Abstract
Resistance to targeted therapeutics is a key issue limiting the long-term utility of these medications in the management of molecularly selected subsets of cancer patients, including patients with non-small cell lung cancer harboring oncogenic alterations affecting EGFR, ALK and other genes. Bypass resistance mediated by activation of MET kinase has emerged as a frequent, validated and pivotal resistance mechanism in multiple types of cancers. Biochemical understanding is accumulating to explain the unique role of MET in such bypass pathways, providing alternate downstream activation opportunities and intricate interactions during epithelial-mesenchymal transitions. Multiple diagnostic testing platforms have become available for selecting appropriate patients for MET targeting in a variety of settings. Importantly, in light of the failures of several earlier clinical studies of MET targeting agents, a large array of recent and current MET-focused trials are incorporating stricter patient selection and more robust predictive biomarkers providing hope for validation of MET targeting as a clinically impactful strategy.
Keywords: MET, EGFR, lung cancer, targeted therapeutics, tyrosine kinase, resistance
Introduction
The current review focuses on the role of the MET proto-oncogene receptor tyrosine kinase (MET)/hepatocyte growth factor (HGF) pathway as a key participant in resistance mechanisms for targeted therapies. We will first introduce the paradigm of resistance mechanisms through the case of epidermal growth factor receptor (EGFR) inhibition in EGFR-mutated lung adenocarcinoma, providing a model for many key aspects that are integral to overcoming drug resistance. Then, the involvement of MET signaling pathways in targeted therapy resistance in a variety of settings will be addressed and biochemical and cell-level mechanisms of the MET pathway will be highlighted. Lastly, we will discuss emerging biomarkers for patient selection as well as provide a survey of relevant ongoing and completed clinical studies focusing on MET inhibition in the context of targeted therapeutic resistance.
Primary (de novo) and acquired resistance to targeted therapy
With rapid advancements in detecting genomic alterations, it is increasingly possible to select specific targeted therapies a priori for patients whose tumors have a high probability of responding to initial treatment, often by identifying molecularly defined subsets associated with oncogene addicted tumors. Prominent examples of such include BCR-ABL1 positive chronic myeloid leukemia, KIT-mutated gastrointestinal stromal tumors and EGFR-mutated, anaplastic lymphoma receptor tyrosine kinase (ALK) gene rearrangement-positive, and ROS proto-oncogene 1 (ROS1) gene rearrangement-positive non-small cell lung cancers (NSCLC). However, even in patients whose tumors dramatically and deeply respond to a rationally chosen targeted therapy, acquired resistance develops in nearly all patients, invariably so in advanced lung cancer with actionable mutations (1). For a given targeted therapy, tumors that do not respond to initial treatment are defined as having primary, or de novo, resistance. Of the subset of tumors that do respond to initial treatment, tumors that eventually stop responding to targeted therapy are said to have acquired resistance. It remains an urgent priority of molecular research to address the difficult challenge of overcoming primary and acquired resistance in order to achieve the goal of durable clinical responses with targeted therapy approaches in oncogene addicted lung cancer and other malignancies.
Acquired resistance via EGFR-dependent pathways in EGFR-mutated lung cancer
In the realm of thoracic malignancies, the great promise and success of targeted therapy and the potential options of overcoming acquired resistance are most developed in EGFR-mutated lung cancer. Mutations in the EGFR gene are present in approximately 10–20% of all NSCLCs with higher incidence in females, non-smokers, younger patients, and adenocarcinoma histology, occurring as frequently as 30–50% in never-smokers with lung adenocarcinoma (2). Since the introduction of small molecule adenosine triphosphate (ATP)-mimetic EGFR tyrosine kinase inhibitors (TKI) and the subsequent discovery of recurrent and actionable EGFR mutations defining a subset of advanced NSCLC patients with dramatic and durable responses to these drugs, research in the EGFR inhibitor space has increased exponentially. While up to 60–80% of patients with EGFR-mutated tumors initially respond to first- and second-generation EGFR TKIs with significant improvement in both response rates and progression-free survival compared to upfront chemotherapy, resistance typically develops after 8–14 months of treatment (3).
Over the past decade, extensive research has identified a multitude of primary and acquired resistance mechanisms. One type of common genomic alteration involves mutations that prevent targeted therapies from effectively inhibiting their respective molecular targets. A prominent example of this involves point mutations resulting in a threonine-to-methionine amino acid substitution at the gatekeeper position 790 of EGFR, which confers resistance to first-generation EGFR TKIs (4). This EGFR T790M mutation has also rarely been reported as a germline alteration leading to a high risk of lung cancer (5). It is also the most common mechanism of acquired resistance developing after treatment with first-generation EGFR TKIs, with 50–65% of tumors harboring such alterations at the time of disease progression (6). Interestingly, recent research suggests that certain tumors may harbor minute subclones with the T790M mutation, while in other tumors the mutation develops de novo. These different evolutionary paths that tumors can take depend on the presence or absence of T790M at disease onset (7). Novel third-generation T790M-directed EGFR inhibitors have demonstrated tremendous success in the clinic for T790M-mediated resistance and one lead compound, osimertinib, is now approved in this setting (8). Examples of similar second site mutations are numerous in other actionable kinases (9-12) and in these cases, cancer cells appear to maintain their original dominant dependence on the integrity of the native oncogenic pathway for continued cell proliferation and survival, principally via activation of cyclin D-dependent proliferative pathways and downregulation of the key pro-apoptotic protein, Bim (13,14).
MET pathway activation as a bypass resistance mechanism
Bypass resistance refers to a type of genomic alteration that results in the activation of alternative ligands and/or receptors that sustain signaling of key downstream pathways, despite successful inhibition of the specific molecular target of a given targeted therapy. The first established model of such bypass pathway activation involves the MET pathway, including MET amplification and overexpression of the MET receptor’s sole cognate ligand hepatocyte growth factor (HGF), which acts as a multi-functional cytokine primarily on cells of epithelial origin (15).
The MET oncogene
MET, which belongs to the MET/RON family, is a proto-oncogene located on chromosome 7q31.2 that encodes for a receptor tyrosine kinase (RTK) (16,17). The binding of HGF to the MET receptor favors its dimerization and its autophosphorylation on two tyrosine residues in its catalytic domain, Y1234 and Y1235. Subsequently, other tyrosine residues are phosphorylated, including Y1003 in the juxtamembrane domain and Y1349 and Y1356 at the C-terminus tail, leading to the avid binding of many effectors. In particular, MET-related phosphorylation and activation of Gab1-mediated downstream pathways involving SHP2, p85-PI3K, PLCγ, and the adapter protein, Crk are more protracted than EGFR-related Gab1 phosphorylation and activation. These events principally result in downstream signaling through the RAS-RAF-MEK-ERK pathway and the PI3K-AKT-mTOR pathway, playing an important role in cellular proliferation and cell survival (18). In addition, MET can associate with many cell membrane proteins including integrins, CD44v6 isoform, plexin type receptors and it can also interact with other RTKs such as EGFR. Even in the absence of HGF, MET can be transactivated by EGFR and simultaneous activation of MET and EGFR is synergistic. In malignant cells, aberrant signaling through the MET receptor promotes cellular invasion, migration, angiogenesis, and eventual metastasis (19,20).
Since it was first discovered in 1984, mutations in the MET gene have been directly associated with papillary renal cell carcinoma and MET aberrations frequently occur in hepatocellular carcinoma as well as head and neck squamous cell carcinomas (21). With regards to NSCLC, MET overexpression, high MET gene copy number, MET gene amplification, and high HGF levels due to overexpression via transcriptional upregulation, activating gene mutations, and alternative splicing have all been significantly associated with negative prognosis (18). MET is also intimately implicated in the processes of tissue remodeling and morphogenic differentiation within the context of transient epithelial-mesenchymal transition (EMT), which is characterized by loss of epithelial differentiation, cell dispersal, cell migration, and degradation of the epithelial matrix (22). In MET-driven cancer, the loss of tight regulation of these events leads to invasion and metastasis. Invasion can also be driven by hypoxia, which induces HGF and MET expression via HIF-1α, rendering cells more sensitive to further HGF stimulation and MET overexpression (23,24).
Initially, the search for biomarkers defining cancers with MET pathway activation focused on MET overexpression, which is frequent and readily testable using routine assays such as immunohistochemistry (IHC). However, it does appear that genetic alterations in the MET pathway provide more robust biomarkers for pathway activation and targeting. MET amplifications are found in 2–4% of untreated NSCLC tumors and are also found in other cancer types, such as gastric adenocarcinoma. Multiple case reports and case series suggest that a subset of MET high-amplified tumors can respond to MET inhibition. In addition, infrequent and recurrent MET gene mutations can occur in the semaphorin extracellular domain, juxtamembrane region, and the kinase domain (25). Mutations that involve the juxtamembrane domain, which is encoded by exons 14 and 15 and is necessary for MET receptor degradation via a critical tyrosine residue (Y1003), can cause tumorigenesis. More recently, recurrent mutations leading to MET exon 14 skipping have been reported to be the most common actionable MET alteration occurring in approximately 3–4% of NSCLCs (26,27) with a higher frequency reported in pulmonary sarcomatoid lung cancer, a rare, very aggressive and treatment-refractory subtype of lung cancer (28).
MET in EGFR TKI resistance
As for the role of the MET pathway in EGFR TKI resistance, the first report by Engelman and colleagues in 2007 provided experimental evidence that focal amplification of MET drives ErbB3-dependent activation of PI3K, conferring resistance to the first-generation EGFR TKI gefitinib (29). By exposing cell line models to increasing concentrations of gefitinib for extended periods of time, an in vitro model of drug resistance was developed. In this model, resistance to gefitinib could be overcome by combined treatment with gefitinib and a MET inhibitor but not with MET inhibitor monotherapy, suggesting a fundamental switch to co-dependence on the activity of both the EGFR pathway and MET pathway where the sustained downstream signaling of just one of these key kinase pathway is sufficient for cellular survival (29). In addition, MET overexpression caused by genomic amplification was noted in patient samples at the time of acquired resistance (29). Interestingly, in another preclinical study the converse situation has also been reported in which a MET-positive gastric carcinoma cell line became resistant to MET inhibitors through bypass activation of the EGFR pathway, resulting in sustained downstream signaling of the RAS-RAF-MEK-ERK and PI3K-AKT-mTOR pathways (30).
Initial reports suggested a fairly high frequency of MET amplification-mediated bypass resistance; however, this frequency was found to be substantially lower with the use of more precise detection techniques. For example, while an initial study noted MET amplification detected by IHC in 4 out of 18 (22%) lung cancer specimens from patients that had developed resistance to EGFR TKIs, a later study from the same group using stricter fluorescence in situ hybridization (FISH) criteria detected 4 out of 37 cases (11%) cases with genomic MET amplification (6). The thoracic oncology group at Memorial Sloan Kettering Cancer Center reported 9 of 43 patients (21%) of MET amplification in their series utilizing array comparative genomic hybridization (31); however, it was noted that only one case involved a high level gene amplification and three of the cases also harbored EGFR T790M. Lower prevalence, as low as 5%, has been reported with the use of more stringent criteria for defining MET amplification, including MET/CEP7 ratio by FISH (32). In summary, MET amplification is a recurrent and definable event occurring in approximately 5–10% of EGFR-mutated tumors that have acquired resistance to EGFR inhibitors, representing the second most common validated EGFR resistance alteration.
While it has been less carefully studied, it appears that MET activation can also serve as a primary resistance mechanism to EGFR-targeted therapy in EGFR-mutated lung cancer. The best evidence for this comes from the recent case report by Gainor and colleagues reporting on a 73-year-old never-smoker patient presenting with a malignant effusion from lung adenocarcinoma. Molecular testing demonstrated a classic EGFR L858R mutation as well as high level MET amplification of at least 30 copies, with a MET/CEP7 ratio greater than 15 in the pre-treatment diagnostic specimen (33). The patient was started on erlotinib therapy as per guideline recommendations, but rapidly progressed 4 weeks into therapy. Based on the patient’s MET amplification crizotinib was added, leading to a dramatic clinical and radiographic response. While the rate of de novo MET amplification is anticipated to be no more than 3%, cases like this argue for broader upfront mutation profiling in EGFR-mutated lung cancer.
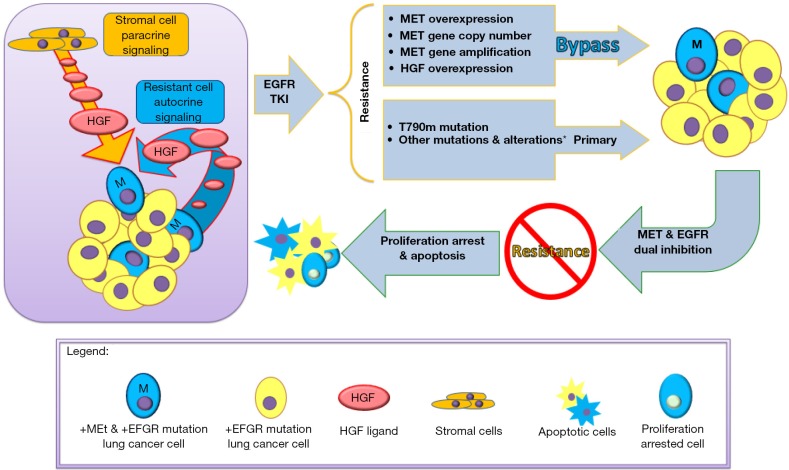
MET gene amplification can also act as a pre-existing resistance mechanism in certain cases (18). Using in vitro resistance models with a pan-ErbB kinase inhibitor, PF00299804, resistant clones of HCC827 were noted to recurrently carry MET amplifications, leading to MET overexpression-dependent EGFR TKI resistance that could be overcome by subsequent MET inhibition. All clones that emerged carried at least four-fold amplification of MET, and closer examination of the chromosome 7 amplicon suggested that all clones shared the same clonal origin. In addition, short-term HGF exposure rescued cells from gefitinib-induced death through Gab1 adaptor protein signaling instead of through an ErbB3-dependent mechanism. In contrast, long-term HGF exposure led to the emergence of MET-amplified clones, resulting in a more typical resistance pattern. High-throughput FISH studies demonstrated that parental HCC827 cells harbor minor subfractions of cells with MET copy gains (6 out of 4,237, 0.14%). HGF exposure alone did not lead to the emergence of MET-amplified clones; the presence of EGFR inhibition was required, which may provide a unique growth advantage to a subset of cells with MET overexpression. In human correlative studies, MET amplification was identified in 4 out of 27 (15%) post-treatment specimens with the use of an IHC assay. HGF expression by IHC was also higher in some post-treatment specimens. In all four MET-amplified specimens, rare tumor cells with MET amplifications could be identified in the pre-treatment specimens, while only 1 out of 8 post-treatment non-MET amplified specimens harbored cells with detectable MET amplifications. An interesting clinical observation demonstrating the waxing and waning of a MET amplified subclone in a patient dependent on EGFR TKI therapy corroborates these findings (34). Overall, these studies strongly suggest that specific mechanisms of drug resistance that develop as a result of drug exposure may be predetermined and occur as a result of drug selective pressure. In the case of MET pathway activation, resistance can emerge over time in the context of long-term drug exposure due to the presence of MET-amplified subclones and both paracrine and autocrine HGF stimulation, leading to enrichment and clonal selection (Figure 1).
Figure 1.
Schematic illustration of the emergence of drug resistance to EGFR tyrosine kinase inhibitors highlighting the role of autocrine and paracrine MET/HGF pathway alterations and the preexistence of subclonal MET amplified drug-resistant cell subsets.
MET in resistance to third-generation EGFR TKIs
More recently, EGFR T790M-directed third-generation EGFR inhibitors have demonstrated great success in the management of EGFR T790M-mediated resistance. Two lead compounds, rociletinib and osimertinib, showed excellent response rates in this context and osimertinib has been approved by the FDA for this indication (8); however, the development of rociletinib has been suspended since. In vitro studies with HCC827 cell line models demonstrated that MET activation via MET amplification can serve as a resistance mechanism not only to first-generation EGFR inhibitors but also to third-generation EGFR inhibitors (35). In these cell models, a combination of osimertinib and the MET/ALK TKI crizotinib overcame resistance, leading to enhanced Bim stability and augmentation of apoptosis; regaining sensitivity was closely correlated with full suppression of ErbB3 phosphorylation. Since acquired resistance uniformly develops with the use of third-generation EGFR inhibitors as well, case reports and case series are emerging identifying analogous mechanisms of secondary acquired resistance with novel EGFR mutations (C797S) that disallow covalent binding of third-generation EGFR inhibitors as well as bypass resistance via MET pathway activation (36).
For example, in a recent case report of an EGFR-mutated T790M lung cancer patient treated with the third-generation EGFR TKI osimertinib, post-treatment tumor biopsies revealed high levels (30 copies) of MET amplification. Interestingly, crizotinib provided transient symptomatic and clinical benefit, lending credence to the idea of investigating combined EGFR and MET inhibition (37). In a cohort of 43 patients on rociletinib, CAPP-Seq circulating tumor DNA (ctDNA) was utilized to monitor emerging resistance mechanisms and MET copy number gains were noted in 11 patients (26%) (38). High-level MET amplification was also demonstrated in correlative mouse xenograft studies upon rociletinib exposure that could be overcome with the addition of crizotinib.
HGF as paracrine mediator of resistance
HGF overexpression represents another MET pathway-related mechanism of resistance to EGFR inhibitors. Excessive autocrine signaling of HGF through overexpression activates MET in a Gab1 signaling-dependent manner and accelerates the development of MET amplification in vitro and in vivo, leading to resistance to gefitinib (39). This represents an important example of ligand-mediated drug resistance, with HGF overexpression having the ability to independently rescue both PI3K-AKT-mTOR and RAS-RAF-MEK-ERK signaling in the presence of first-generation EGFR tyrosine kinase inhibitors (40). Studies by Yano and colleagues demonstrated that in 23 EGFR-mutated NSCLCs with acquired resistance where post-resistance biopsy specimens were available, 14 had high levels of HGF expression (61%), 12 had EGFR T790M (52%), and two had MET amplification (9%) (41). As HGF is produced by lung cancer cells as well as stromal and fibroblast cells in the tumor microenvironment, resistance can be based on both autocrine and paracrine pathways and can synergize with MET amplification. HGF production by the stroma may also partially explain why clinical resistance might emerge preferentially in some organ sites and tissues. In addition, changes in serum HGF levels may be able to serve as predictive biomarkers of acquired resistance (42).
Other bypass signaling and alternative resistance mechanisms
Similar bypass signaling pathways have been repeatedly identified in preclinical, animal, and human studies, including the activation of ErbB2, AXL and IGF1R pathways; however, these mechanisms are not well validated yet in the clinic (43,44). Other known acquired resistance mechanisms include epithelial-mesenchymal transitions, which to some extent overlap with MET/AXL-activated tumor subsets, and rare transformations of NSCLC into small cell lung cancer which are sensitive to standard small cell lung cancer therapies, highlighting the need to repeatedly assess the genetics and histology of lung cancers throughout their clinical course (6). Additional mechanisms include different pharmacokinetic and pharmacodynamic examples, such as central nervous system (CNS) specific progression related to poor drug distribution of several agents into the CNS compartment. The field is further complicated by the fact that these resistance mechanisms can co-exist, shift and change in a temporally and spatially dynamic manner over the course of the disease dependent on multiple factors, most critically the relevant ongoing selection pressure of targeted therapies.
MET resistance mechanisms in other malignancies
EGFR inhibition in colorectal carcinomas
Multiple studies have demonstrated that MET amplification can occur in the setting of anti-EGFR monoclonal antibody therapy of advanced KRAS wild-type colorectal carcinoma (CRC). This setting differs from EGFR-mutated lung cancer in that the efficacy of anti-EGFR monoclonal antibodies in advanced CRC is limited, with progression usually occurring in 3–12 months. Additionally, there is no specific genetic defect being targeted in this setting. For example, Van Emburgh and colleagues reported that in vitro stimulation of MET by HGF led to resistance in CRC cells and xenografts to cetuximab and panitumumab and 3 out of 7 patients with acquired resistance to EGFR-directed therapy were noted to have MET amplification (45). Pharmacological silencing of the MET pathway in preclinical models led to reconstitution of sensitivity.
ErbB2-positive malignancies
Limited data also suggest potential involvement of MET-mediated resistance with ErbB2-directed therapies. In a study conducted Minuti and colleagues, high gene copy numbers of MET and HGF correlated with poor outcomes and resistance to trastuzumab in HER2-positive breast cancers (46). Shattuck and colleagues demonstrated that trastuzumab-resistant primary cell lines and primary tumors also exhibited elevated expression of MET and HGF (47). Lastly, in ErbB2-positive gastric cancer models, MET activation and downstream ERK and AKT activation has been demonstrated to lead to resistance to the dual EGFR/ErbB2 inhibitor, lapatinib (48). Physical association of MET and ErbB has also been demonstrated (49).
FGFR-driven cancers
In FGFR1-amplified lung cancer cell lines H1581 and DMS114, clones resistant to FGFR inhibitors AZD4547 and BAY116387 were established and RTK arrays showed overexpression and activation of MET with genomic MET amplification identified in some but not all clones (50). Resistance appeared to be mediated via ErbB3 activation. Combination therapy with crizotinib or MET siRNA re-sensitized cells while ectopic MET expression in H1581 cells led to resistance. Resistant clones also appeared to undergo epithelial-mesenchymal transitions with enhanced migration and invasion.
BRAF-mutated tumors
MET activation via stromal HGF secretions was identified by two groups as one of the alternate pathways associated with resistance to the BRAF inhibitor vemurafenib in melanoma (51,52). In addition, Krepler and colleagues reported on 12 patient-derived tumor xenograft (PDX) models established from BRAF-mutated melanomas upon progression on BRAF inhibitor therapy (53). Among other changes detected by next-generation sequencing (Foundation One), MET amplification (copy number ranging from 9–63) was noted in 3 PDX models, and increased phosphorylated MET signal was detected in at least one of the PDX models in which capmatinib MET inhibitor therapy demonstrated success. Lastly, in a recent study of patients with BRAF-mutated CRC who initially responded to combined EGFR/BRAF inhibition with panitumumab and vemurafenib, MET amplification was noted in specimens from resistant tumors and the corresponding pre-treatment specimens were found to harbor pre-existing cells with MET amplification, suggestive of clonal expansion after therapy exposure (54). In CRC cell line models, ectopic MET expression led to resistance to BRAF inhibition that could be overcome by combination therapy. One index patient with MET amplification on crizotinib and vemurafenib had a rapid and marked clinical response, confirming the clinical impact of these findings. In the post-treatment sample of this patient, MET IHC showed strong expression in 90% of cells while MET copy numbers by FISH ranged from 1–30 (mean 8.5) with an overall MET/CEP7 ratio of 2.62 with at least 75% of cells harboring MET amplification.
VEGF pathway targeting
In glioblastoma multiforme models, Jahangiri and colleagues conducted gene expression studies suggesting that in bevacizumab-resistant tumors, MET is one of the most overexpressed genes and that genetic ablation could reverse resistance and reduce tumor cell invasion and tumor cell survival (55). Other studies suggested that dual VEGFR and MET blockade reduces metastasis and improves survival in several preclinical models (56).
Overall, these various studies illustrate that activation of the MET/HGF axis is emerging as a key resistance mechanism in a large variety of clinical settings.
Biochemical mechanisms permitting MET to serve as a key bypass resistance kinase
It is important to note that MET kinase activation functions in a broad variety of settings as a bypass resistance mechanism. Physiologically, the binding of HGF to MET induces its activation, driving a complex biologic program of invasive growth from the promotion of cell proliferation and cell invasion, and protection from apoptosis (16). MET-driven invasive growth is part of a physiological program that occurs during embryonic development and during adulthood within the context of tissue regeneration. MET-mediated signaling results from pathways activated by the MET receptor but it can also be modulated by crosstalk between MET and different membrane receptors in complex interacting networks. The strength and duration of MET-induced signals are regulated by a network of co-receptors, such as adhesive receptors, death receptors, class B plexins, and other tyrosine kinase receptors that physically associate with MET. When it occurs, constitutive MET activation can contribute to several aspects of tumor progression by inducing neoplastic cells to disaggregate from the tumor mass, eroding basement membranes, infiltrating stromal matrices and eventually colonizing new tissues to form metastases (17).
The tumor microenvironment plays an essential role in sustaining resistance to targeted therapies as paracrine and autocrine tumor microenvironment-derived ligands can activate signals sufficient to overcome drug inhibition in tumor cells, especially in the case of HGF, the cognate ligand for MET. Gusenbauer and colleagues set out to investigate the underlying mechanism of HGF-induced EGFR TKI resistance in 12 distinct carcinoma cell lines derived from breast, kidney, liver and tongue (57). They demonstrated that HGF inhibits classical tyrosine kinase activity-dependent EGFR activation in a wide variety of carcinoma cell lines and induces EGFR to directly interact with and stabilize multiple potentially oncogenic proteins in multiprotein complexes, including the RTKs Axl and EphA2 as well as CUB domain-containing protein 1 (CDCP1), integrin β4, and JAK1; all these putative EGFR binding partners were confirmed to co-immunoprecipitate with EGFR by immunoblotting. Within this cell surface multiprotein complex, EGFR signaling no longer depends on its kinase function and downstream signaling activation is independent of ATP binding, preventing inhibition by small molecule EGFR TKI monotherapy. This unique ability of MET to engage other cell surface oncogenic kinases and to yield alternative ways for downstream activation of oncogenic pathways can explain many of the observations noted in the EGFR-mutated lung cancer setting, the association of drug resistance with epithelial-mesenchymal transitions, and the wide involvement of MET bypass signaling in multiple cancer types. In addition, pretreatment with the MET-directed TKI AMG-458 fully restored gefitinib sensitivity and blocked HGF-induced cell scattering, further highlighting the role of HGF paracrine signaling and the HGF/MET axis in EGFR TKI resistance.
To more comprehensively address the role of diverse cognate ligands for various oncogenic RTKs on TKI resistance, Harbinski and colleagues utilized high-throughput secretome screening in which secreted proteins collected from cell media supernatants of various cell lines transfected with a library of 3,432 cDNAs were assessed for their ability to rescue kinase-dependent cancer cells that were simultaneously treated with a corresponding kinase inhibitor (58). For example, HEK293T cells and MKN-45 cells were treated with JAA120, a MET inhibitor, and RT-112 cells were treated with BGJ398, a FGFR inhibitor. Interestingly, among multiple cytokines tested, HGF and FGF emerged as uniquely potent in rescuing cells from drug-induced apoptosis, including HGF for FGFR-sensitive cells and FGF and NRG2 for MET-sensitive cells, suggesting that kinase receptor bypass signaling is a broadly common mechanism of drug resistance. There was also simultaneous and synergistic activity of two RTKs observed, specifically MET and FGFR in combination. In a set of primary xenograft mouse models with MET and FGFR1 co-activation, the combination of capmatinib, an investigational MET inhibitor, and BGJ398, a pan FGFR kinase inhibitor, led to statistically significant and substantial tumor regression, suggesting that simultaneously blocking multiple distinct oncogenic kinase pathways is a rational approach for overcoming resistance to single agent targeted therapy.
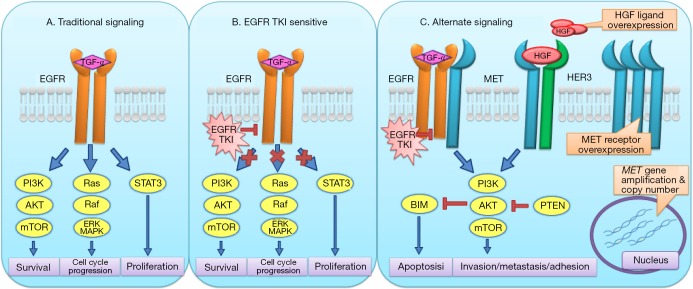
In summary, these data demonstrating involvement of MET in EMT-mediated resistance, potential to lead to RTK uncoupling via the generation of cell surface receptor aggregates and recognition as a key resistance signaling pathway in multiple model systems suggests a key role via unique mechanisms of broad plasticity, receptor engagement and alternative signaling for the MET/HGF axis in bypass resistance (Figure 2).
Figure 2.
Schematic illustration of the unique biochemical aspects of MET-mediated bypass signaling driven by cell surface RTK aggregates leading to de-coupling of EGFR oncogenic function from its tyrosine kinase activity (alternate signaling).
Diagnostic biomarkers for MET pathway activation/targeting
As the role of MET pathway activation in the development of acquired resistance to targeted therapies becomes more fully appreciated, there is an increasingly urgent need for validated diagnostic biomarkers to effectively detect MET pathway activation. Given the dramatic clinical responses seen in selected patients treated with MET-directed therapy, detection of actionable MET alterations needs to be seamlessly incorporated into the clinical management of advanced NSCLC for upfront treatment selection and monitoring for the development of acquired resistance. Multiple methods exist for the detection of MET expression, MET activation, gene copy number changes, and genomic alterations and may be able to serve as biomarkers for sensitivity to MET-targeted agents, facilitating prospective patient selection in the clinic (19). We will review the most relevant assays in the resistance setting below.
Immunohistochemistry (IHC)
While MET IHC is a readily available and easily adoptable assay, its utility for MET targeting has been questioned by a series of recent studies, including multiple negative clinical studies which will be reviewed in detail below. The reason for the poor performance of MET IHC to predict treatment benefit might be related to limited correlation with genetic markers, such as MET exon 14 skipping or MET amplification which have emerged as much more robust biomarkers (59).
Copy number assessment
For copy number changes, both fluorescence in situ hybridization (FISH) and real-time polymerase chain reaction (PCR) can detect increased copy numbers of MET reliably; however, real-time PCR lacks the ability to distinguish true genomic amplification from polysomy (18). Case reports suggest significant benefit of MET inhibition in tumors with high level MET gene amplification (60,61). Recent studies are starting to provide guidance for relevant MET FISH cutoffs to define patients with true sensitivity for MET inhibition. For example, in a study by Camidge and colleagues, 13 patients with MET-amplified NSCLC were treated with crizotinib and 6 patients out of the 13 had high level amplification (MET/CEP7 ratio greater than 5). For low, intermediate, and high MET/CEP7 ratios, objective response rates were 0%, 17%, and 67% respectively, suggesting that a MET/CEP7 ratio greater than or equal to 5 effectively identifies patients with high sensitivity for MET inhibition. Accordingly, the median duration of response was 15.7 weeks for patients with intermediate-level amplification compared to 73.6 weeks for patients with high-level amplification.
Next-generation sequencing (NGS)
NGS has become a very powerful diagnostic tool in our diagnostic armamentarium for MET gene alterations, especially given the broad spectrum of mutations affecting the 5’ and 3’ splice sites of the MET gene that are hard to capture using other methodologies (62,63). NGS has the advantage of providing comprehensive information on mutations, insertions/deletions, copy number changes, and gene fusions. Given the wide range of possible MET genomic alterations, NGS should be viewed as the leading and preferred diagnostic method currently.
Plasma circulating tumor DNA (ctDNA) testing
ctDNA testing has become a valuable tool with validated utility for the detection of oncogenic and resistant EGFR mutations. Several test platforms also allow for NGS-based detection of multiple targets, including MET mutations and amplifications, and several ongoing studies have incorporated plasma ctDNA testing-based biomarkers for patient selection (38,64-66). While ctDNA testing has good sensitivity for point mutations, such as MET exon 14 skipping variants, it has less sensitivity for copy number changes. However, for high level amplifications, ctDNA testing might still be sufficient for relatively reliable detection and further studies in this rapidly developing area are eagerly awaited.
Other biomarkers
As shown in multiple preclinical models, activation of MET can be assessed by the phosphorylation status of tyrosine residues in the kinase domain and/or the C-terminus tail. In pre- and post-treatment biopsies, changes in Met phosphorylation were observed in response to multiple distinct MET tyrosine kinase inhibitors, including tivantinib. Additionally, the phosphorylation of downstream effectors such as Gab1 and ERK can be used as surrogate pharmacodynamic indicators. Plasma biomarkers also hold promise, including HGF and shed-Met (shedding of the Met extracellular domain), VEGF, and soluble VEGFR2, though more prospective clinical trials are needed for validation (16).
Pre-treatment biopsies will be a top priority for prospective clinical trials for MET-directed therapies, as archived biopsies are less likely to capture critical information about MET status after treatment with drugs directed at other molecular targets (67). Although frequent biopsies, including pre- and post-treatment biopsies, are difficult to obtain in the context of clinical studies, they will be necessary to direct more rationally chosen targeted approaches (16). ctDNA testing will provide a convenient compliment to such biomarker assays.
Clinical studies assessing MET inhibition to potentiate the efficacy of targeted therapy
Prior experience
Despite tremendous excitement about the utility of targeting MET for the treatment of NSCLC in a variety of settings, initial efforts were hampered by a series of major setbacks. Reviewing these failures provides context for the challenges of clinical study designs in this area. For example, there was a great deal of optimism for onartuzumab, a monovalent monoclonal antibody designed to specifically target the MET receptor, based on intriguing results of a preceding prominent phase II trial (68). This randomized phase II study demonstrated improved progression-free survival (2.9 vs. 1.5 months; HR =0.53; P=0.04) and overall survival (12.6 vs. 3.8 months; HR =0.37; P=0.002) for the onartuzumab plus erlotinib arm compared to the erlotinib plus placebo arm in the subset of patients with IHC-confirmed MET-positive non-small cell lung cancer patients (69). Unfortunately, the subsequent phase III METLung trial failed to show clinically meaningful efficacy for IHC-confirmed MET-positive patients and further development of onartuzumab in this setting was halted. However, key shortcomings of this study included a molecularly unselected patient population where erlotinib is now understood to have very little clinical utility and the use of a poor biomarker, IHC for MET targeting, without consideration of more robust genetic biomarkers in treatment selection.
Tivantinib, another novel MET receptor TKI, was featured in a phase III multinational, randomized, double-blinded, and placebo-controlled study in patients with previously treated locally advanced or metastatic NSCLC. This study was also founded on encouraging randomized phase II study data (70). The tivantinib plus erlotinib combination modestly improved progression-free survival compared to the placebo plus erlotinib arm, but did not improve overall survival in the nonsquamous NSCLC population (71). Post-hoc subgroup analyses suggested an improvement in overall survival in patients with high MET expression, but statistical significance was not reached and further development of tivantinib in this setting is not currently being pursued. Similar to the phase II onartuzumab study, the lack of biomarker selection for EGFR targeting might have been a major flaw in retrospect and questions have been raised as to whether tivantinib is actually a clinically relevant MET inhibitor.
MET TKI studies
Learning from these drastic failures, more recent studies are much more focused on careful patient selection, enrolling only patients with EGFR-mutated NSCLCs with carefully defined acquired resistance to primary therapy. However, these scientifically cleaner studies suffer from much smaller patient numbers that are relevant for accrual based on stringent selection and the significant tissue needs for study eligibility. Currently, there are a multitude of clinical studies assessing MET inhibition with approved tyrosine kinase inhibitors with activity against MET such as crizotinib and cabozantinib, and with novel MET tyrosine kinase inhibitors. We have highlighted a few select studies below and have provided a more comprehensive list in Table 1.
Table 1. Summary of ongoing studies with MET inhibitors in the drug resistance setting.
| Study ID | Experimental drug | Drug class | Treatment combination | Phase | Patient population | Biomarker selection targets |
|---|---|---|---|---|---|---|
| NCT02468661 | Capmatinib | MET TKI | Alone or with Erlotinib | II | Advanced EGFR-mutated NSCLC | EGFRm+, MET GCN >6 |
| NCT02205398 | Capmatinib | MET TKI | Cetuximab | I/II | Advanced Squamous Cell Carcinoma of Head and Neck, Metastatic Colorectal Cancer | K-Ras/N-Ras- CRC, cMet-positive (combination of IHC and FISH) |
| NCT01982955 | Tepotinib (MSC2156119J) | MET TKI | Gefitinib | I/II | Advanced EGFR mutation+ NSCLC, failed prior EGFR TKI | EGFRm+, (ph II limited to MET IHC+, T790M-) |
| NCT02143466 | Volitinib (AZD6094) | MET TKI | AZD9291 | I/II | Advanced EGFR-mutation+ NSCLC, failed prior EGFR TKI | EGFRm+ (MET+ and T790M- for ph II) |
| NCT02260531 | Cabozantinib | Multi-targeted TKI | Trastuzumab (if ErbB2+) | II | Breast cancer, metastatic brain disease | No biomarker selection |
| NCT01644773 | Crizotinib | Multi-targeted TKI | Dasatinib | I | Diffuse intrinsic pontine glioma, high-grade glioma | No biomarker selection |
| NCT01900652 | LY2875358 | Anti-MET monoclonal antibody | Alone or with erlotinib | II | Advanced EGFR-mutation+ NSCLC, failed prior EGFR TKI | MET IHC+ |
| NCT02318368 | Ficlatuzumab | Anti-HGF monoclonal antibody | Erlotinib | II | Advanced EGFR-mutation+ NSCLC, EGFR TKI-naive | EGFRm+ and BDX004 positive |
Emerging clinical data suggest that MET TKIs in combination with an EGFR TKI may have a favorable benefit-risk ratio for the treatment of cMET-amplified, EGFR T790M negative advanced NSCLC with acquired resistance to prior EGFR TKI. INC280 (capmatinib) is an orally bioavailable, highly potent and selective cMET inhibitor (IC50 =0.13 nM) with greater than 10,000-fold selectivity for MET compared to other tested human kinases (72). Preliminary clinical activity of INC280 in combination with gefitinib was observed in adult patients with locally advanced or metastatic EGFR-mutated, cMET-dysregulated NSCLC who have progressed after EGFR TKI treatment. In particular, patients with a cMET gene copy number greater than or equal to 5 demonstrated an overall response rate of 40% (8/20) and 7 of these responders had a GCN greater than 6 (73). Based on these promising preliminary data, a randomized three-arm phase II study has been launched to compare INC280 versus erlotinib plus INC280 versus standard doublet chemotherapy in patients with EGFR-mutated NSCLC (NCT02468661). Key selection criteria include MET GCN greater than 6 for study entry.
A combination of INC280 and cetuximab is also being explored in cMET-positive metastatic colorectal cancer and head and neck squamous cell carcinoma following progression on prior EGFR monoclonal antibody therapy with cetuximab or panitumumab (NCT02205398). This phase IB dose escalation study will assess the toxicity and then efficacy of the combination at the expanded MTD/RDE dose level. Selection criteria include cMET positivity as defined by IHC and FISH (similar to ErbB2 scoring criteria) as well as KRAS and NRAS wild type status for CRC patients.
Another ongoing study in this area assesses the safety and utility of the highly potent and selective MET TKI inhibitor, tepotinib (MSC2156119J) along with gefitinib in patients with MET-positive advanced EGFR-mutated NSCLC who failed prior gefitinib (NCT01982955). The phase IB portion of the study was presented at ASCO 2016 demonstrating good tolerability (no DLTs noted in two dose levels) and encouraging initial activity (4/18 partial responses) of the combination (74) and the recommended phase II dose of tepotinib at 500 mg PO once daily with gefitinib at 250 mg PO once daily is now being studied in the randomized phase II portion limited to MET-positive and EGFR T790M-negative patients with cisplatin/pemetrexed chemotherapy serving as the control arm. c-Met positivity (2+ or 3+) in this study is determined by IHC using an anti-c-Met monoclonal antibody (SP44; Ventana/Roche).
Another very potent and selective MET inhibitor in active development is volitinib (HMPL-504), an orally bioavailable and potent small molecule ATP-competitive inhibitor with single nanomolar activity and at least 200-fold selectivity for MET kinase inhibition over 247 tested kinases. One ongoing study in China (NCT02374645) is assessing the safety of the combination of gefitinib and volitinib in advanced EGFR-mutated NSCLC. The TATTON clinical study (NCT02143466) is exploring different combinations of the T790M-targeting EGFR inhibitor osimertinib in different combinations, including volitinib, in patients with EGFR-mutated NSCLC upon progression on prior EGFR TKI therapy. In a rolling phase IB dose escalation design, the combination of osimertinib and volitinib appeared to have encouraging activity and was escalated to phase 2 following treatment of 12 patients at two dose levels of volitinib (600 and 800 mg PO once daily) with a set dose of osimertinib of 80 mg PO once daily (75). Partial responses were reported in 6 of 11 evaluable patients with 4 of the responses confirmed, including a major response in a 32-year-old female with an EGFR exon 19 deletion positive NSCLC with documented high MET-amplification positive disease progression.
A phase I study of glesatinib (MGCD265), a potent and highly selective tyrosine kinase inhibitor with predominant activity against MET and AXL has been completed (76). Subsequent phase Ib data revealed clinical activity with durable confirmed partial responses in 3 out of 11 NSCLC patients and tumor regressions in 10 out of 11 NSCLC patients, but bioavailability and tolerability issues prompted the use of a new formulation of glesatinib for the currently ongoing phase II trial (NCT02544633). One of the reported responses to single agent MGCD265 involved a patient with a MET amplification positive, EGFR-mutated advanced NSCLC.
A study focusing on breast cancer patients with CNS metastases is assessing the utility of MET inhibition with cabozantinib in this setting alone or in combination with trastuzumab in ErbB2-positive patients (NCT02260531), utilizing a range of correlative biomarkers including levels and changes in serum MET as well as MET and phosphorylated MET by IHC in available archival tissues (NCT02260531).
Another intriguing study is exploring the combination of PDGFRA and MET inhibition in pediatric brain tumors based on frequent genetic alterations affecting both pathways as well as recent data highlighting genetic heterogeneity in clonal subpopulations suggestive of synergistic value to such combinations. This phase I study assessing the combination of crizotinib and dasatinib found that the combination was too toxic at twice daily dosing and now is accruing patients at once daily dosing cohorts (NCT01644773).
Anti-MET monoclonal antibodies
Another relevant agent being developed in this space is the humanized, bivalent, IgG4 monoclonal antibody LY2875358. It is unique by not exhibiting any functional agonist activity, and it potently blocks HGF binding to MET and HGF-induced MET phosphorylation and cell proliferation. In addition, it also induces MET receptor internalization and degradation and has shown activity in both HGF-dependent and HGF-independent (such as MET-amplified) model systems (77). Initial studies showed good tolerability of this antibody and a randomized phase II study that recently completed accrual will assess single agent LY2875358 and a combination of erlotinib and LY2875358 in patients with advanced EGFR-mutated NSCLC with MET biomarker positive tumors upon progression on prior EGFR TKI therapy (NCT01900652). Results presented in abstract form suggest limited activity of single-agent anti-MET antibody therapy or the combination for the overall population of MET diagnostic positive patients, defined by MET IHC positivity. Since close to 90% of enrolled patients were found to be positive by MET IHC, further results are awaited in more rigorously defined MET-positive patient subsets (78).
Anti-HGF monoclonal antibodies
Ficlatuzumab (AV-299) is a potent, high affinity and selective HGF-inhibitory antibody developed to inhibit HGF/MET-driven biological activities (79). A phase II study of ficlatuzumab and erlotinib versus placebo and erlotinib (FOCAL study, NCT02318368) is ongoing to assess the value of combined inhibition in patients with EGFR-mutated, EGFR TKI-naïve advanced NSCLC. One key eligibility criteria is positivity for the serum proteomic marker, BDX004. Similar to Veristrat, this defines a patient population with poor prognosis.
Summary
Overall, the emerging data puts the HGF/MET signaling axis front and center as a leading example of bypass resistance to a broad spectrum of targeted therapeutics in a variety of clinical settings. Activation of MET signaling represents a unique set of resistance mechanisms to targeted therapies because it can occur through multiple and varied molecular events including genomic amplification and ligand overexpression, leading to rapid evolution of drug resistance. It also provides a paradigm of clonal evolution and selection. The ability of overexpressed MET to form cell surface tyrosine kinase aggregates is a key mechanism for its role in EMT as well as drug resistance via bypass and alternative signaling pathways. In the case of EGFR-mutated lung adenocarcinoma, a wide range of models from preclinical studies to well-annotated case reports suggest true actionability of MET amplification as a resistance mechanism. MET amplification detected by a variety of platforms might be the leading biomarker for ongoing EGFR-mutated NSCLC studies. A wide range of clinical studies exploring small molecule as well as monoclonal antibody MET inhibition approaches in diverse clinical settings should be greatly supported by our larger oncology community to enable us to fully validate MET inhibition in these clinical contexts.
Acknowledgements
None.
Footnotes
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- 1.Chong CR, Janne PA. The quest to overcome resistance to EGFR-targeted therapies in cancer. Nat Med 2013;19:1389-400. 10.1038/nm.3388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.D'Angelo SP, Pietanza MC, Johnson ML, et al. Incidence of EGFR exon 19 deletions and L858R in tumor specimens from men and cigarette smokers with lung adenocarcinomas. J Clin Oncol 2011;29:2066-70. 10.1200/JCO.2010.32.6181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Camidge DR, Pao W, Sequist LV. Acquired resistance to TKIs in solid tumours: learning from lung cancer. Nat Rev Clin Oncol 2014;11:473-81. 10.1038/nrclinonc.2014.104 [DOI] [PubMed] [Google Scholar]
- 4.Kobayashi S, Boggon TJ, Dayaram T, et al. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N Engl J Med 2005;352:786-92. 10.1056/NEJMoa044238 [DOI] [PubMed] [Google Scholar]
- 5.Bell DW, Gore I, Okimoto RA, et al. Inherited susceptibility to lung cancer may be associated with the T790M drug resistance mutation in EGFR. Nat Genet 2005;37:1315-6. 10.1038/ng1671 [DOI] [PubMed] [Google Scholar]
- 6.Sequist LV, Waltman BA, Dias-Santagata D, et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med 2011;3:75ra26. 10.1126/scitranslmed.3002003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hata AN, Niederst MJ, Archibald HL, et al. Tumor cells can follow distinct evolutionary paths to become resistant to epidermal growth factor receptor inhibition. Nat Med 2016;22:262-9. 10.1038/nm.4040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jänne PA, Yang JC, Kim DW, et al. AZD9291 in EGFR inhibitor-resistant non-small-cell lung cancer. N Engl J Med 2015;372:1689-99. 10.1056/NEJMoa1411817 [DOI] [PubMed] [Google Scholar]
- 9.Gainor JF, Dardaei L, Yoda S, et al. Molecular Mechanisms of Resistance to First- and Second-Generation ALK Inhibitors in ALK-Rearranged Lung Cancer. Cancer Discov 2016;6:1118-33. 10.1158/2159-8290.CD-16-0596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Katayama R, Shaw AT, Khan TM, et al. Mechanisms of acquired crizotinib resistance in ALK-rearranged lung Cancers. Sci Transl Med 2012;4:120ra17. 10.1126/scitranslmed.3003316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bauer S, Joensuu H. Emerging Agents for the Treatment of Advanced, Imatinib-Resistant Gastrointestinal Stromal Tumors: Current Status and Future Directions. Drugs 2015;75:1323-34. 10.1007/s40265-015-0440-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O'Hare T, Deininger MW, Eide CA, et al. Targeting the BCR-ABL signaling pathway in therapy-resistant Philadelphia chromosome-positive leukemia. Clin Cancer Res 2011;17:212-21. 10.1158/1078-0432.CCR-09-3314 [DOI] [PubMed] [Google Scholar]
- 13.Costa DB, Halmos B, Kumar A, et al. BIM mediates EGFR tyrosine kinase inhibitor-induced apoptosis in lung cancers with oncogenic EGFR mutations. PLoS Med 2007;4:1669-79; discussion 1680. [DOI] [PMC free article] [PubMed]
- 14.Kobayashi S, Shimamura T, Monti S, et al. Transcriptional profiling identifies cyclin D1 as a critical downstream effector of mutant epidermal growth factor receptor signaling. Cancer Res 2006;66:11389-98. 10.1158/0008-5472.CAN-06-2318 [DOI] [PubMed] [Google Scholar]
- 15.Corso S, Giordano S. Cell-autonomous and non-cell-autonomous mechanisms of HGF/MET-driven resistance to targeted therapies: from basic research to a clinical perspective. Cancer Discov 2013;3:978-92. 10.1158/2159-8290.CD-13-0040 [DOI] [PubMed] [Google Scholar]
- 16.Maroun CR, Rowlands T. The Met receptor tyrosine kinase: a key player in oncogenesis and drug resistance. Pharmacol Ther 2014;142:316-38. 10.1016/j.pharmthera.2013.12.014 [DOI] [PubMed] [Google Scholar]
- 17.Ma PC, Maulik G, Christensen J, et al. c-Met: structure, functions and potential for therapeutic inhibition. Cancer Metastasis Rev 2003;22:309-25. 10.1023/A:1023768811842 [DOI] [PubMed] [Google Scholar]
- 18.Gelsomino F, Facchinetti F, Haspinger ER, et al. Targeting the MET gene for the treatment of non-small-cell lung cancer. Crit Rev Oncol Hematol 2014;89:284-99. 10.1016/j.critrevonc.2013.11.006 [DOI] [PubMed] [Google Scholar]
- 19.Furlan A, Kherrouche Z, Montagne R, Copin MC, Tulasne D. Thirty years of research on met receptor to move a biomarker from bench to bedside. Cancer Res 2014;74:6737-44. 10.1158/0008-5472.CAN-14-1932 [DOI] [PubMed] [Google Scholar]
- 20.Christensen JG, Burrows J, Salgia R. c-Met as a target for human cancer and characterization of inhibitors for therapeutic intervention. Cancer Lett 2005;225:1-26. 10.1016/j.canlet.2004.09.044 [DOI] [PubMed] [Google Scholar]
- 21.Ma PC, Tretiakova MS, MacKinnon AC, Ramnath N, Johnson C, Dietrich S, et al. Expression and mutational analysis of MET in human solid cancers. Genes Chromosomes Cancer 2008;47:1025-37. 10.1002/gcc.20604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rastogi I, Rajanna S, Webb A, et al. Mechanism of c-Met and EGFR tyrosine kinase inhibitor resistance through epithelial mesenchymal transition in non-small cell lung cancer. Biochem Biophys Res Commun 2016;477:937-44. 10.1016/j.bbrc.2016.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kitajima Y, Ide T, Ohtsuka T, et al. Induction of hepatocyte growth factor activator gene expression under hypoxia activates the hepatocyte growth factor/c-Met system via hypoxia inducible factor-1 in pancreatic cancer. Cancer Sci 2008;99:1341-7. 10.1111/j.1349-7006.2008.00828.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pennacchietti S, Michieli P, Galluzzo M, et al. Hypoxia promotes invasive growth by transcriptional activation of the met protooncogene. Cancer Cell 2003;3:347-61. 10.1016/S1535-6108(03)00085-0 [DOI] [PubMed] [Google Scholar]
- 25.Ma PC, Jagadeeswaran R, Jagadeesh S, et al. Functional expression and mutations of c-Met and its therapeutic inhibition with SU11274 and small interfering RNA in non-small cell lung cancer. Cancer Res 2005;65:1479-88. 10.1158/0008-5472.CAN-04-2650 [DOI] [PubMed] [Google Scholar]
- 26.Awad MM, Oxnard GR, Jackman DM, et al. MET Exon 14 Mutations in Non-Small-Cell Lung Cancer Are Associated With Advanced Age and Stage-Dependent MET Genomic Amplification and c-Met Overexpression. J Clin Oncol 2016;34:721-30. 10.1200/JCO.2015.63.4600 [DOI] [PubMed] [Google Scholar]
- 27.Paik PK, Drilon A, Fan PD, et al. Response to MET inhibitors in patients with stage IV lung adenocarcinomas harboring MET mutations causing exon 14 skipping. Cancer Discov 2015;5:842-9. 10.1158/2159-8290.CD-14-1467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu X, Jia Y, Stoopler MB, et al. Next-Generation Sequencing of Pulmonary Sarcomatoid Carcinoma Reveals High Frequency of Actionable MET Gene Mutations. J Clin Oncol 2016;34:794-802. 10.1200/JCO.2015.62.0674 [DOI] [PubMed] [Google Scholar]
- 29.Engelman JA, Zejnullahu K, Mitsudomi T, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science 2007;316:1039-43. 10.1126/science.1141478 [DOI] [PubMed] [Google Scholar]
- 30.Qi J, McTigue MA, Rogers A, et al. Multiple mutations and bypass mechanisms can contribute to development of acquired resistance to MET inhibitors. Cancer Res 2011;71:1081-91. 10.1158/0008-5472.CAN-10-1623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bean J, Brennan C, Shih JY, et al. MET amplification occurs with or without T790M mutations in EGFR mutant lung tumors with acquired resistance to gefitinib or erlotinib. Proc Natl Acad Sci U S A 2007;104:20932-7. 10.1073/pnas.0710370104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu HA, Arcila ME, Rekhtman N, et al. Analysis of tumor specimens at the time of acquired resistance to EGFR-TKI therapy in 155 patients with EGFR-mutant lung cancers. Clin Cancer Res 2013;19:2240-7. 10.1158/1078-0432.CCR-12-2246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gainor JF, Niederst MJ, Lennerz JK, et al. Dramatic Response to Combination Erlotinib and Crizotinib in a Patient with Advanced, EGFR-Mutant Lung Cancer Harboring De Novo MET Amplification. J Thorac Oncol 2016;11:e83-5. 10.1016/j.jtho.2016.02.021 [DOI] [PubMed] [Google Scholar]
- 34.Womack JP, Varella-Garcia M, Camidge DR. Waxing and Waning of MET Amplification in EGFR-Mutated NSCLC in Response to the Presence and Absence of Erlotinib Selection Pressure. J Thorac Oncol 2015;10:e115-8. 10.1097/JTO.0000000000000642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shi P, Oh YT, Zhang G, et al. Met gene amplification and protein hyperactivation is a mechanism of resistance to both first and third generation EGFR inhibitors in lung cancer treatment. Cancer Lett 2016;380:494-504. 10.1016/j.canlet.2016.07.021 [DOI] [PubMed] [Google Scholar]
- 36.Thress KS, Paweletz CP, Felip E, et al. Acquired EGFR C797S mutation mediates resistance to AZD9291 in non-small cell lung cancer harboring EGFR T790M. Nat Med 2015;21:560-2. 10.1038/nm.3854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ou SH, Agarwal N, Ali SM. High MET amplification level as a resistance mechanism to osimertinib (AZD9291) in a patient that symptomatically responded to crizotinib treatment post-osimertinib progression. Lung Cancer 2016;98:59-61. 10.1016/j.lungcan.2016.05.015 [DOI] [PubMed] [Google Scholar]
- 38.Chabon JJ, Simmons AD, Lovejoy AF, et al. Circulating tumour DNA profiling reveals heterogeneity of EGFR inhibitor resistance mechanisms in lung cancer patients. Nat Commun 2016;7:11815. 10.1038/ncomms11815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Turke AB, Zejnullahu K, Wu YL, et al. Preexistence and clonal selection of MET amplification in EGFR mutant NSCLC. Cancer Cell 2010;17:77-88. 10.1016/j.ccr.2009.11.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Donev IS, Wang W, Yamada T, et al. Transient PI3K inhibition induces apoptosis and overcomes HGF-mediated resistance to EGFR-TKIs in EGFR mutant lung cancer. Clin Cancer Res 2011;17:2260-9. 10.1158/1078-0432.CCR-10-1993 [DOI] [PubMed] [Google Scholar]
- 41.Yano S, Wang W, Li Q, et al. Hepatocyte growth factor induces gefitinib resistance of lung adenocarcinoma with epidermal growth factor receptor-activating mutations. Cancer Res 2008;68:9479-87. 10.1158/0008-5472.CAN-08-1643 [DOI] [PubMed] [Google Scholar]
- 42.Tanaka H, Kimura T, Kudoh S, et al. Reaction of plasma hepatocyte growth factor levels in non-small cell lung cancer patients treated with EGFR-TKIs. Int J Cancer 2011;129:1410-6. 10.1002/ijc.25799 [DOI] [PubMed] [Google Scholar]
- 43.Zhang Z, Lee JC, Lin L, et al. Activation of the AXL kinase causes resistance to EGFR-targeted therapy in lung cancer. Nat Genet 2012;44:852-60. 10.1038/ng.2330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Takezawa K, Pirazzoli V, Arcila ME, et al. HER2 amplification: a potential mechanism of acquired resistance to EGFR inhibition in EGFR-mutant lung cancers that lack the second-site EGFRT790M mutation. Cancer Discov 2012;2:922-33. 10.1158/2159-8290.CD-12-0108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Van Emburgh BO, Sartore-Bianchi A, Di Nicolantonio F, et al. Acquired resistance to EGFR-targeted therapies in colorectal cancer. Mol Oncol 2014;8:1084-94. 10.1016/j.molonc.2014.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Minuti G, Cappuzzo F, Duchnowska R, et al. Increased MET and HGF gene copy numbers are associated with trastuzumab failure in HER2-positive metastatic breast cancer. Br J Cancer 2012;107:793-9. 10.1038/bjc.2012.335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shattuck DL, Miller JK, Carraway KL, 3rd, et al. Met receptor contributes to trastuzumab resistance of Her2-overexpressing breast cancer cells. Cancer Res 2008;68:1471-7. 10.1158/0008-5472.CAN-07-5962 [DOI] [PubMed] [Google Scholar]
- 48.Chen CT, Kim H, Liska D, et al. MET activation mediates resistance to lapatinib inhibition of HER2-amplified gastric cancer cells. Mol Cancer Ther 2012;11:660-9. 10.1158/1535-7163.MCT-11-0754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Agarwal S, Zerillo C, Kolmakova J, et al. Association of constitutively activated hepatocyte growth factor receptor (Met) with resistance to a dual EGFR/Her2 inhibitor in non-small-cell lung cancer cells. Br J Cancer 2009;100:941-9. 10.1038/sj.bjc.6604937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim SM, Kim H, Yun MR, et al. Activation of the Met kinase confers acquired drug resistance in FGFR-targeted lung cancer therapy. Oncogenesis 2016;5:e241. 10.1038/oncsis.2016.48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wilson TR, Fridlyand J, Yan Y, et al. Widespread potential for growth-factor-driven resistance to anticancer kinase inhibitors. Nature 2012;487:505-9. 10.1038/nature11249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Straussman R, Morikawa T, Shee K, et al. Tumour micro-environment elicits innate resistance to RAF inhibitors through HGF secretion. Nature 2012;487:500-4. 10.1038/nature11183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Krepler C, Xiao M, Sproesser K, et al. Personalized Preclinical Trials in BRAF Inhibitor-Resistant Patient-Derived Xenograft Models Identify Second-Line Combination Therapies. Clin Cancer Res 2016;22:1592-602. 10.1158/1078-0432.CCR-15-1762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pietrantonio F, Oddo D, Gloghini A, et al. MET-Driven Resistance to Dual EGFR and BRAF Blockade May Be Overcome by Switching from EGFR to MET Inhibition in BRAF-Mutated Colorectal Cancer. Cancer Discov 2016;6:963-71. 10.1158/2159-8290.CD-16-0297 [DOI] [PubMed] [Google Scholar]
- 55.Jahangiri A, De Lay M, Miller LM, et al. Gene expression profile identifies tyrosine kinase c-Met as a targetable mediator of antiangiogenic therapy resistance. Clin Cancer Res 2013;19:1773-83. 10.1158/1078-0432.CCR-12-1281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nakagawa T, Matsushima T, Kawano S, et al. Lenvatinib in combination with golvatinib overcomes hepatocyte growth factor pathway-induced resistance to vascular endothelial growth factor receptor inhibitor. Cancer Sci 2014;105:723-30. 10.1111/cas.12409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gusenbauer S, Vlaicu P, Ullrich A. HGF induces novel EGFR functions involved in resistance formation to tyrosine kinase inhibitors. Oncogene 2013;32:3846-56. 10.1038/onc.2012.396 [DOI] [PubMed] [Google Scholar]
- 58.Harbinski F, Craig VJ, Sanghavi S, et al. Rescue screens with secreted proteins reveal compensatory potential of receptor tyrosine kinases in driving cancer growth. Cancer Discov 2012;2:948-59. 10.1158/2159-8290.CD-12-0237 [DOI] [PubMed] [Google Scholar]
- 59.Borczuk A, Paucar D, Halmos B. Has MET met its match? Ann Transl Med 2016;4:97. 10.21037/atm.2016.01.15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ou SH, Kwak EL, Siwak-Tapp C, et al. Activity of crizotinib (PF02341066), a dual mesenchymal-epithelial transition (MET) and anaplastic lymphoma kinase (ALK) inhibitor, in a non-small cell lung cancer patient with de novo MET amplification. J Thorac Oncol 2011;6:942-6. 10.1097/JTO.0b013e31821528d3 [DOI] [PubMed] [Google Scholar]
- 61.Schwab R, Petak I, Kollar M, et al. Major partial response to crizotinib, a dual MET/ALK inhibitor, in a squamous cell lung (SCC) carcinoma patient with de novo c-MET amplification in the absence of ALK rearrangement. Lung Cancer 2014;83:109-11. 10.1016/j.lungcan.2013.10.006 [DOI] [PubMed] [Google Scholar]
- 62.Frampton GM, Ali SM, Rosenzweig M, et al. Activation of MET via diverse exon 14 splicing alterations occurs in multiple tumor types and confers clinical sensitivity to MET inhibitors. Cancer Discov 2015;5:850-9. 10.1158/2159-8290.CD-15-0285 [DOI] [PubMed] [Google Scholar]
- 63.Schrock AB, Frampton GM, Suh J, et al. Characterization of 298 Patients with Lung Cancer Harboring MET Exon 14 Skipping Alterations. J Thorac Oncol 2016;11:1493-502. 10.1016/j.jtho.2016.06.004 [DOI] [PubMed] [Google Scholar]
- 64.Schwaederle M, Husain H, Fanta PT, et al. Detection rate of actionable mutations in diverse cancers using a biopsy-free (blood) circulating tumor cell DNA assay. Oncotarget 2016;7:9707-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Villaflor V, Won B, Nagy R, et al. Biopsy-free circulating tumor DNA assay identifies actionable mutations in lung cancer. Oncotarget 2016. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sorber L, Zwaenepoel K, Deschoolmeester V, et al. Circulating cell-free nucleic acids and platelets as a liquid biopsy in the provision of personalized therapy for lung cancer patients. Lung Cancer 2016. pii: S0169-5002(16)30312-9. [DOI] [PubMed]
- 67.Ma PC. MET receptor juxtamembrane exon 14 alternative spliced variant: novel cancer genomic predictive biomarker. Cancer Discov 2015;5:802-5. 10.1158/2159-8290.CD-15-0769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Spigel DR, Edelman MJ, Mok T, et al. Treatment Rationale Study Design for the MetLung Trial: A Randomized, Double-Blind Phase III Study of Onartuzumab (MetMAb) in Combination With Erlotinib Versus Erlotinib Alone in Patients Who Have Received Standard Chemotherapy for Stage IIIB or IV Met-Positive Non-Small-Cell Lung Cancer. Clin Lung Cancer 2012;13:500-4. 10.1016/j.cllc.2012.05.009 [DOI] [PubMed] [Google Scholar]
- 69.Spigel DR, Ervin TJ, Ramlau RA, et al. Randomized phase II trial of Onartuzumab in combination with erlotinib in patients with advanced non-small-cell lung cancer. J Clin Oncol 2013;31:4105-14. 10.1200/JCO.2012.47.4189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sequist LV, von Pawel J, Garmey EG, et al. Randomized phase II study of erlotinib plus tivantinib versus erlotinib plus placebo in previously treated non-small-cell lung cancer. J Clin Oncol 2011;29:3307-15. 10.1200/JCO.2010.34.0570 [DOI] [PubMed] [Google Scholar]
- 71.Scagliotti G, von Pawel J, Novello S, et al. Phase III Multinational, Randomized, Double-Blind, Placebo-Controlled Study of Tivantinib (ARQ 197) Plus Erlotinib Versus Erlotinib Alone in Previously Treated Patients With Locally Advanced or Metastatic Nonsquamous Non-Small-Cell Lung Cancer. J Clin Oncol 2015;33:2667-74. 10.1200/JCO.2014.60.7317 [DOI] [PubMed] [Google Scholar]
- 72.Liu X, Wang Q, Yang G, et al. A novel kinase inhibitor, INCB28060, blocks c-MET-dependent signaling, neoplastic activities, and cross-talk with EGFR and HER-3. Clin Cancer Res 2011;17:7127-38. 10.1158/1078-0432.CCR-11-1157 [DOI] [PubMed] [Google Scholar]
- 73.Wu YL, Yang JC, Kim DW, et al. Safety and efficacy of INC280 in combination with gefitinib (gef) in patients with EGFR-mutated (mut), MET-positive NSCLC: A single-arm phase lb/ll study. J Clin Oncol 2014;35:abstr 8017.
- 74.Wu YL, Soo RA, Kim DW, et al. Tolerability, efficacy and recommended phase II dose (RP2D) of tepotinib plus gefitinib in Asian patients with c-Met-positive/EGFR-mutant NSCLC: Phase Ib data. J Clin Oncol 2016;34:abstr e20501.
- 75.Oxnard GR, Ramalingam SS, Ahn MJ, et al. Preliminary results of TATTON, a multi-arm phase Ib trial of AZD9291 combined with MEDI4736, AZD6094 or selumetinib in EGFR-mutant lung cancer. J Clin Oncol 2015;33:abstr 2509.
- 76.Kollmannsberger CK, Sharma S, Shapiro G, et al. Phase I study of receptor tyrosine kinase (RTK) inhibitor, MGCD265, in patients (pts) with advanced solid tumors. J Clin Oncol 2015;33:abstr 2589.
- 77.Yoh K, Doi T, Ohmatsu H, et al. A phase I dose-escalation study of LY2875358, a bivalent MET antibody, given as monotherapy or in combination with erlotinib or gefitinib in Japanese patients with advanced malignancies. Invest New Drugs 2016;34:584-95. 10.1007/s10637-016-0370-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Camidge DR, Moran T, Demedts I, et al. A randomized, open-label, phase 2 study of emibetuzumab plus erlotinib (LY+E) and emibetuzumab monotherapy (LY) in patients with acquired resistance to erlotinib and MET diagnostic positive (MET Dx+) metastatic NSCLC. J Clin Oncol 2016;34:abstr 9070.
- 79.Patnaik A, Weiss GJ, Papadopoulos KP, et al. Phase I ficlatuzumab monotherapy or with erlotinib for refractory advanced solid tumours and multiple myeloma. Br J Cancer 2014;111:272-80. 10.1038/bjc.2014.290 [DOI] [PMC free article] [PubMed] [Google Scholar]