Abstract
The objective of this study was to investigate the antimicrobial resistance, Tn1546 transposon variability and plasmid diversity among Polish vancomycin-resistant Enterococcus faecium (VREfm) isolates of VanA phenotype in the context of their clonal structure. Two hundred sixteen clinical VREfm isolates collected between 1997 and 2010 were studied by antimicrobial susceptibility testing, MLST, MLVA and detection of IS16, esp Efm, pilA, intA and plasmid-specific genes by PCR. Tn1546 structure was revealed by overlapping PCR and sequencing. Selected isolates were subjected to PFGE-S1 and Southern hybridization analyses. The vast majority of the isolates (95.8 %) belonged to lineages 17/18 (during the whole study period 1997–2010) and 78 (mostly in 2006–2010) of hospital-adapted meroclone of E. faecium. All isolates displayed a multi-drug resistance phenotype. Twenty-eight Tn1546 types (including 26 novel ones) were associated with eight different ISs (IS1216, IS1251, ISEfa4, ISEfa5, ISEfm2, ISEf1, IS3-like, ISEfm1-like). The vanA-determinant was typically located on plasmids, which most commonly carried rep2pRE25, rep17pRUM, rep18pEF418, rep1pIP501, ω-ε-ζ and axe-txe genes. VanA isolates from 1997–2005 to 2006–2010 differed in clonal composition, prevalence of gentamicin- and tetracycline-resistance and plasmidome. Our analysis revealed high complexity of Tn1546-type transposons and vanA-plasmids, and suggested that diverse genetic events, such as conjugation transfer, recombination, chromosomal integration and DNA mutations shaped the structure of these elements among Polish VREfm.
Electronic supplementary material
The online version of this article (doi:10.1007/s10096-016-2804-8) contains supplementary material, which is available to authorized users.
Introduction
In the past 20 years, vancomycin-resistant enterococci (VRE) have emerged as nosocomial pathogens worldwide. In Poland, the first VRE outbreak due to Enterococcus faecium (VREfm) of VanA phenotype started in December 1996 in the Gdańsk Medical University [1]. The vast majority of VREfm observed worldwide belongs to a specific hospital meroclone, initially described as clonal complex 17 (CC17), later divided into three distinct lineages 17, 18 and 78 based on multilocus sequence typing (MLST) analyses [2, 3]. Recently, the approach called Bayesian Analysis of Population Structure (BAPS), applied to the E. faecium MLST data delimited two groups within the hospital meroclone, 2–1 and 3–3, corresponding to lineages 78 and 17/18, respectively [4]. Strains belonging to the hospital meroclone are ciprofloxacin- and ampicillin-resistant, enriched in putative virulence traits, and show a distinct genetic repertoire, including cell surface protein genes (fms), regulatory genes, putative pathogenicity islands, plasmids, insertion sequences (IS) and integrated phages, which promote their adaptation [5–7]. The presence of IS16 and the E. faecium-specific esp gene (esp Efm), carried on the integrative conjugative element ICEEfm1, together with the intA integrase gene, are proven molecular markers of hospital-associated E. faecium [8–10].
Several glycopeptide-resistance phenotypes have been described so far, with VanA and VanB being the most common in enterococci isolated from hospital infections [11]. The vanA gene cluster is carried on Tn1546-type transposons [12], which show a significant degree of heterogeneity, associated with presence of point mutations, deletions and presence of various ISs [13, 14]. A few studies demonstrated the location of Tn1546 on Inc18, pRUM-like, pMG1-like, and pLG1 plasmids [15, 16]; however, the knowledge of vanA-plasmids and their epidemiology is still far from being satisfactory and may differ significantly among countries.
In Poland, hospital VRE isolates are continuously submitted for confirmation and further analyses to the National Reference Centre for Susceptibility Testing (NRCST), located at the National Medicines Institute in Warsaw. The aim of this study was to characterize E. faecium VanA isolates collected by the NRCST since 1997 until the end of 2010, focusing on the Tn1546 transposon variability and vanA-plasmid diversity in the context of the clonal structure of VREfm isolates to provide the country-wide picture of these important hospital pathogens.
Materials and methods
Bacterial isolates and susceptibility testing
The study comprised 216 consecutive, non-repetitive (1 isolate per patient) VREfm VanA isolates received by the NRCST from 42 hospitals in 24 cities in Poland over the period 1997–2010. Part of the isolates analyzed in this work correspond to strains partially tested in previous surveillance studies, including: 108 VanA representatives of the VREfm collection from 1997 to 2005 [17] and 20 representative isolates of a E. faecium VanA outbreak in 2009 [18]. The majority of isolates (n = 137) were derived from 11 VanA outbreaks and the remaining 79 isolates were reported as single isolations. Of the 216 isolates, 211 (97.7 %) were from hospitalized patients and five (2.3 %) were from the hospital environment. Among the isolates from hospitalized patients, a total of 37 isolates (17.5 %) were from invasive infections (31 isolates from blood and 6 from other sources); 52 isolates (24.6 %) were from non-invasive infections (21 from urine, 18 from wounds, and 13 from other sources) and 122 (57.8 %) represented faecal carriage. Antimicrobial susceptibility of 88 isolates, not investigated previously, was tested by the Etest method (bioMérieux, Marcy l’Etoile, France) for daptomycin, teicoplanin and vancomycin and by a broth microdilution method [19] for the remaining compounds (Table 1). Multidrug-resistant (MDR) isolates were defined as recommended [20]. Vancomycin-resistance determinants were detected by PCR as described previously [21] with the E. faecium BM4147 and E. faecalis V583 strains as positive vanA and vanB controls, respectively.
Table 1.
MIC values for E. faecium VanA isolated in Poland during the period 1997–2010
| Compound/phenotype | 1997–2010 N = 216 |
1997–2005 N = 128 |
2006–2010 N = 88 |
MIC breakpoints/ECOFF (μg/ml) | |||||
|---|---|---|---|---|---|---|---|---|---|
| Number (%) non-susceptible | Number (%) non-susceptible | MIC 50 (mg/l) | MIC90 (mg/l) | Number (%) non-susceptible | MIC 50 (mg/l) | MIC90 (mg/l) | S ≤ | R > | |
| Vancomycina | 216 (100) | 128 (100) | 512 | >512 | 88 (100) | >256 | >256 | 4 | 4 |
| Teicoplanina | 216 (100) | 128 (100) | 64 | 128 | 88 (100) | 48 | >256 | 2 | 2 |
| Ampicillina | 215 (99.5) | 128 (100) | >128 | >256 | 87 (98.8) | 128 | >256 | 4 | 8 |
| HLGRa | 172 (79.6) | 118 (92.2) | >1024 | >1024 | 54 (64.4) | >1024 | >1024 | 128 | 128 |
| HLSRa | 167 (77.3) | 112 (87.5) | >1024 | >2048 | 55 (62.5) | >1024 | >2048 | 512 | 512 |
| HLAR 28 | 147 (68.1) | 110 (86) | >1024 | >1024 | 37 (42) | >1024 | >2048 | - | - |
| Quinupristin/dalfopristina | 2 (0.9) | 1 (0.8) | 1 | 2 | 1 (1.1) | 1 | 1.5 | 1 | 4 |
| Linezolida | 1 (0.5) | 0 (0) | 1 | 4 | 1 (1.1) | 1 | 2 | 4 | 4 |
| Tigecyklinea | 0 (0) | 0 (0) | 0.06 | 0.19 | 0 (0) | 0.06 | 0.25 | 0.25 | 0.5 |
| Tetracyclineb | 135 (62.2) | 89 (68.9) | 64 | 128 | 46 (52.3) | 8 | 128 | 4 | 4 |
| Ciprofloxacinb | 215 (99.5) | 127 (99.2) | 128 | >256 | 88 (100) | 128 | 256 | 4 | 4 |
| Daptomycinb | 0 (0) | 0 (0) | 2 | 3 | 0 (0) | 2 | 3 | 4 | 4 |
| Chloramphenicolc | 53 (24.4) | 30 (23.2) | 8 | 16 | 23(26.1) | 8 | 16 | 8 | ≥32c |
| MDR28 | 216 (100) | 128 (100) | nc | nc | 88 (100) | nc | nc | – | – |
nc not calculated, n number of isolates
The results were interpreted following the European Committee on Antimicrobial Susceptibility Testing (EUCAST)-approved breakpoints [53] and the Ecological Cut-Off (ECOFF) values for compounds without defined breakpoints (http://mic.eucast.org/Eucast2/, last accessed 20th July 2015). For chloramphenicol the Clinical and Laboratory Standards Institute (CLSI) breakpoints were used [19]a Interpretation according to the EUCAST clinical breakpoint valueb Interpretation according to the EUCAST Ecological Cut-off (ECOFF) valuec Interpretation according to CLSI breakpoint value
DNA isolation and genotyping of isolates
Total DNA of isolates was extracted using Genomic DNA Prep Plus kit (A&A Biotechnology, Gdansk, Poland). Multilocus VNTR analysis (MLVA), MLST, and detection of 19 rep families and the unique rep pMG1 gene were performed as described [22–24]. Sequence types (STs) were grouped to CCs by the comparative eBURST analysis performed against the whole E. faecium MLST database. PCR detection of IS16, esp Efm, fms21 (pilA), rep pLG1, plasmid addiction systems, relaxase genes, and intA ICEEfm1 was performed as described (Supplementary Table 1 and references therein). DNA of enterococcal isolates from our laboratory collection [17, 18, 25] served as positive controls.
Plasmid profiling, hybridization analyses, Tn1546 typing and statistical analysis
DNA in agarose plugs was obtained as described [21], treated with S1 nuclease (Takara Bio, Japan) and separated by PFGE with Lambda Ladder PFG marker (New England Biolabs, Beverly, MA) [26] followed by blotting onto the Hybond membrane (GE Healthcare, Buckinghamshire, UK) by capillary transfer. Hybridization was carried out using the Amersham ECL Random-Prime Labelling and Detection System (GE Healthcare, Buckinghamshire, UK). Tn1546 transposon was investigated by PCR mapping and sequencing (Supplementary Table 1 and references therein) of selected regions encompassing 7571 bp out of 10851 bp (i.e., ∼70 % of the transposon, Fig. 1). The Tn1546 sequence of E. faecium BM4147 (GenBank acc. no.: M97297) [12] was used as a reference. The nomenclature of Tn1546-type transposons in the present study was based on the following alphanumeric code: the ‘A’ types (A1-A6) referred to transposon variants of the wild type (wt) Tn1546 structure (A1) not interrupted by insertion sequences; the ‘B’ types contained 1–3 copies of IS1216 (B, BB, BBB types); the C, D, E, F, G, H and I types carried IS1251, ISEfa5, ISEfa4, ISEfm2, ISEf1, ISEfm1-like and IS3-like elements, respectively. Transposons with more than one IS type were described by a two-, three- or four-letter code (e.g., ‘BC’ with both IS1216 and IS1251). The Arabic numerals indicated differences in the presence of particular point mutations as well as the orientation of ISs and the localization of their insertion sites (e.g., B1-B4). The novel ISEfm1-like sequence was submitted to GenBank (KT719407). Chi-square test was used to assess the differences of distributions, with p ≤ 0.05 considered significant.
Fig. 1.
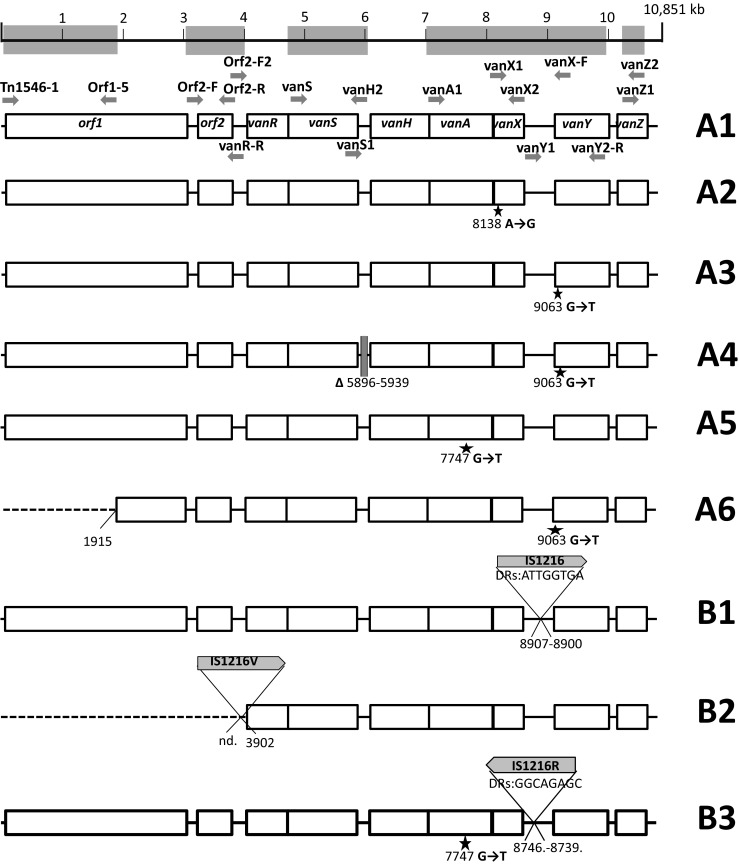
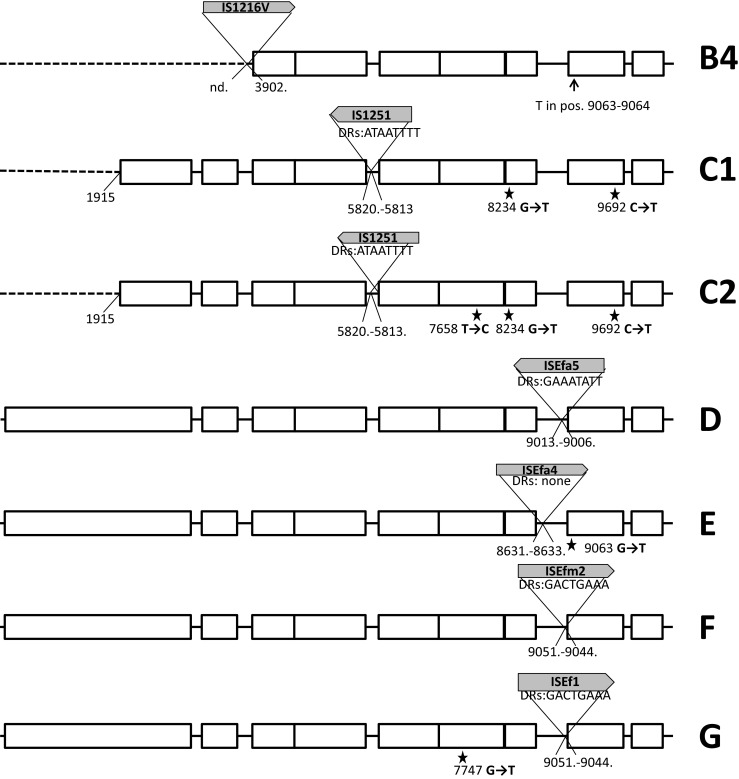
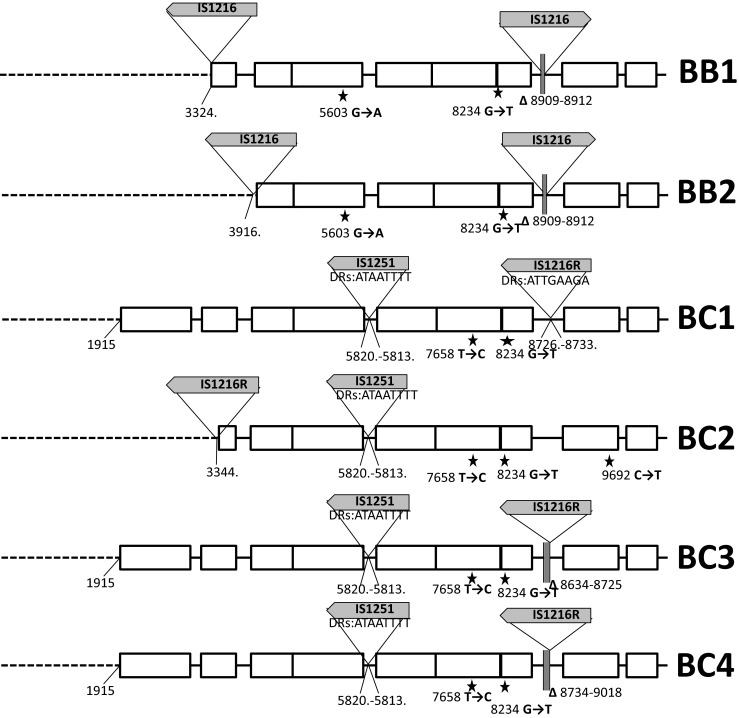
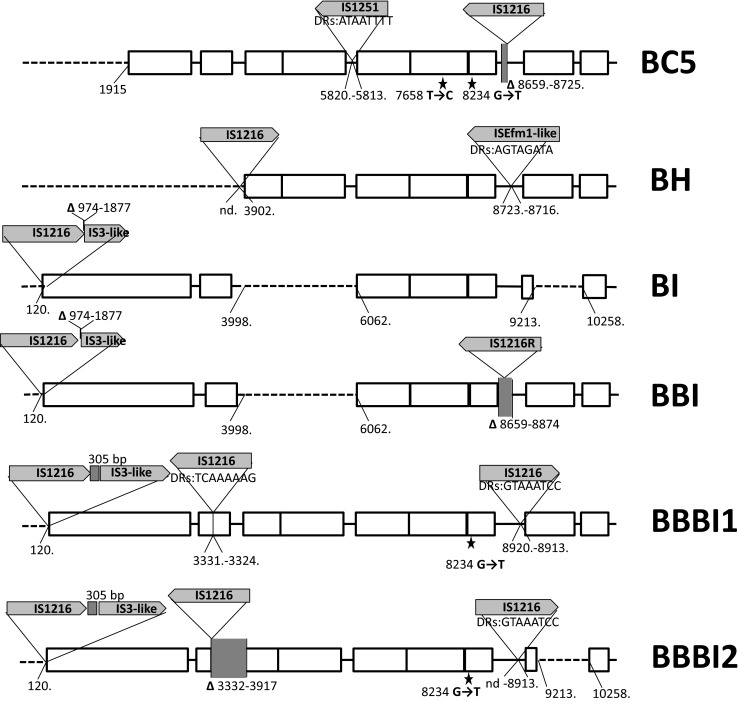
Diversity of Tn1546 transposon types among E. faecium VanA isolates. Position of primers used in PCR mapping and sequencing indicated by arrows with primer names; open rectangles, transposon genes; stars, positions of point mutations; analyzed areas of the transposon shadowed; dashed lines, deletions in the left arm of the transposon; filled rectangles, deletions within the transposon; vertical arrow, triangles with arrows, the IS positions; single-nucleotide insertion in vanY
Results
Susceptibility to antimicrobial agents
All isolates were resistant to vancomycin and teicoplanin (Table 1) and carried vanA. Resistance to ampicillin, ciprofloxacin, tetracycline, chloramphenicol, gentamicin and streptomycin (high level) was prevalent or highly prevalent and all isolates showed the MDR phenotype. A significant decline in the prevalence of both tetracycline-resistance (from 68.9 to 52.3 %, p = 0.01) and high-level gentamicin resistance (from 92.1 to 64.4 %, p < 0.0001) was found between the 1995–2005 and 2006–2010 periods. A single isolate was resistant to linezolid and two isolates to quinupristin/dalfopristin. All isolates were susceptible to tigecycline and daptomycin.
MLVA, MLST, IS16 and virulence markers detection
MLVA was performed for 196 isolates and these results were analysed together with data obtained earlier for 20 isolates from the 2009 outbreak [18]. Among 216 isolates, 37 different MLVA types (MTs) and three incomplete profiles (due to lack of VNTR7 amplification) were observed, that included 207 and nine isolates, respectively (Supplementary Table 2). MT1, MT159, MT25 and MT13 were most prevalent, with 36, 34, 26 and 20 isolates, respectively. All MT159 isolates except one were isolated in 2006–2010 (p ≤ 0.0001), in contrast to isolates of MT25, which all except one were isolated in 1997–2005 (p = 0.0001). The MT1 isolates showed a similar frequency over the whole study period (13.2 % vs 21.6 %, p = 0.07). In the present study, STs and the presence of IS16, esp Efm, intA and pilA were determined for 88 isolates not encompassed by the previous studies (i.e., 68 isolates from 2006 to 2010 and 20 isolates from 2001 to 2002), and these results were analysed with the data obtained previously for the remaining 128 isolates. Altogether, 40 different STs were found, with 18 STs (45.0 %) represented by single isolates. The vast majority of the STs, i.e. 36 STs representing 207 isolates (95.8 %), belonged to the hospital-adapted meroclone of E. faecium; lineage 17/18 included 31 STs with 165 isolates and was present during the whole period 1997–2010, while lineage 78 included five STs characteristic for 42 isolates, occurring mostly in 2006–2010 (p ≤ 0.0001). IS16 was present in 207 (95.8 %) isolates and 186 (86.1 %) isolates harboured the esp Efm gene. The integrase gene intA and the pilA gene were found in 181 (83.8 %) and 214 (99.1 %) isolates, respectively.
Structural diversity of Tn1546 transposons
The structure of Tn1546-type transposons was determined for 187 isolates while for 20 isolates the structure of the transposon (representing types A1 and G, Fig. 1) had been published earlier [18]. In the case of nine isolates the structure of the transposon could not be determined in spite of repeated attempts due to lack of PCR products for some parts of the transposon, which might have been caused by sequence polymorphism(s) within PCR primers annealing sites, and discrepancies of sequencing results in certain regions, likely associated with the presence of more than one transposon in a single isolate. Twenty-eight transposon types, including 26 new ones, were discerned in the analyzed group (Fig. 1). The most predominant types, including C1 (40 isolates), B2 (n = 38), A3 (n = 36), G (n = 25), E (n = 14), A1 (n = 13) and D (n = 7), were associated with several STs and typically showed a multicenter distribution. Eight different ISs were detected within Tn1546, including IS1216, IS1251, ISEfa4, ISEfa5, ISEfm2, ISEf1, IS3-like and ISEfm1-like. The most common IS1216 was present in all 16 B-type transposons, both in the direct and reversed orientations, with five different types of 8-bp direct repeats. These B-type transposons were found in Gdańsk and Warsaw, as well as in 14 other cities. IS1251 was associated with seven Tn1546 types (C1-C2, BC1-BC5) and present in 53 isolates (25.6 %), which mainly originated from Kraków and Warsaw. In these isolates, IS1251 was always inserted in the vanS-vanH intergenic region of the transposon at the position 5813. The D, E, F and G types of transposon were characterized by the presence of ISEfa5, ISEfa4, ISEfm2 and ISEf1 in the vanX-vanY intergenic region, respectively. These types were generally limited to one or two centres, with the exception of the G type, which apart from the outbreak in two Warsaw hospitals [18] occurred in seven other centres. An insertion of an IS in 98 % identical to ISEfm1 (GenBank no. AF138282) in the vanX-vanY intergenic region resulted in the BH-type of transposon.
The variability of Tn1546 was additionally associated with the presence of deletions, insertions and point mutations. In the A4 type, a 44 nt deletion in the vanS-vanH intergenic region (nt 5896–5939) was observed. Other deletions, located in orf2, vanX and the vanX-vanY intergenic region coincided with the presence of ISs (six types: BB1, BB2, BC4, BC5, BBI, BBBI2). Seven different point mutations were detected, including four known previously (T7658C, G7747T, G8234T, C9692T) [18, 27, 28] and three new ones (G5603A, A8138G and G9063T). The G5603A mutation resulted in the A80T change in VanS and the G9063T mutation in the L4F change in VanY. The B4 type, found in two independent isolates, demonstrated the presence of a novel single-nucleotide T insertion between nt 9063–9064 within the vanY gene, resulting in translational frameshift and a truncated VanY. Nevertheless, these two isolates showed high MIC values for vancomycin (>512 mg/L in both cases) and teicoplanin (64 and >128 mg/L).
Plasmid gene content among VREfm and diversity of vanA-plasmids
PCR-based typing of plasmid replication initiator genes (rep) was performed for 196 isolates and these results were combined with the data for 20 isolates, published previously [18]. Altogether, ten rep-types were observed among VREfm-VanA (Fig. 2). Isolates carried from one up to seven different rep genes, with 4.7 rep genes per isolate on average. Isolates positive for rep1 pIP501, rep7 pT181 and rep pMG1 appeared mainly in the period 1997–2005, while rep11 pEF1071 gene was typical for isolates obtained in 2006–2010. The plasmid stabilization systems axe-txe and rep17 pRUM were present in the majority of isolates. Another system, ω-ε-ζ, was also quite common, predominantly among rep2 pRE25-carrying strains, and occurred mainly in the period of 2006–2010. Two additional systems, mazEF and relBE, were observed only in 2003 for six and two isolates, respectively. The rel pCIZ2 and rel pEF1 relaxase genes were prevalent, and additionally rel pHTß and rel pRE25 were detected. The majority of rel pRE25-positive isolates (n = 22, 91.7 %) were also rep2 pRE25-positive, however, most of 169 rep2 pRE25-positive isolates lacked this relaxase. Isolates carrying rel pHTß dominated in 1997–2005 (40 out of 45 rel pHTß-positive isolates) and the majority of rel pHTß -positive isolates also harboured rep pMG1 (n = 43, 95.5 %).
Fig. 2.
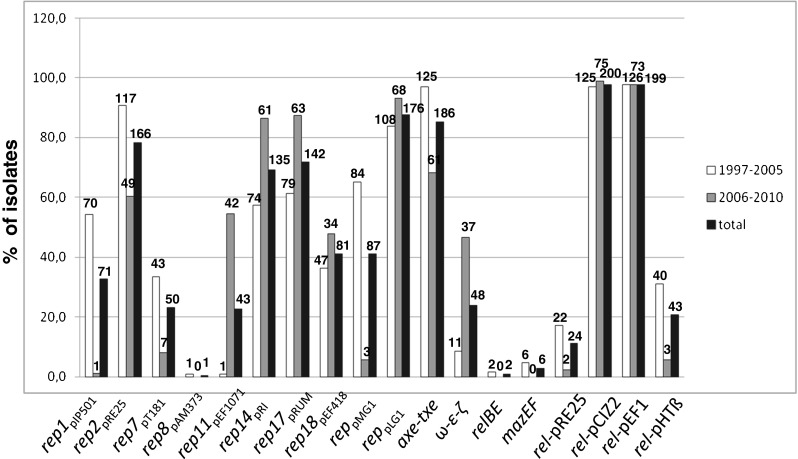
Plasmid-associated gene distribution among Polish VREfm VanA. Number of isolates with a particular gene given above the graph bars
Fifty-two isolates, obtained from 24 medical centres over the whole study period and representing 26 different STs and 21 Tn1546 types were selected for PFGE of S1-digested DNA and hybridization analyses. Additionally, the results obtained previously for three isolates from the 2009 outbreak [18] were included for comparative purposes. Investigated isolates showed the presence from one up to 11 plasmid bands per isolate in PFGE-S1 analyses. Subsequent hybridization with the vanA probe revealed the presence of 86 vanA-plasmids with up to four such plasmids in an isolate, and two cases of chromosomal localization of vanA (Table 2). Further hybridization studies showed the co-localization of vanA determinants with all six tested rep types, including rep2 pRE25, rep17 pRUM, rep18 pEF418, rep1 pIP501, rep pLG1 and rep pMG1 that accounted for 40.7 % (n = 35), 40.7 % (n = 35), 24.4 % (n = 21), 19.8 % (n = 17), 5.8 % (n = 5) and 1.2 % (n = 1) of vanA-plasmids, respectively. The vanA-plasmids with rep1 pIP501 were limited to isolates from 1997 to 2005, circulating in two hospitals in Poznań. These plasmids differed by size (from ca. 30 to ca. 265 kb) and presence of other rep and toxin-antitoxin genes, and carried four different types of Tn1546, with A3 being predominant (8 out of 13 isolates harbouring vanA-plasmids with rep1 pIP501). The vanA-plasmids with rep2 pRE25 , rep17 pRUM and rep18 pEF418 genes showed a multicentre distribution and occurred during the whole study period. In total, 37 (43 %) vanA-plasmids were associated with more than a single rep type and 21 vanA-plasmids (24.4 %), present in 11 isolates, did not hybridize with any of the tested rep genes. With a single exception, these latter plasmids were obtained during 2006–2010 (p = 0.001). Five of the isolates with these unknown replicons concomitantly carried three vanA-plasmids, ca. 30, 160 and 380 kb in size, which did not hybridize with any probes of toxin-antitoxin and relaxase genes tested. All these isolates carried B2 transposons, but belonged to diverse STs and MTs, and originated from four different medical centres over 2006–2010. Two toxin-antitoxin systems, ω-ε-ζ and axe-txe were commonly carried by vanA-plasmids (35 and 32 plasmids, respectively). The ω-ε-ζ system was characteristic for rep2 pRE25 plasmids and axe-txe for rep17 pRUM plasmids (71.4 and 62.9 % of the respective vanA-plasmids). The gene specifying pEF1-relaxase was located on 11 vanA-plasmids (12.8 %), with various rep types. Some of the presumed genetic events, that could be inferred on the basis of these analyses, include examples of transposon evolution within an enterococcal strain, Tn1546 transposition among plasmids, conjugative transfer of plasmids, and their changes such as recombination or chromosomal integration as proposed in Table 2.
Table 2.
Characteristics of the VanA plasmidome of 55 selected VREfm isolates
| Strain ID/year of isolation | Code of medical centrea | Tn1546 type | MLST type (lineage) | Number of VanA plasmid bands | Hypothetical genetic event b | VanA plasmid replicon types and stabilization systems (approximate size in kb)c | ||
|---|---|---|---|---|---|---|---|---|
| 1639/1997 | Gd-a | BBBI1 | 407(17/18) | 1 | rep17 TA1 (45) | |||
| 1641/1997 | Gd-a | BBBI2 | 408(17/18) | 1 | Chromosomal integration of 50-kb plasmid | rep17 rep18 TA1 (50) | rep2 rep17 rep18 TA1 (chr) | |
| 3132/1998 | Gd-p | A1 | 18(17/18) | 2 | Transfer of 40-kb plasmid between ST18 and ST411 strains, followed by plasmid recombination or transposition of A1 | rep2 rep17 TA1 (35) | rep2 TA2 (40) | |
| 3136/1998 | Gd-p | A1 | 411 (singleton) | 2 | rep2 TA2 (40) | rep2 TA2 (270) | ||
| 7952/1999 | Gd-p | nt | 381(17/18) | 3 | rep2 rep17 TA1 (35) | rep17 TA1 (170) | rep17 TA1 (320) | |
| 2509/2000 | Po-1 | A3 | 386(17/18) | 2 | Chromosomal integration of 40-kb plasmid | rep2 (30) | rep2 TA1 TA2 (40) | rep1 rep2 TA1 TA2 (chr) |
| 2524/2000 | Po-1 | A3 | 382(17/18) | 2 | rep17 TA2 (40) | rep1 TA2 relpEF1(140) | ||
| 2712/2000 | Po-1 | A3 | 385(17/18) | 3 | Transposition of A3 or plasmid recombination in ST385 strain | TA2 (40) | rep1 TA2 (140) | rep1 reppLG1 TA2 relpEF1 (265) |
| 1409/2002 | Po-4 | A3 | 385(17/18) | 1 | rep2 reppMG1 TA1 TA2 (250) | |||
| 2506/2000 | Po-1 | A4 | 385(17/18) | 2 | Derivative of A3 in ST385 strain, with concomitant change of plasmid backbone | rep2 rep18 TA2 (<30) | rep1 rep18 TA2 (265) | |
| 291/2002 | Po-2 | A3 | 117(17/18) | 1 | rep1 rep17 TA1 TA2 (145) | |||
| 1714/2003 | Po-2 | A3 | 117(17/18) | 1 | Clonal spread of ST117 with 265-kb plasmid, followed by transposition of A3 among plasmids or plasmid recombination | rep1 (265) | ||
| 710/2004 | Po-2 | A3 | 117(17/18) | 4 | rep1 (265) | rep1 rep2 rep18 TA1 TA2 (255,310,360) | ||
| 914/2002 | Po-2 | A3 | 17(17/18) | 1 | rep1 rep17 TA1 (155) | |||
| 2039/2003 | Po-2 | A3 | 410(17/18) | 1 | rep1 (145) | |||
| 3010/2003 | Po-2 | A3 | 192(78) | 1 | rep1 rep2 rep18 (145) | |||
| 655/2007 | Po-4 | A6 | 549(78) | 1 | rep2 TA1 TA2 (40) | |||
| 3003/2003 | Po-2 | E | 117(17/18) | 1 | Transposition of E or plasmid recombination in ST117 strain | rep1 rep2 rep18 reppLG1 TA1 (165) | ||
| 2127/2004 | Po-2 | E | 117(17/18) | 1 | rep1 reppLG1 (165) | |||
| 2512/2000 | Po-1 | D | 17(17/18) | 1 | rep1 TA2 (30) | |||
| 1156/2002 | Po-2 | D | 382(17/18) | 1 | rep1 rep2 TA2 (50) | |||
| 714/2003 | Kr-1 | C1 | 117(17/18) | 1 | rep2 rep18 rep17 reppLG1 TA1 TA2 (450) | |||
| 756/2003 | Kr-1 | C1 | 17(17/18) | 1 | rep17 rep18 TA1 TA2 relpEF1 (70) | |||
| 1679/2003 | Kr-1 | C1 | 18(17/18) | 1 | Clonal spread of ST18 strain and concomitant change of plasmid size by a presumable deletion (loss of relpEF1) | rep2 rep18 TA1 TA2 relpEF1 (70) | ||
| 3779/2004 | Kr-4 | C1 | 18(17/18) | 1 | rep2 rep18 TA1 TA2 (65) | |||
| 4002/2005 | Kr-3 | C1 | 387(17/18) | 1 | Plasmid transfer and ∼15-kb deletion | rep2 rep18 TA1 TA2 relpEF1 (55) | ||
| 1332/2003 | Kr-1 | C1 | 132(17/18) | 1 | Clonal spread of ST132 strain with 45-kb plasmid harbouring C1 transposon | rep17 TA1 (45) | ||
| 1336/2003 | Kr-1 | C1 | 132(17/18) | 1 | rep17 TA1 (45) | |||
| 2981/2003 | Mi | ntd | 132(17/18) | 1 | Evolution of C1 transposon within the same ST132 strain and 45-kb plasmid backbone | rep17 TA1 (45) | ||
| 2216/2005 | Kr-1 | C1 | 388(17/18) | 1 | rep2 rep17 rep18 TA1 TA2 relpEF1 (75) | |||
| 84/2010 | Gdy | C2 | 17(17/18) | 1 | rep2 rep17 TA1 TA2 (50) | |||
| 3552/2009e | Wa-10 | A1 | 18(17/18) | 2 | Ancestor for B2 transposon, associated with plasmids of unknown rep-type(s) | (<30) | (170) | |
| 3240/2006 | Po-5 | B2 | 17(17/18) | 3 | Concomitant transfer of three ∼30-, 160- and 380-kb plasmids with unknown rep-type(s), carrying B2 transposon, into diverse clonal backgrounds | (30) | (160) | (380) |
| 1930/2007 | Wa-1 | B2 | 64(17/18) | 3 | (30) | (160) | (380) | |
| 4285/2008 | Wa-1 | B2 | 192(78) | 3 | (30) | (160) | (380) | |
| 5151/2008 | Osw | B2 | 18(17/18) | 3 | (30) | (160) | (380) | |
| 1767/2010 | Wa-3 | B2 | 780(17/18) | 3 | (<30) | (160) | (380) | |
| 5009/2009 | Wa-2 | B2 | 230(78) | 3 | Transfer of 160-kb plasmid of unknown rep-type, followed by transposition of B2 among plasmids or plasmid recombination | rep17 TA1 (<30), rep17 (45) | (160) | |
| 3238/2006 | Sk | B2 | 279(17/18) | 2 | Transfer of 30-kb plasmid of unknown rep-type, followed by transposition of B2 among plasmids or plasmid recombination | (30) | rep18 reppLG1 (180) | |
| 2546/2008 | Gr | BH | 202(17/18) | 3 | BH derivative of B2 transposon on ∼370-kb plasmid of unknown rep-type, followed by transposition of BH among plasmids or plasmid recombination | rep17 rep18 TA1 (30) | rep17 rep18 TA1 (155) | (370) |
| 8744/2010 | Wa-2 | B2 | 561(17/18) | 1 | rep2 rep17 TA2 relpEF1(35) | |||
| 484/2010 | Ke | B2 | 17(17/18) | 3 | Transfer of <30-kb plasmid among strains of ST877 and ST17 | rep2 rep17 rep18 TA1 (<30) | rep2 (160) | rep2 rep17 rep18 TA1 (320) |
| 9363/2010 | Sw | B2 | 877(17/18) | 1 | rep2 rep17 rep18 TA1 (<30) | |||
| 3856/2005 | Wa-1 | B4 | 78(78) | 1 | rep2 TA2 (50) | |||
| 991/2009e | Wa-4 | G | 18(17/18) | 1 | Recombination events or transposition of G among rep17 plasmids. | rep17 (100) | ||
| 3554/2009e | Wa-10 | G | 192(78) | 1 | rep17 (50) | |||
| 2944/2009 | Ko | G | 18(17/18) | 1 | rep2 rep17 TA2 (35) | |||
| 3392/2009 | Ost | G | 78(78) | 1 | rep17 relpEF1 (45) | |||
| 726/2010 | In | G | 17(17/18) | 1 | rep17 relpEF1 (100) | |||
| 3322/2007 | Wa-2 | BC1 | 412(78) | 1 | Evolution of BC transposons within the same ST412 strain in the ∼35-kb plasmid backbone | rep2 rep17 TA2 (35) | ||
| 107/2010 | Wa-2 | BC5 | 412(78) | 1 | rep2 rep17 TA2 (35) | |||
| 3948/2010 | Wa-2 | BC4 | 412(78) | 1 | rep2 rep17 TA2 (35) | |||
| 1901/2005 | Lo | F | 279(17/18) | 1 | rep17 rep18 TA2 (50) | |||
| 8034/2010 | Kr-5 | B3 | 341(78) | 2 | rep2 rep17 TA1 TA2 relpEF1(50) | rep2 rep17 TA2 relpEF1(65) | ||
| 8628/2010 | Ka | BBI | 202(17/18) | 1 | (115) | |||
nt non-typeable
a By Bydgoszcz, Gd-a Gdańsk, adult hemathology ward; Gd-p Gdańsk, paediatric haematology ward; Gdy Gdynia, Gr Grodzisk Mazowiecki, In Inowrocław, Ka Katowice, Ke Kętrzyn; Ko Konin, Kos Kościerzyna, Kr Kraków, Lo Łódź, Mi Mielec, Op Opole, Os Ostrów Mazowiecki, Osw Ostrów Wielkopolski, Ost Ostrzeszów, Ot Otwock, Pi Pisz, Pl Płock, Po Poznań, Rz Rzeszów, Sk Skierniewice, Sw Świdnica, Wa Warszawa, Wr Wrocław, Zi Zielona Góra; the city abbreviation is followed by the centre number b Shadowed boxes indicate presumable associations among isolates; c rep1, rep2, rep17, rep18 – plasmid replicon families according to Jensen et al., 2010 [24]; TA1, TA2- axe-txe and ω-ε-ζ stabilization systems specific for pRUM and pRE25, respectively; d Presumably C1 transposon type, however no amplification of the region containing IS1251 could be obtained, in spite of several attempts
eResults from Wardal et al., 2014 [18]
Discussion
Currently, VREfm play an increasingly important role in nosocomial infections and are considered alert pathogens [29], with vanA as a main determinant of this phenotype within many countries [30]. In Poland VRE remain less prevalent than in the United States or some European countries, e.g. our recent study revealed 7 % vancomycin-resistance among invasive E. faecium collected during 2010–2011 [31]. Although we observed an increasing prevalence of VanB E. faecium [17], VanA is still most frequent among Polish VREfm ([31] and NRCST unpublished observations). In the present study we aimed at the characterization of clonality of VanA-VREfm and genetic elements associated with this resistance determinant. Numerous reports show that in the case of human nosocomial infections vancomycin resistance is almost exclusively acquired by the hospital-adapted meroclone of E. faecium, now widespread all over the world [2, 3] and prevalent among invasive E. faecium in Polish hospitals [31]. In this study, the vanA determinant was carried by representatives of this meroclone with only a few exceptions limited to the 1997–2005 period. These isolates might represent intermediates, by which glycopeptides resistance determinants were introduced into hospitals. All isolates belonging to hospital meroclone, as expected, were resistant to both ampicillin and ciprofloxacin, and enriched in putative virulence traits / markers such as IS16, esp Efm, intA Efm and pili genes. The population structure determined for Polish VREfm VanA closely resembled these of hospital-associated E. faecium in other countries. High diversity of STs/MTs is consistent with the presence of polyclonal hospital population of E. faecium that subsequently acquires vancomycin resistance determinants [13, 14]. The vast majority of isolates grouped into hospital lineage 17/18, mostly represented by STs 17, 117, 18, 132, 202 and lineage 78, which included STs 78, 192 and 412. In contrast to several other countries, where ST203 and ST16 constituted a significant proportion of hospital E. faecium [3, 14, 32, 33], in our population only one representative of ST16 was found and ST203 was completely absent. The characteristic change in the proportion of isolates belonging to lineage 17/18 and lineage 78 was observed since the year 2005 when lineage 78 started to be significantly more frequent in Poland. Our results are in agreement with observations made in other studies, suggesting waves of successful E. faecium, first from lineage 17/18 and followed by lineage 78 strains [4, 34]. This population shift, apparent in MLST and MLVA, was additionally associated with a change in plasmidome composition and observed decreased resistance levels to tetracycline and aminoglycosides.
Diversity of Tn1546 in VREfm is typical for this transposon, as reported by others [13, 14]. Nevertheless, in the present study we observed several new variants of Tn1546. VanA transposons indistinguishable from the Tn1546 A1 prototype [12] were frequently encountered in Europe, especially in the late 1990s and 2000s [14, 27]. This type and its mutational derivatives (A2-A6) were ubiquitous among early VREfm in our study. Single-nucleotide T insertion between nt 9063–9064 in the vanY gene of B4 type of transposon resulted in a translational frameshift and a truncated translation product. This change, however, did not abolish the glycopeptide resistance. VanY is a membrane-associated D,D-carboxypeptidase that hydrolyses the C-terminal D-Ala or D-Lac residue of peptidoglycan precursors but lacks transpeptidase activity. VanY, together with VanZ, represent accessory proteins, which are not required for the expression of glycopeptide resistance but increase its level [35]. Isolates with a deletion of vanY gene showed lower resistance levels to teicoplanin, likely due to the diminished transcription of vanZ while point mutations in vanY, observed so far were not associated with a loss of protein function [27, 36].
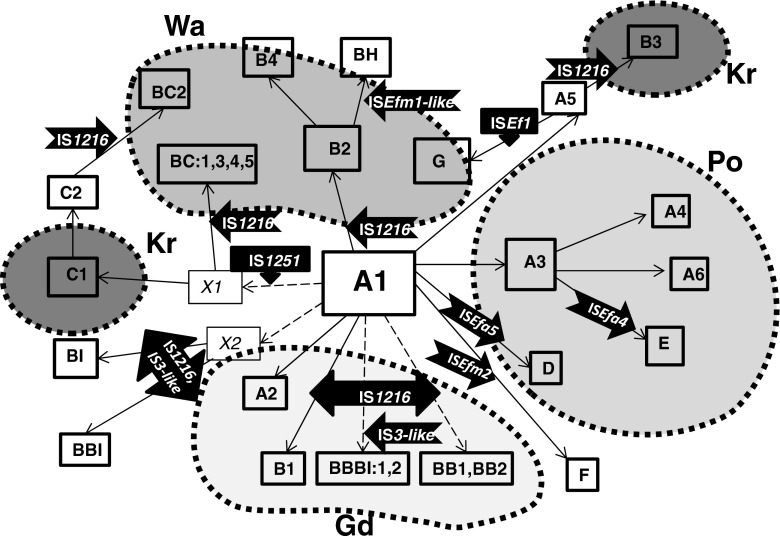
Activity of various ISs represented a very important factor, contributing to the formation of several novel transposon types. IS1216, the most common IS in our study, characteristic for B-types, was detected at various positions of the transposon, and its insertion often resulted in deletions of adjacent sequences in ORF2, vanX and the vanX-vanY intergenic region, as observed by others [13, 27]. The BI, BBI, BBBI types, apart from IS1216, exhibited the concomitant presence of a IS1216V-IS3-like element, originally reported in 1995 [37]. Since then, this element was described in several studies, which reported intact as well as a 5′-truncated IS3-like sequence [27, 38], both of which were also detected in our study. The C-type, harbouring IS1251, was relatively frequent and the integration sites of this IS were identical to those published by others [14, 37, 39]. Sequencing analysis allowed discerning the C1-type, specific for Krakow hospitals and the C2-type, found in other cities. The D type transposon, containing ISEfa5, to our knowledge, represents the first example of this variant outside of South America [34]. Type E represents the first insertion of ISEfa4 in the vanX-vanY intergenic region. This IS was described earlier in orf2-vanR and vanS-vanR intergenic regions [40, 41], as well as within IS1542 [42]. The F type harboured ISEfm2, representative of the IS256 family. Thus far, in enterococci this IS has been solely observed inserted between orf13 and tetS within CTn6000 transposon [43]. In our study, we report for the first time the insertion of ISEfm2 into Tn1546. The G type transposon with ISEf1, reported earlier for the VREfm outbreak in two neighbouring Warsaw medical centres in 2009 [18] was additionally detected in isolates from 2009 to 2010 derived from seven different cities, which may indicate its multicenter spread. Integration of the ISEfm1-like element, belonging to the IS982 family in the BH type, represents yet another novel insertion event in Tn1546. The presence of ISEfm1 was described previously in the vanX-vanY intergenic region [44], as well as within the vanD operon [45]. Complex analysis of transposon structures described in our study revealed the potential scheme of their hypothetical evolution among Polish VREfm. In this scenario, we propose the A1 type as presumable ancestor variant, with remaining types being its direct or indirect derivatives (Fig. 3).
Fig. 3.
Hypothetical evolution of Tn1546 structures among Polish VREfm VanA. Type A1, found in different cities, is a presumable ancestor variant with remaining types being its direct or indirect derivatives. Types A2, A3 and A5 developed by point mutations in wt type. A4 developed from A3 through single deletion events in the vanS-vanH intergenic region. A6 is an A3 derivative that lacks ca. 1900 bps in the 5′ end. The E type transposon, a third potential derivative of A3, arose through acquisition of ISEfa4 between vanX and vanY. A3 and its derivatives were typical for Poznań (Po) medical centres. B3 and G variants presumably developed from A5 after insertion events of IS1216 and ISEf within vanX-vanY intergenic region in Kraków (Kr) and Warsaw (Wa), respectively. The ubiquitous B2 type, typical for Warsaw, probably emerged from a single insertion event of IS1216 within the A1 type with a concomitant deletion of the 5′ end of the transposon. The B4 (additional single nucleotide insertion within vanY) and BH (ISEfm1-like insertion between vanX and vanY) types represent possible derivatives of B2. The B1, D and F transposon types are potential derivatives of A1 formed by IS1216, ISEfa5 and ISEfm2 insertions, respectively. Another group of transposon variants, encompassing types BI, BBI, BBBI1, BBBI2, BB1 and BB2 emerged through complex insertion and deletion events in different regions of wt transposon promoted mostly by IS1216 elements. This group was detected mainly in Gdańsk (Gd). The activity of another insertion sequence, IS1251, followed by IS1216 insertions and several point mutations resulted in the formation of C- and BC-types in Kraków and Warsaw, respectively
The vanA determinants are almost exclusively located on plasmids and these elements play a very important role in the spread of glycopeptides resistance [46]. Our results show that the Polish VREfm population is enriched in plasmid replicons of different families including megaplasmids, Inc18-, pRUM- and pMG1/pHT-like plasmids, encountered in VanA-VREfm in other countries [16, 46, 47]. Size variation of vanA-plasmids, even within the same family indicates their flexibility, and identification of multiple rep types in a single plasmid suggests a common presence of plasmid cointegrates. We observed an interesting change in the VanA-associated plasmidome between early (1997–2005) and more recent (2006–2010) isolates. In particular, rep1-vanA replicons were quite abundant among early isolates and typically located on plasmids over 140 bp in size. Together with rep2-vanA replicons they represent the Inc18 family, associated with Tn1546 elements among clinical E. faecium in Europe [16, 47]. Another shift in plasmidome composition between early and recent isolates was shown for pMG1 replicons, present exclusively among early VREfm. High prevalence of pMG1-like elements was observed among VREfm in the United States and Japan, where they contributed to the spread of both aminoglycoside and glycopeptide resistance [46]. Apart from Inc18 plasmids, the pRUM derivatives constitute the second main carrier of vancomycin resistance among the contemporary E. faecium isolates [16, 46]. Plasmids with the pRUM-like rep can be divided into two groups, one with axe-txe genes and mob regions from the staphylococcal pC223 plasmid and the other with relaxase from pEF1 and lacking axe-txe [15, 16, 46]. Our results indicate that representatives of both these groups are present among Polish VREfm. Additionally, we observed plasmids with unknown rep types among isolates obtained since 2006, which suggests the appearance of a new vanA-plasmid type(s), not included in the available classification scheme [24] and which will be a subject to further studies.
Finally, the analysis of PFGE-S1 hybridization results in the context of epidemiological information, determined Tn1546 types and the clonal background of the isolates, which revealed a high complexity of genetic events involving VREfm with VanA phenotype and resulting in the dissemination of this type of resistance. As these 52 isolates were pre-selected for a maximal representation of the collection diversity, only a few examples of clonal spread were observed, such as dissemination of ST132 strain with a 45-kb plasmid harbouring C1 transposon in a Krakow hospital (Table 2). The role of VREfm clonal spread in Poland, however, had been demonstrated before in our outbreak studies [18, 39, 48]. Particular types of transposons in the analysed group were frequently associated with various plasmid vectors. This situation may have resulted from transposition of Tn1546 among plasmids [12], promoted by integration ‘hot-spots’ [49] and from the recombination processes among enterococcal plasmids [15, 50]. The present study also provided examples of involvement of plasmids as vectors of vancomycin resistance, by demonstrating the presence of plasmids of the same size and with the same transposon types and plasmid-specific genes in different strains, as found in other studies [15, 51]. Occasionally, vanA-plasmids appeared to be integrated into bacterial chromosome, in agreement with other observations [52]. Further detailed studies employing extensive sequencing are indispensable to fully elucidate the events involving genetic elements engaged in the dissemination of the vanA gene cluster in the population of Polish VREfm.
In conclusion, the VREfm of the VanA phenotype collected in our country over the period 1997–2010 represent a highly variable group in the respect of their clonal composition, plasmid content and structures of Tn1546, a direct carrier of vanA genes. High genetic plasticity of these organisms, together with a rapid global spread of successful hospital-adapted enterococcal clones constitute a significant and continuously increasing epidemiological threat for human health. Thus, both epidemiological situation concerning VREfm as well as genetic elements and strains associated with VanA vancomycin resistance warrant further studies.
Electronic supplementary material
Below is the link to the electronic supplementary material.
(DOCX 20 kb)
(DOCX 58 kb)
(DOCX 16 kb)
Acknowledgments
We thank all Polish microbiologists who sent VREm isolates to our laboratory, Janetta Top for assigning new alleles, STs and MTs, and Kenneth Van Horn for critical reading of the manuscript. This publication made use of the Enterococcus faecium MLST website (http://efaecium.mlst.net/) hosted at Imperial College of the University of Oxford and funded by the Wellcome Trust.
Compliance with ethical standards
Funding
This work was supported by the grant N N401588540 from the Narodowe Centrum Nauki (NCN), Poland, by the MIKROBANK funding from the Ministry of Science and Higher Education, Poland, and by a statutory funding from the Ministry of Science and Higher Education, Poland.
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval and informed consent
Isolates were obtained as a part of routine activity of the NRCST and were analysed anonymously in a retrospective manner. Ethical approval and informed consent were thus not required.
References
- 1.Hryniewicz W, Szczypa K, Bronk M, Samet A, Hellmann A, Trzcinski K. First report of vancomycin-resistant Enterococcus faecium isolated in Poland. Clin Microbiol Infect. 1999;5(8):503–505. doi: 10.1111/j.1469-0691.1999.tb00181.x. [DOI] [PubMed] [Google Scholar]
- 2.Willems RJ, Top J, van Santen M, Robinson DA, Coque TM, Baquero F, Grundmann H, Bonten MJ. Global spread of vancomycin-resistant Enterococcus faecium from distinct nosocomial genetic complex. Emerg Infect Dis. 2005;11(6):821–828. doi: 10.3201/1106.041204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Willems RJ, Hanage WP, Bessen DE, Feil EJ. Population biology of gram-positive pathogens: high-risk clones for dissemination of antibiotic resistance. FEMS Microbiol Rev. 2011;35(5):872–900. doi: 10.1111/j.1574-6976.2011.00284.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Willems RJ, Top J, van Schaik W, Leavis H, Bonten M, Siren J, Hanage WP, Corander J. Restricted gene flow among hospital subpopulations of Enterococcus faecium. MBio. 2012;3(4):e00151–00112. doi: 10.1128/mBio.00151-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hendrickx AP, Bonten MJ, van Luit-Asbroek M, Schapendonk CM, Kragten AH, Willems RJ. Expression of two distinct types of pili by a hospital-acquired Enterococcus faecium isolate. Microbiology. 2008;154(Pt 10):3212–3223. doi: 10.1099/mic.0.2008/020891-0. [DOI] [PubMed] [Google Scholar]
- 6.Heikens E, van Schaik W, Leavis HL, Bonten MJ, Willems RJ. Identification of a novel genomic island specific to hospital-acquired clonal complex 17 enterococcus faecium isolates. Appl Environ Microbiol. 2008;74(22):7094–7097. doi: 10.1128/AEM.01378-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.7. Lebreton F, van Schaik W, McGuire AM, Godfrey P, Griggs A, Mazumdar V, Corander J, Cheng L, Saif S, Young S, Zeng Q, Wortman J, Birren B, Willems RJ, Earl AM, Gilmore MS (2013) Emergence of epidemic multidrug-resistant Enterococcus faecium from animal and commensal strains. MBio 4(4). doi:10.1128/mBio.00534-13 [DOI] [PMC free article] [PubMed]
- 8.Top J, Sinnige JC, Majoor EA, Bonten MJ, Willems RJ, van Schaik W. The recombinase IntA is required for excision of esp-containing ICEEfm1 in Enterococcus faecium. J Bacteriol. 2011;193(4):1003–1006. doi: 10.1128/JB.00952-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Willems RJL, Homan W, Top J, van Santen-Verheuvel M, Tribe D, Manzioros X, Gaillard C, Vandenbroucke-Grauls CMJE, Mascini EM, van Kregten E, van Embden JDA, Bonten MJM. Variant esp gene as a marker of a distinct genetic lineage of vancomycinresistant Enterococcus faecium spreading in hospitals. Lancet. 2001;357(9259):853–855. doi: 10.1016/S0140-6736(00)04205-7. [DOI] [PubMed] [Google Scholar]
- 10.Werner G, Fleige C, Geringer U, van Schaik W, Klare I, Witte W. IS element IS16 as a molecular screening tool to identify hospital-associated strains of Enterococcus faecium. BMC Infect Dis. 2011;11:80. doi: 10.1186/1471-2334-11-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Courvalin P. Genetics of glycopeptide resistance in gram-positive pathogens. Int J Med Microbiol. 2005;294(8):479–486. doi: 10.1016/j.ijmm.2004.10.002. [DOI] [PubMed] [Google Scholar]
- 12.Arthur M, Molinas C, Depardieu F, Courvalin P. Characterization of Tn1546, a Tn3-related transposon conferring glycopeptide resistance by synthesis of depsipeptide peptidoglycan precursors in Enterococcus faecium BM4147. J Bacteriol. 1993;175(1):117–127. doi: 10.1128/jb.175.1.117-127.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Talebi M, Pourshafie MR, Katouli M, Mollby R. Molecular structure and transferability of Tn1546-like elements in Enterococcus faecium isolates from clinical, sewage, and surface water samples in Iran. Appl Environ Microbiol. 2008;74(5):1350–1356. doi: 10.1128/AEM.02254-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Werner G, Klare I, Fleige C, Witte W. Increasing rates of vancomycin resistance among Enterococcus faecium isolated from German hospitals between 2004 and 2006 are due to wide clonal dissemination of vancomycin-resistant enterococci and horizontal spread of vanA clusters. Int J Med Microbiol. 2008;298(5–6):515–527. doi: 10.1016/j.ijmm.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 15.Freitas AR, Novais C, Tedim AP, Francia MV, Baquero F, Peixe L, Coque TM. Microevolutionary events involving narrow host plasmids influences local fixation of vancomycin-resistance in Enterococcus populations. PLoS One. 2013;8(3):e60589. doi: 10.1371/journal.pone.0060589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rosvoll TC, Pedersen T, Sletvold H, Johnsen PJ, Sollid JE, Simonsen GS, Jensen LB, Nielsen KM, Sundsfjord A. PCR-based plasmid typing in Enterococcus faecium strains reveals widely distributed pRE25-, pRUM-, pIP501- and pHTbeta-related replicons associated with glycopeptide resistance and stabilizing toxin-antitoxin systems. FEMS Immunol Med Microbiol. 2010;58(2):254–268. doi: 10.1111/j.1574-695X.2009.00633.x. [DOI] [PubMed] [Google Scholar]
- 17.Sadowy E, Sienko A, Gawryszewska I, Bojarska A, Malinowska K, Hryniewicz W. High abundance and diversity of antimicrobial resistance determinants among early vancomycin-resistant Enterococcus faecium in Poland. Eur J Clin Microbiol Infect Dis. 2013;32(9):1193–1203. doi: 10.1007/s10096-013-1868-y. [DOI] [PubMed] [Google Scholar]
- 18.Wardal E, Markowska K, Zabicka D, Wroblewska M, Giemza M, Mik E, Polowniak-Pracka H, Wozniak A, Hryniewicz W, Sadowy E. Molecular analysis of vanA outbreak of Enterococcus faecium in two Warsaw hospitals: the importance of mobile genetic elements. Biomed Res Int. 2014;2014:575367. doi: 10.1155/2014/575367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.CLSI (2015) Clinical and Laboratory Standards Institute (CLSI) (2015) Performance standards for antimicrobial susceptibility testing; 25th informational supplement. CLSI document M100-S20. CLSI, Wayne, PA
- 20.Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, Harbarth S, Hindler JF, Kahlmeter G, Olsson-Liljequist B, Paterson DL, Rice LB, Stelling J, Struelens MJ, Vatopoulos A, Weber JT, Monnet DL. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18(3):268–281. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 21.Clark NC, Cooksey RC, Hill BC, Swenson JM, Tenover FC. Characterization of glycopeptide-resistant enterococci from U.S. hospitals. Antimicrob Agents Chemother. 1993;37(11):2311–2317. doi: 10.1128/AAC.37.11.2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Top J, Schouls LM, Bonten MJ, Willems RJ. Multiple-locus variable-number tandem repeat analysis, a novel typing scheme to study the genetic relatedness and epidemiology of Enterococcus faecium isolates. J Clin Microbiol. 2004;42(10):4503–4511. doi: 10.1128/JCM.42.10.4503-4511.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Homan WL, Tribe D, Poznanski S, Li M, Hogg G, Spalburg E, Van Embden JD, Willems RJ. Multilocus sequence typing scheme for Enterococcus faecium. J Clin Microbiol. 2002;40(6):1963–1971. doi: 10.1128/JCM.40.6.1963-1971.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jensen LB, Garcia-Migura L, Valenzuela AJ, Lohr M, Hasman H, Aarestrup FM. A classification system for plasmids from enterococci and other Gram-positive bacteria. J Microbiol Methods. 2010;80(1):25–43. doi: 10.1016/j.mimet.2009.10.012. [DOI] [PubMed] [Google Scholar]
- 25.Wardal E, Gawryszewska I, Hryniewicz W, Sadowy E. Abundance and diversity of plasmid-associated genes among clinical isolates of Enterococcus faecalis. Plasmid. 2013;70(3):329–342. doi: 10.1016/j.plasmid.2013.07.003. [DOI] [PubMed] [Google Scholar]
- 26.Barton BM, Harding GP, Zuccarelli AJ. A general method for detecting and sizing large plasmids. Anal Biochem. 1995;226(2):235–240. doi: 10.1006/abio.1995.1220. [DOI] [PubMed] [Google Scholar]
- 27.Willems RJ, Top J, van den Braak N, van Belkum A, Mevius DJ, Hendriks G, van Santen-Verheuvel M, van Embden JD. Molecular diversity and evolutionary relationships of Tn1546-like elements in enterococci from humans and animals. Antimicrob Agents Chemother. 1999;43(3):483–491. doi: 10.1128/aac.43.3.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jensen LB, Ahrens P, Dons L, Jones RN, Hammerum AM, Aarestrup FM. Molecular analysis of Tn1546 in Enterococcus faecium isolated from animals and humans. J Clin Microbiol. 1998;36(2):437–442. doi: 10.1128/jcm.36.2.437-442.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boucher HW, Talbot GH, Bradley JS, Edwards JE, Gilbert D, Rice LB, Scheld M, Spellberg B, Bartlett J. Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clin Infect Dis. 2009;48(1):1–12. doi: 10.1086/595011. [DOI] [PubMed] [Google Scholar]
- 30.Werner G, Coque TM, Hammerum AM, Hope R, Hryniewicz W, Johnson A, Klare I, Kristinsson KG, Leclercq R, Lester CH, Lillie M, Novais C, Olsson-Liljequist B, Peixe LV, Sadowy E, Simonsen GS, Top J, Vuopio-Varkila J, Willems RJ, Witte W, Woodford N (2008) Emergence and spread of vancomycin resistance among enterococci in Europe. Euro Surveill 13(47) [PubMed]
- 31.Gawryszewska I, Zabicka D, Bojarska K, Malinowska K, Hryniewicz W, Sadowy E (2016) Invasive enterococcal infections in Poland: the current epidemiological situation. Eur J Clin Microbiol Infect Dis 35(5):847–856. doi:10.1007/s10096-016-2607-y [DOI] [PMC free article] [PubMed]
- 32.Johnson PD, Ballard SA, Grabsch EA, Stinear TP, Seemann T, Young HL, Grayson ML, Howden BP. A sustained hospital outbreak of vancomycin-resistant Enterococcus faecium bacteremia due to emergence of vanB E. faecium sequence type 203. J Infect Dis. 2010;202(8):1278–1286. doi: 10.1086/656319. [DOI] [PubMed] [Google Scholar]
- 33.Zheng B, Tomita H, Xiao YH, Wang S, Li Y, Ike Y. Molecular characterization of vancomycin-resistant enterococcus faecium isolates from mainland China. J Clin Microbiol. 2007;45(9):2813–2818. doi: 10.1128/JCM.00457-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Khan MA, Northwood JB, Loor RG, Tholen AT, Riera E, Falcon M, Paraguayan Antimicrobial N, van Belkum A, van Westreenen M, Hays JP. High prevalence of ST-78 infection-associated vancomycin-resistant Enterococcus faecium from hospitals in Asuncion, Paraguay. Clin Microbiol Infect. 2010;16(6):624–627. doi: 10.1111/j.1469-0691.2009.02898.x. [DOI] [PubMed] [Google Scholar]
- 35.Arthur M, Depardieu F, Reynolds P, Courvalin P. Quantitative analysis of the metabolism of soluble cytoplasmic peptidoglycan precursors of glycopeptide-resistant enterococci. Mol Microbiol. 1996;21(1):33–44. doi: 10.1046/j.1365-2958.1996.00617.x. [DOI] [PubMed] [Google Scholar]
- 36.Khan SA, Sung K, Layton S, Nawaz MS. Heteroresistance to vancomycin and novel point mutations in Tn1546 of Enterococcus faecium ATCC 51559. Int J Antimicrob Agents. 2008;31(1):27–36. doi: 10.1016/j.ijantimicag.2007.08.007. [DOI] [PubMed] [Google Scholar]
- 37.Handwerger S, Skoble J. Identification of chromosomal mobile element conferring high-level vancomycin resistance in Enterococcus faecium. Antimicrob Agents Chemother. 1995;39(11):2446–2453. doi: 10.1128/AAC.39.11.2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jung WK, Hong SK, Lim JY, Lim SK, Kwon NH, Kim JM, Koo HC, Kim SH, Seo KS, Ike Y, Tanimoto K, Park YH. Phenotypic and genetic characterization of vancomycin-resistant enterococci from hospitalized humans and from poultry in Korea. FEMS Microbiol Lett. 2006;260(2):193–200. doi: 10.1111/j.1574-6968.2006.00311.x. [DOI] [PubMed] [Google Scholar]
- 39.Kawalec M, Kedzierska J, Gajda A, Sadowy E, Wegrzyn J, Naser S, Skotnicki AB, Gniadkowski M, Hryniewicz W. Hospital outbreak of vancomycin-resistant enterococci caused by a single clone of Enterococcus raffinosus and several clones of Enterococcus faecium. Clin Microbiol Infect. 2007;13(9):893–901. doi: 10.1111/j.1469-0691.2007.01774.x. [DOI] [PubMed] [Google Scholar]
- 40.Gu L, Cao B, Liu Y, Guo P, Song S, Li R, Dai H, Wang C. A new Tn1546 type of VanB phenotype-vanA genotype vancomycin-resistant Enterococcus faecium isolates in mainland China. Diagn Microbiol Infect Dis. 2009;63(1):70–75. doi: 10.1016/j.diagmicrobio.2008.08.018. [DOI] [PubMed] [Google Scholar]
- 41.Depardieu F, Reynolds PE, Courvalin P. VanD-type vancomycin-resistant Enterococcus faecium 10/96A. Antimicrob Agents Chemother. 2003;47(1):7–18. doi: 10.1128/AAC.47.1.7-18.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cha JO, Yoo JI, Kim HK, Kim HS, Yoo JS, Lee YS, Jung YH. Diversity of Tn1546 in vanA-positive Enterococcus faecium clinical isolates with VanA, VanB, and VanD phenotypes and susceptibility to vancomycin. J Appl Microbiol. 2013;115(4):969–976. doi: 10.1111/jam.12300. [DOI] [PubMed] [Google Scholar]
- 43.Novais C, Freitas AR, Silveira E, Baquero F, Peixe L, Roberts AP, Coque TM. Different genetic supports for the tet(S) gene in enterococci. Antimicrob Agents Chemother. 2012;56(11):6014–6018. doi: 10.1128/AAC.00758-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Novais C, Freitas AR, Sousa JC, Baquero F, Coque TM, Peixe LV. Diversity of Tn1546 and its role in the dissemination of vancomycin-resistant enterococci in Portugal. Antimicrob Agents Chemother. 2008;52(3):1001–1008. doi: 10.1128/AAC.00999-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Boyd DA, Conly J, Dedier H, Peters G, Robertson L, Slater E, Mulvey MR. Molecular characterization of the vanD gene cluster and a novel insertion element in a vancomycin-resistant Enterococcus isolated in Canada. J Clin Microbiol. 2000;38(6):2392–2394. doi: 10.1128/jcm.38.6.2392-2394.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lanza VF, Tedim AP, Martinez JL, Baquero F, Coque TM (2015) The plasmidome of firmicutes: impact on the emergence and the spread of resistance to antimicrobials. Microbiol Spectr 3 (2):PLAS-0039-2014. doi:10.1128/microbiolspec.PLAS-0039-2014 [DOI] [PubMed]
- 47.Sletvold H, Johnsen PJ, Wikmark OG, Simonsen GS, Sundsfjord A, Nielsen KM. Tn1546 is part of a larger plasmid-encoded genetic unit horizontally disseminated among clonal Enterococcus faecium lineages. J Antimicrob Chemother. 2010;65(9):1894–1906. doi: 10.1093/jac/dkq219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kawalec M, Gniadkowski M, Hryniewicz W. Outbreak of vancomycin-resistant enterococci in a hospital in Gdask, Poland, due to horizontal transfer of different Tn1546-like transposon variants and clonal spread of several strains. J Clin Microbiol. 2000;38(9):3317–3322. doi: 10.1128/jcm.38.9.3317-3322.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Garcia-Migura L, Hasman H, Svendsen C, Jensen LB. Relevance of hot spots in the evolution and transmission of Tn1546 in glycopeptide-resistant Enterococcus faecium (GREF) from broiler origin. J Antimicrob Chemother. 2008;62(4):681–687. doi: 10.1093/jac/dkn265. [DOI] [PubMed] [Google Scholar]
- 50.Teuber M, Schwarz F, Perreten V. Molecular structure and evolution of the conjugative multiresistance plasmid pRE25 of Enterococcus faecalis isolated from a raw-fermented sausage. Int J Food Microbiol. 2003;88(2–3):325–329. doi: 10.1016/S0168-1605(03)00195-8. [DOI] [PubMed] [Google Scholar]
- 51.Garcia-Migura L, Liebana E, Jensen LB. Transposon characterization of vancomycin-resistant Enterococcus faecium (VREF) and dissemination of resistance associated with transferable plasmids. J Antimicrob Chemother. 2007;60(2):263–268. doi: 10.1093/jac/dkm186. [DOI] [PubMed] [Google Scholar]
- 52.Paulsen IT, Banerjei L, Myers GS, Nelson KE, Seshadri R, Read TD, Fouts DE, Eisen JA, Gill SR, Heidelberg JF, Tettelin H, Dodson RJ, Umayam L, Brinkac L, Beanan M, Daugherty S, DeBoy RT, Durkin S, Kolonay J, Madupu R, Nelson W, Vamathevan J, Tran B, Upton J, Hansen T, Shetty J, Khouri H, Utterback T, Radune D, Ketchum KA, Dougherty BA, Fraser CM. Role of mobile DNA in the evolution of vancomycin-resistant Enterococcus faecalis. Science. 2003;299(5615):2071–2074. doi: 10.1126/science.1080613. [DOI] [PubMed] [Google Scholar]
- 53.EUCAST (2015) European Committee on Antimicrobial Susceptibility Testing (EUCAST) (2015) Breakpoint tables for interpretation of MICs and zone diameters. Version 5.0, EUCAST, Basel, Switzerland
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 20 kb)
(DOCX 58 kb)
(DOCX 16 kb)