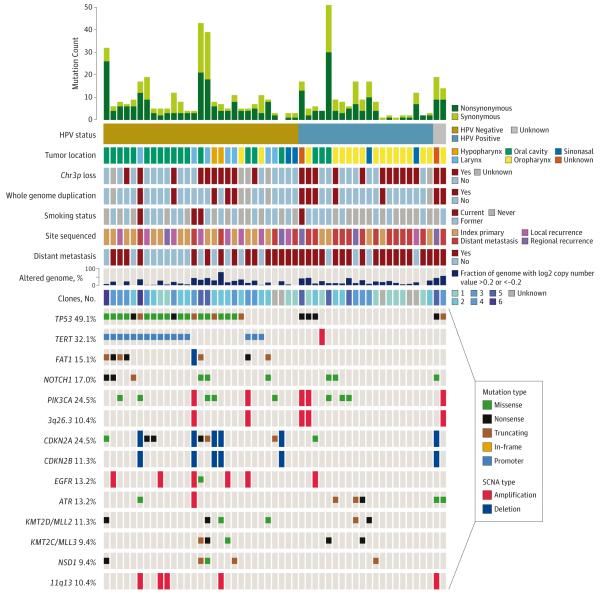
The most frequent single nucleotide variants and copy number variants in HNSCC are depicted in Figure 2 and eTable 4 in Supplement 2. Whole-genome duplication was present in 12 of 48 cases (25%) with complete copy number data (Figure 2) (eFigures 1 and 2 in Supplement 1).17
Figure 2. The Molecular Landscape of Recurrent and/or Metastatic Head and Neck Squamous Cell Carcinoma.
Genetic alterations and clinical data for 53 recurrent and/or metastatic head and neck squamous cell carcinomas. Copy number values are based on 48 cases with informative copy number data; 11q13 includes CCND1, FGF3, FGF4, and FGF19; 3q26.3 includes PIK3CA, SOX2, and DCUN1D1. Clones indicates number of subclonal populations per tumor; HPV, human papillomavirus; SCNA, somatic copy number alterations.
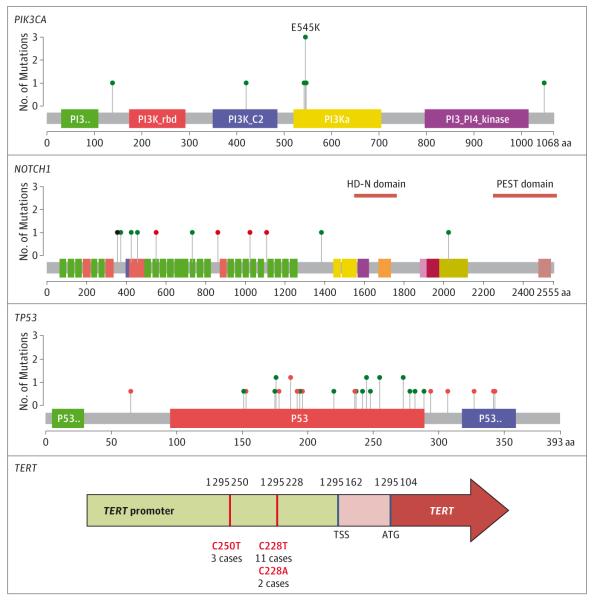
The promoter region of TERT harbored mutations in 16 of 30 HPV-negative tumors (53%) (Figure 3). In this recurrent and metastatic population, mutations in the TERT promoter were significantly more frequent than rates reported in primary HPV-negative HNSCCs18 (12 of 70 [17%]) (odds ratio (OR), 5.5; P < .001), indicating enrichment for this mutation in advanced cancers, although we note that deep sequencing may be more sensitive for single nucleotide variants. Remarkably, 10 of 11 (91%) HPV-negative tongue SCCs had TERT mutations. TERT promoter mutations have been linked with poorer survival.19 There were no TERT promoter mutations in HPV-positive HNSCC, consistent with the ability of the E6 oncoprotein to activate telomerase.20 Therefore, TERT mutation and HPV infection may represent parallel mechanisms of telomerase activation in HNSCC.
Figure 3. Mutation Distributions in PIK3CA, NOTCH1, TP53 and TERT in Head and Neck Squamous Cell Carcinomas.
Green circles indicate missense mutations; red, truncating; black, in-frame insertions or deletions.
Four tumors had mutations in mismatch repair genes, including 1 truncating mutation in MLH1 (in the highly mutated HPV-positive case) and missense mutations in MSH2, MSH6, and POLD1 (HPV-negative cases). The average mutation count in these tumors was significantly elevated (17.3 vs 4.5; P < .001).
After NGS, 17 of 53 patients with HNSCC (32%) were enrolled in clinical trials. Treatment was guided by molecular data in 13 patients (25%). In 7 patients (13%), NGS nominated targeted therapies, similar to the rate in another recent HNSCC precision oncology study (10%).21 Four patients with alterations in PIK3CA or PTEN were treated on PI3K inhibitor trials, 1 HRAS mutation on a farnesyl transferase inhibitor trial, 1 MAPK1 mutation with an ERK inhibitor on a single-patient IND (Investigational New Drug) application, and 1 SMARCB1 deletion on an EZH2 inhibitor trial. In 6 patients (12%), the lack of actionable alterations on MSK-IMPACT prompted the decision to instead enroll on immunotherapy trials.
HPV-Positive and HPV-Negative HNSCC
Comparing HPV-positive tumors (n = 21 [39.6%]) with HPV-negative tumors (n = 30), current and former tobacco use was slightly more common in the HPV-negative population (18 patients [60%] vs 10 patients [48%]) (eTable 3 in Supplement 2). In contrast to prior studies of primary tumors,2,3,6 recurrent and metastatic HPV-positive and HPV-negative tumors had similar mutation counts (4.4 vs 5.9; P = .42) and copy number–altered fraction of genome (0.19 vs 0.18; P = .78) (Figure 2).
Several genes were more frequently altered in recurrent or metastatic HPV-negative tumors compared with HPV-positive tumors, including TP53 (21 [72%] vs 3 [15%]; P < .001), TERT promoter (16 [55%] vs 0 [0%]; P = 9 × 10−5), and CDKN2A mutation and deletion (11 [37%] vs 1 [5%]; P = .016). A correlation network of altered genes revealed clusters altered primarily in HPV-positive or HPV-negative tumors (eFigure 14 in Supplement 1). Gene ontology annotations of the HPV-positive clusters were strongly enriched for tyrosine kinase signaling (P = 3 × 10−9) (eTable 5 in Supplement 2).
We used FACETS to infer levels of intratumor heterogeneity.14 In multivariable analyses, HPV-positive tumors had fewer subclonal populations (P = .03). Recurrent and metastatic sites trended toward having more subclonal mutations than primary sites (P = .08) (eTables 6 and 7 in the Supplement 2).
Alterations Enriched in Recurrent and Metastatic HPV-Positive HNSCC
The vast majority of HPV-positive HNSCCs are cured with standard therapy.22 The genetic alterations in the minority of these tumors developing recurrence and/or metastasis are largely undefined. We examined alterations that were enriched in recurrent and metastatic HPV-positive HNSCC sequenced with MSK-IMPACT, in comparison to primary HPV-positive tumors sequenced by TCGA.3 These 2 cohorts had similar clinical and demographic profiles, including smoking history (50% vs 61%; P = .57).
We first examined variant profiles and determined the degree of similarity between each advanced HPV-positive tumor sequenced with MSK-IMPACT compared with HPV-positive and HPV-negative primary tumors sequenced by TCGA. We used a support vector machine algorithm to categorize tumors as more similar to HPV-positive or HPV-negative profiles based on mutations and copy number variants. The classifier was first trained and cross-validated in the TCGA cohort, then applied to 53 sequenced MSK-IMPACT samples in 51 patients with known HPV status. The classifier correctly categorized 29 of 31 (91%) HPV-negative MSK-IMPACT cases, but only 12 of 21 (57%) HPV-positive cases (P = .007) (eFigure 15 in Supplement 1). The classifier had significantly higher confidence probabilities for HPV-negative tumors (P = 4.4 × 10−5) (eFigure 3A in Supplement 1). Therefore, in this cohort, 43% of advanced HPV-positive HNSCC tumors had genotypes that resembled primary HPV-negative tumors (eFigure 3B in Supplement 1). These “HPV-negative-like” HPV-positive tumors trended toward poorer survival (hazard ratio [HR], 2.3; P = .19) although this comparison was limited by small sample size (eFigure 3C in Supplement 1).
The mutational profile of recurrent and metastatic HPV-positive tumors sequenced by MSK-IMPACT appeared intermediate between the HPV-positive and HPV-negative primary tumors in the TCGA. In comparison, the mutational profile of HPV-negative recurrent and metastatic tumors closely matched the primary HPV-negative cohort (P = 1.3 × 10−4) (eFigure 4 and eFigure 16 in Supplement 1).
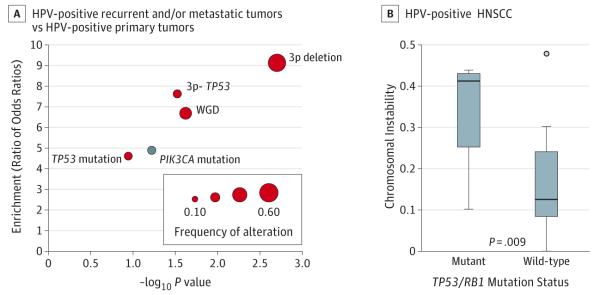
We examined enrichment for specific genetic alterations, focusing on the most frequent: whole-genome duplication, 3p deletion, TP53 mutation, and PIK3CA mutation. To allow for potential differences between platforms, we determined enrichment by comparing HPV-negative:HPV-positive ORs between data sets (Figure 4A) (eTable 8 in Supplement 2). In the recurrent and metastatic HPV-positive tumors, there was significant enrichment for whole-genome duplication (6.7-fold; P = .024), 3p deletion (9.1-fold; P = .002), and 3p deletion-TP53 mutation (7.6-fold; P = .03). Compared with primary HPV-positive tumors, recurrent and metastatic HPV-positive tumors had higher rates of TP53 mutation (3 of 20 tumors [15%] vs 1 of 36 tumors [3%]; enrichment 4.6; P = .11) and lower rates of PIK3CA mutation (2 of 20 tumors [10%] vs 13 of 36 tumors [36%]; enrichment 0.21, P = .06).
Figure 4. Genetic Alterations Enriched in Recurrent and/or Metastatic HPV-Positive HNSCC.
A, Enrichment analysis for common alterations, comparing recurrent and/or metastatic HPV-positive tumors to primary HPV-positive tumors. Red dots indicate alterations enriched in recurrent and/or metastatic tumors, and blue dots indicate alterations enriched in primary tumors. For these comparisons, recurrent and/or metastatic tumors are MSK-IMPACT cases and primary tumors are The Cancer Genome Atlas cases.3 B, Differences in chromosomal instability, defined as proportion of genome copy number-altered, in TP53/RB1-mutated or wild-type HPV-positive tumors in MSK-IMPACT (Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets). Boxes represent interquartile ranges and whiskers represent maxima and minima. HPV indicates human papillomavirus. HNSCC, head and neck squamous cell carcinoma.
We sought to confirm these findings in additional data sets of recurrent and metastatic HPV-positive HNSCC. While there are no comparable cohorts limited to recurrent and metastatic disease, the data sets of Chung et al23 and Chau et al21 included mutational data for HPV-positive tumors, a proportion of which were recurrent or metastatic. Consistent with our data, each of the HPV-positive cohorts in the studies by Chung et al and Chau et al exhibited higher frequencies of TP53 mutation (5%-13%) and lower frequencies of PIK3CA mutation (13%-20%), compared with observed rates in HPV-positive primary tumors.2,3,5,6 For additional confirmation, we analyzed TP53 status in primary uterine cervical cancers (TCGA, n = 170) and recurrent and metastatic HPV-positive cervical cancers (MSK-IMPACT, n = 16). The frequency of TP53 mutation was markedly higher in metastatic cases than primary cases (4 of 16 tumors [25%] vs 5 of 170 [3%] tumors; OR, 8.3; P = .008) (eTable 9 in Supplement 2).
TP53 mutations are uncommon (0%-3%) in HPV-positive primary HNSCC.2,3,5,6 All TP53 mutations were truncating (eTable 4 in Supplement 2). One patient was a current smoker (35 pack-year), and 2 were former oligosmokers (1 and 5 pack-year). Although p53 is inactivated by the HPV E6 oncoprotein,24 in vitro data reveal that complete inactivation of p53 in HPV-positive HNSCC cell lines induces resistance to radiotherapy and more aggressive tumor behavior.25 Interestingly, TP53 mutations in HPV-positive tumors were much more likely to be subclonal than in HPV-negative tumors (3 of 4 mutations [75%] vs 4 of 27 mutations [15%]; P = .03) (eTable 7 in Supplement 2). Our hypothesis generating data suggest that TP53 mutation in a subset of HPV-positive tumors may be associated with tobacco use, occur later in tumor evolution, and reflect poorer prognosis.
Similarly, alterations (mutations or deletions) in RB1 were present in 3 of 20 (15%) recurrent and metastatic HPV-positive tumors. The degree of chromosomal instability in TP53 or RB1 mutated HPV-positive tumors was significantly higher than in TP53/RB1 wildtype tumors (P = .009). This suggests that complete inactivation of p53 or RB has functional sequelae beyond those resulting from incomplete degradation by E6/E7 (Figure 4B).
Mutational Signatures in HNSCC
The tobacco signature was strongest in current smokers and was most evident in laryngeal cancers, similar to findings in TCGA exome data.26 (eFigure 5 in Supplement 1) There was no difference in APOBEC scores between HPV-positive and HPV-negative tumors (1.68 vs 1.36; P = .29), in contrast to data from primary tumors, where the APOBEC signature is associated with HPV-positivity.27 This is consistent with the overall trend of molecular differences between HPV-positive and HPV-negative disease being attenuated in advanced cancers. The tumors with the strongest APOBEC signatures were either sinonasal or oropharynx primaries, mostly HPV-positive (eFigure 5 in Supplement 1).
Adenoid Cystic Carcinomas
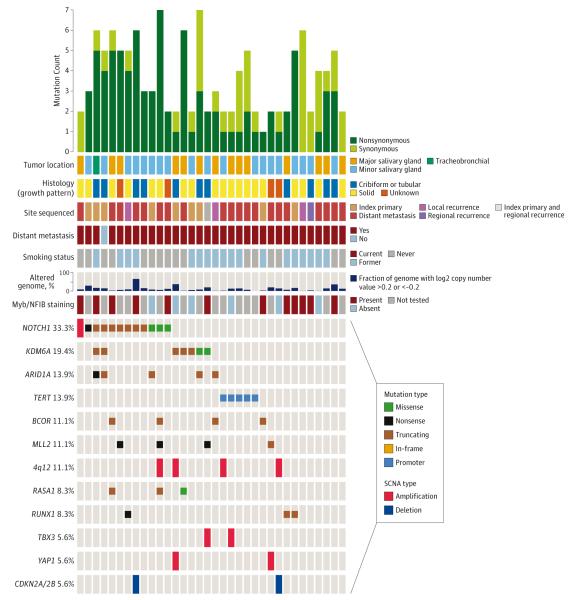
Of 36 recurrent and metastatic ACCs, mutational load (mean, 2.3) and fraction of copy number-altered genome (mean, 0.12) were low. The most frequently altered gene was NOTCH1 (12 of 33 [33%]). In 8 of 36 cases (22.2%), the alterations (mutation or amplification) were consistent with activating events (Figure 5) (eFigure 17 in Supplement 1). Three samples had recurrent truncating alterations at serine 2467, which are activating mutations in leukemia. Comparing recurrent and metastatic ACCs to primary ACCs previously sequenced by our group,9 we identified significant enrichment of NOTCH1 activating alterations (8 of 36 tumors [22.2%] vs 2 of 60 tumors [3.3%]; OR, 8.3; P = .005) (eFigure 6 in Supplement 1), which have been linked to aggressive tumor behavior in ACC.28 Activating NOTCH1 mutations confer sensitivity to γ-secretase inhibitors,29 nominating these drugs as potentially active in NOTCH1-mutated ACCs. TERT promoter mutations were also identified in 5 ACCs (14%). TERT promoter mutations have not been previously identified in salivary cancers (eFigure 17 in Supplement 1 and eTable 4 in Supplement 2).
Figure 5. Mutational Landscape of Recurrent and/or Metastatic Adenoid Cystic Carcinoma.
Genetic and clinical data for 36 recurrent and/or metastatic adenoid cystic carcinomas. SCNA indicates somatic copy number alterations.
Twenty (56%) patients with ACCs were enrolled on clinical trials. In 4 patients (11%), protocol selection was guided by NGS: 1 patient with FGFR3 mutation was treated with an FGFR inhibitor, 1 patient with a PIK3CA mutation and 1 patient with a PIK3R1 mutation were treated with a PI3K inhibitor, and 1 patient with MDM2 amplification was treated with an MDM2 inhibitor. Although not guided by NGS, 4 patients treated with the multikinase inhibitor regorafenib were subsequently found to have PDGFRA/KDR/KIT amplification, justifying the appropriateness of this protocol.
Other Salivary Cancers
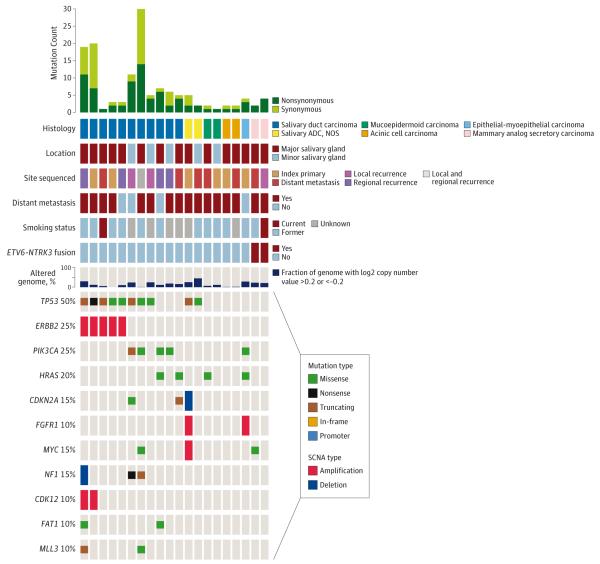
There were 20 advanced salivary cancers of other histologies: 11 salivary duct carcinomas, 2 acinic cell carcinomas, 2 mammary analog secretory carcinomas (MASC), 2 mucoepidermoid carcinomas, 2 salivary adenocarcinomas, and 1 epithelial-myoepithelial carcinoma (Figure 6) (eTable 4 in Supplement 2).
Figure 6. Mutational Landscape of Recurrent and/or Metastatic Other Salivary Carcinoma Types.
Genetic and clinical data for 20 recurrent and/or metastatic other salivary carcinoma types. ADC indicates adenocarcinoma; SCNA, somatic copy number alterations.
Salivary duct carcinomas harbored numerous potentially actionable alterations (including PIK3CA, HRAS, ERBB2, and AR) and several interesting structural variants in NF1 and ROS1 (eTable 2) (eFigure 7 in Supplement 1). One mucoepidermoid tumor with an HRAS G13V mutation was enrolled on a farnesyl transferase inhibitor trial.
One tumor was initially diagnosed as acinic cell carcinoma. Sequencing identified an ETV6-NTRK3 fusion (eFigure 8 in Supplement 1), changing the diagnosis to MASC.30 The patient was then enrolled on a TRK inhibitor basket study, with a clinical near-complete response.31 A second acinic cell carcinoma was rediagnosed as MASC and enrolled on a TRK inhibitor basket study, also with a similar major response.
Cutaneous Carcinomas
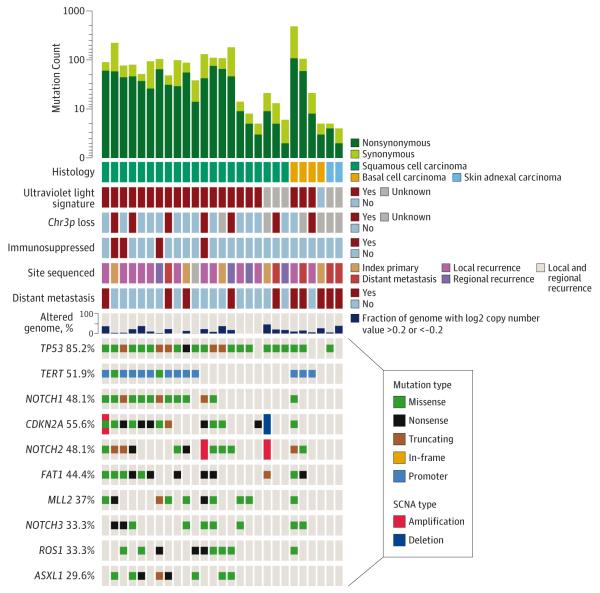
Twenty-seven advanced nonmelanoma skin carcinomas of the head and neck were sequenced: 21 squamous, 2 skin adnexal, and 4 basal cell carcinomas (BCCs) (Figure 7) (eFigure 18 in Supplement 1 and eTable 4 in Supplement 2). A predominating UV light mutational signature was evident in 18 of 21 (86%) cutaneous SCCs and was associated with higher average mutation counts than the cutaneous SCCs lacking a UV light mutational signature (37.9 mutations vs 4.3 mutations; P = .008). Deletion of 3p in combination with TP53 mutation is a strong negative prognostic factor in mucosal HNSCC,32 but these co-occurring alterations have not been described in cutaneous SCC. We found 3p deletion and TP53 mutation in 6 of 19 cases (32%).
Figure 7. Mutational Landscape of Advanced Cutaneous Carcinomas of the Head and Neck.
Genetic and clinical data for 27 advanced cutaneous head and neck cancers. SCNA indicates somatic copy number alterations
Three of 4 advanced BCCs exhibited the UV light mutational signature, including 1 hypermutated tumor with 122 mutations. In this case, the finding of high mutational load led to a decision to treat with immunotherapy.
One BCC lacking the UV light signature had only 2 non-synonymous mutations: a patient who had scalp irradiation in childhood. Three of four BCCs responded initially to hedgehog pathway inhibitors (prior to NGS) and were subsequently confirmed to have PTCH1 mutations.
Nasopharyngeal Carcinomas
Eight recurrent and metastatic nasopharyngeal carcinomas were sequenced (eFigure 19 in Supplement 1 and eTable 4 in Supplement 2). Four patients with nasopharyngeal carcinoma (50%) were enrolled on clinical trials, of which one (13%) was NGS-guided: a PIK3CA mutation led to enrollment on a PI3K inhibitor trial.
Odontogenic Carcinomas
Two odontogenic carcinomas were profiled. Unexpectedly, EWSR1 gene fusions were found in both (eFigure 9 in Supplement1).One case, initially diagnosed as ameloblastic carcinoma, had an EWSR1-FLI1 fusion, confirmed with a polymerase chain reaction assay. Fluorescence in situ hybridization analysis performed earlier had not identified rearrangement of EWSR1, indicating the superior sensitivity of NGS in this case. This changed the diagnosis to Ewing sarcoma with epithelial differentiation and ameloblastic morphology. This information was used to guide chemotherapy with a sarcoma regimen.
The second case was a metastatic clear cell odontogenic carcinoma of the maxilla, with an EWSR1-ATF1 fusion, which has been seen in clear cell sarcomas and hyalinizing clear cell carcinoma of salivary glands, but not in (morphologically similar) odontogenic tumors.
Other Rare Cancers
The findings in 2 olfactory neuroblastomas and 3 head and neck neuroendocrine carcinomas are detailed in eTable 4 in Supplement 2.
Germline Analysis
One HNSCC patient had a germline heterozygous deletion in FANCA (eFigure 10 in Supplement 1) and a tumor with a somatic FANCA stopgain mutation. While homozygous loss of FANCA in the germline causes Fanconi anemia, this double-hit affecting FANCA is an interesting finding in HNSCC,33 similar to a report in a prostate cancer, where it conferred sensitivity to cisplatin.34 The ability of NGS to profile both somatic and germline alterations is an additional source of potentially actionable data.
Complete genomic and clinical data from this study are available in searchable form at http://www.cbioportal.org/study?id=hnc_mskcc_2016.
Discussion
Critical gaps in knowledge must be addressed to rationally devise new therapies for head and neck cancers. To date, genomics studies have been limited to cohorts of primary tumors. The molecular landscape of the most clinically challenging tumors—treatment-resistant, recurrent and metastatic disease—has not been defined. Furthermore, many uncommon cancer types have not been molecularly profiled.
Here, we review our implementation of precision oncology for patients with advanced head and neck cancers. This effort has been useful in 3 respects: (1) illuminating the unique molecular landscape of advanced or understudied cancers; (2) refining diagnoses of rare cancers; and (3) guiding treatment based on molecular alterations.
In the initial 2 years of this approach, NGS guided therapy in 14% of patients (25% of HNSCC patients). A major limitation has been the low frequency (21%) of targetable alterations. This underscores the importance of profiling these tumors, and the potential benefits of basket studies for diseases such as head and neck cancer. In several cases, such as the enrollment of metastatic MASC tumors on TRK inhibitor trials, NGS facilitated dramatic therapeutic responses.
Focused analyses of recurrent and metastatic tumors can provide valuable biologic insights, identifying alterations that are associated with advanced cancers.35 For example, we identified a high rate of activating NOTCH1 mutations in metastatic ACC. We also identified genetic alterations defining recurrent and metastatic HPV-positive cancers: many tumors had genetic profiles closer to HPV-negative tumors, a finding that has potential significance to current studies of targeted therapies and chemoradiotherapy deintensification in this population. An important caveat is that these findings are based on a small cohort, and when comparing results to other data sets, it is possible that differences in patient characteristics, prior therapy, sequencing or analytic methodologies may contribute to differences observed. These results require further confirmation.
Many head and neck cancers are understudied due to their rarity. We report the first molecular data in mucoepidermoid carcinomas, acinic cell carcinomas, epithelialmyoepithelial carcinomas, MASC, salivary adenocarcinoma, skin adnexal carcinomas, odontogenic carcinomas, and head and neck neuroendocrine carcinomas. While sample sizes were too limited to draw broad conclusions, many potentially actionable findings were identified and several uncommon diagnoses refined.
Finally, this analysis demonstrated the potential of targeted NGS to measure intratumor heterogeneity and identify certain mutational signatures: those of carcinogenesis driven by UV light, tobacco, APOBEC, and mismatch repair deficiency.15,36,37 Emerging data implicate each of these features as potential predictive biomarkers for immunotherapy: ITH, mutational load, and the tobacco and UV light mutational signatures have each been associated with response to immune checkpoint inhibitors in lung, skin, and colorectal cancers.38-42
Conclusions
To advance precision head and neck oncology, a necessary first step is describing the complete catalog of molecular alterations in incurable and rare cancers. As demonstrated here, the molecular profile of recurrent and metastatic tumors is quite distinct from primary tumors. In these understudied diseases, no single institution will be able to profile a large number of patients. Pooling genomic data will be critical to devising rational therapies. Finally, a major therapeutic limitation in head and neck cancer has been the lack of matched trial options. This shows the value of basket studies, which accounted for half of clinical trial enrollments in this cohort. As understanding of molecular drivers and biomarkers expands, we anticipate the clinical value added by routine NGS will continue to increase.
Supplementary Material
Key Points.
Question
Can clinical implementation of next-generation sequencing of tumors help guide treatment and expand our understanding of the biology of advanced head and neck cancer?
Findings
Molecular profiling of advanced (nearly all recurrent and metastatic) head and neck cancers was able to identify actionable alterations and guide treatment in a portion of patients, refine diagnoses in certain rare cancers, and discover unique genetic findings that are enriched in advanced cancers.
Meaning
Implementation of precision oncology is beginning to guide treatment and improve our knowledge of molecular alterations relevant to advanced, treatment-resistant head and neck cancers.
Acknowledgments
Funding/Support: This work was supported by NIH P30 CA008748 (to the Memorial Sloan Kettering Cancer Center), the Damon Runyon Cancer Research Foundation, the National Institutes of Health (grant No. NIH K08 DE024774), and the Society of MSK (to Dr Morris).
Role of the Funder/Sponsor: The funders/sponsors had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; and decision to submit the manuscript for publication.
Footnotes
Supplemental content at jamaoncology.com
Author Contributions: Dr Morris had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Concept and design: Morris, Chan, Hyman, Solit, Ho.
Acquisition, analysis, or interpretation of data: Morris, Chandramohan, West, Zehir, Chakravarty, Pfister, Wong, Sherman, Baxi, Ganly, Singh, Shah, Shaha, Patel, Roman, Barker, McBride, Chan, Dogan, Hyman, Berger, Solit, Riaz, Ho.
Drafting of the manuscript: Morris, Chan, Solit.
Critical revision of the manuscript for important intellectual content: Morris, Chandramohan, West, Zehir, Chakravarty, Pfister, Wong, Sherman, Baxi, Ganly, Singh, Shah, Shaha, Boyle, Patel, Roman, Barker, McBride, Chan, Dogan, Hyman, Berger, Solit, Riaz, Ho.
Statistical analysis: Morris, Chandramohan, Berger, Riaz.
Obtaining funding: Morris, Chan, Solit.
Administrative, technical, or material support: Morris, Sherman, Baxi, Singh, Shaha, Roman, Barker, Chan, Hyman, Solit, Ho.
Study supervision: Morris, Chan, Dogan, Berger, Solit, Ho.
Other: Morris, West.
Conflict of Interest Disclosures: None reported.
Additional Contributions: We thank our patients and acknowledge our colleagues from the Head and Neck Cancer Disease Management Team and the Center for Molecular Oncology.
REFERENCES
- 1.Duvvuri U, Myers JN. Cancer of the head and neck is the sixth most common cancer worldwide. Curr Probl Surg. 2009;46(2):114–117. doi: 10.1067/j.cpsurg.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 2.Agrawal N, Frederick MJ, Pickering CR, et al. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science. 2011;333(6046):1154–1157. doi: 10.1126/science.1206923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cancer Genome Atlas Network Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 2015;517(7536):576–582. doi: 10.1038/nature14129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.India Project Team of the International Cancer Genome Consortium Mutational landscape of gingivo-buccal oral squamous cell carcinoma reveals new recurrently-mutated genes and molecular subgroups. Nat Commun. 2013;4:2873. doi: 10.1038/ncomms3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Seiwert TY, Zuo Z, Keck MK, et al. Integrative and comparative genomic analysis of HPV-positive and HPV-negative head and neck squamous cell carcinomas. Clin Cancer Res. 2015;21(3):632–641. doi: 10.1158/1078-0432.CCR-13-3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stransky N, Egloff AM, Tward AD, et al. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011;333(6046):1157–1160. doi: 10.1126/science.1208130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lin DC, Meng X, Hazawa M, et al. The genomic landscape of nasopharyngeal carcinoma. Nat Genet. 2014;46(8):866–871. doi: 10.1038/ng.3006. [DOI] [PubMed] [Google Scholar]
- 8.Pickering CR, Zhou JH, Lee JJ, et al. Mutational landscape of aggressive cutaneous squamous cell carcinoma. Clin Cancer Res. 2014;20(24):6582–6592. doi: 10.1158/1078-0432.CCR-14-1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ho AS, Kannan K, Roy DM, et al. The mutational landscape of adenoid cystic carcinoma. Nat Genet. 2013;45(7):791–798. doi: 10.1038/ng.2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stephens PJ, Davies HR, Mitani Y, et al. Whole exome sequencing of adenoid cystic carcinoma. J Clin Invest. 2013;123(7):2965–2968. doi: 10.1172/JCI67201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hyman DM, Solit DB, Arcila ME, et al. Precision medicine at Memorial Sloan Kettering Cancer Center: clinical next-generation sequencing enabling next-generation targeted therapy trials. Drug Discov Today. 2015;20(12):1422–1428. doi: 10.1016/j.drudis.2015.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheng DT, Mitchell TN, Zehir A, et al. Memorial Sloan Kettering-integrated mutation profiling of actionable cancer targets (MSK-IMPACT): a hybridization capture-based next-generation sequencing clinical assay for solid tumor molecular oncology. J Mol Diagn. 2015;17(3):251–264. doi: 10.1016/j.jmoldx.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schrader KA, Cheng DT, Joseph V, et al. Germline Variants in Targeted Tumor Sequencing Using Matched Normal DNA. JAMA Oncol. 2016;2(1):104–111. doi: 10.1001/jamaoncol.2015.5208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shen R, Seshan V. FACETS: allele-specific copy number and clonal heterogeneity analysis tool for high-throughput DNA sequencing. Nucleic Acids Res. 2016 Jun 7;:pii:gkw520. doi: 10.1093/nar/gkw520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roberts SA, Lawrence MS, Klimczak LJ, et al. An APOBEC cytidine deaminase mutagenesis pattern is widespread in human cancers. Nat Genet. 2013;45(9):970–976. doi: 10.1038/ng.2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pfeifer GP, You YH, Besaratinia A. Mutations induced by ultraviolet light. Mutat Res. 2005;571(1-2):19–31. doi: 10.1016/j.mrfmmm.2004.06.057. [DOI] [PubMed] [Google Scholar]
- 17.Zack TI, Schumacher SE, Carter SL, et al. Pan-cancer patterns of somatic copy number alteration. Nat Genet. 2013;45(10):1134–1140. doi: 10.1038/ng.2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Killela PJ, Reitman ZJ, Jiao Y, et al. TERT promoter mutations occur frequently in gliomas and a subset of tumors derived from cells with low rates of self-renewal. Proc Natl Acad Sci U S A. 2013;110(15):6021–6026. doi: 10.1073/pnas.1303607110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qu Y, Dang S, Wu K, et al. TERT promoter mutations predict worse survival in laryngeal cancer patients. Int J Cancer. 2014;135(4):1008–1010. doi: 10.1002/ijc.28728. [DOI] [PubMed] [Google Scholar]
- 20.Liu X, Dakic A, Zhang Y, Dai Y, Chen R, Schlegel R. HPV E6 protein interacts physically and functionally with the cellular telomerase complex. Proc Natl Acad Sci U S A. 2009;106(44):18780–18785. doi: 10.1073/pnas.0906357106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chau NG, Li YY, Jo VY, et al. Incorporation of next-generation sequencing into routine clinical care to direct treatment of head and neck squamous cell carcinoma [published online January 13, 2016] Clin Cancer Res. 2016 doi: 10.1158/1078-0432.CCR-15-2314. [DOI] [PubMed] [Google Scholar]
- 22.Ang KK, Harris J, Wheeler R, et al. Human papillomavirus and survival of patients with oropharyngeal cancer. N Engl J Med. 2010;363(1):24–35. doi: 10.1056/NEJMoa0912217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chung CH, Guthrie VB, Masica DL, et al. Genomic alterations in head and neck squamous cell carcinoma determined by cancer gene-targeted sequencing. Ann Oncol. 2015;26(6):1216–1223. doi: 10.1093/annonc/mdv109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Werness BA, Levine AJ, Howley PM. Association of human papillomavirus types 16 and 18 E6 proteins with p53. Science. 1990;248(4951):76–79. doi: 10.1126/science.2157286. [DOI] [PubMed] [Google Scholar]
- 25.Kimple RJ, Smith MA, Blitzer GC, et al. Enhanced radiation sensitivity in HPV-positive head and neck cancer. Cancer Res. 2013;73(15):4791–4800. doi: 10.1158/0008-5472.CAN-13-0587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pickering CR, Zhang J, Neskey DM, et al. Squamous cell carcinoma of the oral tongue in young non-smokers is genomically similar to tumors in older smokers. Clin Cancer Res. 2014;20(14):3842–3848. doi: 10.1158/1078-0432.CCR-14-0565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Henderson S, Chakravarthy A, Su X, Boshoff C, Fenton TR. APOBEC-mediated cytosine deamination links PIK3CA helical domain mutations to human papillomavirus-driven tumor development. Cell Rep. 2014;7(6):1833–1841. doi: 10.1016/j.celrep.2014.05.012. [DOI] [PubMed] [Google Scholar]
- 28.Ferrarotto R, Mitani Y, Cai Y, Diao L. Notch1 mutations to define a subgroup of adenoid cystic carcinoma (ACC): Tumor stage, propensity to bone and liver metastasis, risk of relapse, and overall survival. ASCO Annual Meeting Abstract 6081. J Clin Oncol. 2015;33(Suppl) abstract 6081. [Google Scholar]
- 29.Stoeck A, Lejnine S, Truong A, et al. Discovery of biomarkers predictive of GSI response in triple-negative breast cancer and adenoid cystic carcinoma. Cancer Discov. 2014;4(10):1154–1167. doi: 10.1158/2159-8290.CD-13-0830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Skàlovà A, Vanecek T, Sima R, et al. Mammary analogue secretory carcinoma of salivary glands, containing the ETV6-NTRK3 fusion gene: a hitherto undescribed salivary gland tumor entity. Am J Surg Pathol. 2010;34(5):599–608. doi: 10.1097/PAS.0b013e3181d9efcc. [DOI] [PubMed] [Google Scholar]
- 31.Drilon A, Li G, Dogan S, et al. What hides behind the MASC: clinical response and acquired resistance to entrectinib after ETV6-NTRK3 identification in a mammary analogue secretory carcinoma (MASC) Ann Oncol. 2016;27(5):920–926. doi: 10.1093/annonc/mdw042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gross AM, Orosco RK, Shen JP, et al. Multi-tiered genomic analysis of head and neck cancer ties TP53 mutation to 3p loss. Nat Genet. 2014;46(9):939–943. doi: 10.1038/ng.3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mathew CG. Fanconi anaemia genes and susceptibility to cancer. Oncogene. 2006;25(43):5875–5884. doi: 10.1038/sj.onc.1209878. [DOI] [PubMed] [Google Scholar]
- 34.Beltran H, Eng K, Mosquera JM, et al. Whole-Exome Sequencing of Metastatic Cancer and Biomarkers of Treatment Response. JAMA Oncol. 2015;1(4):466–474. doi: 10.1001/jamaoncol.2015.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Robinson D, Van Allen EM, Wu YM, et al. Integrative clinical genomics of advanced prostate cancer. Cell. 2015;161(5):1215–1228. doi: 10.1016/j.cell.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alexandrov LB, Nik-Zainal S, Wedge DC, et al. Australian Pancreatic Cancer Genome Initiative. ICGC Breast Cancer Consortium. ICGC MMML-Seq Consortium. ICGC PedBrain Signatures of mutational processes in human cancer. Nature. 2013;500(7463):415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cleaver JE, Crowley E. UV damage, DNA repair and skin carcinogenesis. Front Biosci. 2002;7:d1024–d1043. doi: 10.2741/A829. [DOI] [PubMed] [Google Scholar]
- 38.Rizvi NA, Hellmann MD, Snyder A, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348(6230):124–128. doi: 10.1126/science.aaa1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Goh G, Walradt T, Markarov V, et al. Mutational landscape of MCPyV-positive and MCPyV-negative Merkel cell carcinomas with implications for immunotherapy. Oncotarget. 2016;7(3):3403–3415. doi: 10.18632/oncotarget.6494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nghiem P, Bhatia S, Daud A, et al. Activity of PD-1 blockade with pembrolizumab as first systemic therapy in patients with advanced Merkel cell carcinoma (abstract). Paper presented at: Proceedings of the European Cancer Congress 2015; Vienna, Austria. September 25-29; 2015.p. 22LBA. [Google Scholar]
- 41.McGranahan N, Furness AJ, Rosenthal R, et al. Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science. 2016;351(6280):1463–1469. doi: 10.1126/science.aaf1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morris LG, Riaz N, Desrichard A, et al. Pan-cancer analysis of intratumor heterogeneity as a prognostic determinant of survival. Oncotarget. 2016;7(9):10051–10063. doi: 10.18632/oncotarget.7067. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.