Abstract
Drug-induced liver injury (DILI) remains a leading cause of drug withdrawal from human clinical trials or the marketplace. Due to species-specific differences in liver pathways, predicting human-relevant DILI using in vitro human liver models is critical. Microfabrication tools allow precise control over the cellular microenvironment towards stabilizing liver functions for weeks. These tools are used to engineer human liver models with different complexities and throughput using cell lines, primary cells, and stem cell-derived hepatocytes. Including multiple human liver cell types can mimic cell-cell interactions in specific types of DILI. Finally, organ-on-a-chip models demonstrate how drug metabolism in the liver affects multi-organ toxicities. In this review, we survey engineered human liver platforms within the needs of different phases of drug development.
Keywords: liver-on-a-chip, microfabrication, micropatterned co-cultures, drug-induced liver injury, primary human hepatocytes
The Drug Development Pipeline
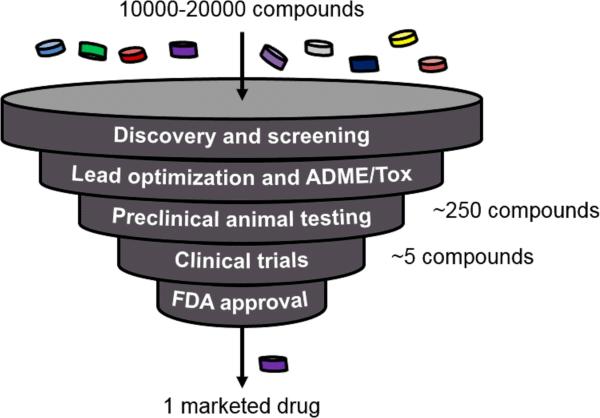
Drug development begins when a compound library (~10-20 thousand compounds) is tested in a high-throughput format for activity (e.g., inhibition) against a protein implicated in a disease [1]. If the target protein is unidentified, the compounds are tested to alleviate a diseased phenotype in cells (this process is called phenotypic drug discovery) [2]. The lead “hit” compound and its backups then undergo ADME/Tox (see Glossary) characterization in vitro and in animals before entering human clinical trials (Fig. 1). Despite the large amount of money (~$3-5B) and time (12-15 years) invested during drug development, ~90% of compounds fail during clinical trials, with one-third of such failures attributed to organ toxicity [3]. The liver is especially susceptible to such toxicity due to its central role in drug metabolism. Drug-induced liver injury (DILI) is a leading cause of drug attrition in preclinical and clinical testing, black-box warnings on marketed drugs, the withdrawal of previously approved drugs, and acute liver failures [4].
Figure 1. The drug development pipeline.
Starting with a set of between 10,000 and 20,000 compounds created via combinatorial chemistry, early stages of the pipeline greatly reduce the number of potential compounds for further development. Eventually only one drug gets launched into the marketplace after $3-5B and 12-15 years of going through the pipeline. The discovery/screening, lead optimization and ADME/Tox (absorption, distribution, metabolism, excretion, and toxicity), and preclinical animal testing are the phases where engineered human liver models can make the greatest impact to reduce and in some cases replace the usage of animals and prevent harm to patients in clinical trials and in the marketplace.
Animal models are not always good predictors of human-relevant DILI due to significant species-specific differences in drug metabolism pathways [5]. Thus, in vitro models of the human liver are important to understand human drug metabolism and toxicity prior to clinical trials [6]. Such models can also aid in phenotypic drug discovery against liver diseases (i.e. hepatitis B/C viral infections, fatty liver disease, fibrosis, carcinoma) [7]. Primary human hepatocytes (PHHs) are ideal for creating human liver models; however, their liver-specific functions (see Box 1) rapidly decline in conventional culture formats, which leads to a low (<50%) sensitivity for DILI prediction [6,8]. Therefore, engineers have developed tools that can control the cellular microenvironment towards enabling higher and more stable PHH functions for several weeks [6]. The ability to test drugs chronically on stable PHHs has led to significant improvements in sensitivities for DILI detection, as well as better modeling of liver diseases. The latest efforts towards organ-on-a-chip platforms are geared towards understanding how drug metabolism in the liver affects efficacy and/or toxicity in other tissues [9]. In this review, we discuss the design features and utility of engineered liver models and organ-on-a-chip devices within the context of different phases of drug development.
High-Throughput Hepatic Systems
In early drug development, when many compounds need to be tested and the amount of compounds is limiting, culture platforms should be high-throughput, cost relatively little, and provide actionable data quickly (within 24-48 hours). In the context of the liver, metabolic stability, major metabolites, and toxicity of compounds are important parameters to evaluate [6]. To reduce cost, cancerous hepatic cell lines (i.e. THLE-2, HepG2, Hep3B, HepaRG) can provide an initial assessment of drug toxicity; however, for proper metabolism, the use of PHHs becomes important because cell lines downregulate levels of drug metabolism enzymes (messenger-RNA and protein) in the human liver [10]. In this section, we discuss how engineering has helped to advance high-throughput culture platforms that are often compatible with multiple liver cell sources.
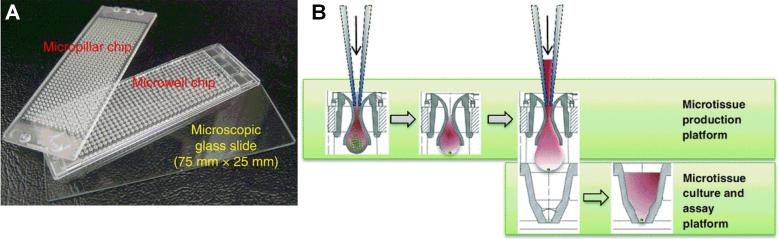
Kwon et al. designed a microchip platform for transducing 3D liver cell cultures with genes for drug metabolism enzymes [11] (Fig. 2A). The platform features 532 reaction vessels (micropillars and corresponding microwells) on a 75 mm by 25 mm outline. Cells are suspended in a Matrigel™ droplet (~60 nL), which is spotted on a micropillar. The micropillar is then placed into a corresponding microwell containing recombinant adenoviruses. THLE-2 cells were transduced with adenoviruses to manipulate the expression of human drug metabolism enzyme genes. A single microarray was used to create 84 combinations of metabolic gene expressions, which provided information on which enzyme combinations led to drug toxicity in cells. In another example, a 3D Hep3B microarray was coupled with a microarray containing various combinations of recombinant drug metabolism enzymes to evaluate the metabolism-mediated toxicity of drugs [12]. Ultimately, PHHs will better capture complex mechanisms of DILI, where the ratios of many enzymes and transporters may be important. Therefore, adapting the aforementioned platforms to PHHs (in addition to cell lines) will be important, while continuing to reduce cell numbers and drug amounts in the droplets, which is desirable for early drug development.
Figure 2. High-throughput hepatic systems.
(A) 532-well micropillar and microwell plate combination that can contain as little as 60 nL in each well. Hepatic cells (i.e. THLE-2) suspended in a Matrigel™ droplet are spotted on each micropillar, which are then placed in a microwell containing recombinant adenoviruses. The adenoviruses are engineered to transduce the cells with different combinations of drug metabolism genes towards determining enzymes involved in the observed toxicity of a drug. Adapted from [11] with permission from Nature Publishing Group. (B) Hanging-drop method for generating liver spheroids of controlled diameters. Briefly, a cell mixture is seeded into the wells of a specialized 96-well plate that allows the spheroid to form and mature in a hanging drop. The spheroids are then transferred to another culture plate for drug dosing. Adapted from [13] with permission from Springer.
Other platforms can also form 3D spheroids of uniform sizes, as opposed to spheroids of heterogeneous sizes that can become necrotic in their cores if the diameter becomes too large (>200 μm) for adequate oxygen diffusion. Hanging drops of uniform sizes containing HepG2 or PHHs can be cultured in a specialized plate, which causes the formation of spheroids of controlled diameters (253 ± 7.4 μm) within 4 days [13]. These spheroids are then transferred to another multi-well plate for drug screening (Fig. 2B). Miyamoto et al. developed a tapered cell seeding device containing 400 microwells with a top aperture (500 μm × 500 μm) and a bottom aperture (300 μm diameter circle) per microwell to form HepG2 spheroids of uniform diameters [14]. Fukuda and Nakazawa designed a spheroid microarray that allowed stable immobilization of 100 μm rat hepatocyte spheroids in microwells for probing cytochrome P450 (CYP450) activities following drug treatment [15].
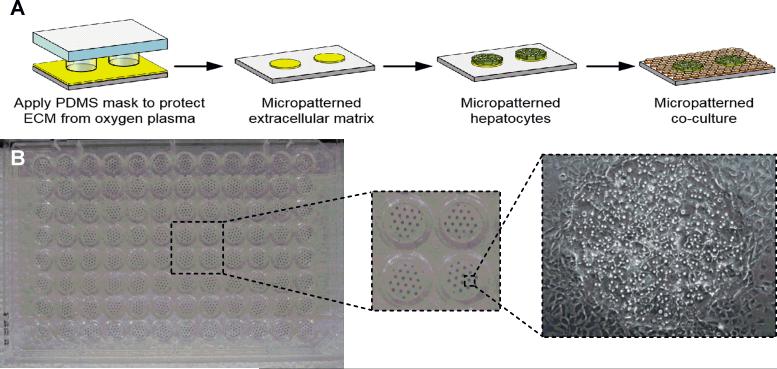
Spheroidal cultures are not always compatible with high content screening (HCS), in which multiplexed fluorescent readouts are used to screen for DILI liabilities at the organelle level in individual cells (i.e. mitochondria, reactive oxygen species or ROS). On the other hand, monolayer cultures are well-suited for HCS [8]. For instance, in micropatterned co-cultures (MPCCs) of PHHs and supportive nonparenchymal cells (NPCs), the PHH fluorescent signals can be computationally separated from the NPC signals [16]. In this model, multi-well plates are subjected to soft lithography utilizing polydimethylsiloxane (PDMS) stamps to micropattern collagen islands on which PHHs selective attach and are subsequently surrounded by NPCs [17] (Fig. 3A). The diameter/spacing of the PHH domains have been optimized to enable optimal cell-cell interactions between PHHs and NPCs, which leads to higher and more stable (4-6 weeks) liver functions than randomly distributed co-cultures. MPCCs are routinely cultured in a 96-well plate format (Fig. 3B), but more recently, the Bhatia group has adapted MPCCs to a 384-well plate.
Figure 3. Micropatterned co-cultures.
(A) Tissue culture polystyrene (or glass) can be uniformly coated with extracellular matrix protein (ECM) such as collagen and protected with a polydimethylsiloxane (PDMS) stamp. Exposed areas of ECM are ablated under oxygen plasma, leaving micropatterned ECM islands that match the geometry of the PDMS stamp. Hepatocytes selectively attach to ECM islands, and nonparenchymal cells (NPCs) fill in the surrounding area. (B) An industry standard 96-well plate showing uniform hepatocyte islands micropatterned using the process in panel A. The NPCs used in this example are 3T3-J2 murine embryonic fibroblasts surrounding the primary human hepatocyte colonies. Reprinted from [6] with permission from Sage Publications. More recently, MPCCs have been adapted to a 384-well plate format by the Bhatia group at the Massachusetts Institute of Technology.
MPCCs have been validated using well-annotated compounds for applications in drug development. Wang et al. detected a significantly greater number of clinically-relevant drug metabolites in MPCCs than short-term PHH suspensions [18]. Lin et al. showed that MPCCs predicted 73%, 92%, and 96% of clearance rates for 26 drugs within 2-fold, 3-fold, and 4-fold of in vivo rates, respectively [19]. On the other hand, suspension hepatocytes and conventional monolayers predicted only 30-50% of the clearance rates for tested drugs within 4-fold of in vivo rates. Additionally, MPCCs recapitulated drug-drug interactions that can affect drug clearance in patients [20]. In another study, MPCCs were dosed with clinical hepatotoxins for 9 days, which improved the sensitivity for DILI detection to ~66% versus ~29% in short-term monocultures [21]. MPCCs have also been used to model liver diseases towards utility in phenotypic drug discovery. Specifically, MPCCs can be infected with several pathogens, such as hepatitis B virus [22], hepatitis C virus [23], and malaria [24], and they can be used to screen for glucose-lowering diabetes drugs [25].
The availability of cryopreserved PHHs allows the construction of on-demand cultures for drug screening. However, PHHs are a limited resource for sustainable drug screening using the same donor(s) and are limited in the genetic diversity available to predict inter-individual differences in DILI outcomes, which are relevant in the clinic [4]. On the other hand, induced pluripotent stem cells (iPSCs) can be derived from many patients and are a nearly infinite cell source for drug screening. Protocols to differentiate iPSCs into hepatocyte-like cells (iPSC-HHs) use growth factors inspired from embryology; however, liver functions remain very low (<10%) relative to adult PHHs [26]. Engineering tools can be used to further mature iPSC-HHs [27]. Takayama et al. created a nanopillar plate for the spheroidal culture of iPSC-HHs, which helped to further improve liver functions [28]. Berger et al. showed that the MPCC technique further matures and stabilizes iPSC-HH functions for 4 weeks [29]. Furthermore, such ‘iMPCCs’ dosed with 37 hepatotoxins for 6 days yielded a sensitivity (65%) that was similar to the sensitivity obtained in PHH-based MPCCs (70%) using the same drug set, while the specificity in both models was 100% using 10 non-liver-toxic drugs [30]. These results suggest that iPSC-HHs in an engineered platform may be suitable for an initial drug toxicity screen; however, mechanistic inquiries into drug responses as in later stages of drug development will require detailed comparisons of molecular pathways affected by drugs in iPSC-HHs and PHHs.
Engineered Liver Co-Cultures
As a lead candidate compound progresses through drug development, its effects on tissue types need to be investigated at the mechanistic level in a more physiological context where high-throughput and rapid screening are not always required given the significant reduction in the number of compounds for testing relative to early drug development (Fig. 1). While hepatic cell lines provide cheap and abundant cell sources for high-throughput systems deployed during early drug development, they are not suitable for accurately modeling complex physiological outcomes during lead optimization because they are limited to single donors and down-regulate drug metabolism pathways [10]. In contrast, PHHs can mitigate such limitations of cell lines. Besides PHHs, NPCs resident in the liver (see Box 2) are known to either experience toxicity to drugs directly and/or secrete molecules that regulate PHH response [31]. Thus, investigators have incorporated one or more liver NPCs alongside hepatocytes into engineered liver models as discussed below; however, it is not yet clear how to incorporate cholangiocytes for bile drainage into a separate compartment as in vivo, which could determine drug disposition in the bile. Initial designs of devices are often tested with rat liver cells or cell lines, though adaptation to primary human liver cells is progressing rapidly. In this section, we discuss the latest engineered platforms to improve co-culture functions and their initial validation with annotated drugs.
Static platforms
The MPCC platform is modular in that the NPC population can be modified without significantly affecting the PHH homotypic interactions on the micropatterned domains for the proper formation of bile canaliculi between the hepatocytes. Nguyen et al. first allowed MPCCs to stabilize for ~1 week and then seeded human liver Kupffer macrophages (KMs) atop the monolayer [32]. Stimulating KMs with bacterial lipopolysaccharide (LPS) led to cytokine-mediated down-regulation of hepatic CYP450s, which can affect drug outcomes. Current studies are focused on creating MPCCs with other liver NPCs, while maintaining the multi-well format for screening several drugs or conditions.
Other groups have created PHH-NPC co-cultures in 3D configurations. Kostadinova et al. seeded a mixture of liver NPCs (liver sinusoidal endothelial cells or LSECs, hepatic stellate cells or HSCs, and KMs; see Box 2) onto a porous nylon scaffold in a 24-well format followed by seeding of PHHs ~1 week later. PHHs displayed CYP450 activities for ~3 months, while the NPCs expressed prototypical markers [33]. Drug toxicity studies showed the ability of the co-cultures to predict clinical outcomes. In a scaffold-free example, the hanging drop culture method discussed above was used to create spheroids of PHHs, KMs, and endothelia [13]. Trovafloxacin-induced hepatotoxicity was exacerbated when KMs were activated via LPS.
In contrast to a random orientation of cell types in the aforementioned spheroids, bioprinting can be used to position liver NPCs relative to PHHs to create a compartmentalized architecture that can provide insights into how the relative positions of the cell types changes upon drug treatment. Organovo developed bioprinted co-cultures containing PHHs, HSCs, and LSECs in a 24-well plate format. Ma et al. bioprinted iPSC-HHs, endothelial cells, and adipose-derived stem cells in a hexagonal architecture embedded in a hydrogel that mimics the liver lobule [34].
Perfused liver-on-a-chip platforms
In comparison to static models, perfusion of cultures allows for continuous nutrient exchange, better oxygen delivery, and physiologic shear stress; however, complexities in liquid handling are introduced along with reduced throughput relative to multi-well plates. Miniaturized microfluidic bioreactors can further allow control over the soluble microenvironment of cells at a scale similar to a tissue's functional unit [9] (Fig. 4A). The Griffith group engineered a device in which preformed hepatic aggregates adhered to the collagen-coated walls of an array of micro-channels were perfused, which led to higher hepatic functions than static controls [35]. This platform was later adapted to co-cultures between rat hepatocytes and LSECs [36]. Toh et al. designed a microfluidic chip to generate linear gradients of drug concentrations and establish dose-response curves for drugs incubated on rat hepatocytes [37]. Similar concentration gradients were established in another biochip designed to mimic the liver sinusoid [38]. The Leclerc group has designed biochips for characterizing the metabolic profiles of rat hepatocytes [39,40], evaluating drug metabolism in PHHs [41,42], and investigating drug-induced ROS formation and glutathione depletion in rat hepatocytes [43] and HepG2/C3a cells [44]. Chao et al. cultured PHH monolayers in a microfluidic platform for evaluating drug clearance [45], which was extended by Novik et al. to PHH-NPC co-cultures [46]. The co-cultures under flow were capable of clearing compounds with better correlation to in vivo clearance values than the static controls. More modern spheroidal cultures feature microfluidic connections that can enable continuous perfusion of medium. Kim et al. used such a platform for monitoring rat hepatic spheroids over time [47], while Frey et al. used a similar method for evaluating toxicity of the anti-cancer drug 5-fluorouracil [48].
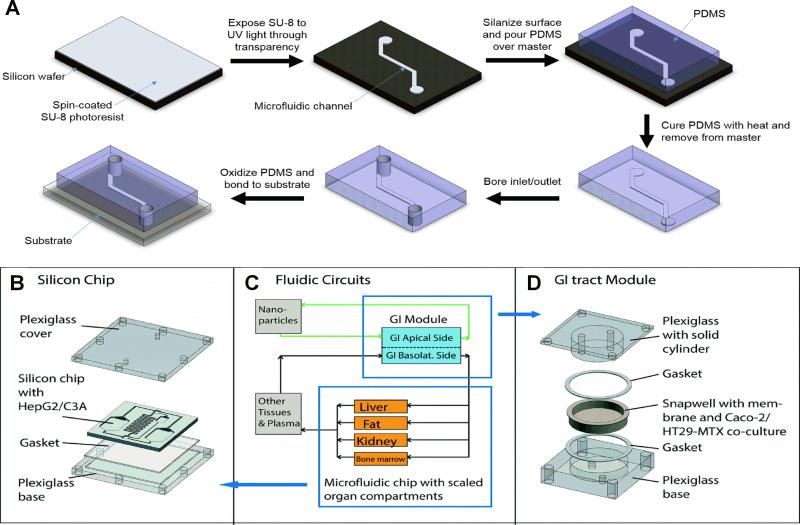
Figure 4. Liver-on-a-chip platforms.
(A) Liver-on-a-chip platforms are often manufactured with soft lithography techniques, whereby a design is transferred to SU-8 photoresist using ultraviolet (UV) light through a high-resolution mask such as a transparency. Polydimethylsiloxane (PDMS) is then poured over the patterned SU-8 to construct the microfluidic channel. After curing and boring the inlet and outlet ports, the PDMS is oxidized and bonded to the device substrate. (B) A liver-on-a-chip platform featuring HepG2/C3A cells is established on a silicon chip sandwiched between plexiglass layers. (C) Using a physiologically-based pharmacokinetic (PBPK) model, the fluidic circuit is designed to mimic the interactions between the liver and gastrointestinal (GI) compartments. (D) The liver on-a-chip platform is connected with a GI platform consisting of a co-culture of Caco-2 and HT29-MTX cells. Reprinted from [65] with permission from the Royal Society of Chemistry.
Several groups have created co-cultures of PHHs and endothelial cells in microfluidic devices, although primary human LSECs are not always used due to limited availability. Bovine aortic endothelial cells were used in a biochip that separates hepatocytes from endothelia with either an extracellular matrix (ECM) protein layer or a microporous membrane [49]. Bovine pulmonary microcapillary endothelial cells were incorporated into a layered model to induce the formation of capillary-like structures [50]. Two groups used EA.hy926 (transformed endothelial cells) in lieu of primary cells in their microfluidic devices, which also contained LX-2 cells (immortalized HSC line), U937 monocytes (cell line from lung lymphoma), and PHHs in a layered sinusoid-like architecture [51,52]. However, the aforementioned readily available cell types cannot fully mimic human liver physiology. To eliminate the endothelial cells altogether, some groups have developed devices that simulate the endothelial fenestrations with artificial barrier slits [37,53,54]; however, such devices are devoid of PHH-LSEC crosstalk, which is important in liver diseases [55].
Microfluidic devices are difficult to set up and handle relative to multi-well plates. Therefore, to aid ease of usability and allow rapid assessment of drug effects, real-time monitoring of toxicity biomarkers is being incorporated into microfluidic devices. Bavli et al. developed a biochip with a computer-controlled microfluidic switchboard that can simultaneously monitor mitochondrial respiration, glucose, and lactate in HepG2/C3A aggregates in real-time [56]. A shift from oxidative mitochondrial respiration to glycolysis was then shown after treatment with two hepatotoxins. Rennert et al. established a liver biochip consisting of human umbilical vein endothelial cells and monocyte-derived macrophages in the vascular plane that was separated from the hepatic plane containing HepaRG and LX-2 cells with a membrane mimicking the space of Disse [57]. Luminescent-based sensor spots were integrated in the chip for real-time measurement of oxygen consumption. Vernetti et al. created a platform in which PHHs, EA.hy926, U937 monocytes, and LX-2 cells were sequentially layered in a microfluidic device that had fluorescent protein biosensors inside ~20% of the PHHs to detect apoptosis and ROS following drug exposure [51]. Finally, other groups have developed microfluidic devices with incorporated antibody-based biosensors for monitoring transforming growth factor (TGF)-β1, an activator of HSCs [58,59].
Oxygen gradients can modulate hepatic CYP450 expression along the length of the sinusoid (zonation), which can cause zonal patterns of DILI [60]. Allen et al. generated oxygen gradients across rat hepatocyte-fibroblast co-cultures by cell-mediated depletion of oxygen dissolved in the culture medium from the inlet to the outlet of a bioreactor [61]. The oxygen gradient led to an in vivo-like higher expression of CYP450s and greater acetaminophen toxicity in the hepatocytes subjected to the low-oxygen regions (outlet) as compared to the high-oxygen regions (inlet). Sato et al. also developed a biochip for providing a continuous oxygen gradient to cultured mouse hepatocytes [62]. Peng et al. designed a 16-well device with channels underneath the cell culture chamber to introduce oxygen scavenging chemicals and induce hypoxic conditions [63].
Organ-On-A-Chip Platforms
Drug metabolism in the liver can affect the efficacy and/or toxicity of drugs in other organ systems. Thus, organ-on-a-chip platforms with microfluidic perfusion are being developed to investigate interactions between tissue types following drug exposure [9]. The Shuler lab designed an early organ-on-a-chip model in which cell lines were used to model lung, liver, and fat compartments that were linked with microfluidic flow to investigate drug biodistribution [64]. Using cell lines is common in current organ-on-a-chip designs to show proof-of-concept prior to transitioning to primary cells, which can be more difficult to source and culture in complex devices.
The liver and the intestine are connected in vivo by the portal circulation and the common bile duct. Often drugs and their metabolites can undergo enterohepatic recirculation between the liver and the intestine, which can increase exposure to both organs. Esch et al. constructed a liver-intestine chip to investigate nanoparticle toxicity (Fig. 4B-D) [65]. The liver compartment was modeled with HepG2/C3A cells cultured in a silicon chip between plexiglass layers, while the intestine was represented with a co-culture of Caco-2 (colon carcinoma) and HT29-MTX (mucus-secreting colon epithelia) cells. When only the liver compartment was exposed to nanoparticles, the HepG2/C3A cells released more aspartate aminotransferase (cellular injury marker) than the vehicle-only control. This phenomenon was exacerbated when the liver and intestine compartments were connected via microfluidic channels. Bricks et al. coupled polycarbonate cell culture inserts and microfluidic biochips to create a platform in which the liver compartment and intestinal compartments were mimicked by HepG2-C3A and Caco-2 cells, respectively [66]. The transport of phenacetin through the intestinal barrier and its metabolism into acetaminophen by the liver compartment were demonstrated. Besides the intestine, interactions of the liver with other organs are also important for evaluating drug effects. Chouca-Snouber et al. created a microfluidic biochip that combined HepG2/C3A or HepaRG liver and MDCK (Madin-Darby canine kidney) cell lines [67]. Ifosfamide was metabolized by HepaRG cells but not HepG2/C3A cells into a metabolite that was toxic to MDCK cells. Materne et al. created a biochip consisting of liver spheroids with HepaRG and primary HSCs, and differentiated NT2 cell neurospheres [68]. After 2 weeks of dosing with 2,5-hexanedione (neurotoxin), the co-cultures were more sensitive than the single-tissue cultures in the biochip. Similar techniques have been used with liver-tumor interactions for studying endothelial permeability [69]. Other groups have used biochips to study liver-skin interactions in troglitazone-induced toxicity [70] and topical substance exposure [71]. Physiologically based pharmacokinetic (PBPK) modeling is a powerful application for organs-on-a-chip devices since it can provide insights into key clinical parameters of the drug (i.e. maximum plasma concentration and half-life). Kimura et al. developed a liver-lung-intestine model for PBPK analysis. While the cell lines HepG2, A549, and Caco-2 were used, they were still able to model the distribution of three anti-cancer drugs [72]. Similarly, Sung et al. modeled liver-tumor-marrow interactions using a microfluidic chip with cell lines and derived a PK model for the distribution of 5-fluorouracil [73]. Their results showed that when compared to static cultures, the microfluidic device was able to more accurately reproduce liver metabolism of the cancer prodrug tegafur to 5-fluorouracil, which was toxic to the tumor model.
Finally, a few groups have created four-organ-chips. Maschmeyer et al. combined intestine, skin, liver, and kidney modules onto a chip and assessed functionality of the cells over 28 days [74]. Oleaga et al. combined cardiac, muscle, neuronal, and liver modules and evaluated the toxicities of doxorubicin, atorvastatin, valproic acid, acetaminophen, and N-acetyl-m-aminophenol [75]. With increased research funding from federal agencies (i.e. National Institutes of Health and Defense Advanced Research Projects Agency), development of multi-organ chips with greater automation and better physiological relevance (i.e. using primary cells) will continue to grow.
Concluding Remarks
Engineered culture platforms can keep human liver cells highly functional for several weeks in vitro towards better predicting clinical drug outcomes than is possible with declining monolayers and animal models. Some common trends in such platforms have emerged. First, the functional stabilization of PHHs for several weeks in vitro is of paramount importance even in the absence of mimicking the organization of the human liver. In particular, static disorganized spheroids and micropatterned co-cultures with murine embryonic fibroblasts have been shown to stabilize PHH functions for at least 4 weeks [6]. Second, optimizing cell-cell interactions between PHHs and surrounding NPCs in both 2D (micropatterned) and 3D (bioprinted) formats can enhance and stabilize PHH functions reproducibly across many donors as opposed to an inherent phenotypic instability in randomly distributed co-cultures. Third, certain types of DILI in which inflammation and fibrosis are important can be more accurately modeled by including Kupffer macrophages and hepatic stellate cells in co-culture with PHHs, respectively. Fourth, a single liver platform doesn't satisfy the requirements for all of the phases of drug development. The choice of the culture model therefore depends on the complexity of the hypotheses being tested, the acceptable level of sensitivity/specificity, throughput required, and the cost-benefit analysis. Finally, liver-on-a-chip models are being coupled with models of other tissues on organ-on-a-chip to determine the effects of liver drug metabolism on drug efficacy and/or toxicity in other tissues.
Even with considerable developments in engineering of human liver platforms, some key issues need to be addressed in next-generation platforms. First, gene expression/functions of multiple liver cell types in an engineered platform over time needs to be compared to freshly-isolated cells prior to plating towards elucidating those pathways that return to physiological levels in culture while others that deviate considerably and could serve as benchmarks for iterative device optimizations. Second, comparing 2D and 3D engineered liver platforms (as opposed to using declining monolayers as controls) with the same drug sets will determine potential improvements in sensitivity for predicting clinical liver-drug interactions in the 3D models. Third, consensus needs to be reached on the use of specific biomarkers to validate the utility of engineered liver platforms for drug screening. Finally, iPSC-derived human liver cells need to be further differentiated towards the adult liver phenotype using engineering tools in order to better utilize these cells for determining mechanisms underlying inter-individual variations in DILI outcomes (‘idiosyncratic’ or unpredictable). Ultimately, personalized drug screening on mature iPSC-derived human liver cells could potentially allow custom drug therapies for different patients.
In conclusion, various engineered human liver models can serve the biological and throughput needs of different phases of drug development within budgetary constraints. While the exact future of engineered human liver models is uncertain (see Outstanding Questions), we anticipate that continued advances in microfabrication, liver cell culture, and cell sourcing will reduce drug attrition, prevent harm to patients, and enable discovery of novel drugs.
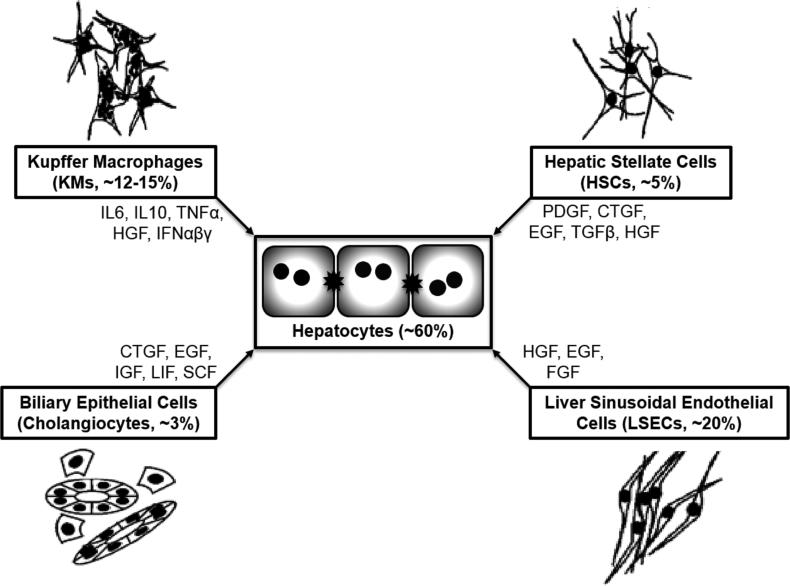
Figure I. Parenchymal and nonparenchymal cell types of the liver.
Kupffer macrophages (KMs), hepatic stellate cells (HSCs), liver sinusoidal endothelial cells (LSECs), and biliary epithelial cells are the major nonparenchymal cells (NPCs) in the liver. By secreting the indicated molecules, these NPCs impact the phenotype of primary human hepatocytes (PHHs). Percentages represent the relative number of each cell type in the human liver. Adapted from [6].
Box 1: The Liver And Its Functions.
The liver is the largest internal organ of the body (~1.5 kg in a 70 kg human). It is responsible for over 500 functions that include synthesis of proteins (i.e. albumin, transferrin) and clotting factors; metabolism of fats, proteins, and carbohydrates; storage of glycogen and lipids; detoxification of endogenous (i.e. bilirubin: by-product of red blood cell breakdown) and exogenous (i.e. drugs) substrates; and production of bile that aids in the digestion and absorption of fats in the small intestine. Primary human hepatocytes (PHHs) perform the majority of the functions in the liver. PHHs form between each other membrane-bound structures called bile canaliculi, which can drain specific drugs/metabolites and bile into the biliary duct that connects to the gall bladder.
The functional longevity/health of PHHs in engineered liver models is assessed over time using a subset of the aforementioned PHH phenotypic biomarkers [6], including
PHH morphology (e.g., polygonal shape, distinct nuclei/nucleoli, formation of bile canaliculi) because it correlates with retention of some functions.
Albumin, a major plasma protein responsible for maintaining osmotic pressure in the blood. Albumin secretion can be measured with an enzyme-linked immunosorbent assay (ELISA) in culture supernatants.
Urea, a by-product of nitrogen metabolism that can be measured in culture supernatants using a colorimetric assay [17].
Cytochrome P450 enzymes: the activities of specific isoforms can be evaluated via in situ optical readouts (luminescence, fluorescence) or incubation with isozyme-specific substrates and measuring the metabolite in culture supernatants with liquid chromatography–mass spectrometry.
Other liver-specific functions (e.g., glucose metabolism) can be measured depending on the hypotheses.
Non-hepatocyte-specific biomarkers can also be evaluated using optical readouts, including adenosine triphosphate (ATP, the major energy-carrying molecule in a cell) and glutathione in cell lysates.
For an in vitro drug toxicity screen, drug dose- and time-dependent decreases in the levels of phenotypic biomarkers over time are compared to biomarker levels in drug-free controls, which contain the organic solvent (e.g., dimethylsulfoxide) the drug was dissolved in to account for the effects of that solvent on hepatic functions. One typical method to classify a compound as “toxic” is to interpolate within the drug dose range tested a TC50 value, defined as the drug concentration that reduces one of the tested biomarkers to 50% of the drug-free control. Drugs can then be rank-ordered based on the TC50 values. Non-toxic drugs do not display interpolated TC50 values for any of the tested biomarkers [21,30].
Box 2: Liver Nonparenchymal Cells.
Besides PHHs (see Box 1), the liver contains sinusoidal endothelial cells (LSECs), Kupffer macrophages (KMs), hepatic stellate cells (HSCs), and cholangiocytes that communicate with PHHs via the secretion of specific molecules [55] (Fig. I). LSECs with pores for transport (fenestrae) line the liver capillaries (sinusoids), thereby separating PHHs from flowing blood. A thin ECM layer (space of Disse) separates the LSECs and PHHs. Additionally, LSECs participate in the metabolism of macromolecules (i.e. glycoproteins, lipoproteins). KMs are exposed to gut-derived bacterial endotoxins (i.e. lipopolysaccharide), which can activate these cells and cause secretion of cytokines. Quiescent HSCs store vitamin A in lipid droplets; however, liver injury can activate HSCs into myofibroblasts that secrete excessive collagen and cause fibrosis. Cholangiocytes line the bile ducts that drain the contents of the bile canaliculi into the gall bladder.
Outstanding.
To what extent are cancerous cell lines (both hepatocytes and liver nonparenchymal cells) representative of primary cells depending on the particular application in a specific phase of drug development?
Can iPSC-derived human hepatocyte-like cells in engineered platforms replace cell lines and primary human hepatocytes in the early stages of drug development?
How important is the inclusion of liver nonparenchymal cells, such as liver sinusoidal endothelial cells, Kupffer macrophages, and hepatic stellate cells, in high-throughput liver systems designed for screening drug toxicity in the early stages of drug development?
Are 3D engineered liver platforms, such as spheroids of controlled dimensions and bioprinted organoids, better predictive of clinical outcomes than 2D engineered liver platforms, such as micropatterned co-cultures?
Is perfusion necessary to maintain functions of liver cell types for prolonged times in vitro?
How should comparisons across different liver models be standardized across laboratories, with respect to both measured endpoints and annotated drug sets, so that it becomes easier to determine advances and rooms for further improvement?
How should the relative sizes of different organ types be modeled in organ-on-a-chip devices?
What technological advances are necessary for better ease of use (i.e., automation) and higher throughput of microfluidic culture platforms?
What impact will improvements in physiologically-based pharmacokinetic modeling (PBPK) have on advancing organ-on-a-chip devices?
Can in vitro engineered human liver platforms eventually replace the need for testing drug-induced liver injury (DILI) liabilities in animal models?
Can iPSC-derived hepatocytes from many individuals be useful to predict idiosyncratic DILI (occurs unpredictably in a small number of patients) across different patients?
Trends.
Early phases of drug development need lower cost/higher throughput, while later phases need a better physiological context for human liver models.
Multiple cell sources (i.e. cell lines, primary cells) can be used in liver platforms designed for different phases of drug development.
High-throughput systems feature cellular microarrays, spheroids of controlled diameters, and micropatterned co-cultures (MPCCs). Spheroids and MPCCs can stabilize hepatic functions for several weeks for chronic drug dosing.
Lower throughput systems include co-cultures of multiple human liver cell types in static (bioprinted) and perfused (liver-on-a-chip) formats.
Organ-on-a-chip platforms allow investigations into how liver drug metabolism affects other tissues.
Continued functional maturation of iPSC-derived hepatocytes in engineered platforms could enable personalized drug screening in the future.
ACKNOWLEDGMENTS
Funding was provided by the National Science Foundation (CAREER award CBET-1351909 to S.R.K.) and the National Institute of Biomedical Imaging and Bioengineering (7R03EB019184-03 to S.R.K.). We acknowledge helpful discussions with Matthew Davidson and Christine Lin.
GLOSSARY
- Absorption, distribution, metabolism, excretion, and toxicity (ADME/Tox)
Processes by which a drug gets absorbed into the blood stream from the small intestine, gets distributed to various organs, and eventually gets metabolized and excreted out of the body. A drug and/or its metabolites can cause toxicity to one or more organs.
- Cytochrome P450 (CYP450) enzymes
Drug metabolizing enzymes (oxidoreductases) in the liver with major human isoforms such as CYP1A2, 2C9, 2C19, 2D6, 2E1, and 3A4.
- Enterohepatic recirculation
The movement of bile acids, bilirubin, drugs, and other biochemicals between the liver and small intestine. The hepatocytes in the liver excrete molecules into the small intestine via the bile, and the enterocyte in the small intestine can send some or all of those same molecules back to the liver through the portal vein (i.e., recirculation).
- Enzyme-linked immunosorbent assay (ELISA)
A quantitative colorimetric method of quantifying a specific protein in a sample via the binding of antibody to the protein/antigen of interest.
- Glutathione (GSH)
An antioxidant that is synthesized in the body from amino acids L-cysteine, L-glutamic acid, and glycine, and can detoxify reactive compounds such as drug metabolites.
- Induced pluripotent stem cell-derived human hepatocytes (iPSC-HHs)
Somatic cells are first converted into induced pluripotent stem cells (iPSCs), which are then differentiated into hepatocyte-like cells via one of several in vitro protocols that mimic different phases of liver development.
- Lipopolysaccharide (LPS)
Biomolecule consisting of a lipid and polysaccharide that is a component of bacterial membranes and can trigger an inflammatory response in the liver and other organs.
- Nonparenchymal cells (NPCs)
Supporting cells that help a parenchymal (major) cell type of an organ/tissue execute its functions. In the liver, NPCs include liver sinusoidal endothelial cells (LSECs), Kupffer macrophages (KMs), hepatic stellate cells (HSCs), and cholangiocytes, which help support parenchymal cells called hepatocytes.
- Physiologically based pharmacokinetic (PBPK) modeling
The use of in silico methods for predicting the absorption, distribution, metabolism, and excretion of a drug throughout a patient's body. This technique is important for predicting key clinical parameters of a drug, such as half-life and maximum plasma concentration.
- Polydimethylsiloxane (PDMS)
A silicone-based polymer often used in microfluidic devices due to its optical clarity (for microscopy), chemical inertness, and biocompatibility (i.e. non-toxic to cells).
- Sensitivity
The portion of clinically toxic drugs correctly identified as such with an in vitro assay. High sensitivity in a preclinical assay ensures that ‘toxic’ drugs do not enter clinical trials.
- Specificity
The portion of clinically non-toxic drugs correctly identified as such with an in vitro assay. High specificity ensures that a non-toxic drug is not incorrectly eliminated from the drug development pipeline when it could be helping patients in clinical trials and the marketplace by being efficacious against the targeted disease.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Kaitin KI. Deconstructing the drug development process: the new face of innovation. Clin. Pharmacol. Ther. 2010;87:356–361. doi: 10.1038/clpt.2009.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mullard A. The phenotypic screening pendulum swings. Nat Rev Drug Discov. 2015;14:807–809. doi: 10.1038/nrd4783. [DOI] [PubMed] [Google Scholar]
- 3.Kola I, Landis J. Can the pharmaceutical industry reduce attrition rates? Nat Rev Drug Discov. 2004;3:711–715. doi: 10.1038/nrd1470. [DOI] [PubMed] [Google Scholar]
- 4.Kaplowitz N. Idiosyncratic drug hepatotoxicity. Nat Rev Drug Discov. 2005;4:489–499. doi: 10.1038/nrd1750. [DOI] [PubMed] [Google Scholar]
- 5.Olson H, et al. Concordance of the toxicity of pharmaceuticals in humans and in animals. Regul. Toxicol. Pharmacol. 2000;32:56–67. doi: 10.1006/rtph.2000.1399. [DOI] [PubMed] [Google Scholar]
- 6.Khetani SR, et al. Microengineered liver tissues for drug testing. J Lab Autom. 2015;20:216–250. doi: 10.1177/2211068214566939. [DOI] [PubMed] [Google Scholar]
- 7.Lin C, et al. The application of engineered liver tissues for novel drug discovery. Expert Opin Drug Discov. 2015;10:519–540. doi: 10.1517/17460441.2015.1032241. [DOI] [PubMed] [Google Scholar]
- 8.Xu JJ, et al. Cellular Imaging Predictions of Clinical Drug-Induced Liver Injury. Toxicological Sciences. 2008;105:97–105. doi: 10.1093/toxsci/kfn109. [DOI] [PubMed] [Google Scholar]
- 9.Huh D, et al. From 3D cell culture to organs-on-chips. Trends Cell Biol. 2011;21:745–754. doi: 10.1016/j.tcb.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wilkening S, et al. Comparison of primary human hepatocytes and hepatoma cell line Hepg2 with regard to their biotransformation properties. Drug Metabolism and Disposition. 2003;31:1035–1042. doi: 10.1124/dmd.31.8.1035. [DOI] [PubMed] [Google Scholar]
- 11.Kwon SJ, et al. High-throughput and combinatorial gene expression on a chip for metabolism-induced toxicology screening. Nat Commun. 2014;5:3739. doi: 10.1038/ncomms4739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee DW, et al. Application of the DataChip/MetaChip technology for the evaluation of ajoene toxicity in vitro. Arch Toxicol. 2014;88:283–290. doi: 10.1007/s00204-013-1102-9. [DOI] [PubMed] [Google Scholar]
- 13.Messner S, et al. Multi-cell type human liver microtissues for hepatotoxicity testing. Arch Toxicol. 2013;87:209–213. doi: 10.1007/s00204-012-0968-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miyamoto Y, et al. Spheroid Formation and Evaluation of Hepatic Cells in a Three-Dimensional Culture Device. Cell Med. 2015;8:47–56. doi: 10.3727/215517915X689056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fukuda J, Nakazawa K. Hepatocyte spheroid arrays inside microwells connected with microchannels. Biomicrofluidics. 2011;5:22205. doi: 10.1063/1.3576905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Trask OJ, Jr, et al. A Micropatterned Hepatocyte Cocucolture Model for Assessment of Liver Toxicity Using High-Content Imaging Analysis. Assay Drug Dev Technol. 2014;12:16–27. doi: 10.1089/adt.2013.525. [DOI] [PubMed] [Google Scholar]
- 17.Khetani SR, Bhatia SN. Microscale culture of human liver cells for drug development. Nat Biotechnol. 2008;26:120–126. doi: 10.1038/nbt1361. [DOI] [PubMed] [Google Scholar]
- 18.Wang WW, et al. Assessment of a Micropatterned Hepatocyte Coculture System to Generate Major Human Excretory and Circulating Drug Metabolites. Drug Metabolism and Disposition. 2010;38:1900–1905. doi: 10.1124/dmd.110.034876. [DOI] [PubMed] [Google Scholar]
- 19.Lin C, et al. Prediction of Drug Clearance and Drug-Drug Interactions in Microscale Cultures of Human Hepatocytes. Drug Metab Dispos. 2016;44:127–136. doi: 10.1124/dmd.115.066027. [DOI] [PubMed] [Google Scholar]
- 20.Prueksaritanont T, et al. Drug-drug interaction studies: regulatory guidance and an industry perspective. AAPS J. 2013;15:629–645. doi: 10.1208/s12248-013-9470-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khetani SR, et al. Use of micropatterned cocultures to detect compounds that cause drug-induced liver injury in humans. Toxicol Sci. 2013;132:107–117. doi: 10.1093/toxsci/kfs326. [DOI] [PubMed] [Google Scholar]
- 22.Shlomai A, et al. Modeling host interactions with hepatitis B virus using primary and induced pluripotent stem cell-derived hepatocellular systems. Proceedings of the National Academy of Sciences. 2014;111:12193–12198. doi: 10.1073/pnas.1412631111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ploss A, et al. Persistent hepatitis C virus infection in microscale primary human hepatocyte cultures. Proceedings of the National Academy of Sciences. 2010;107:3141–3145. doi: 10.1073/pnas.0915130107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.March S, et al. A microscale human liver platform that supports the hepatic stages of Plasmodium falciparum and vivax. Cell Host Microbe. 2013;14:104–115. doi: 10.1016/j.chom.2013.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Davidson MD, et al. Hormone and Drug-Mediated Modulation of Glucose Metabolism in a Microscale Model of the Human Liver. Tissue Eng Part C Methods. 2015;21:716–725. doi: 10.1089/ten.tec.2014.0512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schwartz RE, et al. Pluripotent stem cell-derived hepatocyte-like cells. Biotechnol. Adv. 2014;32:504–513. doi: 10.1016/j.biotechadv.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Davidson MD, et al. Stem cell-derived liver cells for drug testing and disease modeling. Discov Med. 2015;19:349–358. [PMC free article] [PubMed] [Google Scholar]
- 28.Takayama K, et al. 3D spheroid culture of hESC/hiPSC-derived hepatocyte-like cells for drug toxicity testing. Biomaterials. 2013;34:1781–1789. doi: 10.1016/j.biomaterials.2012.11.029. [DOI] [PubMed] [Google Scholar]
- 29.Berger DR, et al. Enhancing the functional maturity of induced pluripotent stem cell-derived human hepatocytes by controlled presentation of cell-cell interactions in vitro. Hepatology. 2015;61:1370–1381. doi: 10.1002/hep.27621. [DOI] [PubMed] [Google Scholar]
- 30.Ware BR, et al. Prediction of Drug-Induced Liver Injury in Micropatterned Cocultures Containing iPSC-Derived Human Hepatocytes. Toxicol Sci. 2015;145:252–262. doi: 10.1093/toxsci/kfv048. [DOI] [PubMed] [Google Scholar]
- 31.LeCluyse EL, et al. Organotypic liver culture models: meeting current challenges in toxicity testing. Crit. Rev. Toxicol. 2012;42:501–548. doi: 10.3109/10408444.2012.682115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nguyen TV, et al. Establishment of a hepatocyte-kupffer cell coculture model for assessment of proinflammatory cytokine effects on metabolizing enzymes and drug transporters. Drug Metab Dispos. 2015;43:774–785. doi: 10.1124/dmd.114.061317. [DOI] [PubMed] [Google Scholar]
- 33.Kostadinova R, et al. A long-term three dimensional liver co-culture system for improved prediction of clinically relevant drug-induced hepatotoxicity. Toxicol Appl Pharmacol. 2013;268:1–16. doi: 10.1016/j.taap.2013.01.012. [DOI] [PubMed] [Google Scholar]
- 34.Ma X, et al. Deterministically patterned biomimetic human iPSC-derived hepatic model via rapid 3D bioprinting. Proceedings of the National Academy of Sciences. 2016;113:2206–2211. doi: 10.1073/pnas.1524510113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sivaraman A, et al. A microscale in vitro physiological model of the liver: predictive screens for drug metabolism and enzyme induction. Curr Drug Metab. 2005;6:569–591. doi: 10.2174/138920005774832632. [DOI] [PubMed] [Google Scholar]
- 36.Domansky K, et al. Perfused multiwell plate for 3D liver tissue engineering. Lab Chip. 2010;10:51–58. doi: 10.1039/b913221j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Toh Y-C, et al. A microfluidic 3D hepatocyte chip for drug toxicity testing. Lab Chip. 2009;9:2026–2035. doi: 10.1039/b900912d. [DOI] [PubMed] [Google Scholar]
- 38.Shih M-C, et al. A microfluidic device mimicking acinar concentration gradients across the liver acinus. Biomed Microdevices. 2013;15:767–780. doi: 10.1007/s10544-013-9762-z. [DOI] [PubMed] [Google Scholar]
- 39.Legendre A, et al. Metabolic characterization of primary rat hepatocytes cultivated in parallel microfluidic biochips. J Pharm Sci. 2013;102:3264–3276. doi: 10.1002/jps.23466. [DOI] [PubMed] [Google Scholar]
- 40.Baudoin R, et al. Evaluation of a liver microfluidic biochip to predict in vivo clearances of seven drugs in rats. J Pharm Sci. 2014;103:706–718. doi: 10.1002/jps.23796. [DOI] [PubMed] [Google Scholar]
- 41.Prot JM, et al. A cocktail of metabolic probes demonstrates the relevance of primary human hepatocyte cultures in a microfluidic biochip for pharmaceutical drug screening. Int J Pharm. 2011;408:67–75. doi: 10.1016/j.ijpharm.2011.01.054. [DOI] [PubMed] [Google Scholar]
- 42.Baudoin R, et al. Evaluation of seven drug metabolisms and clearances by cryopreserved human primary hepatocytes cultivated in microfluidic biochips. Xenobiotica. 2013;43:140–152. doi: 10.3109/00498254.2012.706725. [DOI] [PubMed] [Google Scholar]
- 43.Leclerc E, et al. Integration of pharmacokinetic and NRF2 system biology models to describe reactive oxygen species production and subsequent glutathione depletion in liver microfluidic biochips after flutamide exposure. Toxicol In Vitro. 2014;28:1230–1241. doi: 10.1016/j.tiv.2014.05.003. [DOI] [PubMed] [Google Scholar]
- 44.Choucha-Snouber L, et al. Metabolomics-on-a-chip of hepatotoxicity induced by anticancer drug flutamide and Its active metabolite hydroxyflutamide using HepG2/C3a microfluidic biochips. Toxicol Sci. 2013;132:8–20. doi: 10.1093/toxsci/kfs230. [DOI] [PubMed] [Google Scholar]
- 45.Chao P, et al. Evaluation of a microfluidic based cell culture platform with primary human hepatocytes for the prediction of hepatic clearance in human. Biochem Pharmacol. 2009;78:625–632. doi: 10.1016/j.bcp.2009.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Novik E, et al. A microfluidic hepatic coculture platform for cell-based drug metabolism studies. Biochem Pharmacol. 2010;79:1036–1044. doi: 10.1016/j.bcp.2009.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim J-Y, et al. 96-well format-based microfluidic platform for parallel interconnection of multiple multicellular spheroids. J Lab Autom. 2015;20:274–282. doi: 10.1177/2211068214564056. [DOI] [PubMed] [Google Scholar]
- 48.Frey O, et al. Reconfigurable microfluidic hanging drop network for multi-tissue interaction and analysis. Nat Commun. 2014;5:4250. doi: 10.1038/ncomms5250. [DOI] [PubMed] [Google Scholar]
- 49.Kang YBA, et al. Liver sinusoid on a chip: Long-term layered co-culture of primary rat hepatocytes and endothelial cells in microfluidic platforms. Biotechnol Bioeng. 2015;112:2571–2582. doi: 10.1002/bit.25659. [DOI] [PubMed] [Google Scholar]
- 50.Kasuya J, et al. Spatio-temporal control of hepatic stellate cell-endothelial cell interactions for reconstruction of liver sinusoids in vitro. Tissue Eng Part A. 2012;18:1045–1056. doi: 10.1089/ten.TEA.2011.0351. [DOI] [PubMed] [Google Scholar]
- 51.Vernetti LA, et al. A human liver microphysiology platform for investigating physiology, drug safety, and disease models. Exp. Biol. Med. (Maywood) 2016;241:101–114. doi: 10.1177/1535370215592121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Prodanov L, et al. Long-term maintenance of a microfluidic 3D human liver sinusoid. Biotechnol Bioeng. 2016;113:241–246. doi: 10.1002/bit.25700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lee PJ, et al. An artificial liver sinusoid with a microfluidic endothelial-like barrier for primary hepatocyte culture. Biotechnol Bioeng. 2007;97:1340–1346. doi: 10.1002/bit.21360. [DOI] [PubMed] [Google Scholar]
- 54.Nakao Y, et al. Bile canaliculi formation by aligning rat primary hepatocytes in a microfluidic device. Biomicrofluidics. 2011;5:22212. doi: 10.1063/1.3580753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kmieć Z. Cooperation of liver cells in health and disease. Adv Anat Embryol Cell Biol. 2001;161:III–XIII. 1–151. doi: 10.1007/978-3-642-56553-3. [DOI] [PubMed] [Google Scholar]
- 56.Bavli D, et al. Real-time monitoring of metabolic function in liver-on-chip microdevices tracks the dynamics of mitochondrial dysfunction. Proceedings of the National Academy of Sciences. 2016;113:E2231–40. doi: 10.1073/pnas.1522556113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rennert K, et al. A microfluidically perfused three dimensional human liver model. Biomaterials. 2015;71:119–131. doi: 10.1016/j.biomaterials.2015.08.043. [DOI] [PubMed] [Google Scholar]
- 58.Matharu Z, et al. Detecting transforming growth factor-β release from liver cells using an aptasensor integrated with microfluidics. Anal Chem. 2014;86:8865–8872. doi: 10.1021/ac502383e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhou Q, et al. Liver injury-on-a-chip: microfluidic co-cultures with integrated biosensors for monitoring liver cell signaling during injury. Lab Chip. 2015;15:4467–4478. doi: 10.1039/c5lc00874c. [DOI] [PubMed] [Google Scholar]
- 60.Anundi I, et al. Zonation of acetaminophen metabolism and cytochrome P450 2E1-mediated toxicity studied in isolated periportal and perivenous hepatocytes. Biochem Pharmacol. 1993;45:1251–1259. doi: 10.1016/0006-2952(93)90277-4. [DOI] [PubMed] [Google Scholar]
- 61.Allen JW, et al. In vitro zonation and toxicity in a hepatocyte bioreactor. Toxicol Sci. 2005;84:110–119. doi: 10.1093/toxsci/kfi052. [DOI] [PubMed] [Google Scholar]
- 62.Sato A, et al. An in vitro hepatic zonation model with a continuous oxygen gradient in a microdevice. Biochem Biophys Res Commun. 2014;453:767–771. doi: 10.1016/j.bbrc.2014.10.017. [DOI] [PubMed] [Google Scholar]
- 63.Peng C-C, et al. A microfluidic cell culture array with various oxygen tensions. Lab Chip. 2013;13:3239–3245. doi: 10.1039/c3lc50388g. [DOI] [PubMed] [Google Scholar]
- 64.Viravaidya K, et al. Development of a microscale cell culture analog to probe naphthalene toxicity. Biotechnol. Prog. 2004;20:316–323. doi: 10.1021/bp0341996. [DOI] [PubMed] [Google Scholar]
- 65.Esch MB, et al. Body-on-a-chip simulation with gastrointestinal tract and liver tissues suggests that ingested nanoparticles have the potential to cause liver injury. Lab Chip. 2014;14:3081–3092. doi: 10.1039/c4lc00371c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bricks T, et al. Development of a new microfluidic platform integrating co-cultures of intestinal and liver cell lines. Toxicol In Vitro. 2014;28:885–895. doi: 10.1016/j.tiv.2014.02.005. [DOI] [PubMed] [Google Scholar]
- 67.Choucha-Snouber L, et al. Investigation of ifosfamide nephrotoxicity induced in a liver-kidney co-culture biochip. Biotechnol Bioeng. 2013;110:597–608. doi: 10.1002/bit.24707. [DOI] [PubMed] [Google Scholar]
- 68.Materne E-M, et al. A multi-organ chip co-culture of neurospheres and liver equivalents for long-term substance testing. J. Biotechnol. 2015;205:36–46. doi: 10.1016/j.jbiotec.2015.02.002. [DOI] [PubMed] [Google Scholar]
- 69.Zervantonakis IK, et al. Three-dimensional microfluidic model for tumor cell intravasation and endothelial barrier function. Proceedings of the National Academy of Sciences. 2012;109:13515–13520. doi: 10.1073/pnas.1210182109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wagner I, et al. A dynamic multi-organ-chip for long-term cultivation and substance testing proven by 3D human liver and skin tissue co-culture. Lab Chip. 2013;13:3538–3547. doi: 10.1039/c3lc50234a. [DOI] [PubMed] [Google Scholar]
- 71.Materne E-M, et al. The multi-organ chip--a microfluidic platform for long-term multi-tissue coculture. J Vis Exp. 2015 doi: 10.3791/52526. DOI: 10.3791/52526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kimura H, et al. An on-chip small intestine-liver model for pharmacokinetic studies. J Lab Autom. 2015;20:265–273. doi: 10.1177/2211068214557812. [DOI] [PubMed] [Google Scholar]
- 73.Sung JH, et al. A microfluidic device for a pharmacokinetic-pharmacodynamic (PKPD) model on a chip. Lab Chip. 2010;10:446–455. doi: 10.1039/b917763a. [DOI] [PubMed] [Google Scholar]
- 74.Maschmeyer I, et al. A four-organ-chip for interconnected long-term co-culture of human intestine, liver, skin and kidney equivalents. Lab Chip. 2015;15:2688–2699. doi: 10.1039/c5lc00392j. [DOI] [PubMed] [Google Scholar]
- 75.Oleaga C, et al. Multi-Organ toxicity demonstration in a functional human in vitro system composed of four organs. Sci Rep. 2016;6:20030. doi: 10.1038/srep20030. [DOI] [PMC free article] [PubMed] [Google Scholar]