Abstract
Tumor-repopulating cells are a tumorigenic sub-population of cancer cells that drives tumorigenesis. We have recently reported that soft fibrin matrices maintain tumor-repopulating cell growth by promoting histone 3 lysine 9 (H3K9) demethylation and Sox2 expression and that Cdc42 expression influences H3K9 methylation. However, the underlying mechanisms of how soft matrices induce H3K9 demethylation remain elusive. Here we find that tumor-repopulating cells exhibit lower focal adhesion kinase (FAK) and H3K9 methylation levels in soft fibrin matrices than control melanoma cells on 2D rigid substrates. Silencing FAK in control melanoma cells decreases H3K9 methylation, whereas overexpressing FAK in tumor-repopulating cells enhances H3K9 methylation. Overexpressing Cdc42 or RhoA in the presence of FAK knockdown restores H3K9 methylation levels. Importantly, silencing FAK, Cdc42, or RhoA promotes Sox2 expression and proliferation of control melanoma cells in stiff fibrin matrices, whereas overexpressing each gene suppresses Sox2 expression and reduces growth of TRCs in soft but not in stiff fibrin matrices. Our findings suggest that low FAK mediated by soft fibrin matrices downregulates H3K9 methylation through reduction of Cdc42 and RhoA and promotes growth of tumor-repopulating cells.
Keywords: focal adhesion kinase, cancer, tumor repopulating cell, growth, matrix stiffness
INTRODUCTION
Cancer Stem Cells (CSCs) are a subpopulation of cancer cells within a tumor that possesses self-renewing capability and drives tumor progression and metastasis. It has been demonstrated that TICs (tumor-initiating cells) or CSC like cells exist in several types of cancer, including blood [1], brain [2, 3], skin [4, 5], colon [6], and intestine [7]. These tumorigenic cells are resistant to conventional chemotherapy and radiotherapy and speculated to be key players in cancer relapse [8, 9]. Therefore, understanding the molecular mechanisms underlying TIC’s high tumorigenicity is essential to achieve complete tumor eradication. We previously mechanically selected tumorigenic B16 tumor-repopulating cells (TRCs) by culturing single cancer cells from cancer cell lines or primary tumors in three-dimensional (3D) soft fibrin matrices [4]. The selected TRCs exhibit high tumorigenic potential as they can efficiently repopulate tumors locally and in distant organs in wild-type syngeneic and non-syngeneic mice. These CSC like TRCs express the same levels of surface stem cell marker CD133 as the unselected B16 cells. Therefore, these cells are different from CSCs. These TRCs are also different from tumor-initiating cells (TICs) that have 3 distinct subtypes of transient, delayed contributing, and long-term self-renewing. Soft fibrin matrices regulate TRC growth by promoting histone 3 lysine residue 9 (H3K9) demethylation that is associated with Cdc42 downregulation and Sox2 expression [10]. However, the underlying mechanisms of how soft fibrin matrices regulate H3K9 demethylation remain elusive. Here we demonstrate that focal adhesion kinase (FAK) regulates H3K9 methylation via Cdc42 and RhoA. Soft fibrin matrices maintain TRC growth by downregulating FAK, which in turn lowers Cdc42 and RhoA to mediate H3K9 demethylation and Sox2 upregulation.
MATERIALS AND METHODS
Cell culture
Murine melanoma cell line B16-F1 was purchased from American Type Culture Collection. Human breast cancer cell line MCF-7 and human lung cancer cell line A549 were obtained from Chinese Type Culture Collection. Briefly, cells were cultured on rigid dishes in cell culture medium supplemented with 10% fetal bovine serum (Invitrogen), 23mM L-glutamine (Invitrogen), 13mM sodium pyruvate and 0.13mM penicillin/streptomycin at 373°C with 5% CO2.
Polyacrylamide and 3D fibrin gel preparation
Polyacrylamide gels were prepared following the protocol reported previously [11]. 3D fibrin gels were prepared as described previously [4, 10]. Briefly, fibrinogen was diluted with T7 buffer. Fibrinogen and single cell solution mixture was made by mixing the same volume. 250 μl cell/fibrinogen mixture was seeded into each well of 24-well plate and mixed well with pre-added 53μl thrombin (1003U/ml). The cell culture plate was then incubated in 373°C cell culture incubator for 103min. Finally, 13ml of cell medium was added.
qPCR analysis
Total mRNA was isolated from cells using the RNeasy Mini Kit (QIAGEN) according to the supplier’s instruction. qPCR was performed using the SsoAdvanced™ Universal Probes Supermix (Bio-Rad). The data were normalized against mouse GAPDH. The sequences of all the primers were listed in the Supplemental Table 1.
Western blotting assay
To quantify the expressions of FAK, H3K9 di- and tri-methylation, cells were lysed with RIPA lysis buffer (Beyotime). 20 μl of each sample was separated by 8–15% SDS-PAGE, blocked with 5% BSA overnight at 4 °C and incubated with primary antibodies to FAK (Rabbit, 1:1000, Abcam, ab40794), FAK-Y397 (Rabbit, 1:1000, Abcam, ab81298), H3K9 di-methylation (Rabbit, 1:300, Millipore, 17-648) and tri-methylation (Rabbit, 1:300, Millipore, 17-625), Histone H3 (Rabbit, 1:500, CST, #D1H2), RhoA (Rabbit, 1:500, Abcam, ab187027), and GAPDH (Mouse, 1:1000, Abcam, ab8245). Primary antibodies were detected with goat anti-Rabbit IgG-HRP (1:2000, Santa Cruz, sc-2004) or anti-Mouse IgG-HRP (1:2000, Santa Cruz, sc-2005). The blots were developed using SuperSignal West Pico chemiluminescent substrate (Millipore).
Chromatin immunoprecipitation assay
We performed ChIP assay following the manufacturer’s instructions (EZ-ChIP kit, Millipore). Briefly, cells were subjected to cross-linking with 1% formaldehyde in medium for 10 min at 373°C and then lysed on ice. Chromatin was sonicated to shear DNA to an average length of 0.2–1.0 kb. Antibodies to di- and tri- methyl-histone H3 (Lys9) (Abcam) were used for immunoprecipitation. Normal mouse IgG (Santa Cruz) was used as negative control. The immunoprecipitation was heated to reverse the formaldehyde cross-linking and the DNA fragments in the precipitates was purified for qPCR analysis.
FRET imaging
The Lyn-FAK and H3K9 biosensors used in this study were reported elsewhere. The specificity of Lyn-FAK [10, 12] and H3K9 FRET biosensors [13] were determined using appropriate controls and point-mutations. The biosensors were transfected into cells. A Leica inverted fluorescence microscope integrated with Dual-View MicroImager system (Optical Insights) was used to capture CFP and YFP (YPet) emission images. For FRET imaging, each CFP and YFP image were simultaneously captured on the same screen by using a charge-coupled device camera (C4742-95-12ERG; Hamamatsu). A customized Matlab (Mathworks) program was used to analyze YPet/CFP (for H3K9 methylation) or CFP/YFP (for FAK activity) emission ratios.
RNA interference
Cells were transfected with siRNAs and complementary DNA using Lipofectamine 2000 (Invitrogen) following the manufacturer’s protocol. The construct sequences were shown in Supplemental Table 2. Knocking down or overexpression efficiency of the protein levels was shown in Supplemental Fig. 1.
Statistical analysis
Two-tailed Student’s t-test was used to conduct all the statistics.
RESULTS
Soft matrices promote TRC growth via low FAK
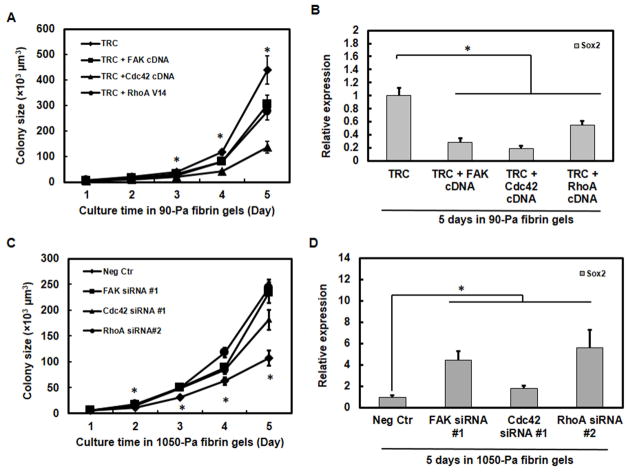
We have reported that soft fibrin matrices decrease H3K9 methylation, increase Sox2 (a self-renewal gene) expression, and promote TRC’s high tumorigenicity [4, 10]. However, the early cascade of matrix-cell mechanical signaling remains elusive. It is known that cells sense and respond to extracellular matrices mainly through integrin-mediated adhesion [14–16]. As one of the first proteins that interacts with cytoplasmic tails of integrins, FAK is abnormally expressed in several types of cancer and involved in tumor progression and metastasis [17–19]. We hypothesized that FAK might play a critical role in regulating TRC growth. To demonstrate the functional roles of FAK and the downstream molecules Cdc42 and RhoA in TRC growth, we examined their effects on the colony growth in 3D fibrin matrices. Melanoma cells expressed higher FAK, Cdc42, RhoA, and methyltransferases and lower Sox2 in stiff (1050-Pa) than in soft (90-Pa) 3D fibrin gels (Supplemental Fig. 2A, B). Overexpressing FAK or Cdc42 or transfecting a constitutively active construct RhoA V14 in TRCs suppressed the colony growth in soft (Fig. 1A) but not in stiff fibrin matrices (Supplemental Fig. 3A). This finding is supported by the results that overexpressing these genes inhibited TRC growth via suppression of Sox2 gene expression in soft (Fig. 1B) but not in stiff fibrin gels (Supplemental Fig. 3B), possibly because Sox2 is already very low in stiff fibrin gels [10]. On the other hand, silencing FAK, Cdc42, or RhoA in control melanoma cells increased colony growth in stiff fibrin matrices (Fig. 1C), possibly due to upregulation of Sox2 expression (Fig. 1D). Interestingly, silencing FAK or Cdc42 but not RhoA promoted colony growth in soft fibrin matrices (Supplemental Fig. 3C) without upregulating Sox2 gene expression (Supplemental Fig. 3D), likely because Sox2 expression was already very high in the soft matrices and thus could not be elevated further. These findings suggest that FAK and its downstream molecules Cdc42 and RhoA may negatively regulate the growth of TRCs and that local matrix stiffness influences tumor growth by modulating Sox2 expression through these genes.
Fig. 1. FAK and Cdc42 and RhoA regulate growth of tumor-repopulating cells.
Overexpression of FAK, Cdc42, or RhoA inhibits the colony growth (A) and Sox2 expression (B) of TRCs in soft fibrin matrices. Inhibition of FAK, Cdc42, or RhoA promotes the colony growth (C) and Sox2 expression (D) of control melanoma cells in stiff fibrin matrices. Colony growth was monitored from day 1 to day 5 (n=30 colonies per condition). In (A), significant differences between TRC and TRC+FAK cDNA, TRC+Cdc42 cDNA, or TRC+RhoA V14 from day 3 to day 5. In (C), significant differences between Neg Ctr (negative control) and FAK siRNA #1, Cdc42 siRNA #1, or RhoA siRNA from day 2 to day 5. After 5 days, the mRNAs were extracted for analysis of Sox2 expression by qPCR in (B) and (D) (n=3 independent experiments). *P<0.05 in all subfigures.
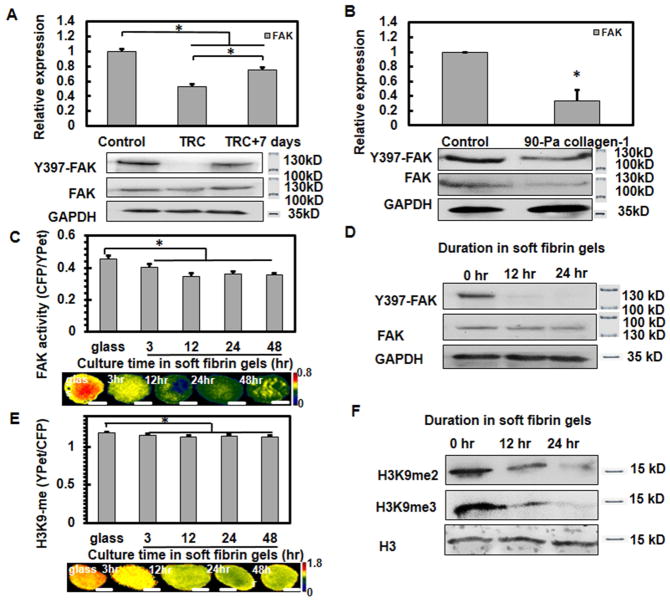
Next we examined whether matrix rigidity might influence FAK in melanoma cells. In comparison to control melanoma cells, TRCs expressed lower FAK mRNA (Fig. 2A, top panel), lower level of FAK protein and substantially lower Y-397 phosphorylation of FAK (Fig. 2A, bottom panel,), suggested that both of total FAK protein amount and FAK activity decreased in TRCs. Because TRCs could maintain their mechanical memory on rigid glass for 5 days [10], we re-plated TRCs back onto 2D rigid glass for 7 days and found that FAK mRNAs and activity were significantly increased toward the level of the control B16 cells (Fig. 2A). We further assayed colony growth in stiff fibrin matrices after inhibiting FAK activity pharmacologically with PF573228, and found that inhibiting FAK activity without changing FAK amounts also significantly increased colony growth in stiff fibrin matrices (Supplemental Fig. 4), suggesting that both of the FAK protein amount and activity contribute to TRC growth. To further explore if FAK downregulation in TRCs was due to lack of β1 integrin attachment, we cultured B16 cells on 90 Pa collagen-1 gels surface and compared FAK activity with control cells on collagen-1 coated rigid plastic. FAK mRNAs, protein levels, and activity all significantly decreased when compared with control cells (Fig. 2B), suggesting that low FAK levels and low FAK activity are a result of the low substrate stiffness and not due to difference in integrin subtypes. The findings of low FAK amount and activity in B16 melanoma cells in 90-Pa fibrin gels were confirmed in human breast cancer MCF-7 cells and human lung cancer A549 cells cultured in 90 Pa fibrin gels (Supplemental Fig. 2C, D), suggesting that low FAK in growing TRCs in soft matrices is not limited to murine melanoma cells.
Fig. 2. FAK activity and H3K9 methylation are tightly associated.
(A) TRCs exhibit low FAK gene expression (top panel) and activity (bottom panel). (B) FAK activity decreases in cell cultured on 90-Pa collagen-1 gels surface. Control melanoma cells decrease FAK activity (C, D) and H3K9 methylation (E, F) after culture in soft fibrin matrices. Control cells were transfected with FAK or H3K9 biosensors on rigid plastic and then cultured in soft fibrin matrices. n=35 cells per condition; *P<0.05. Representative FRET images were shown in the bottom panels of (C, E). Scale bars: 10 μm. The proteins of these cells were extracted for analysis of FAK and H3K9 methylation by western blots. Representative images of 3 independent experiments.
Association of low FAK activity with low H3K9 methylation
Next, we examined how FAK influenced Sox2 expression. B16 TRCs exhibited lower gene expressions of H3K9 methyltransferases EHMT2 and SUV39h1 and lower H3K9 di- and tri-methylation [10]. To quantify FAK activity and H3K9 methylation at the single cell level, we used a FAK biosensor [12] and a H3K9 methylation biosensor [10, 13], both of which are based on the principle of fluorescence resonance energy transfer (FRET). To better understand the underlying mechanisms, we set out to determine the dynamic changes of FAK activity and H3K9 methylation of melanoma cells at early time points (up to 48 hrs) after being cultured in soft fibrin gels. To rule out the effect of apoptosis, only cells with no detectable morphological signs of apoptosis (e.g., no shrinkage of cell volume and no blebbing) were chosen for experiments. Indeed, after FRET measurements, these same cells were able to continue to proliferate in the next few days. Our data show that both FAK activity and H3K9 methylation started to decrease as early as 3 hrs and stayed at the reduced levels up to 48 hrs after culture in soft (90-Pa) fibrin gels (Fig. 2C, E), which was further confirmed by western blotting assayed after 12 and 24 hrs in the soft gels (Fig. 2D, F). We recently has shown that B16 TRCs lose Sox2 expression on rigid substrates [10]. Here we found that FAK mRNA expression and activity (Supplemental Fig. 5A and 6A), EHMT2 and SUV39h1 mRNA expression (Supplemental Fig. 5B), and H3K9 methylation levels (Supplemental Fig. 6B) increased with culture time when TRCs were plated back (up to 7 days) onto the rigid glass. These results demonstrate that FAK activation levels and H3K9 methylation levels are closely associated in melanoma cells.
FAK regulates H3K9 methylation through small GTPases
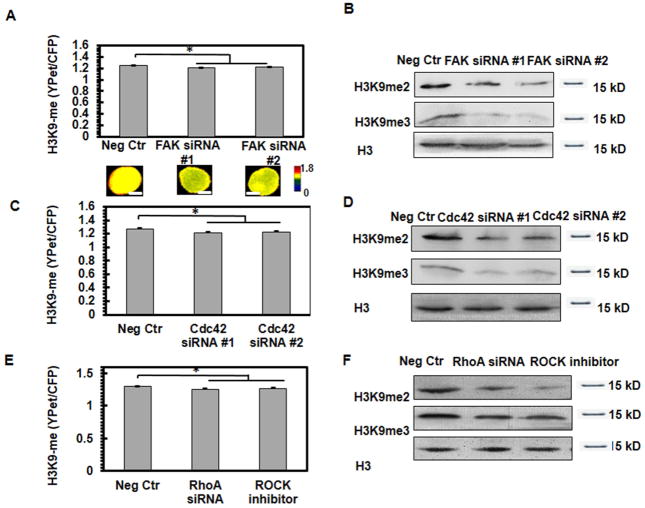
Since FAK is a cytoplasmic protein tyrosine kinase that serves as a critical mechanosensor in integrin-mediated mechanotransduction [15, 16, 20, 21], we wondered whether soft fibrin matrices induce H3K9 demethylation in the nucleus through FAK. Silencing FAK significantly decreased H3K9 methylation in control melanoma cells on the rigid glass (Fig. 3A and 3B; Supplemental Fig. 8A). Melanoma cells elevated their H3K9 methylation on 2D stiff substrates (8-kPa) or in 3D stiff fibrin gels (1050-Pa) compared to cells on soft substrates (0.15-kPa) or to cells in 3D soft (90-Pa) fibrin gels, which was abolished after FAK inhibition (Supplemental Fig. 7A and 7B, Supplemental Fig. 8B). Overexpressing FAK in melanoma cells increased H3K9 methylation in 3D soft fibrin gels but not on the rigid glass (Supplemental Fig. 7C and 7D). These results suggest that FAK regulates H3K9 methylation.
Fig. 3. FAK regulates H3K9 methylation through small GTPases.
Silencing FAK (A, B), Cdc42 (C, D) or RhoA (E, F) decreases H3K9 methylation. Control melanoma cells were transfected with H3K9 biosensors and siRNAs or treated with 10 μM ROCK inhibitor Y-27632. n=35 cells per condition; *P<0.05. Representative FRET images were shown in the bottom panels of (A). Scale bars, 5 μm. The proteins of these cells were extracted for analysis of H3K9 methylation by western blots. Representative images of 3 independent experiments.
As the downstream proteins of FAK and key cytoskeleton regulators[19, 22–24], Cdc42 and RhoA gene expression levels were downregulated in TRCs compared to control melanoma cells and both were increased after plating TRCs back to the rigid glass (Supplemental Fig. 5C, D). We have shown previously that silencing Cdc42 significantly decreased H3K9 methylation of control melanoma cells [10], which is supported by the data at the single-cell level on the rigid glass (Fig. 3C, D and Supplemental Fig. 8C). On the other hand, knocking down RhoA or inhibiting Rho kinase (ROCK) with 10 μM Y-27632 in control melanoma cells significantly decreased H3K9 methylation (Fig. 3E, F and Supplemental Fig. 8D. These data suggest that both Cdc42 and RhoA regulate H3K9 methylation. To determine if FAK regulates H3K9 methylation through Cdc42 and RhoA, we knocked down FAK but overexpressed Cdc42 or RhoA or transfected RhoA V14 in control melanoma cells and then cultured them on 2D substrates. Overexpressing Cdc42 or RhoA or transfecting RhoA-V14 restored H3K9 methylation levels even when FAK was silenced (Supplemental Fig. 7E and 7F). Interestingly, when RhoA was silenced, overexpressing Cdc42 restored H3K9 methylation levels (Supplemental Fig. 9A and 9B), suggesting that there might be an interplay between Cdc42 and RhoA in regulating H3K9 methylation.
Low FAK mediates H3K9 demethylation and Sox2 upregulation
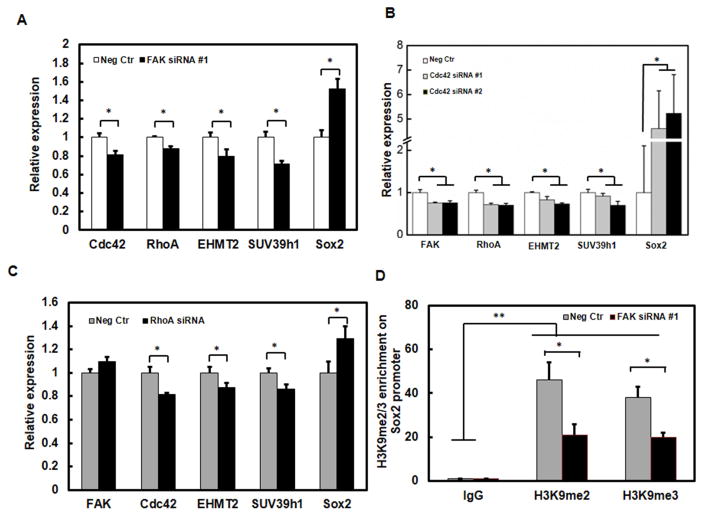
We have recently reported that H3K9 demethylation increases Sox2 expression and growth of TRCs [10]. We further asked whether and how low levels of FAK promote Sox2 expression in TRCs. In control melanoma cells, knocking down FAK inhibited the gene expressions of Cdc42, RhoA, EHMT2, and SUV39h1 and increased Sox2 gene expression (Fig. 4A); silencing Cdc42 decreased the gene expressions of FAK, RhoA, EHMT2, and SUV39h1 and increased Sox2 gene expression (Fig. 4B); inhibiting RhoA had no effect on FAK expression but significantly inhibited the gene expressions of Cdc42, EHMT2, and SUV39h1 and increased Sox2 expression (Fig. 4C). Our previous chromatin immunoprecipitation (ChIP) data showed that EHMT2 and SUV39h1 methylated H3K9 at the Sox2 promoter site block Sox2 expression and that silencing EHMT2 or SUV39h1 significantly increased Sox2 expression. Importantly, EHMT2 and SUV39h1 are important in regulating tumor cell growth in soft fibrin gels [10]. To further explore if FAK-Cdc42/RhoA-H3K9 methylation pathway regulate Sox2 directly by binding to its promoter region, we performed ChIP assay to test the H3K9 methylation levels on Sox2 promoter region after silencing FAK. We found that H3K9 methylation levels on Sox2 promoter region were significantly downregulated after silencing FAK (Fig. 4D). These findings suggest that low FAK induces H3K9 demethylation at the Sox2 promoter region and thus Sox2 upregulation via downregulation of Cdc42 and RhoA.
Fig. 4. Sox2 expression is regulated by low FAK mediated H3K9 demethylation via Cdc42 and RhoA.
(A) Knockdown of FAK decreases the expressions of Cdc42, RhoA, EHMT2, and SUV39h1 and increases Sox2 expression. (B) Knockdown of Cdc42 decreases the expressions of FAK, RhoA, EHMT2, and SUV39h1 and increases Sox2 expression. (C) Knockdown of RhoA decreases the expressions of Cdc42, EHMT2, and SUV39h1 and increases Sox2 expression. Control melanoma cells were transfected with siRNAs. The mRNAs were then extracted for analysis of the indicated genes by qPCR. (D) H3K9 di- (me2) and tri-methylation (me3) levels in the promoter region of Sox2 quantified by the ChIP assay. Relative enrichment was determined by qPCR. Mean ± s.e.m; n=3; **P<0.01; *P<0.05.
DISCUSSION
In the current study, we have found that compared to control melanoma cells on the rigid glass, CSC like TRCs exhibit low amount and low Y397 phosphorylation of FAK and high Sox2 expression. Recently we have demonstrated that TRCs are undifferentiated cells since they express low levels of Mitf and high levels of Sox2 [10, 25]. Importantly, we find that silencing FAK in control melanoma cells increases Sox2 expression in stiff but not in soft fibrin matrices and promotes growth in both soft and stiff fibrin matrices, whereas overexpressing FAK in TRCs decreases Sox2 expression and suppresses growth in soft but not in stiff fibrin matrices.
It has been reported that FAK is overexpressed in several types of cancer and responsible for high tumorigenicity and metastatic potential of tumor cells [26–28]. These findings have led to the development of FAK inhibitors as a potential anticancer therapy. The discrepancy between previous reports and our current study may result from the fact that most of the total population of tumor cells are probably differentiated tumor cells in those previous reports whereas only undifferentiated TRCs are analyzed in our present study [10]. Our results are consistent with the previous findings that the reduction in FAK activity promotes growth of stem cells [29] while the elevation in FAK activity facilitates their differentiation [30]. These findings may explain why FAK inhibition may not block tumorigenic cell proliferation as the current strategy in clinical trials of the candidate drugs to inhibit FAK does not work [31]. I Nevertheless, we acknowledge that our current findings are limited only to a few types of tumor cells and whether it can be extended to other types of cancer or to in vivo condition should be interrogated rigorously.
Based on our current findings, we propose a working model for the signaling pathways of soft-fibrin induced TRC’s growth (Supplemental Fig. 10). Cells sense mechanical forces from soft fibrin matrices via integrin subset αvβ3, which is the receptor of fibrin/fibrinogen [4]. FAK, one of the first molecules downstream of integrin, serves as a mechanosensor and mediates force transduction into the cells. Soft fibrin matrices downregulate FAK that lowers Cdc42 and RhoA. The reduced levels of Cdc42 and RhoA reorganize the cytoskeletal structure, disassemble the stress fibers, and decrease the cytoskeletal tension [32]. However, it is still not clear how Cdc42 and RhoA regulate H3K9 methylation in the nucleus. One possibility is that LINC (Liner of Nucleoskeleton of Cytoskeleton complex) couples the propagation of low forces into the nucleus [33]. Low forces further induce H3K9 demethylation at the region of Sox2 promoter [10], which promotes Sox2 expression and TRC growth. In addition, it is well-documented that histone modification including H3K9 methylation regulates nuclear architecture and chromatin assembly and condensation [34]. Nevertheless, the exact signaling pathways of matrix-softness-dependent tumorigenicity remain to be elucidated in the future.
Supplementary Material
Highlights.
FAK activity, mRNA and protein are low in highly tumorigenic cells.
FAK is tightly associated with H3K9 methylation and negatively related to the growth of tumor-repopulating cells, a subpopulation of melanoma cells.
Silencing or inhibiting FAK promotes rather than blocks the growth of highly tumorigenic cells. Overexpressing FAK downregulates the growth of the tumorigenic cells.
Matrix stiffness (rigidity) is a key player in regulating the response of the melanoma cells to FAK modulation via RhoA and Cdc42.
Acknowledgments
We thank Dr. Yingxiao Wang of UCSD for kindly providing the FAK biosensor.
Funding
This work was supported by National Institutes of Health grant GM072744, Ministry of Science and Technology of China grant 2016YFA0101100, and by Huazhong University of Science and Technology.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
N.W. conceived the project; Y.T., A.R.W., Q.J., and N.W. designed the experiments; Y.T., A.R.W., and Q.J. carried out most of the experiments and analyzed the data; W.Z., J.L., F.Y., J.W.C., and J.J.C. performed the western blotting assay and RT-PCR; Jian S. performed some experiments of qPCR; J.S., A.T., and R.S. assisted and commented on the project; N.W., Y.T., and Q.J. wrote the manuscript with inputs from other authors.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lapidot T, Sirard C, Vormoor J, et al. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature. 1994;367:645–8. doi: 10.1038/367645a0. [DOI] [PubMed] [Google Scholar]
- 2.Singh SK, Hawkins C, Clarke ID, et al. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 3.Chen J, Li Y, Yu TS, et al. A restricted cell population propagates glioblastoma growth after chemotherapy. Nature. 2012;488:522–6. doi: 10.1038/nature11287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu J, Tan Y, Zhang H, et al. Soft fibrin gels promote selection and growth of tumorigenic cells. Nat Mater. 2012;11:734–41. doi: 10.1038/nmat3361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Driessens G, Beck B, Caauwe A, et al. Defining the mode of tumour growth by clonal analysis. Nature. 2012;488:527–30. doi: 10.1038/nature11344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schepers AG, Snippert HJ, Stange DE, et al. Lineage tracing reveals Lgr5+ stem cell activity in mouse intestinal adenomas. Science. 2012;337:730–5. doi: 10.1126/science.1224676. [DOI] [PubMed] [Google Scholar]
- 7.Ricci-Vitiani L, Lombardi DG, Pilozzi E, et al. Identification and expansion of human colon-cancer-initiating cells. Nature. 2007;445:111–5. doi: 10.1038/nature05384. [DOI] [PubMed] [Google Scholar]
- 8.Dean M, Fojo T, Bates S. Tumour stem cells and drug resistance. Nat Rev Cancer. 2005;5:275–84. doi: 10.1038/nrc1590. [DOI] [PubMed] [Google Scholar]
- 9.Baumann M, Krause M, Hill R. Exploring the role of cancer stem cells in radioresistance. Nat Rev Cancer. 2008;8:545–54. doi: 10.1038/nrc2419. [DOI] [PubMed] [Google Scholar]
- 10.Tan Y, Tajik A, Chen J, et al. Matrix softness regulates plasticity of tumour-repopulating cells via H3K9 demethylation and Sox2 expression. Nat Commun. 2014;5:4619. doi: 10.1038/ncomms5619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang YL, Pelham RJ. Preparation of a flexible, porous polyacrylamide substrate for mechanical studies of cultured cells. Methods Enzymol. 1998;298:489–496. doi: 10.1016/s0076-6879(98)98041-7. [DOI] [PubMed] [Google Scholar]
- 12.Seong J, Ouyang M, Kim T, et al. Detection of focal adhesion kinase activation at membrane microdomains by fluorescence resonance energy transfer. Nat Commun. 2011;2:406. doi: 10.1038/ncomms1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin CW, Jao CY, Ting AY. Genetically encoded fluorescent reporters of histone methylation in living cells. J Am Chem Soc. 2004;126:5982–3. doi: 10.1021/ja038854h. [DOI] [PubMed] [Google Scholar]
- 14.Chen HC, Guan JL. Association of focal adhesion kinase with its potential substrate phosphatidylinositol 3-kinase. Proc Natl Acad Sci. 1994;91:10148–10152. doi: 10.1073/pnas.91.21.10148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Geiger B, Spatz JP, Bershadsky AD. Environmental sensing through focal adhesions. Nat Rev Mol Cell Biol. 2009;10:21–33. doi: 10.1038/nrm2593. [DOI] [PubMed] [Google Scholar]
- 16.Wang HB, Dembo M, Hanks SK, et al. Focal adhesion kinase is involved in mechanosensing during fibroblast migration. Proc Natl Acad Sci U S A. 2001;98:11295–300. doi: 10.1073/pnas.201201198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao J, Guan JL. Signal transduction by focal adhesion kinase in cancer. Cancer Metastasis Rev. 2009;28:35–49. doi: 10.1007/s10555-008-9165-4. [DOI] [PubMed] [Google Scholar]
- 18.Itoh S. Role of Expression of Focal Adhesion Kinase in Progression of Hepatocellular Carcinoma. Clin Cancer Res. 2004;10:2812–2817. doi: 10.1158/1078-0432.ccr-1046-03. [DOI] [PubMed] [Google Scholar]
- 19.McLean GW, Carragher NO, Avizienyte E, et al. The role of focal-adhesion kinase in cancer - a new therapeutic opportunity. Nat Rev Cancer. 2005;5:505–15. doi: 10.1038/nrc1647. [DOI] [PubMed] [Google Scholar]
- 20.Wu Y, Zhang K, Seong J, et al. In-situ coupling between kinase activities and protein dynamics within single focal adhesions. Sci Rep. 2016;6:29377. doi: 10.1038/srep29377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iskratsch T, Wolfenson H, Sheetz MP. Appreciating force and shape — the rise of mechanotransduction in cell biology. Nat Rev Mol Cell Biol. 2014;15:825–833. doi: 10.1038/nrm3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Han JW, Lee HJ, Bae GU, et al. Promyogenic function of Integrin/FAK signaling is mediated by Cdo, Cdc42 and MyoD. Cell Signal. 2011;23:1162–9. doi: 10.1016/j.cellsig.2011.03.001. [DOI] [PubMed] [Google Scholar]
- 23.Myers JP, Robles E, Ducharme-Smith A, et al. Focal adhesion kinase modulates Cdc42 activity downstream of positive and negative axon guidance cues. J Cell Sci. 2012;125:2918–29. doi: 10.1242/jcs.100107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nobes CD, Hall A. Rho, Rac, Cdc42 GTPases regulate the assembly of multimolecular focal complexes associated with actin stress fibers, lamellipodia, and filopodia. Cell. 1995;81:53–62. doi: 10.1016/0092-8674(95)90370-4. [DOI] [PubMed] [Google Scholar]
- 25.Hemesath TJ, Steingrimsson E, McGill G, et al. Microphthalmia, a critical factor in melanocyte development, defines a discrete transcription factor family. Genes Dev. 1994;8:2770–80. doi: 10.1101/gad.8.22.2770. [DOI] [PubMed] [Google Scholar]
- 26.Ashton GH, Morton JP, Myant K, et al. Focal adhesion kinase is required for intestinal regeneration and tumorigenesis downstream of Wnt/c-Myc signaling. Dev Cell. 2010;19:259–69. doi: 10.1016/j.devcel.2010.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lahlou H, Sanguin-Gendreau V, Zuo D, et al. Mammary epithelial-specific disruption of the focal adhesion kinase blocks mammary tumor progression. Proc Natl Acad Sci. 2007;104:20302–20307. doi: 10.1073/pnas.0710091104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fan H, Zhao X, Sun S, et al. Function of focal adhesion kinase scaffolding to mediate endophilin A2 phosphorylation promotes epithelial-mesenchymal transition and mammary cancer stem cell activities in vivo. J Biol Chem. 2013;288:3322–33. doi: 10.1074/jbc.M112.420497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wrighton PJ, Klim JR, Hernandez BA, et al. Signals from the surface modulate differentiation of human pluripotent stem cells through glycosaminoglycans, integrins. Proc Natl Acad SciU SA. 2014;111:18126–31. doi: 10.1073/pnas.1409525111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murray P, Prewitz M, Hopp I, et al. The self-renewal of mouse embryonic stem cells is regulated by cell-substratum adhesion and cell spreading. Int J Biochem Cell Biol. 2013;45:2698–705. doi: 10.1016/j.biocel.2013.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kaiser J. The cancer stem cell gamble. Science. 2015;347:226–9. doi: 10.1126/science.347.6219.226. [DOI] [PubMed] [Google Scholar]
- 32.Sit ST, Manser E. Rho GTPases and their role in organizing the actin cytoskeleton. J Cell Sci. 2011;124:679–83. doi: 10.1242/jcs.064964. [DOI] [PubMed] [Google Scholar]
- 33.Tajik A, Zhang Y, Wei F, et al. Transcription upregulation via force-induced direct stretching of chromatin. Nat Mater. 2016;15:1287–1296. doi: 10.1038/nmat4729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Towbin BD, González-Aguilera C, Sack R, et al. Step-wise methylation of histone H3K9 positions heterochromatin at the nuclear periphery. Cell. 2012;150:934–47. doi: 10.1016/j.cell.2012.06.051. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.