Abstract
The BXD family of mice were generated by crossing and inbreeding ethanol-preferring C57BL/6J and ethanol-avoiding DBA/2J strains that differ greatly in genome sequence and other behaviors. This study evaluated variations in the level of voluntary ethanol intake in a cohort of 42 BXD strains and both progenitor strains using a model of alcohol dependence and relapse drinking. A total of 119 BXDs (85 males, 34 females) (n ~ 4 per genotype; 1/genotype/sex/group) were evaluated along with males from both progenitor strains (n = 14–15/genotype). Mice were evaluated for intake using limited access (2 hr/day) 2-bottle (15% v/v ethanol vs. water) model for 6 weeks (baseline intake). Each animal received 4 weekly cycles of chronic intermittent ethanol (CIE) vapor exposure (CIE group) or air control exposure (CTL group) (16 hr/day × 4 days) interleaved by 5-day drinking test cycles. Blood ethanol concentrations (BEC) ranged from 150–300 mg/dl across genotypes. Baseline intake varied greatly among cases—from ~0.8 to ~2.9 g/kg. As expected, CIE exposure induced a significant increase in ethanol drinking in C57BL/6J relative to baseline as well as air controls that remained relatively stable over the four test cycles. In contrast, DBA/2J cases did not show a significant increase in consumption. Heritability of variation in baseline consumption, calculated from C57BL/6J and DBA/2J strains is about 54% but this increases following treatment to 60–80%. As expected from the marked difference between progenitors, ethanol intake and level of escalation varied greatly among BXDs after exposure (~-1.3 to ~2.6 g/kg). Interestingly, the magnitude and direction of changes in ethanol intake did not relate to BEC values of the preceding CIE exposure cycle. Overall, these data indicate significant variation in consumption and even escalation, much of it under genetic control, following repeated CIE treatment.
Keywords: Ethanol dependence, voluntary ethanol intake, recombinant inbred BXD mice
Introduction
There is considerable evidence regarding genetic influence on alcohol use disorders (Cloninger, 1987; Dick & Foroud, 2002; Kendler, Aggen, Prescott, Crabbe, & Neale, 2012; Merikangas et al., 1998). Understanding the basis of individual differences in response to alcohol effects, tolerance or sensitivity, and risk for development of dependence are critical to better understand alcoholism and to implement preventive measures and develop appropriate treatment. For example, sensitivity to adverse (e.g., sedative) effects of alcohol intoxication can influence the amount of alcohol consumed and the risk of developing future alcohol abuse and dependence. Subjects with a positive family history for alcoholism have been found to show more subjective stimulation and less sedation that can also predict later alcohol problems (King, Houle, de Wit, Holdstock, & Schuster, 2002; Quinn & Fromme, 2011; Schuckit & Smith, 2000).
Animal models have been extremely valuable in advancing the study of the role of genetic factors in alcoholism. Some studies have used lines of rats or mice that, starting with outbred subjects, were created through selected breeding based on their alcohol (ethanol) preference (P, NP rats), level of intake (HAD, LAD rats; HAP, LAP mice), acute response to ethanol (FAST, SLOW mice), drinking to intoxication (HDID mice), ethanol withdrawal sensitivity (WSP, WSR mice), to name a few (Crabbe, Harris, & Koob, 2011; Crabbe, Phillips, & Belknap, 2010). Interestingly, in some cases the selection for a particular trait, for example high ethanol preference, also results in a change in other parameters that favor ethanol intake such as tolerance to alcohol’s sedative effects (Crabbe et al., 2010). This suggests that genetic influences for various alcohol-related phenotypes may be related, thereby providing clues about common mechanisms underlying increased risk for alcohol abuse (Crabbe et al., 2012; Crabbe et al., 2010; Cunningham et al., 1991; Metten et al., 1998; Risinger, Malott, Prather, Niehus, & Cunningham, 1994). An alternative approach is the direct manipulation of a gene or groups of genes of interest to evaluate its impact on alcohol intake (Crabbe et al., 2010; Crabbe, Phillips, Harris, Arends, & Koob, 2006).
The use of inbred strains of mice has also helped evaluate different genetic components of ethanol effects and intake. While C57BL/6J has been characterized as an ethanol preferring strain, the DBA/2J strain has been characterized as ethanol avoiding based on their voluntary ethanol intake levels (Belknap, Crabbe, & Young, 1993; Crabbe, Young, & Kosobud, 1983). These mouse strains also differ in other ethanol-related effects. For example, DBA/2J mice are more sensitive than C57BL/6J mice to behavioral and physiological manifestations of ethanol withdrawal (Metten et al., 1998). The BXD recombinant inbred strains were generated by inbreeding F2 generation subjects obtained by crossing ethanol-preferring C57BL/6J (B) and ethanol avoiding DBA/2J (D), The BXD progeny strain inherits stretches of DNA from either the B or D parent, and each strain has its own unique and fixed pattern of B and D genotypes across the entire genome. The BXDs were originally developed in the 1970s with around 30 RI lines and more recently expanded to ~ 150 RI strains (Peirce, Lu, Gu, Silver, & Williams, 2004; Taylor, 1978; Wang et al., 2016), and are now used to map and define gene variants that control a wide range of traits (Houtkooper et al., 2013; Wang et al., 2016). BXD strains have been evaluated along their progenitor strains for voluntary ethanol intake (Gill, Liu, & Deitrich, 1996; Phillips, Crabbe, Metten, & Belknap, 1994; Rodriguez, Plomin, Blizard, Jones, & McClearn, 1994, 1995), ethanol-induced conditioned place preference (Risinger & Cunningham, 1998), and acute response to ethanol and withdrawal from chronic ethanol exposure (Buck, Rademacher, Metten, & Crabbe, 2002; Metten et al., 1998; Philip et al., 2010; Phillips et al., 1994; Putman et al., 2016). Thus, this resource has been very valuable for examining genetic contributions to various pharmacological effects of ethanol and motivation effects of ethanol (Crabbe et al., 1983; Philip et al., 2010; Phillips et al., 1994).
Studies conducted with rodents have demonstrated that dependence results in escalation of voluntary ethanol drinking (Becker, 2013). In most studies, dependence was produced via chronic intermittent ethanol (CIE) and ethanol consumption was measured in the home cage. Using this approach, a mouse model of ethanol dependence and relapse drinking that involves repeated cycles of CIE exposure and demonstrates escalation of voluntary ethanol drinking has been developed using male C57BL/6J mice (Becker & Lopez, 2004; Griffin, Lopez, & Becker, 2009; Lopez & Becker, 2005). This escalation of drinking produced more than a 2-fold increase in blood ethanol levels and brain ethanol concentrations (Becker & Lopez, 2004; Griffin, Lopez, Yanke, Middaugh, & Becker, 2009). Further, increased consumption appears specific to ethanol because CIE exposure did not alter intake of sucrose or saccharin (Becker & Lopez, 2004; Lopez, Griffin, Melendez, & Becker, 2012). CIE exposed mice also show tolerance to ethanol’s aversive effects (Lopez et al., 2012) and are less sensitive to devaluation of ethanol’s reinforcing effect (Lopez, Becker, & Chandler, 2014), which can help maintaining higher levels of voluntary ethanol intake.
Few studies have examined genetic factors that may influence this CIE-induced escalation of voluntary ethanol intake. Indeed, most of the above mentioned studies were conducted using the C57BL/6J strain. The study presented here is a first attempt to explore the influence of genetic background on ethanol consumption both prior to and during the course of CIE exposure. Specifically, a panel of BXD strains were used to evaluate baseline ethanol intake and changes in intake after repeated cycles of CIE (or air control) exposure under a limited access free-choice drinking paradigm. The study also included male C57BL/6J mice that served as a positive control, since this model of ethanol dependence and relapse drinking was developed using these mice. This initial foray into the genetics of CIE escalation was designed in collaboration with researchers of the NIAAA-funded INIA-Stress consortium to generate not only critical phenotype data (ethanol intake, blood ethanol levels during CIE exposure), but also to provide tissue to generate endocrine, neurochemical and genetic/genomic parameters for these BXD strains.
We have intentionally limited resampling within single strains to a bare minimum (usually two cases per strain) in the interest of screening larger numbers of diverse genotypes. This is the polar extreme of studying only one or two strains (C57BL/6J and DBA/2J) in great depth. Our approach provides a good view of the range of variation across a genetically diverse family but of course, does not provide precise estimate of strain averages. In contrast, the analysis of a single strain provides much more accurate data for one genotype, but results will often not generalize well. In this study, we generally do not make claims about phenotypes of individual BXD stains, but we can compute heritability of the consequences of CIE using the well replicated data from the progenitor strains and we can also estimate general effects of sex on CIE-associated traits.
Methods
Subjects
A total of 119 mice (85 males and 34 females) were used in this study. Mice representing 42 BXD genotypes, D2B6F1 hybrids (DBA/2J females crossed with C57BL/6J males) as well as both progenitor strains (C57BL/6 and DBA/2) were included in the design of the study. All of these mice were obtained from University of Tennessee (UT) and were 12–16 weeks old upon arrival. The mice were kept in quarantine for 3 weeks before use. Adult (10 weeks old upon arrival) C57BL/6J and DBA/2J stock obtained from The Jackson Laboratory (Bar Harbor, ME) served as positive and negative progenitor control (N = 8/group; see Table 1). The general study design typically involved the use of 4–8 more cases per genotype, and 1–2 cases per experimental cell as defined by genotype, sex, and group (CIE, CTL). This design does not allow us to accurately define means per strain and treatment (except for the progenitors). We used data from both progenitor strains and both sexes to estimate heritability under precisely the same conditions used to treat all BXD cases. It is reasonable to assume the heritabilities computed from contrast of the progenitors will also apply to their inbred progeny. Mice were individually housed with free access to food (Harland Teklad, Madison, WI) and water throughout all phases of the experiments. Body weights were recorded weekly during ethanol drinking weeks or daily during chronic intermittent ethanol (CIE) or air exposure (detailed below). Mice were housed in a temperature and humidity-controlled animal facility under a reversed 12-hr light/dark cycle (lights on at 0200 hr). Mice were not food or water deprived at any time during the study. All procedures were approved by the Institutional Animal Care and Use Committee and followed the NIH Guide for the Care and Use of Laboratory Animals (8th edition, National Research Council, 2011).
Table 1.
Left: Number of mice per strain and sex that were included in the study, were evaluated for baseline ethanol intake, and were later distributed in CIE and CTL conditions. Right: Number of CIE mice distributed based on the last CIE cycle completed. Whenever a CIE-exposed subject was lost, the corresponding CTL mouse from the same strain and sex was removed from the analysis as well.
| Last CIE exposure cycle | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Strain | Total N | Males | Females | CTL | CIE | 1 | 2 | 3 | 4 | 5 | |
| 1 | BXD5 | 2 | 2 | 0 | 1 | 1 | 1 | ||||
| 2 | BXD12 | 2 | 2 | 0 | 1 | 1 | 1 | ||||
| 3 | BXD14 | 3 | 3 | 0 | 1 | 2 | 1 | 1 | |||
| 4 | BXD16 | 2 | 2 | 0 | 1 | 1 | 1 | ||||
| 5 | BXD19 | 2 | 2 | 0 | 1 | 1 | 1 | ||||
| 6 | BXD24 | 2 | 2 | 0 | 1 | 1 | 1 | ||||
| 7 | BXD32 | 2 | 2 | 0 | 1 | 1 | 1 | ||||
| 8 | BXD34 | 2 | 2 | 0 | 1 | 1 | 1 | ||||
| 9 | BXD36 | 2 | 2 | 0 | 1 | 1 | 1 | ||||
| 10 | BXD38 | 2 | 2 | 0 | 1 | 1 | 1 | ||||
| 11 | BXD39 | 3 | 3 | 0 | 1 | 2 | 2 | ||||
| 12 | BXD43 | 3 | 3 | 0 | 1 | 2 | 2 | ||||
| 13 | BXD44 | 2 | 2 | 0 | 1 | 1 | 1 | ||||
| 14 | BXD45 | 2 | 2 | 0 | 1 | 1 | 1 | ||||
| 15 | BXD49 | 2 | 0 | 2 | 1 | 1 | 1 | ||||
| 16 | BXD50 | 2 | 0 | 2 | 1 | 1 | 1 | ||||
| 17 | BXD51 | 2 | 2 | 0 | 1 | 1 | 1 | ||||
| 18 | BXD55 | 3 | 0 | 3 | 1 | 2 | 2 | ||||
| 19 | BXD61 | 2 | 2 | 0 | 1 | 1 | 1 | ||||
| 20 | BXD62 | 2 | 0 | 2 | 1 | 1 | 1 | ||||
| 21 | BXD64 | 2 | 0 | 2 | 1 | 1 | 1 | ||||
| 22 | BXD66 | 2 | 2 | 0 | 1 | 1 | 1 | ||||
| 23 | BXD68 | 2 | 2 | 0 | 1 | 1 | 1 | ||||
| 24 | BXD69 | 2 | 0 | 2 | 1 | 1 | 1 | ||||
| 25 | BXD71 | 2 | 0 | 2 | 1 | 1 | 1 | ||||
| 26 | BXD73 | 2 | 0 | 2 | 1 | 1 | 1 | ||||
| 27 | BXD74 | 3 | 3 | 0 | 1 | 2 | 1 | 1 | |||
| 28 | BXD75 | 3 | 0 | 3 | 1 | 2 | 2 | ||||
| 29 | BXD77 | 2 | 2 | 0 | 1 | 1 | 1 | ||||
| 30 | BXD80 | 3 | 3 | 0 | 1 | 2 | 2 | ||||
| 31 | BXD81 | 2 | 2 | 0 | 1 | 1 | 1 | ||||
| 32 | BXD83 | 2 | 2 | 0 | 1 | 1 | 1 | ||||
| 33 | BXD84 | 2 | 2 | 0 | 1 | 1 | 1 | ||||
| 34 | BXD85 | 3 | 0 | 3 | 1 | 2 | 2 | ||||
| 35 | BXD87 | 2 | 0 | 2 | 1 | 1 | 1 | ||||
| 36 | BXD89 | 2 | 2 | 0 | 1 | 1 | 1 | ||||
| 37 | BXD90 | 2 | 2 | 0 | 1 | 1 | 1 | ||||
| 38 | BXD99 | 3 | 0 | 3 | 1 | 2 | 2 | ||||
| 39 | BXD100 | 2 | 2 | 0 | 1 | 1 | 1 | ||||
| 40 | BXD101 | 2 | 2 | 0 | 1 | 1 | 1 | ||||
| 41 | BXD102 | 2 | 2 | 0 | 1 | 1 | 1 | ||||
| 42 | BXD103 | 2 | 2 | 0 | 1 | 1 | 1 | ||||
| 43 | C57(UT) | 4 | 2 | 2 | 2 | 2 | 1 (F) | 1 (M) | |||
| 44 | D286F1 | 2 | 0 | 2 | 1 | 1 | 1 | ||||
| 45 | DBA/2 (UT) | 4 | 2 | 2 | 2 | 2 | 1 (M) | 1 (F) | |||
| 46 | C57BL/6J | 16 | 16 | 0 | 8 | 8 | 2 | 6 | |||
| 47 | DBA/2J | 16 | 16 | 0 | 8 | 8 | 1 | 7 | |||
| Total | 135 | 101 | 34 | 63 | 72 | 11 | 1 | 7 | 4 | 45 | |
DBA/2J mice were evaluated in a separate study.
Study design
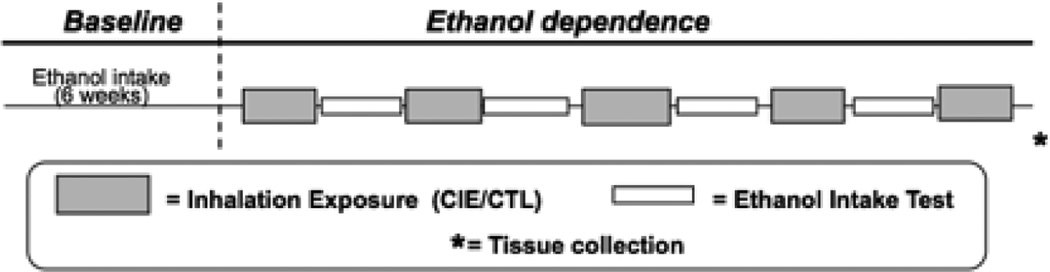
The general study design is summarized in Figure 1. Once within our facilities, all mice were housed individually. Seventy-two hours later, mice were offered ethanol (15% v/v vs. water) using a limited access procedure described below. The study involved first recoding baseline levels of intake for six weeks. Mice representative of each genotype were then separated in paired groups to be exposed to either weekly cycles of CIE exposure (CIE group) or air control (CTL group) exposure as described below. Seventy-two hours after each cycle of CIE (or air control) exposure, mice resumed ethanol drinking for five consecutive days. This pattern of CIE or air control exposure followed by five days of ethanol self-administration was repeated for four cycles. A fifth cycle of CIE (or air) exposure followed the last ethanol intake evaluation and mice were sacrificed for brain and other tissue collection at the 72 hr withdrawal time point. Other research teams within the INIA-stress consortium used the tissue generated in this study for neuroendocrine, neurochemical and genetic analyses (data presented in other articles in this special issue).
Figure 1.
Time line of the scheme used to evaluate voluntary ethanol intake during baseline and after several cycles of CIE (or air for CTL) exposure, and to collect brain and plasma samples used for other studies.
Limited access drinking procedure
Using a procedure previously described, mice were allowed to drink ethanol in the home cage under a daily (Mon-Fri) limited access (2 hr/day) schedule (Becker & Lopez, 2004; Griffin, Lopez, & Becker, 2009; Griffin, Lopez, Yanke, et al., 2009; Lopez & Becker, 2005). The 2 hr daily drinking sessions commenced 30 min before the start of the dark cycle, when mice were presented with a 2-bottle choice to drink ethanol (15% v/v) or tap water as the alternative fluid. The ethanol solution was prepared fresh each day by mixing 95% ethanol with deionized water. The position of the ethanol and water bottles was alternated daily to avoid development of a side preference. The amount of ethanol consumed by each mouse was converted to g/kg based on the milliliters of ethanol consumed (± 0.1 ml) and body weight (± 0.1 g). Water intake was also recorded but it was not included in these analyses. However, these data may be obtained from the full dataset archived in www.genenetwork.org.
Chronic Intermittent Ethanol (CIE) Exposure Procedure
Chronic ethanol vapor exposure was administered in inhalation chambers for 16 hr each day for four days starting on Monday afternoon and finishing Friday morning as detailed previously (Becker & Lopez, 2004; Griffin, Lopez, & Becker, 2009; Griffin, Lopez, Yanke, et al., 2009; Lopez & Becker, 2005). Ethanol concentration in the inhalation chambers was uniformly set for all genotypes and monitored daily to ensure that the inhalation conditions produced stable blood ethanol concentrations (BEC) around 175 mg/dl in male C57BL/6J mice. Blood ethanol concentrations (BEC) were assessed once each week by sampling blood from the retro-orbital sinus immediately upon removal from the chamber. Blood samples were centrifuged for phase separation and 5 µl of plasma was injected into an Analox Instrument analyzer (Lunenburg, MA) for measurement of BEC (Lopez et al., 2014; Lopez et al., 2012). Before each 16 hr ethanol exposure CIE mice were administered ethanol (1.6 g/kg; 8% w/v) combined with the alcohol dehydrogenase inhibitor pyrazole (1 mmol/kg) intraperitoneally (IP) in a volume of 0.02 ml/g body weight. CTL mice were similarly handled, but administered the same pyrazole dose without ethanol (in a saline solution) prior to being placed in control (air) inhalation chambers. Thus, all mice received the same number and timing of pyrazole injections prior to final removal from the inhalation chambers.
Results
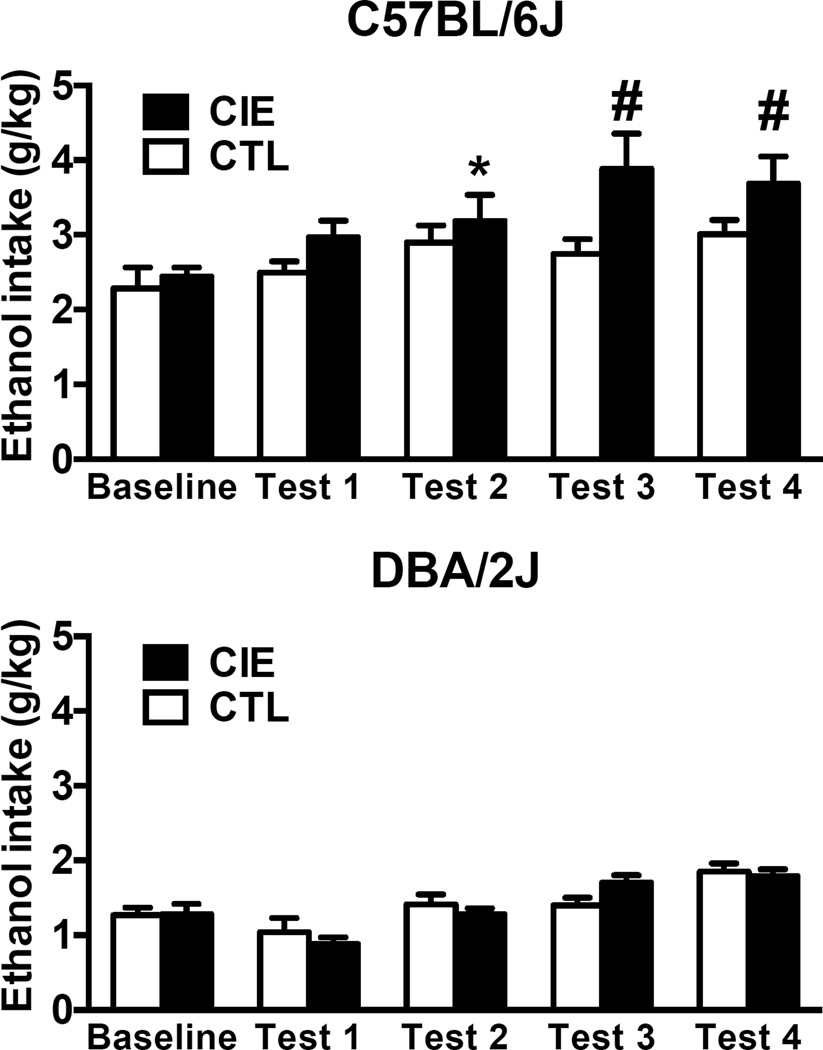
We used all C57BL/6J and DBA/2J cases to compare baseline consumption to that ethanol intake during test cycles 1–4. Data were averaged over days and analyzed by ANOVA with Group (CIE, CTL) as a between-subject factor and Phase (Baseline, Test 1–4) as a repeated measure. ANOVA indicated a significant main effect of Phase [F(4,48)=12.14; p<0.01] due to overall higher intake during the test cycles compared to baseline. Importantly, the ANOVA indicated a significant interaction between Group and Phase [F(4,48)=2.61; p<0.05]. Newman-Keuls post-hoc comparisons indicated that there were no differences in ethanol intake during the last week of baseline between CTL and CIE mice. Furthermore, CTL mice continued to drink ethanol at the same level across the test cycles compared to baseline. As expected, CIE C57BL/6J mice showed a significant increase in ethanol intake during test cycle 2 compared to their own baseline and compared their own baseline and to CTL mice during test cycles 3 and 4 (Figure 2). Male DBA/2J did not show significant changes in voluntary ethanol intake due to CIE exposure (Figure 2). The ANOVA for this dataset indicated only a significant main effect of Phase [F(4,52)=16.05; p<0.01] due to higher intake in test 4 compared to baseline and all the other test cycles, and lower intake in Test 1 compared to baseline. However, ANOVA did not reveal any significant effect of CIE exposure or interaction between CIE exposure and Phase of the study (Figure 2).
Figure 2.
Ethanol intake (g/kg) for male C57BL/6J (top) and DBA/2J mice averaged over the last 5 days of baseline and the five days of ethanol intake test cycles 1–4. * indicates higher intake compared to baseline; # indicates higher intake compared to baseline and CTL group in the same test cycle. Values are mean ± SEM.
Heritability was calculated from a set of 29 progenitors (14 C57BL/6J and 15 DBA/2J, all male) that were treated along with the cohort of BXD animals using identical protocols, personnel, and time of year. With this level of replication and with matched environmental factors we can accurately estimate variance due to a strain main effect, treatment main effect, and interaction between the two. Our main assumption is that heritability estimated from the contrast between the two progenitor strains is a minimally biased estimate of that of BXD progeny strains. Since all of these strains are now fully inbred (no residual heterozygosity), there is no reason to expect a bias in estimates. Heritability is defined here simply as the fraction of the sum of squares attributable to the strain effect using an ordinary least square ANOVA. For example, the total sum of squares for ethanol consumption on Test 4 is 24.2 (g/kg)2, and strain accounts for 16.6 (g/kg)2, or 69% of the variance. The F ratio is 60.8 and the p value is <0.0001. Similar estimates were computed for all test sessions. Strain by treatment interaction effects were also estimated. Based on the analysis of 29 progenitor subjects (14 C57BL/6J and 15 DBA/2J), heritability of the differences in consumption before treatment (baseline) is 54% and this corresponds to roughly a 2-fold difference in consumption (1.28 vs. 2.35 g/kg). In all subsequent cycles, heritability was higher (82% after the first cycle and 69% after the last cycle). As predicted there is a statistically significant strain-by-CIE treatment effect with greater escalation of drinking in C57BL/6J [F(1,25)=4.17]. However, escalation data—the difference of consumption between test cycle 4 and baseline—is significantly noisier than consumption data. Escalation data therefore have a markedly lower heritability in the range of ~15% under these conditions.
The whole dataset obtained in this experiment is available in GeneNetwork (www.genenetwork.org). Table 1 indicates the number of mice per strain and sex that were assigned to the CIE or CTL condition. There were 24 mice that died during the course of CIE exposure (during exposure cycles 1–4). These mice either completed the exposure cycle and died during the 72 hr period before resuming ethanol intake or died during that exposure cycle (Table 1). As indicated above, the ethanol vapor inhalation chambers were set to deliver an amount of ethanol exposure that has been shown to be most effective in producing a reliable increase in ethanol consumption in male C57BL/6J mice (Griffin, Lopez, & Becker, 2009). The BEC registered in each CIE exposure cycle are presented in Table 2 (males) and Table 3 (females). As indicated, there was a considerable range in BEC across strains during the CIE exposure cycles. Some mice did not tolerate the level of intoxication or the chronicity of the exposure to ethanol in this schedule of intermittent exposure and withdrawal. Some mice were removed from the study because of poor response to the CIE procedure. While a few subjects were found dead after ethanol exposure (n = 6), most (n = 18) were removed in consultation with an attending veterinarian familiar with the CIE model. Whenever a CIE-exposed subject was lost, the corresponding CTL mouse from the same strain and sex was removed from the analysis as well. Some mice survived all five CIE exposure cycles despite experiencing BEC noticeably higher than the level registered in C57BL/6J mice (e.g. BXD83 and BXD102).
Table 2.
Blood ethanol concentration (BEC, mg/dl) for male subjects registered in each cycle of CIE exposure. Most values were obtained from a single subject, except for C57BL/6J and DBA/2J.
| Males | CIE Cycle 1 | CIE Cycle 2 | CIE Cycle 3 | CIE Cycle 4 | CIE Cycle 5 |
|---|---|---|---|---|---|
| BXD5 | 144.7 | 86.7 | 196.6 | 230.0 | 123.2 |
| BXD12 | * | 108.7 | 253.8 | 287.1 | 233.1 |
| BXD14 | 104.2 | 150.3 | 190.5 | 195.8 | 210.6 |
| BXD16 | 133.3 | 98.1 | 184.6 | 250.0 | 250.3 |
| BXD19 | 282.7 | 143.2 | 370.0 | ||
| BXD32 | 236.7 | 127.4 | 228.6 | 388.7 | |
| BXD34 | * | 164.3 | 214.0 | 235.2 | 249.0 |
| BXD36 | 225.2 | 241.3 | 149.0 | ||
| BXD39 | 270.4 | 134.8 | 299.7 | 242.2 | |
| BXD43 | 320.5 | 288.8 | 283.6 | 307.1 | 230.5 |
| BXD45 | 145.0 | 99.8 | 157.3 | 254.9 | 345.7 |
| BXD51 | 243.1 | 198.8 | 244.0 | 284.5 | |
| BXD66 | 173.4 | 116.4 | 300.8 | 280.0 | 190.7 |
| BXD74 | 276.2 | 94.1 | 308.9 | 302.9 | 258.3 |
| BXD77 | 180.2 | 64.4 | 303.3 | 257.7 | 227.8 |
| BXD80 | 220.2 | 47.5 | 122.9 | 226.8 | |
| BXD80 | 226.2 | 133.1 | 127.5 | 242.3 | |
| BXD81 | 190.6 | 102.2 | 189.2 | 270.0 | 253.0 |
| BXD83 | 411.6 | 371.2 | 386.4 | 356.3 | 362.9 |
| BXD84 | 125.1 | 104.7 | 300.8 | ||
| BXD89 | 223.4 | 118.9 | 303.2 | 311.3 | |
| BXD100 | 141.9 | 141.6 | 144.2 | 250.7 | 180.1 |
| BXD101 | 341.4 | 256.4 | 245.3 | 318.6 | |
| BXD102 | 154.0 | 161.1 | 349.5 | 365.7 | 358.9 |
| BXD103 | 197.2 | 180.9 | 273.4 | 221.1 | 259.6 |
| DBA/2 (UT) | * | 92.1 | 215.0 | 342.3 | |
| C57 (UT) | 187.6 | 125.4 | 183.4 | 269.0 | 147.0 |
| C57BL/6J | 179.9 | 156.6 | 207.1 | 250.6 | 173.0 |
| DBA/2J | 438.3 | 295.7 | 433.9 | 441.1 |
indicates values that were not recorded due to a technical error.
Table 3.
Blood ethanol concentration (BEC, mg/dl) for female subjects registered in each cycle of CIE exposure
| Females | CIE Cycle 1 | CIE Cycle 2 | CIE Cycle 3 | CIE Cycle 4 | CIE Cycle 5 |
|---|---|---|---|---|---|
| BXD49 | 139.6 | 105.3 | 140.8 | 232.9 | 215.9 |
| BXD50 | 178.8 | 88.0 | 271.3 | 223.9 | 307.3 |
| BXD55 | 72.9 | 78.6 | 141.1 | 166.5 | 112.6 |
| BXD62 | 188.8 | 162.1 | 214.2 | 215.5 | 257.0 |
| BXD64 | 253.7 | 128.4 | 282.1 | 410.0 | |
| BXD69 | 204.7 | 155.8 | 339.2 | ||
| BXD71 | * | 127.2 | 260.3 | 222.9 | 186.8 |
| BXD73 | * | 105.7 | 192.5 | 235.2 | |
| BXD75 | 214.5 | 209.3 | 323.3 | 322.9 | 308.0 |
| BXD85 | 85.4 | 98.9 | 117.6 | 157.2 | 157.0 |
| BXD87 | 227.9 | ||||
| BXD99 | 204.2 | 134.5 | 245.2 | ||
| D2B6F1 | 205.6 | 114.9 | 151.0 | 192.9 | 157.6 |
| DBA/2 (UT) | 138.7 | 160.8 | 273.3 | 274.3 | 374.8 |
indicates values that were not recorded due to a technical error.
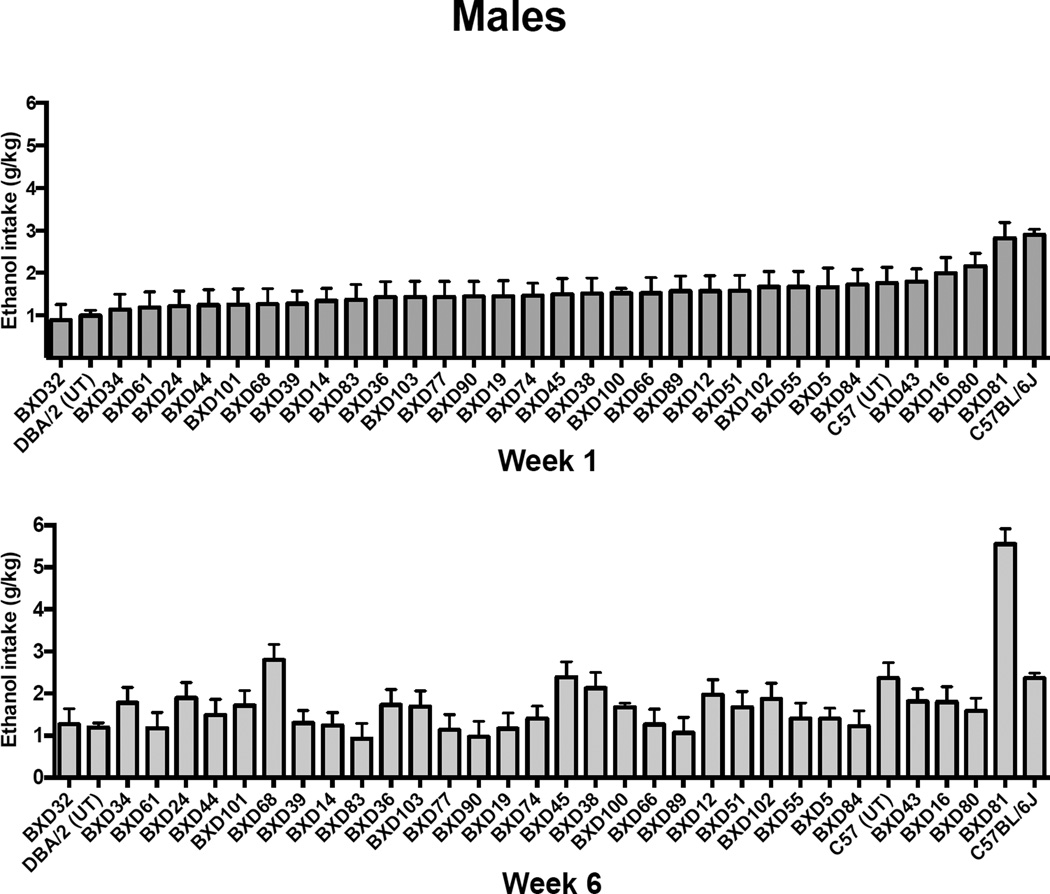
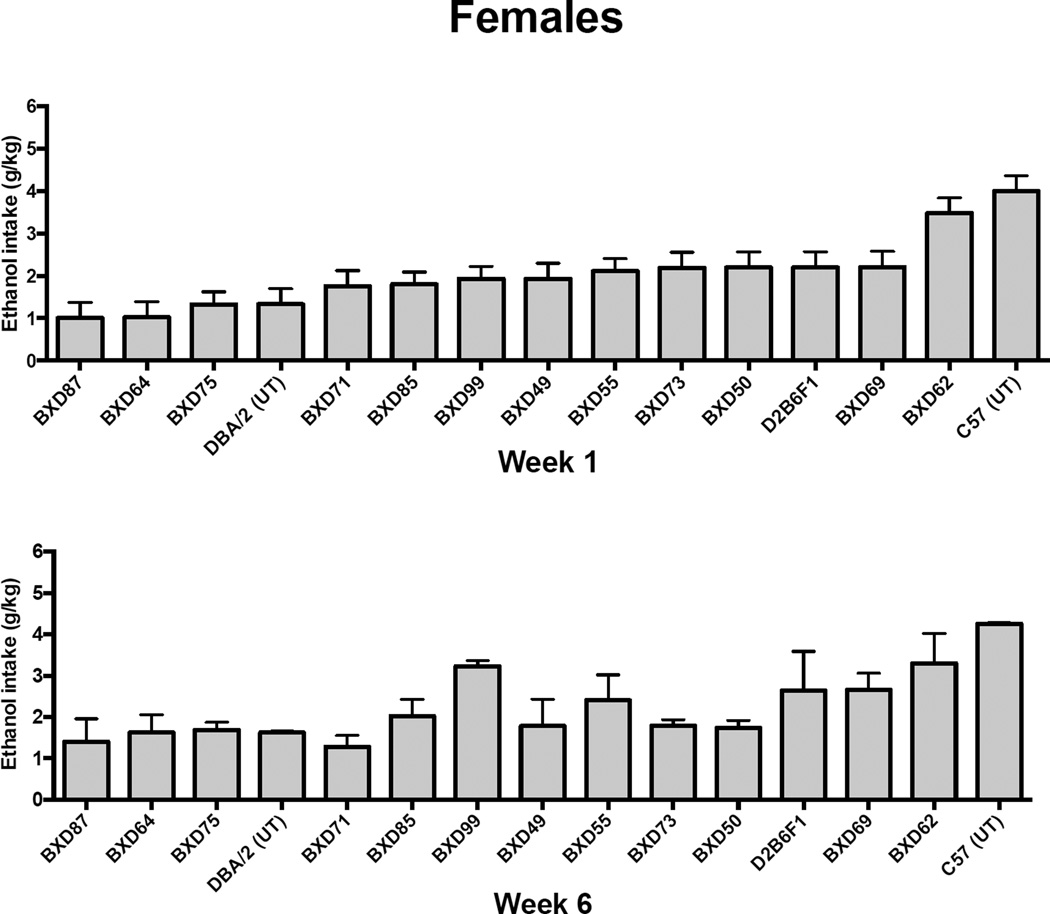
Baseline ethanol intake (averaged for weeks 1 and 6; ranked from low to high) for males (Figure 3) and females (Figure 4) varied between 1 and 3 g/kg across strains in the 2 hr limited access procedure during week 1. A similar range was observed for the last week of baseline (week 6), except that BXD81 males showed a weekly average ethanol intake over 5 g/kg (Figure 3). The amount of ethanol consumed by C57BL/6J mice matched the level observed in previously published studies (Becker & Lopez, 2004; Griffin, Lopez, & Becker, 2009; Griffin, Lopez, Yanke, et al., 2009; Lopez & Becker, 2005). It is also important to note that the rank order of some strains varied from the first to the last week of baseline. For example, BXD68 males showed an increase in ethanol intake from week 1 to week 6 (Figure 3). However, most of the changes in intake were small, and were within 0.5 g/kg difference from week 1 to week 6 for males and females.
Figure 3.
Weekly ethanol intake (g/kg) averaged over the first and last (6) week of baseline drinking for male BXD strains, progenitor strains (from UT), and C57BL/6J mice. Values are ranked from low to high and each bar represent the mean ± SEM.
Figure 4.
Weekly ethanol intake (g/kg) averaged over the first and last (6) week of baseline drinking for female BXD strains and progenitor strains. Values are ranked from low to high and each bar represent the mean ± SEM.
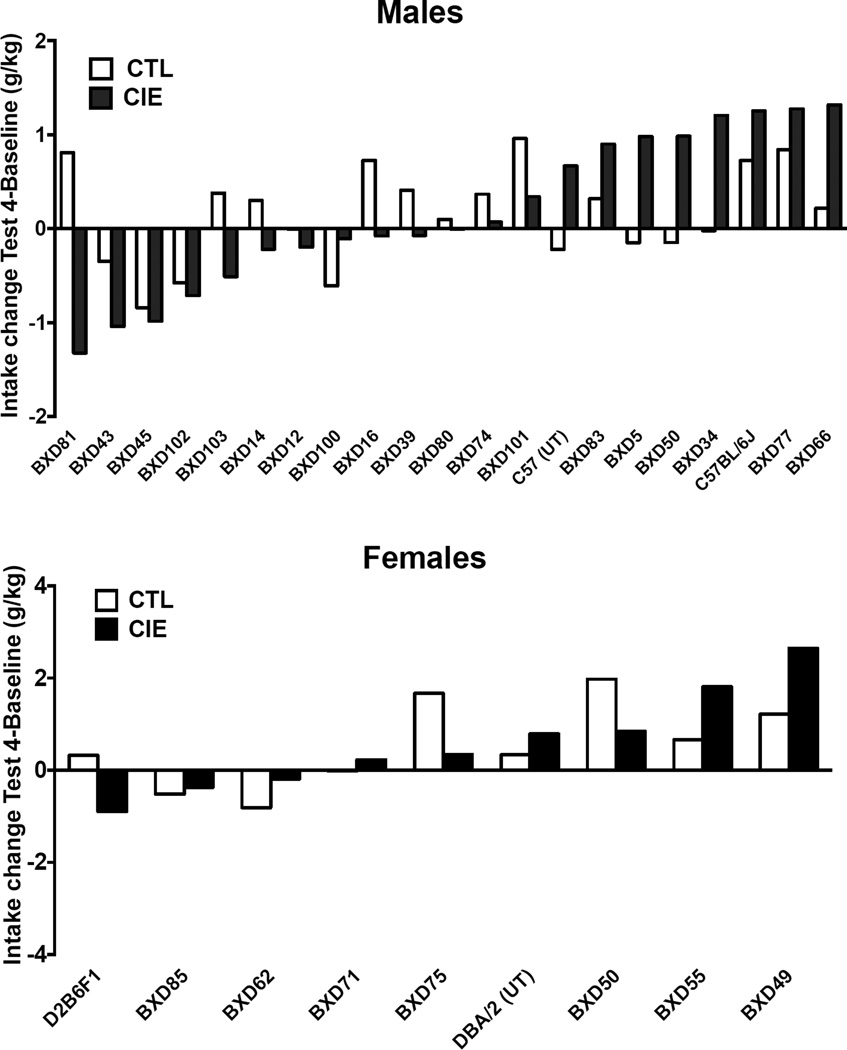
Analysis of ethanol intake from BXD lines included only data obtained from strains that completed all cycles of CIE exposure. Figure 5 shows the absolute change in ethanol intake values (g/kg) from Baseline to Test 4 in male and female mice. The data is ranked based on the magnitude of the change from the biggest decrease to the biggest increase in intake for CIE and CTL mice. In some cases (e.g., males BXD45, BXD77) both CTL and CIE mice showed a change in the same direction, while in others the change was in the opposite direction (e.g., male BXD81) or there was no change in the CTL or the CIE mouse from baseline to Test 4 (Figure 5). Similar results were obtained for test cycles 1–3 (data not shown).
Figure 5.
Change in ethanol intake from baseline to test cycle 4 in g/kg for CTL and CIE, male and female BXD mice, progenitor strains and C57BL/6J.
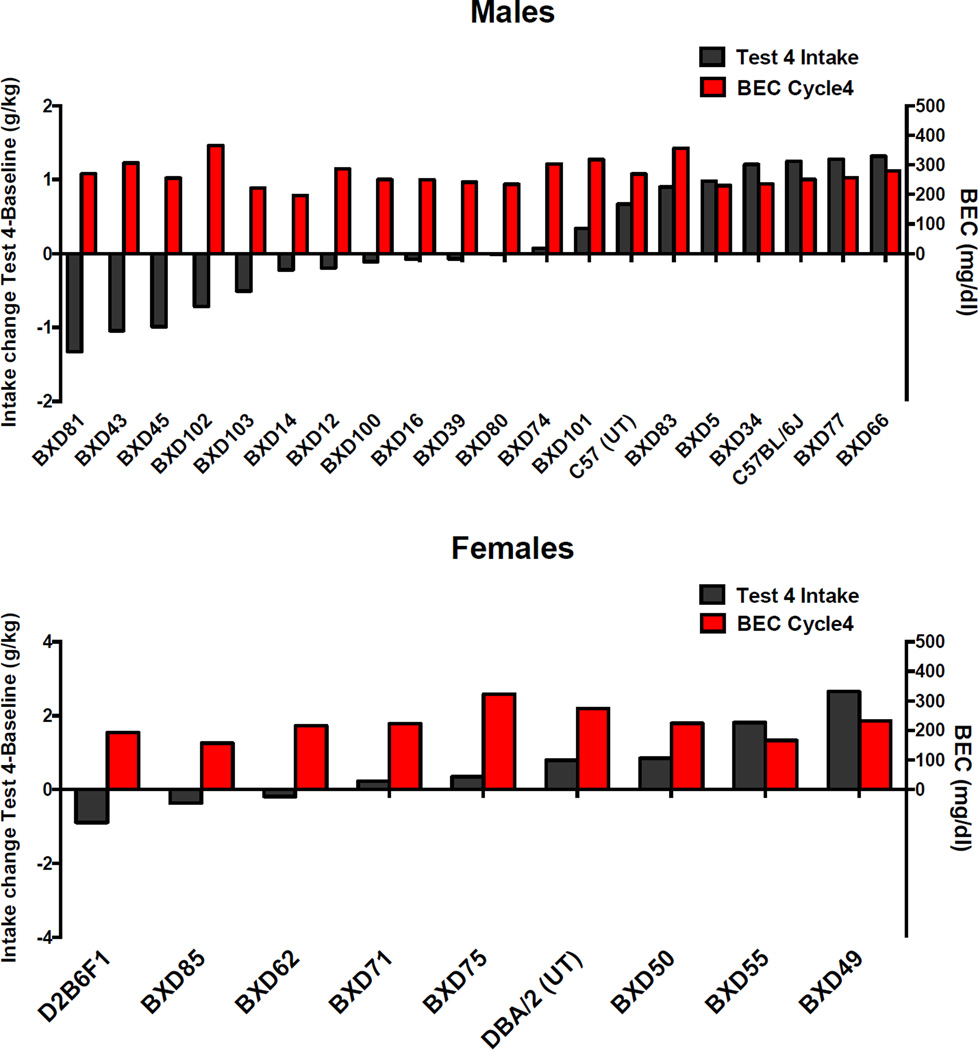
Data presented in Figure 6 depicts the change in ethanol intake (g/kg) from baseline to Test 4 only for CIE mice. The data is ranked as in the previous figure based on the direction of the change for males and female CIE mice. The figure also includes the blood ethanol concentration registered during the fourth cycle of CIE exposure for that particular mouse, or group in the case of C57BL/6J males. As can be observed, there is no relation between the magnitude of the change in ethanol intake and blood ethanol concentrations (BEC) during the previous CIE exposure cycle (R2= 0.0071 for males and 0.013 for females) (Figure 6). Similar results were obtained for test cycles 1–3 (data not shown).
Figure 6.
Change in ethanol intake (g/kg) from baseline to test cycle 4 for CIE, male and female BXD mice, progenitor strains and C57BL/6J and BEC (mg/dl) registered during the fourth cycle of CIE exposure.
Discussion
The present study examined voluntary ethanol intake in a panel of male and female BXD genotypes during a baseline period and after repeated cycles of CIE (or air control) exposure. The study included the evaluation of adult C57BL/6J mice that served as positive controls, since this model of ethanol dependence and relapse drinking was developed using these mice (Becker & Lopez, 2004; Griffin, Lopez, & Becker, 2009; Griffin, Lopez, Yanke, et al., 2009; Lopez & Becker, 2005). While ethanol intake during the baseline phase of the study differed among genotypes tested, most subjects showed baseline levels of intake within 1 and 2 g/kg of ethanol. The rank order of these strains show DBA/2 mice at the low end of the spectrum while C57BL/6 are among the high drinking subjects (see Figures 3 and 4). This was expected since DBA/2 are usually low drinkers while C57BL/6J are high drinkers in various models of ethanol self-administration. Overall, many of the genotypes tested exhibited a change in ethanol consumption during the 6-week baseline phase of the study. That is, rank order of genotypes by amount of ethanol consumed changed for each baseline week of the study. This suggests that known genetic factors that contribute to initial avidity for ethanol may be somewhat distinct from those genetic factors that modulate consumption over a prolonged period of access to ethanol. To our knowledge, this is the first study to examine voluntary ethanol consumption over a substantial period of time (i.e., several weeks) in a large panel of diverse genotypes. Previous studies have evaluated voluntary ethanol intake over 15–20 days and in some cases after a period of forced ethanol intake (single bottle model) (Phillips et al., 1994; Rodriguez et al., 1994, 1995).
It is important to note that in the present study ethanol intake in the BXD lines was evaluated using a limited access procedure with a 15% v/v ethanol solution vs. water. Previous studies have evaluated ethanol intake in BXDs using an initial period of forced exposure to 10% ethanol in the home cage for 24 hr/day (Gill et al., 1996; Rodriguez et al., 1994) or evaluation of 3% or 10% v/v ethanol vs. water also measured every 24 hours (Crabbe et al., 1983; Phillips et al., 1994; Rodriguez et al., 1994, 1995). Thus, differences in the ranking of genotypes in this study compared to previous studies may be related to consumption under limited vs. continuous access situations.
Another issue to consider in comparing baseline intake in the present study to previously reported intake in BXDs is the specific lines selected for analysis. The present study used subjects from both the original BXD set (BXD1 to BXD32) and the newly developed strains (Peirce et al., 2004; Taylor, 1978). However, only seven strains evaluated in this study overlap with strains previously studied for ethanol intake. This allowed for a few, but limited evaluation of rank order correlations between the data obtained in this study and previously published work. In some cases, the correlations were positive and significant, but this was observed in a few cases and varied according to the week of baseline intake from the present study that was selected for analysis (data not shown). A follow up study using a larger number of strains and replicates within strains will be necessary to better compare genotypic differences in free-choice ethanol consumption under limited and continuous access conditions.
Repeated cycles of CIE exposure results in a significant increase in voluntary ethanol intake in male C57BL/6J mice. A set of these mice was included in the present study as positive control for CIE exposure-induced drinking. Results obtained with these mice replicated the results obtained in previous studies (Becker & Lopez, 2004; Dhaher, Finn, Snelling, & Hitzemann, 2008; Finn et al., 2007; Griffin, Lopez, & Becker, 2009; Griffin, Lopez, Yanke, et al., 2009; Lopez & Becker, 2005; Lopez et al., 2012), with CIE mice showing a significant increase in voluntary ethanol intake compared to air-exposed control subjects and their own baseline intake level (see Figure 2). The magnitude of the increase is also comparable to the increase observed in previous studies that resulted in significantly higher blood and brain ethanol levels (Becker & Lopez, 2004; Griffin, Lopez, Yanke, et al., 2009). A set of male DBA/2J mice from a separate study was also included for comparative purpose. Although these mice were not part of the same study, they were exposed to the same concentration of ethanol in the inhalation chamber that was used to evaluate CIE exposure in C57BL/6J and the BXD strains evaluated. Under these experimental parameters, DBA/2J mice consumed less 15% ethanol than C57BL/6J mice and they did not show escalation in voluntary ethanol intake as a consequence of CIE exposure (see Figure 2). The results and estimates of heritable differences in consumption and the vapor chamber protocols using data from the two progenitor strains are more than sufficient to motivate our analysis of the BXD family for their range of responses to CIE and to justify more extensive analysis. The implication of our findings are that obtaining accurate strain means for escalation following treatment will benefit significantly from sample sizes of six or higher, especially if one particular BXD strain is to be used as a model However, QTL mapping, like genetic analysis of human cohorts, benefits far more from analysis of larger numbers of diverse genotypes and fewer replicates. Given that there are now 150 BXD strains, if the goal is strictly to map QTLs, then it is still preferable to sample as many strains as possible, even if this means sampling fewer than four cases per strain (Belknap 1998, Williams & Williams, 2016).
As indicated before, DBA/2J are usually described as ethanol avoiders. However, others have shown that DBA/2J self-administer significantly higher levels of ethanol if ethanol is delivered intragastrically (Fidler et al., 2011) or when ethanol is gradually introduced along with masking its taste with monosodium glutamate (MSG) (McCool & Chappell, 2014, 2015). In this study, the use of a sucrose fading or a MSG fading procedure was omitted to avoid masking the initial intake level of C57BL/6J, DBA/2J and the different BXD strains. Although we did not observe a CIE-related increase in ethanol intake in DBA/2J mice in the present study, others have reported that these mice will self-administer ethanol to alleviate ethanol withdrawal’s effects (Cunningham, Fidler, Murphy, Mulgrew, & Smitasin, 2013; Fidler et al., 2012) and will show a significant increase in ethanol intake after CIE exposure (McCool & Chappell, 2015). Perhaps the use of an alternative route of administration that prevents processing ethanol’s taste (Cunningham et al., 2013) or the use of a prolonged fading procedure to introduce ethanol (McCool & Chappell, 2015) is necessary to observed increased ethanol intake after chronic ethanol exposure in DBA/2J mice. These results regarding DBA/2J mice also impact findings across the BXD strains, suggesting that our data should be considered an admixture of genetic influences on ethanol consumption due to rewarding properties or taste.
As previously noted, a decision was made a priori to evaluate ethanol consumption in all genotypes under the same experimental conditions. As a consequence, all mice were exposed to the same ethanol vapor concentrations in inhalation chambers. Out of the 64 subjects assigned to receive CIE exposure, only 40 were evaluated for voluntary ethanol intake in the last (fourth) ethanol intake test cycle (see Table 1). Since the ethanol concentration in the inhalation chambers was not differentially adjusted for each particular strain, a wide range of BEC levels were recorded across the CIE exposure cycles (see Table 2). Most of the attrition could be attributed to lethal ethanol exposure or its subsequent withdrawal. The large variance in BEC values resulting from the first CIE exposure cycle may be useful in analysis of the genetic contribution to this robust metabolic effect. However, despite the loss of some BXD RI strains and DBA/2 male mice obtained from UT, the majority of genotypes included in the study completed all 4 CIE exposure cycles (see Tables 1 and 2). Moreover, changes in ethanol consumption during test cycles relative to baseline level of intake (increased, unchanged, or decreased) across the panel of BXD RI strains did not appear to vary as a function of BEC values attained during the CIE exposure cycles (see Figure 6). This suggests that genetic factors that contribute to differences in propensity to escalate voluntary ethanol intake as a function of experience with repeated CIE exposure cycles across BXD RI strains are independent from genetic influences on ethanol pharmacokinetics. Additionally, it could be argued that exposure to higher BEC levels in DBA/2J mice compared to C57BL/6J during CIE exposure could have favor a conditioned aversion to alcohol in DBA/2J mice that prevented the escalation on intake expected in dependent subjects. As indicated before, it is possible for DBA/2J mice to show increase ethanol intake after chronic ethanol exposure under different experimental conditions (Cunningham, Fidler, Murphy, Mulgrew, & Smitasin, 2013; Fidler et al., 2012; McCool & Chappell, 2015).
Of particular interest is that ethanol consumption difference from baseline to the last test of ethanol intake (Test 4) varied in a unique manner across the panel of BXD RI strains. This was true for both CIE and CTL subjects (see Figure 5). In some cases there was evidence for escalation of drinking in CIE compared to CTL subjects of a given genotype, while other strains evidenced decreased intake over the 4 test cycles. Some BXD RI strains did not significantly change ethanol consumption during the course of the testing phase of the study. The results presented here are centered on the change in intake from baseline to test 4, but the study design also affords the opportunity to analyze the drinking data for each Test Cycle (comparing CIE vs. CTL conditions).
The results presented here represent a first step towards a more comprehensive analysis of BXD RI strains in this model of CIE exposure and relapse drinking. The goal is to identify genotypes that exhibit robust progressive increases in ethanol consumption across the test cycles as well as genotypes that evidenced decreases in ethanol consumption as a consequence of CIE exposure. This may provide clues about genetic factors that confer greater risk for escalation of drinking and a more dangerous trajectory of addiction. Just as important, identifying genotypes that do not display such escalated drinking may offer insights about genetic factors that confer protection or resilience to such hazardous drinking. Studies conducted to increase the number of subjects assigned to CIE or CTL conditions for a more limited number of BXD RI strains are currently under analysis. These strains were selected based on the results from this study considering two main criteria. First, the level of intake of CTL subjects that did not change from baseline to Test 4. Second, the CIE subject from the same strain completed all CIE exposure cycles and shows a robust increase or decrease in ethanol intake from baseline to Test 4. The outcome of this complementary screening, with a higher number of subjects per experimental condition (defined by strain, sex, and CIE or CTL exposure), will represent a stronger evaluation of BXD RI strains in this model of dependence and ethanol intake.
Highlights.
CIE exposure induces changes in voluntary ethanol intake that vary between C57BL/6J and DBA/2J genotypes of mice.
Heritability of variation is above 50% and increases following treatment.
CIE and air control, exposure induces increases or decreases in ethanol intake across BXD genotypes.
The increase or decrease in intake does not depend on BEC during CIE exposure
Acknowledgments
Supported by NIAAA grants U24 AA020929 (MFL), U01 AA014095 (HCB), P50 AA010761 (HCB), U01 AA016667 (MFM), P50 AA022537 (MFM), U01 AA016662 (RWW), U01 AA013499 (RWW), U24 AA013513 (RWW), U01 AA014425(RWW).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Becker HC. Animal models of excessive alcohol consumption in rodents. Curr Top Behav Neurosci. 2013;13:355–377. doi: 10.1007/7854_2012_203. [DOI] [PubMed] [Google Scholar]
- Becker HC, Lopez MF. Increased ethanol drinking after repeated chronic ethanol exposure and withdrawal experience in C57BL/6 mice. Alcohol Clin Exp Res. 2004;28(12):1829–1838. doi: 10.1097/01.alc.0000149977.95306.3a. [DOI] [PubMed] [Google Scholar]
- Belknap JK. Effect of within-strain sample size on QTL detection and mapping using recombinant inbred mouse strains. Behav Genet. 1998;28(1):29–38. doi: 10.1023/a:1021404714631. [DOI] [PubMed] [Google Scholar]
- Belknap JK, Crabbe JC, Young ER. Voluntary consumption of ethanol in 15 inbred mouse strains. Psychopharmacology (Berl) 1993;112(4):503–510. doi: 10.1007/BF02244901. [DOI] [PubMed] [Google Scholar]
- Buck KJ, Rademacher BS, Metten P, Crabbe JC. Mapping murine loci for physical dependence on ethanol. Psychopharmacology (Berl) 2002;160(4):398–407. doi: 10.1007/s00213-001-0988-8. [DOI] [PubMed] [Google Scholar]
- Cloninger CR. Neurogenetic adaptive mechanisms in alcoholism. Science. 1987;236(4800):410–416. doi: 10.1126/science.2882604. [DOI] [PubMed] [Google Scholar]
- Crabbe JC, Colville AM, Kruse LC, Cameron AJ, Spence SE, Schlumbohm JP, Metten P. Ethanol tolerance and withdrawal severity in high drinking in the dark selectively bred mice. Alcohol Clin Exp Res. 2012;36(7):1152–1161. doi: 10.1111/j.1530-0277.2011.01715.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabbe JC, Harris RA, Koob GF. Preclinical studies of alcohol binge drinking. Ann N Y Acad Sci. 2011;1216:24–40. doi: 10.1111/j.1749-6632.2010.05895.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabbe JC, Phillips TJ, Belknap JK. The complexity of alcohol drinking: studies in rodent genetic models. Behav Genet. 2010;40(6):737–750. doi: 10.1007/s10519-010-9371-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabbe JC, Phillips TJ, Harris RA, Arends MA, Koob GF. Alcohol-related genes: contributions from studies with genetically engineered mice. Addict Biol. 2006;11(3–4):195–269. doi: 10.1111/j.1369-1600.2006.00038.x. [DOI] [PubMed] [Google Scholar]
- Crabbe JC, Young ER, Kosobud A. Genetic correlations with ethanol withdrawal severity. Pharmacol Biochem Behav. 1983;18(Suppl 1):541–547. doi: 10.1016/0091-3057(83)90233-2. [DOI] [PubMed] [Google Scholar]
- Cunningham CL, Fidler TL, Murphy KV, Mulgrew JA, Smitasin PJ. Time-dependent negative reinforcement of ethanol intake by alleviation of acute withdrawal. Biol Psychiatry. 2013;73(3):249–255. doi: 10.1016/j.biopsych.2012.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham CL, Hallett CL, Niehus DR, Hunter JS, Nouth L, Risinger FO. Assessment of ethanol's hedonic effects in mice selectively bred for sensitivity to ethanol-induced hypothermia. Psychopharmacology (Berl) 1991;105(1):84–92. doi: 10.1007/BF02316868. [DOI] [PubMed] [Google Scholar]
- Dhaher R, Finn D, Snelling C, Hitzemann R. Lesions of the extended amygdala in C57BL/6J mice do not block the intermittent ethanol vapor-induced increase in ethanol consumption. Alcohol Clin Exp Res. 2008;32(2):197–208. doi: 10.1111/j.1530-0277.2007.00566.x. [DOI] [PubMed] [Google Scholar]
- Dick DM, Foroud T. Genetic strategies to detect genes involved in alcoholism and alcohol-related traits. Alcohol Res Health. 2002;26(3):172–180. [PMC free article] [PubMed] [Google Scholar]
- Fidler TL, Dion AM, Powers MS, Ramirez JJ, Mulgrew JA, Smitasin PJ, Cunningham CL. Intragastric self-infusion of ethanol in high- and low-drinking mouse genotypes after passive ethanol exposure. Genes Brain Behav. 2011;10(3):264–275. doi: 10.1111/j.1601-183X.2010.00664.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fidler TL, Powers MS, Ramirez JJ, Crane A, Mulgrew J, Smitasin P, Cunningham CL. Dependence induced increases in intragastric alcohol consumption in mice. Addict Biol. 2012;17(1):13–32. doi: 10.1111/j.1369-1600.2011.00363.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn DA, Snelling C, Fretwell AM, Tanchuck MA, Underwood L, Cole M, Roberts AJ. Increased drinking during withdrawal from intermittent ethanol exposure is blocked by the CRF receptor antagonist D-Phe-CRF(12–41) Alcohol Clin Exp Res. 2007;31(6):939–949. doi: 10.1111/j.1530-0277.2007.00379.x. [DOI] [PubMed] [Google Scholar]
- Gill K, Liu Y, Deitrich RA. Voluntary alcohol consumption in BXD recombinant inbred mice: relationship to alcohol metabolism. Alcohol Clin Exp Res. 1996;20(1):185–190. doi: 10.1111/j.1530-0277.1996.tb01063.x. [DOI] [PubMed] [Google Scholar]
- Griffin WC, 3rd, Lopez MF, Becker HC. Intensity and duration of chronic ethanol exposure is critical for subsequent escalation of voluntary ethanol drinking in mice. Alcohol Clin Exp Res. 2009;33(11):1893–1900. doi: 10.1111/j.1530-0277.2009.01027.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin WC, 3rd, Lopez MF, Yanke AB, Middaugh LD, Becker HC. Repeated cycles of chronic intermittent ethanol exposure in mice increases voluntary ethanol drinking and ethanol concentrations in the nucleus accumbens. Psychopharmacology (Berl) 2009;201(4):569–580. doi: 10.1007/s00213-008-1324-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houtkooper RH, Mouchiroud L, Ryu D, Moullan N, Katsyuba E, Knott G, Auwerx J. Mitonuclear protein imbalance as a conserved longevity mechanism. Nature. 2013;497(7450):451–457. doi: 10.1038/nature12188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendler KS, Aggen SH, Prescott CA, Crabbe J, Neale MC. Evidence for multiple genetic factors underlying the DSM-IV criteria for alcohol dependence. Mol Psychiatry. 2012;17(12):1306–1315. doi: 10.1038/mp.2011.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King AC, Houle T, de Wit H, Holdstock L, Schuster A. Biphasic alcohol response differs in heavy versus light drinkers. Alcohol Clin Exp Res. 2002;26(6):827–835. [PubMed] [Google Scholar]
- Lopez MF, Becker HC. Effect of pattern and number of chronic ethanol exposures on subsequent voluntary ethanol intake in C57BL/6J mice. Psychopharmacology (Berl) 2005;181(4):688–696. doi: 10.1007/s00213-005-0026-3. [DOI] [PubMed] [Google Scholar]
- Lopez MF, Becker HC, Chandler LJ. Repeated episodes of chronic intermittent ethanol promote insensitivity to devaluation of the reinforcing effect of ethanol. Alcohol. 2014;48(7):639–645. doi: 10.1016/j.alcohol.2014.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez MF, Griffin WC, 3rd, Melendez RI, Becker HC. Repeated cycles of chronic intermittent ethanol exposure leads to the development of tolerance to aversive effects of ethanol in C57BL/6J mice. Alcohol Clin Exp Res. 2012;36(7):1180–1187. doi: 10.1111/j.1530-0277.2011.01717.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCool BA, Chappell AM. Persistent enhancement of ethanol drinking following a monosodium glutamate-substitution procedure in C57BL6/J and DBA/2J mice. Alcohol. 2014;48(1):55–61. doi: 10.1016/j.alcohol.2013.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCool BA, Chappell AM. Chronic intermittent ethanol inhalation increases ethanol self-administration in both C57BL/6J and DBA/2J mice. Alcohol. 2015;49(2):111–120. doi: 10.1016/j.alcohol.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merikangas KR, Stolar M, Stevens DE, Goulet J, Preisig MA, Fenton B, Rounsaville BJ. Familial transmission of substance use disorders. Arch Gen Psychiatry. 1998;55(11):973–979. doi: 10.1001/archpsyc.55.11.973. [DOI] [PubMed] [Google Scholar]
- Metten P, Phillips TJ, Crabbe JC, Tarantino LM, McClearn GE, Plomin R, Belknap JK. High genetic susceptibility to ethanol withdrawal predicts low ethanol consumption. Mamm Genome. 1998;9(12):983–990. doi: 10.1007/s003359900911. [DOI] [PubMed] [Google Scholar]
- Peirce JL, Lu L, Gu J, Silver LM, Williams RW. A new set of BXD recombinant inbred lines from advanced intercross populations in mice. BMC Genet. 2004;5:7. doi: 10.1186/1471-2156-5-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philip VM, Duvvuru S, Gomero B, Ansah TA, Blaha CD, Cook MN, Chesler EJ. High-throughput behavioral phenotyping in the expanded panel of BXD recombinant inbred strains. Genes Brain Behav. 2010;9(2):129–159. doi: 10.1111/j.1601-183X.2009.00540.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips TJ, Crabbe JC, Metten P, Belknap JK. Localization of genes affecting alcohol drinking in mice. Alcohol Clin Exp Res. 1994;18(4):931–941. doi: 10.1111/j.1530-0277.1994.tb00062.x. [DOI] [PubMed] [Google Scholar]
- Putman AH, Wolen AR, Harenza J, Yordanova RK, Webb BT, Chesler EJ, Miles MF. Identification of Quantitative Trait Loci and Candidate Genes for an Anxiolytic-like Response to Ethanol in BXD Recombinant Inbred Strains. Genes Brain Behav. 2016 doi: 10.1111/gbb.12289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn PD, Fromme K. Subjective response to alcohol challenge: a quantitative review. Alcohol Clin Exp Res. 2011;35(10):1759–1770. doi: 10.1111/j.1530-0277.2011.01521.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risinger FO, Cunningham CL. Ethanol-induced conditioned taste aversion in BXD recombinant inbred mice. Alcohol Clin Exp Res. 1998;22(6):1234–1244. [PubMed] [Google Scholar]
- Risinger FO, Malott DH, Prather LK, Niehus DR, Cunningham CL. Motivational properties of ethanol in mice selectively bred for ethanol-induced locomotor differences. Psychopharmacology (Berl) 1994;116(2):207–216. doi: 10.1007/BF02245064. [DOI] [PubMed] [Google Scholar]
- Rodriguez LA, Plomin R, Blizard DA, Jones BC, McClearn GE. Alcohol acceptance, preference, and sensitivity in mice. I. Quantitative genetic analysis using BXD recombinant inbred strains. Alcohol Clin Exp Res. 1994;18(6):1416–1422. doi: 10.1111/j.1530-0277.1994.tb01444.x. [DOI] [PubMed] [Google Scholar]
- Rodriguez LA, Plomin R, Blizard DA, Jones BC, McClearn GE. Alcohol acceptance, preference, and sensitivity in mice. II. Quantitative trait loci mapping analysis using BXD recombinant inbred strains. Alcohol Clin Exp Res. 1995;19(2):367–373. doi: 10.1111/j.1530-0277.1995.tb01517.x. [DOI] [PubMed] [Google Scholar]
- Schuckit MA, Smith TL. The relationships of a family history of alcohol dependence, a low level of response to alcohol and six domains of life functioning to the development of alcohol use disorders. J Stud Alcohol. 2000;61(6):827–835. doi: 10.15288/jsa.2000.61.827. [DOI] [PubMed] [Google Scholar]
- Taylor BA. Recombinant inbred strains: use in gene mapping. In: Morse HC, editor. Origins of inbred mice. New York: Academic Press; 1978. pp. 423–438. [Google Scholar]
- Wang X, Pandey AK, Mulligan MK, Williams EG, Mozhui K, Li Z, Williams RW. Joint mouse-human phenome-wide association to test gene function and disease risk. Nat Commun. 2016;7:10464. doi: 10.1038/ncomms10464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RW, Williams EG. Resources for systems genetics. In: Schughart K, Williams RW, editors. Methods in Molecular Biology. Humana Press; 2016. in press. [DOI] [PubMed] [Google Scholar]