Abstract
Aminoacyl-tRNA synthetases (ARSs) are ubiquitously expressed, essential enzymes responsible for charging tRNA with cognate amino acids—the first step in protein synthesis. ARSs are required for protein translation in the cytoplasm and mitochondria of all cells. Surprisingly, mutations in 28 of the 37 nuclear-encoded human ARS genes have been linked to a variety of recessive and dominant tissue-specific disorders. Current data sustains that impaired enzyme function is a robust predictor of the pathogenicity of ARS mutations. However, experimental model systems that distinguish between pathogenic and non-pathogenic ARS variants are required for implicating newly identified ARS mutations in disease. Here, we outline strategies to assist in predicting the pathogenicity of ARS variants and urge cautious evaluation of genetic and functional data prior to linking an ARS mutation to a human disease phenotype.
Mutations in nuclear-encoded ARS enzymes cause human inherited disease
Aminoacyl-tRNA synthetases (ARSs) are ubiquitously expressed, essential enzymes that charge tRNA molecules with cognate amino acids in the cytoplasm and mitochondria. The human nuclear genome harbors 37 ARS loci: 17 that encode a cytoplasmic enzyme, 17 that encode a mitochondrial enzyme, and three that encode a bi-functional enzyme that charges tRNA for both cytoplasmic and mitochondrial protein translation [1]. One of the more interesting, albeit perplexing, findings in ARS research is that mutations in many of the genes encoding these enzymes cause myriad tissue-specific human diseases. As such, determining the genetic heterogeneity of ARS-related disease and the pathogenic mechanism of each disease-associated ARS mutation will be important for patient diagnosis, prognosis, and treatment. As the number of ARS mutations implicated in human disease rises, it is important to reflect on our current knowledge of the consequences of ARS mutations so that the pathogenicity of newly identified alleles can be carefully evaluated.
To date, mutations in 28 aminoacyl-tRNA synthetase (ARS) genes have been implicated in a spectrum of inherited, single-gene (Mendelian) human disorders (Table 1)[2]. Genes encoding mitochondrial ARSs have been implicated in recessive syndromes, while those encoding cytoplasmic ARSs have been implicated in both recessive and dominant disorders. Not surprisingly, mutations in genes encoding mitochondrial ARS enzymes often cause phenotypes in tissues with high metabolic demands such as the brain, muscle, and liver, similar to pathogenic mutations in mitochondrial genes and in other nuclear genes that encode mitochondrial proteins. For example, mutations in mitochondrial glutamyl-(EARS2) and aspartyl-tRNA synthetase (DARS2) cause autosomal recessive leukoencephalopathy [3,4], and mutations in mitochondrial phenylalanyl-tRNA synthetase (FARS2) have been associated with liver disease, encephalopathy, and lactic acidosis [5]. However, mutations in other mitochondrial ARS enzymes appear to cause tissue-restricted phenotypes; for example, mutations in mitochondrial histidyl-tRNA synthetase (HARS2) cause autosomal recessive ovarian dysgenesis and sensorineural hearing loss [6].
Table 1.
ARS Loci Implicated in Human Disease
| Gene | Locus | Location of Protein Function | Mode of Inheritance | Disease Phenotype(s) | References |
|---|---|---|---|---|---|
| AARS | 16q22 | Cytoplasm | Autosomal dominant Autosomal recessive |
CMT2N EEIE29 |
[12,34,94] |
| AARS2 | 6p21.1 | Mitochondria | Autosomal recessive | Mitochondrial Infantile CMP Leukoencephalopathy with ovarian failure |
[63,75] |
| CARS2 | 13q34 | Mitochondria | Autosomal recessive | COXPD27 | [45] |
| DARS | 2q21.3 | Cytoplasm | Autosomal recessive | HBSL | [8] |
| DARS2 | 1q25.1 | Mitochondria | Autosomal recessive | LBSL | [4] |
| EARS2 | 16p12.2 | Mitochondria | Autosomal recessive | LTBL COXPD12 |
[3] |
| FARS2 | 6p25.1 | Mitochondria | Autosomal recessive | COXPD14 SPG77 |
[5,95,96] |
| GARS | 7p15 | Cytoplasm & Mitochondria | Autosomal dominant Autosomal recessive |
CMT2D dSMA-V Myalgia, CMP |
[10,97] |
| HARS | 5q31.3 | Cytoplasm | Autosomal dominant Autosomal recessive |
CMT2W Usher Syndrome 3B |
[14,98] |
| HARS2 | 5q31.3 | Mitochondria | Autosomal recessive | Perrault Syndrome 2 | [6] |
| IARS | 9q22.31 | Cytoplasm | Autosomal recessive | Intellectual disability, growth retardation, muscular hypotonia | [47] |
| IARS2 | 1q41 | Mitochondria | Autosomal recessive | CAGSSS; Leigh Syndrome | [77] |
| KARS | 16q23.1 | Cytoplasm & Mitochondria | Autosomal recessive | RI-CMTB DFNB89 Visual impairment, microcephaly, DD, seizures |
[48,99,100] |
| LARS | 5q32 | Cytoplasm | Autosomal recessive | Infantile hepatopathy | [101] |
| LARS2 | 3p21.31 | Mitochondria | Autosomal recessive | Perrault syndrome 4 HLASA |
[50,64] |
| MARS | 12q13.3 | Cytoplasm | Autosomal dominant Autosomal recessive |
CMT2U1 ILLD |
[52,102] |
| MARS2 | 2q33.1 | Mitochondria | Autosomal recessive | Spastic Ataxia 3 COXPD25 |
[53,103] |
| NARS2 | 11q14.1 | Mitochondria | Autosomal recessive | COXPD24 | [79] |
| PARS2 | 3p21.31 | Mitochondria | Autosomal recessive | Alpers syndrome | [78] |
| QARS | 3p21.31 | Cytoplasm & Mitochondria | Autosomal recessive | MSCCA | [54] |
| RARS | 5q34 | Cytoplasm | Autosomal recessive | HLD9 | [7] |
| RARS2 | 6q16.1 | Mitochondria | Autosomal recessive | PCH6 | [55] |
| SARS2 | 19q13.2 | Mitochondria | Autosomal recessive | HUPRA Syndrome | [56] |
| TARS2 | 1q21.2 | Mitochondria | Autosomal recessive | COXPD21 | [57] |
| VARS | 6p21.33 | Cytoplasm | Autosomal recessive | Severe DD, microcephaly, seizures | [104] |
| VARS2 | 6p21.33 | Mitochondria | Autosomal recessive | COXPD20 Encephaloardiomyopathy |
[57,105,106] |
| YARS | 1p35.1 | Cytoplasm | Autosomal dominant Autosomal recessive |
DI-CMTC Multi-system disease, DD, FTT |
[18,107] |
| YARS2 | 12p11.21 | Mitochondria | Autosomal recessive | MLASA2 | [58] |
Notes: CAGSSS: Cataracts, growth hormone deficiency, sensory neuropathy, sensorineural hearing loss, and skeletal dysplasia; CMP: Cardiomyopathy; CMT2D: Charcot Marie Tooth disease type 2D; CMT2N: Charcot Marie Tooth disease type 2N; CMT2U: Charcot Marie Tooth disease type 2U; CMT2W: Charcot Marie Tooth disease type 2W; COXPD12: Combined oxidative phosphorylation deficiency 12; COXPD20: Combined oxidative phosphorylation deficiency 20; COXPD21: Combined oxidative phosphorylation deficiency 21; COXPD24: Combined oxidative phosphorylation deficiency 24; COXPD25: Combined oxidative phosphorylation deficiency 25; DD: Developmental delay; DI-CMT: Dominant-intermediate Charcot Marie Tooth disease; dSMA-V: distal spinal muscular atrophy type V; EEIE29: Epileptic encephalopathy, early infantile, 29; FTT: failure to thrive; HBSL: Hypomyelination with brainstem and spinal cord involvement and leg spasticity; HLASA: Hydrops, lactic acidosis and sideroblastic anemia; HLD9: hypomyelinating leukodystrophy 9; HUPRA: Hyperuricemia, pulmonary hypertension, renal failure, and alkalosis; ILLD: Interstitial lung and liver disease; LBSL: Hypomyelination with brainstem and spinal cord involvement and elevated lactate; LTBL: Leukoencephalopathy with thalamus and brainstem involvement and high lactate; MLASA2: myopathy, lactic acidosis, and sideroblastic anemia; MSCCA: Progressive microcephaly, intractable seizures, and cerebral and cerebellar atrophy; PCH6: pontocerebellar hypoplasia type 6; RI-CMTB: Recessive-intermediate Charcot Marie Tooth disease type B; SPG77: spastic paraplegia 77.
While missense variants in MARS have been identified in patients with CMT, the genetic evidence is not strong enough to conclude, at this point, that this gene is associated with CMT disease.
Similar to mitochondrial ARSs, mutations in genes encoding cytoplasmic ARS enzymes cause a spectrum of recessive syndromes, many of which include neurological phenotypes (Table 1). For example, arginyl-(RARS) and aspartyl-tRNA synthetase (DARS) mutations cause autosomal recessive hypomyelination in the central nervous system [7,8], and homozygosity for Y454S histidyl-tRNA synthetase (HARS) has been implicated in Usher syndrome [9]. Interestingly, only a single dominant phenotype has been associated with heterozygosity for mutations in a cytoplasmic ARS; mutations in glycyl-(GARS), tyrosyl-(YARS), alanyl-(AARS), and histidyl-tRNA synthetase (HARS) cause autosomal dominant peripheral neuropathy [9–14] (Table 1). Also referred to as Charcot-Marie-Tooth (CMT) disease, peripheral neuropathies compose a heterogeneous class of neurodegenerative diseases characterized by muscle weakness and sensory loss in the distal extremities [15]. CMT disease is divided into two major subgroups based on the primary cell type affected, with CMT1 referring to demyelinating neuropathy caused by a defect in Schwann cells and CMT2 referring to axonal neuropathy caused by impaired axon function [16]. To date, CMT-causing ARS mutations have been primarily associated with dominantly inherited CMT2 indicating that the mutations are particularly detrimental to the long axons of the peripheral nervous system.
Since 28 of the 37 human ARS loci have been implicated in disease phenotypes, it is reasonable to predict that mutations in the remaining nine ARS genes may also cause human disease. Furthermore, newly discovered mutations in previously implicated ARS loci are being reported at a rapid rate. As such, each newly identified ARS gene and allele must be carefully evaluated to distinguish bona fide disease-causing mutations from rare, non-pathogenic variants coincidentally identified in a patient with a relevant phenotype. To achieve this goal, we need to utilize all of the current knowledge of well-characterized, disease-associated ARS mutations. Here, we present strategies for assessing the pathogenicity of ARS variants identified in patients with inherited disease and provide considerations for careful interpretation of the data generated from each genetic and functional approach.
ARS variant identification and validation
Linkage analysis
To identify a genetic locus involved in Mendelian disease pathogenesis, investigators must first ensure that the phenotype of interest is both monogenetic (i.e., caused by a single gene) and measurable [17]. When feasible, generating a pedigree that illustrates the familial structure and phenotypic information for each individual allows for interrogation of the inheritance pattern (e.g., dominant, recessive, or X-linked). Clinical data must be carefully collected to ensure that factors such as age of onset, incomplete penetrance, variable expressivity, and phenocopy do not confound the analysis. Furthermore, detailed clinical evaluations are critical for identifying families with similar phenotypes that may have disease-causing mutations in the same gene. For example, CMT-associated YARS mutations were identified through the analysis of two families with a similar dominant, intermediate form of CMT disease [11,18] and GARS mutations were identified through the analysis of unrelated families with a unique, upper-limb predominant form of CMT disease [10,19–23]. Once family and clinical data have been collected, various approaches can be used to identify disease-associated mutations including linkage, candidate gene, and whole-exome sequence (WES) analyses.
Linkage analysis is used to identify loci harboring a pathogenic mutation [24] by assessing for the co-segregation of genetic loci (e.g., polymorphic markers) and the disease phenotype (i.e., a marker of the disease-causing mutation) within pedigrees [17,24]. To perform linkage, affected and unaffected individuals are genotyped using genetic markers (e.g., SNPs or microsatellite repeats) at known locations throughout the genome. Subsequently, a logarithm of the odds of linkage (LOD) score [25] is calculated—log10[(1−θ)NR*θR/(0.5)NR+R]—where θ is the recombination fraction, ‘NR’ is the number of non-recombinant offspring, and ‘R’ is the number of recombinant offspring. A LOD score of +3 is required for linkage with a 5% error rate [26]; however, a LOD score of +3.3 is required to correct for multiple testing in genome-wide analyses [26]. LOD scores are additive across pedigrees, which underscores the importance of having multiple, well-phenotyped families in order to identify a disease-associated locus. Limitations of linkage studies include errors in genotyping and phenotyping individuals (e.g., misdiagnosis, phenocopy, late onset, and incomplete penetrance) and the limited resolution of genotyping platforms (i.e. the number of genetic markers assessed). Once a region of the genome has been linked to a disease phenotype, each gene (as well as non-coding sequences) can be interrogated for causative mutations.
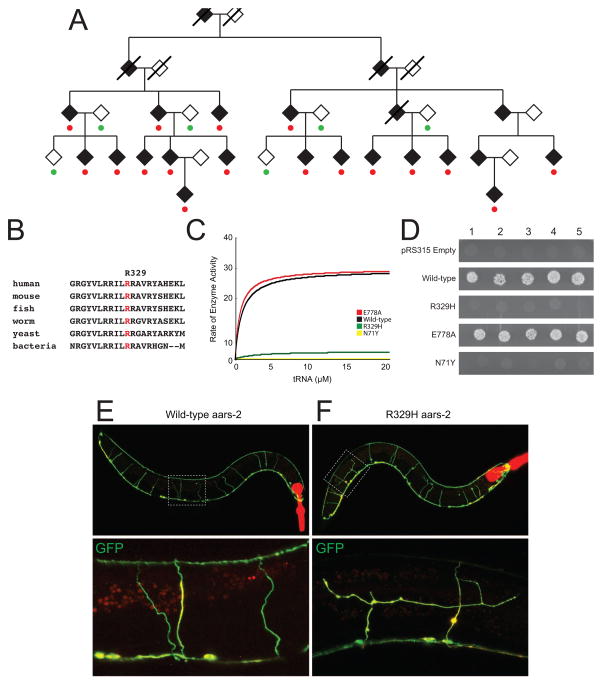
Linkage analysis in multiple, large pedigrees remains the most convincing approach for identifying single-gene disease loci; note that this includes linkage analysis using genome-wide sequencing data [27]. For example, R329H AARS was identified via linkage analysis of 17 affected individuals from a five-generation family with autosomal dominant axonal CMT disease (Figure 1A)[12]. This revealed a region on chromosome 16—containing the alanyl-tRNA synthetase (AARS) gene—with a maximum LOD score of 4.77. Since GARS and YARS mutations were already reported to cause CMT disease [10,11], subsequent mutation screening focused on AARS, which revealed that all affected individuals carry the R329H missense mutation (Figure 1A). Genetic data that further supported the pathogenicity of R329H AARS in CMT disease include: (1) an additional small family with CMT disease carrying this mutation [12]; (2) an additional large, four-generation family with eight affected individuals with CMT disease that carry this mutation [13]; and (3) experimental data showing that R329H AARS is a recurrent mutation that has occurred on multiple, independent haplotypes [13]. Thus, there is strong genetic evidence that implicates R329H AARS in human disease (i.e., dominant CMT2). We will therefore use this mutation as an example in the approaches outlined below.
Figure 1. Characterization of the R329H AARS allele associated with dominant CMT disease.
(A) R329H AARS segregates with dominant CMT disease in a large pedigree. Filled symbols represent affected individuals, empty symbols represent unaffected individuals, and deceased individuals are crossed out. Red circles denote heterozygosity for the R329H AARS allele and green circles represent homozygosity for wild-type AARS. Diamonds were used to protect the identity of the family. (B) A multiple-species sequence alignment at the R329 amino-acid residue illustrates conservation among diverse species. The affected residue is indicated in red. (C) Aminoacylation results for tRNAALA charging for wild-type (red), E778A (black), R329H (green), and N71Y AARS (yellow). The rate of enzyme activity (pmol/min/pmol enzyme) is plotted against tRNA concentration. (D) Yeast complementation results for wild-type, N71Y, R329H, and E778A AARS modeled in the yeast ortholog, ALA1. Five independent cultures for each indicated genotype were grown on solid medium containing 5-FOA. (E and F) Confocal, fluorescence microscopy of animals over-expressing the wild-type (E) or mutant (R329H; F) worm AARS ortholog (aars-2, tagged with TagRFP. In each panel, merged images of GFP-filled GABA motor neurons (Punc-47::GFP) and the TagRFP::aars-2 fusion protein are shown. The bottom frames are high magnification images and boxes in the top frames indicate the zoomed in area. Panel A was adapted from Latour et al., 2010, and Panels C and D were adapted from McLaughlin et al., 2014.
Candidate gene studies
Linkage analysis is often not possible due to, for example, limited access to DNA samples or small pedigree sizes. In these cases, a candidate gene approach can be pursued where genes or gene families known or predicted to cause the observed phenotype are directly assessed for disease-causing mutations. For example, since mutations in three ARS enzymes (GARS, YARS, and AARS) had been implicated in dominantly inherited CMT disease [10–13], it was proposed that mutations in other ARS loci may lead to a similar phenotype[14]. To test this hypothesis, 355 patients with CMT disease and no known disease-causing mutation were screened for variants in the protein-coding sequences of all 37 ARS enzymes by PCR amplification and DNA sequence analysis. The results of these studies included a histidyl-tRNA synthetase variant (R137Q HARS) identified in a single individual with peripheral neuropathy [14]. While functional studies supported the pathogenicity of R137Q HARS (see below), the genetic evidence was insufficient to implicate HARS in the pathogenesis of peripheral neuropathy (demonstrating an important limitation to candidate gene approaches). However, these findings made HARS an attractive candidate gene for further genetic studies in individuals with peripheral neuropathy. Indeed, linkage analysis and whole-exome sequencing subsequently revealed four disease-associated HARS mutations in four families with peripheral neuropathy, which provided the necessary genetic evidence to implicate HARS in CMT disease [9].
Whole-exome sequencing analysis
As the cost of whole-exome (WES) and whole-genome (WGS) sequencing decreases, the use of these technologies to identify disease-causing mutations is becoming more common [28–30]. A major strength of WES and WGS is the ability to directly scrutinize all (or most) relevant nucleotides in patient DNA samples, where WES focuses on protein-coding sequences and WGS provides data from both coding and non-coding sequences (e.g., to include transcriptional regulatory elements). While variant filtering is often employed to prioritize mutations for follow up studies [31], WES and WGS can be combined with linkage analyses to build a statistical argument for pathogenicity [27]. Currently, WES is more commonly used due to the lower cost and the rationale that disease-associated mutations are likely to reside in and around protein-coding sequences. For example WES was used to identify aspartyl-tRNA synthetase (DARS) mutations in patients with recessive hypomyelination of the central nervous system [8]. In this study, the authors computationally prioritized all of the identified variants, which revealed DARS as the sole candidate gene for the disease phenotype. Caveats to WES include an inability to assess non-coding regions of the genome and an inability to capture 100% of protein-coding sequences in the genome during sequence library preparation [31]. However, the nature of the mutations and the identification of multiple DARS mutations in unrelated individuals with a nearly identical recessive phenotype provided strong genetic evidence to implicate DARS in human disease [8].
Considerations for building a genetic argument for pathogenicity
As discussed above, recent advances have increased our ability to identify disease-causing alleles in human populations. These include sequencing technologies that allow rapid and affordable data acquisition as well as a deeper knowledge of human disease genetics, which permits efficient mutation analysis in research and clinical settings. However, two major issues face the field of human genetics. First, erroneous assignment of a gene as disease-associated can occur if rigorous validation and statistical analyses are not performed [32]. Second, once a gene is implicated in disease based on a strong genetic argument, newly identified variants can be erroneously attributed as ‘pathogenic’, which can be overlooked if the variant was coincidentally identified in a patient with the correct disease phenotype. For example, S581L GARS was identified in a patient with peripheral neuropathy and was therefore classified as pathogenic [33]. However, genetic studies revealed that S581L GARS occurs in the general population (see below) and, more importantly, that this mutation does not segregate with CMT disease in two unrelated families, where affected individuals did not carry the variant [34]. These data strongly argue against a pathogenic role for S581L GARS in dominant CMT disease and illustrate the importance of strong genetic evidence before implicating newly identified alleles in a disease phenotype.
To initially assess the pathogenicity of a Mendelian disease-causing variant, three basic approaches have been employed. First, genotyping is performed on all available family members to ensure that the variant segregates with disease with respect to the inheritance pattern (i.e., all affected individuals have the expected genotype while all unaffected individuals do not). For example, all affected individuals in the pedigree with AARS-associated dominant CMT disease are heterozygous for R329H AARS and all unaffected individuals do not carry this mutation (Figure 1A) [12]. In contrast, S581L GARS does not segregate with disease in multiple families (see above) [34], which excludes it as a pathogenic mutation in these cases. There are, however, important caveats to this approach including phenocopy (e.g., certain individuals do not carry the mutation but have a phenotype similar to other family members that is due to non-genetic factors), incomplete penetrance (e.g., certain individuals carry the mutation but never express the disease phenotype, possibly due to environmental or genetic modifiers), and age of onset (e.g., certain individuals carry the mutation but have not reach an age required for disease manifestation).
Second, genotyping is performed on unaffected control individuals (i.e., individuals at the appropriate age that have been clinically assessed for the studied disease) and databases are scrutinized to rule out the presence of the variant in the general population. For example, R329H AARS was not detected in 1,086 chromosomes from individuals without neurological disease and has not been detected (as of the preparation of this manuscript) in the Exome Aggregation Consortium (ExAC) database (http://exac.broadinstitute.org) (doi: http://dx.doi.org/10.1101/030338) nor in the overlapping and updated genome Aggregate Database (gnomAD; http://gnomad.broadinstitute.org). In contrast, S581L GARS is present in both ExAC (16 out of 119,342 alleles) and gnomAD (54 out of 282,216 alleles); note that S581L is equivalent to S635L on an alternate protein isoform. Furthermore, 39 of the 54 S581L GARS alleles in gnomAD are from 10,144 alleles from individuals of Ashkenazi Jewish decent, underscoring the importance of employing ethnically matched control populations. Combined, these data are consistent with S581L GARS being a non-pathogenic allele. There are also caveats to relying on this approach. In addition to the issues discussed for segregation analysis, the presence of a variant in the general population may not exclude it from being pathogenic especially if the phenotype is characterized by a late onset or if the variant is observed in non-disease-associated genotypes (e.g., a heterozygous carrier for a mutation associated with a recessive disease). Indeed, G240R GARS has now been detected in gnomAD (1 out of 252,112 alleles; Table 2) and this mutation was identified via linkage analysis. To adjust for these possibilities, the population frequency of the variant should be compared to the frequency of that variant in patients with the disease phenotype.
Table 2.
Characteristics of ARS variants identified in patients with dominantly inherited disease
| Gene | Variant | Method of Discovery | Enzyme Activity | Yeast Viability | Detection in gnomAD | References |
|---|---|---|---|---|---|---|
| AARS | N71Y | Candidate gene sequencing | Reduced | Lethal | Not present | [13,108] |
| G102R | Candidate gene sequencing | NA | Lethal | Not present | [67] | |
| R326H | Linkage analysis | Reduced | Lethal | Not present | [12,13] | |
| D893N | Candidate gene sequencing | NA | NA | Not present | [109] | |
| GARS | A57V | Candidate gene sequencing | Reduced | NA | Not present | [34,110] |
| E71G | Linkage analysis | Normal | Viable | Not present | [10,65,80] | |
| L129P | Linkage analysis | Reduced | Reduced | Not present | [10,65,80] | |
| D146N | Whole-exome sequencing | Reduced | Reduced | Not present | [34,111] | |
| S211F | Whole-exome sequencing | Reduced | NA | Not present | [34,111] | |
| G240R | Linkage analysis | Reduced | Viable | 1/252,112 | [10,65,80] | |
| P244L | Candidate gene sequencing | Reduced | Lethal | Not present | [34,112] | |
| I280F | Candidate gene sequencing | Reduced | Viable | Not present | [33,34] | |
| H418R | Candidate gene sequencing | Reduced | Lethal | Not present | [34,65,80,113] | |
| D500N | Candidate gene sequencing | Normal | NA | 7/281,982 | [80,114] | |
| G526R | Candidate gene sequencing | Reduced | Lethal | Not present | [10,65,80,115] | |
| S581L1 | Candidate gene sequencing | Normal | NA | 54/282,216 | [33,80] | |
| G598A | Candidate gene sequencing | Reduced | Viable | Not present | [33,34,92] | |
| HARS | T132I | Linkage analysis | NA | Lethal | Not present | [9] |
| P134H | Linkage analysis and candidate gene sequencing | NA | Lethal | Not present | [9] | |
| R137Q | Candidate gene sequencing | NA | Lethal | 20/252,310 | [14] | |
| D175E | Whole-exome sequencing | NA | Reduced | Not present | [9] | |
| D364Y | Linkage analysis and whole-exome sequencing | NA | Lethal | Not present | [9] | |
| MARS3 | R618C | Whole-exome sequencing | NA | Lethal | 3/282,684 | [102] |
| P800T | Whole-exome sequencing | NA | NA | 11/252,290 | [116] | |
| YARS | G41R | Linkage analysis | Reduced | Lethal | Not present | [11,18,42] |
| 153–156Δ VKQV | Candidate gene sequencing | Reduced | NA | Not present | [11,42] | |
| E196K | Linkage analysis | Reduced2 | Reduced | Not present | [11,18,42] | |
| E196Q | Whole-exome sequencing | NA | Reduced | Not present | [60] |
S581L GARS was identified in a patient with axonal neuropathy via candidate gene screening and it is unclear if segregation analysis was performed in the family. Subsequent studies showed that S581L GARS does not segregate with neuropathy in two additional families. Please see text for details.
E196K YARS was reported as having reduced enzyme activity in one study [17] and as having normal activity in another study [41].
While missense variants in MARS have been identified in patients with CMT, the genetic evidence is not strong enough to conclude, at this point, that this gene is associated with CMT disease.
Finally, conservation analysis is performed to determine if the disease-associated variant occurs at a nucleotide, or affects an amino-acid residue, that is conserved among evolutionarily diverse species. For example, R329H AARS affects an arginine that is conserved between human and bacteria, suggesting that this residue is important for enzyme function and consistent with pathogenicity (Figure 1B) [12]. In contrast, S581 GARS is only conserved among vertebrate species [34] indicating that this residue may not be essential for enzyme function. While this approach can be useful for mutations that impair gene function, conservation may not be expected for mutations that cause a gain-of-function effect; however, in this case one would not predict to identify the disease-associated change in another species. Of course, assessing the cis-genomic context at individual loci will provide a more accurate assessment of conservation as it relates to mutation pathogenicity [35].
Functional studies to predict the pathogenicity of ARS variants
Linkage, statistical, and validation studies are required for implicating a mutation in any Mendelian disease. However, functional studies can also be used to assist in predicting, but not proving, pathogenicity. These studies are most useful when the functional consequences of a newly identified variant can be compared to the functional consequences of variants that have been implicated in disease via strong genetic data (e.g., R329H AARS). As mentioned, mutations in genes encoding ARS enzymes have been implicated in recessive and dominant disease phenotypes. In the case of recessive diseases, the inheritance patterns and phenotypes indicate a loss-of-function effect (Table 1). In the case of dominant CMT disease caused by ARS mutations, however, the pathogenic mechanism is less clear. While the majority (if not all) of CMT-causing variants impair ARS function (Table 2) it is not clear how this relates to the CMT disease phenotype [1,36,37]. In this section, we review model systems that have been employed to validate the pathogenicity of ARS mutations implicated in recessive and dominant disease phenotypes. We argue that, regardless of the downstream effect of the mutations, impaired ARS function is a valuable predictor of pathogenicity.
Biochemical studies: pyrophosphate release and aminoacylation assays
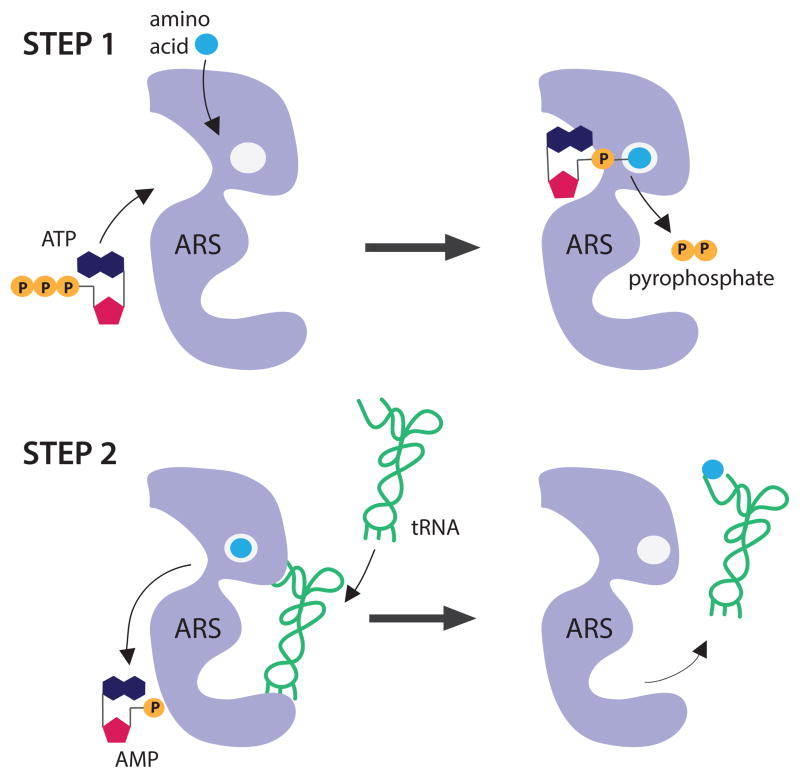
Two approaches have been used to study the effects of ARS mutations on enzyme function: in vitro enzyme kinetic assays and in vivo yeast complementation assays. ARS enzymes charge tRNA with cognate amino acids via a two-step reaction (Figure 2) [38]. The first step involves binding of the ARS to the amino acid and ATP to form the amino-adenylate intermediate, which results in pyrophosphate release. In the second step, the amino-adenylate intermediate recognizes and binds to the appropriate tRNA, which is then covalently linked to the amino acid. Pyrophosphate release and aminoacylation assays are used to determine the ability of mutant ARS enzymes to charge tRNA in vitro compared to the wild-type enzyme [39] with pyrophosphate release assays testing for completion of the first step of the reaction [40,41]. Briefly, purified recombinant ARS enzyme (mutant or wild-type) is incubated with tRNA, ATP, cognate amino acid, and inorganic pyrophosphatase. Pyrophosphate is released upon formation of the amino intermediate, which can be measured by the hydrolysis of PPi by pyrophosphatases. Addition of a colorimetric reagent is used to quantitate the amount of free phosphates and absorbance is proportional to the amount of amino-adenylate intermediate formed. Assays to evaluate the first step of aminoacylation were used to determine the effects of YARS mutations (E196K and G41R) implicated in dominant CMT disease, and HARS2 (L200V and V368L) and FARS2 (I329T) mutations implicated in recessive disease phenotypes [5,6,11]. These efforts revealed that all five mutant proteins have reduced activity compared to the respective wild-type enzyme; however, another study showed that E196K YARS completes the first step of aminoacylation in a manner similar to wild-type YARS (please see below) [42].
Figure 2. Schematic of the two-step aminoacylation reaction.
The aminoacyl-tRNA synthetase (ARS; purple), amino acid (blue), ATP, AMP, pyrophosphate (orange), and tRNA (green) are all indicated. The reaction occurs in two steps, as depicted. First, the ARS binds the amino acid and ATP to form the amino-adenylate intermediate, which releases pyrophosphate. In the second step, the ARS binds the cognate tRNA to facilitate transfer of the amino acid to the tRNA. The tRNA is then released for protein synthesis. The image represents a monomeric enzyme; however, the reaction proceeds similarly for oligomeric enzymes.
Aminoacylation assays test the completion of both steps of the charging reaction and have been more commonly used to assess the effect of human mutations on ARS function. Briefly, purified recombinant ARS enzyme (wild-type or mutant) is incubated with tRNA, ATP, and radiolabeled amino acid. Aliquots of the reaction mixture are collected, spotted on filter paper, and the tRNA is precipitated using TCA to remove any unincorporated amino acid. Radioactivity levels are then assessed to determine the amount of amino acid ligated to tRNA molecules. Steady-state kinetics are calculated by fitting the initial rate of aminoacylation as a function of tRNA concentration (or the concentration of another substrate) to the Michaelis–Menten equation [43]. Aminoacylation assays have been used to demonstrate a loss-of-function effect on tRNA charging for mutations in 15 genes implicated in recessive disease (AARS, CARS2, DARS2, FARS2, IARS, KARS, LARS2, MARS, MARS2, QARS, RARS2, SARS2, TARS2, VARS2, and YARS2) [4,44–59]. Interestingly, aminoacylation assays have also revealed a loss-of-function effect of GARS, YARS, and AARS mutations that cause dominant peripheral neuropathy (Table 2); HARS mutations associated with dominant CMT disease have not been tested. For example, R329H AARS has 1/50 the charging capacity compared to the wild-type AARS protein (Figure 1C) [13]; the disease-associated mutant protein N71Y AARS also has dramatically reduced activity (Figure 1C). In contrast, the rare, non-pathogenic variants E778A AARS and S581L GARS do not affect enzyme activity (Figure 1C and Table 2). These data indicate that tRNA charging assays should be employed to predict the pathogenicity of all newly identified ARS variants, with impaired function supporting a pathogenic role. However, additional functional analysis of non-pathogenic alleles is required to confirm that this assay can distinguish between disease-causing mutations and rare, non-pathogenic ARS variants.
Aminoacylation and pyrophosphate assays are limited in that they may not reflect the activity of ARS enzymes in living cells; for example, post-translational modifications or stabilizing cellular molecules may impact the function of the ARS enzyme in vivo [34]. Furthermore, there are a few notable discrepancies in the literature that question the rationale for using reduced enzyme activity as a predictor of ARS mutation pathogenicity, specifically in dominant CMT disease. For example, two studies assessed the effect of three CMT-associated YARS mutations (G41R, 153-156delVKQV, and E196K) on enzyme function. In one study, G41R and E196K had reduced activity in pyrophosphate exchange assays; 156delVKQV was not evaluated [11]. In the other study, G41R and 156delVKQV had reduced activity while E196K displayed activity similar to wild-type YARS [42]. Based on the normal activity of E196K YARS, the authors of the second study concluded that YARS-associated CMT disease is not caused by altered enzyme function. However, two studies reported that E196K YARS has reduced function when studied in vivo using yeast complementation assays (see below) [11,60] and another loss-of-function variant at this residue (E196Q) has been identified in a family with dominant CMT disease [60]; together, these data support an important role for E196 in YARS enzyme function.
Yeast complementation assays
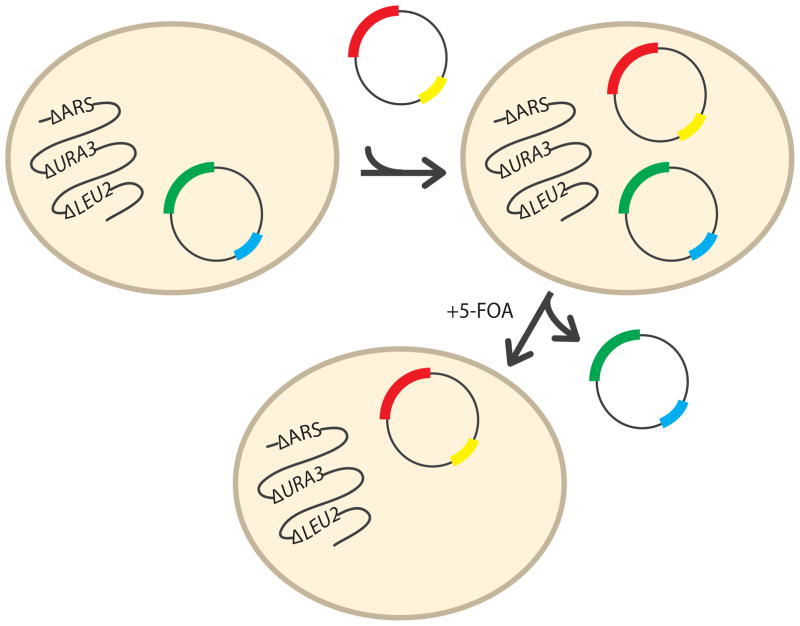
Baker’s yeast (Saccharomyces cerevisiae) is a robust, single-celled eukaryotic organism that is genetically tractable [61] and that shares certain biological features with mammalian cells, including ARS function. The effect of ARS mutations on gene function has been assessed via yeast complementation assays, which evaluate the ability of yeast cells to survive and grow in the presence of only the mutant ARS allele (Figure 3). Here, haploid yeast strains are developed with a non-functional (e.g., deleted) ARS gene. Since ARS proteins are essential for cell survival, a wild-type copy of the ARS gene under study is expressed from a maintenance vector that also encodes the yeast URA3 gene. URA3 is an enzyme required for the biosynthesis of pyrimidines and, importantly, is lethal to yeast cells in the presence of 5-fluoroorotic acid (5-FOA); URA3 converts 5-FOA to a toxic compound. Mutant ARS alleles are then modeled in either the yeast or human ARS ortholog and expressed from a second, experimental vector [62]. This system (Figure 3) allows the use of 5-FOA to select against the presence of the maintenance vector (containing the wild-type ARS locus) and to test for the ability of yeast cells to grow in the presence of only the experimental ARS allele (e.g., wild-type or mutant). Thus, a functional allele on the experimental vector will result in yeast cell growth while a non-functional ARS allele will result in no (or reduced) yeast cell growth; here, cellular growth is used as a proxy for enzyme function.
Figure 3. Overview of the yeast complementation assay.
Haploid yeast disrupted for the endogenous ARS (ΔARS) locus of interest are maintained viable via a maintenance vector bearing a wild-type copy of the respective ARS locus (green) and the URA3 gene (blue) for selection. Wild-type or mutant alleles (red) are introduced via a LEU2-bearing (yellow) vector and resulting strains are selected for in medium lacking uracil and leucine. Subsequent growth on medium containing 5-FOA allows for the selection of cells that have spontaneously lost the maintenance vector and thus growth is correlated to the function of the ARS allele (red) on the experimental vector.
Yeast complementation assays have been used to demonstrate a loss-of-function effect on ARS function for mutations in eight genes implicated in recessive diseases (AARS, AARS2, HARS2, IARS, KARS, LARS2, MARS, VARS2) [6,44,47,48,52,57,63,64]. Similar to aminoacylation assays, in vivo studies in yeast have revealed a loss-of-function effect for GARS, YARS, AARS, and HARS mutations that cause dominant CMT disease (Table 2). For example, R329H AARS modeled in the yeast ortholog ALA1 does not allow yeast cell growth consistent with a loss of enzyme function (Figure 1D). Similarly, the disease-associated N71Y ALA1 allele does not support yeast cell growth while the non-pathogenic E778A ALA1 allele allows growth similar to wild-type ALA1 (Figure 1D) [13]. Likewise, the T132I HARS mutation associated with dominant CMT disease impairs yeast viability while the rare population variant T132S HARS allele sustains viability [9]. Together, these data indicate that yeast complementation assays can be employed to predict the pathogenicity of newly identified ARS variants, with reduced yeast cell growth supporting a pathogenic role. As with aminoacylation assays, the inclusion of non-pathogenic alleles is essential for assessing the efficacy of yeast to predict pathogenicity. Indeed, the results from yeast complementation studies of AARS variants are consistent with the biochemical studies (Figure 1C and 1D) [13] and suggest that these assays can distinguish between disease-causing and benign AARS alleles; however, many more disease-associated mutations and non-pathogenic variants need to be assessed.
There are clear limitations to the utility of yeast complementation studies for assessing the consequences of ARS mutations. First, while all mutations that demonstrate decreased yeast cell growth also show impaired function in aminoacylation assays, the converse is not true (Table 2). For example, G240R GARS has dramatically reduced enzyme activity in vitro but supports yeast cell growth in vivo when modeled in the yeast ortholog GRS1 [34,65]. Explanations for this type of discrepancy include: (1) an inability of the yeast assay to resolve subtle differences in cell growth; (2) the growth of yeast at 30°C compared to the physiological environment afforded the human enzymes in vivo and the temperature at which aminoacylation assays are conducted (37°C); and (3) differences between the human and yeast ARS proteins that may lead to differential effects of mutations on the function of the two orthologs. Some of the above issues have been addressed: growth curves have been assessed to detect more subtle differences in yeast cell growth [11], certain GARS mutations modeled in GRS1 grow the same at 30°C and 37°C [34], and human YARS has been used to show a loss-of-function effect of YARS mutations in yeast [11]. However, the discrepancies outlined above warrant careful interpretation when studying human ARS mutations in a yeast model system. Second, yeast studies (similar to the kinetic assays) evaluate individual alleles in isolation, which is appropriate for assessing impaired function but does not allow an assessment of interactions between the two alleles that exist in a human patient. Finally, yeast are small, single-celled organisms, which do not represent differentiated human cells within a multicellular environment. Indeed, one mutation with strong genetic evidence that has yet to show reduced function in enzyme kinetic or yeast complementation assays (E71G GARS) [34,65] shows a loss-of-function effect in a fly model (see below) [66], warranting a more detailed characterization of the enzyme activity of the mutant protein. In summary, while caution is required when interpreting the data, yeast complementation assays are currently the most effective and efficient in vivo test for predicting the pathogenicity of ARS mutations [67].
Animal models to predict the pathogenicity of ARS variants
Assessing the functional consequences of ARS variants in multi-cellular organisms is important for validating the pathogenicity of ARS variants in human disease. Drosophila is a powerful model organism due to the short life cycle and complex neural network that recapitulates many aspects of the mammalian nervous system. Two fly models were developed to assess the pathogenicity of GARS and YARS mutations identified in patients with dominant CMT disease [66,68]. The mosaic analysis with a repressible marker (MARCM) system in combination with a forward genetic screen revealed a mutation in the Drosophila gars gene (P98L) that causes axonal and dendritic morphological defects in olfactory projection neurons [66]. Neurons homozygous for the P98L gars mutation had normal growth and guidance of axonal and dendritic stalks but were unable to properly arborize the distal axons and dendrites. To determine if the P98L mutation defects were due to loss of GARS function, a neuron-specific gars null allele was generated; neurons homozygous for the gars null allele had an identical phenotype to neurons homozygous for P98L. This model was then used to test the functional effects of human disease-associated GARS mutations (E71G and L129P) on neurons. Specifically, expression of wild-type human GARS rescued the gars−/− neuronal phenotype, while L129P did not and E71G only partially rescued axon growth. This study provided the first Drosophila model of ARS-mediated disease and supported a loss-of-function effect as part of the pathogenic mechanism of GARS-associated CMT disease [66]; however, non-pathogenic variants have yet to be evaluated in this system.
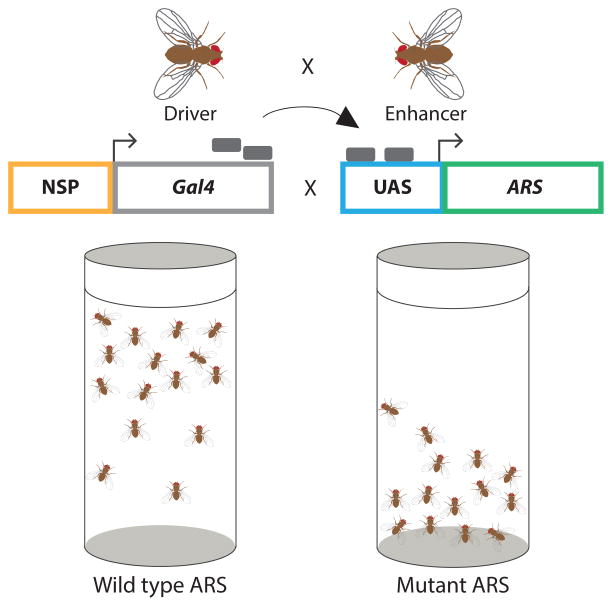
A second Drosophila model was developed to assess YARS mutations implicated in CMT disease for dominant toxicity (Figure 4) [69]. Here, mutant human alleles modeled in the fly ortholog were expressed in neurons from an inducible transgene. All three CMT-associated mutations tested (G41R, 153-156delVKQV, and E196K) caused a dominant climbing defect while wild-type YARS and a non-pathogenic variant (K265N) did not give rise to this phenotype. These data are critical for two reasons: (1) they reveal the fly model as effective in distinguishing between pathogenic alleles and rare, non-pathogenic variants; and (2) they demonstrate a dominant, toxic effect of YARS mutations consistent with the autosomal dominant patient phenotype and previous studies in yeast demonstrating a dominant-negative effect. In summary, informative fly models are available—and should be further employed—to evaluate the pathogenicity of newly identified ARS alleles.
Figure 4. Studying ARS toxicity in Drosophila.
Flies expressing Gal4 (grey rectangles) from a neuron-specific promoter (NSP) are crossed with flies bearing a transgene with the ARS cDNA of interest under control of an upstream activation sequence (UAS). In the progeny, Gal4 binds to the UAS to activate transcription of the ARS cDNA of interest in a neuron-specific fashion. Flies are then evaluated for their ability to climb using a negative geotaxis-climbing assay. Motor neuron function is quantified by the average climbing height of each cohort. The cartoon illustrates climbing defects observed in transgenic animals expressing a mutant (right), but not wild-type (left), ARS.
C. elegans is a nematode (roundworm) that has also proven to be a relevant model to test ARS variants for dominant toxicity. C. elegans has characteristics that make it particularly useful for studying neuron biology [70]. First, it has a short lifespan and can be synchronized to specific developmental stages, which allows the evaluation of gene mutations associated with late-onset diseases. Second, C. elegans is transparent and has a fully defined, non-myelinated nervous system that permits easy visualization of axonal defects in vivo. The worm GABAergic nervous system consists of 26 neurons, including 19 inhibitory (D-type) motor neurons that project commissural axons from nerve cell bodies located in the ventral nerve cord to the dorsal nerve cord [71]. These morphologically simple and largely invariant axonal projections innervate the body wall muscles of the worm and are critical for proper locomotion. Functional or morphological alterations in these axonal projections can lead to abnormal locomotion, which can be assessed using a battery of behavioral assays. For example, the thrash assay quantifies the number of lateral swimming movements an animal makes over time (thrashes min−1) and can be used to examine the fidelity of GABA nervous system connectivity and function [72]. Typically, animals that have abnormal D-type GABA motor neuron function display irregular body bending and a reduction in the number of body bends over time.
A C. elegans model has been developed to study the effects of loss-of-function, CMT-associated missense ARS mutations on neurons (Figure 5) [9,14]. Briefly, wild-type or mutant (R137Q and D364Y) C. elegans HARS cDNA was cloned downstream of a GABAergic neuron-specific promoter (Punc-25) and injected into a C. elegans strain that stably expresses GFP under the GABA-specific vesicular transporter gene (Punc-47) promoter. Offspring were synchronized to the fourth larval stage and aged for 1 to 4 days before axons were visualized using fluorescence microscopy to assess for nervous system abnormalities. Confocal imaging revealed abnormal axonal branching and blebbing, failure of axonal commissures to extend to the dorsal nerve cord, and large dorsal nerve cord gaps that progressed with age in animals expressing mutant but not wild type HARS. Decreased locomotion in thrash assays correlated with abnormal axonal morphology that also progressed with age in animals expressing the mutant proteins [9,14]. Similarly, overexpression of R329H AARS leads to axonal defects in worm, which is not observed upon overexpression of wild-type AARS (Figure 1E and 1F) (Antonellis and Beg, unpublished data). These results have implications similar to the YARS studies in fly. Specifically, they show that: (1) the worm may be useful for distinguishing between pathogenic ARS alleles and non-pathogenic variants; and (2) loss-of-function missense mutations are dominantly toxic in overexpression studies, consistent with the patient phenotype. We therefore suggest that the complementary fly and worm model systems should be further used to predict the pathogenicity of ARS alleles identified in patients with dominantly inherited CMT disease. Importantly, data sets from a broader panel of disease-causing and benign ARS alleles will provide key information on the utility of these model systems.
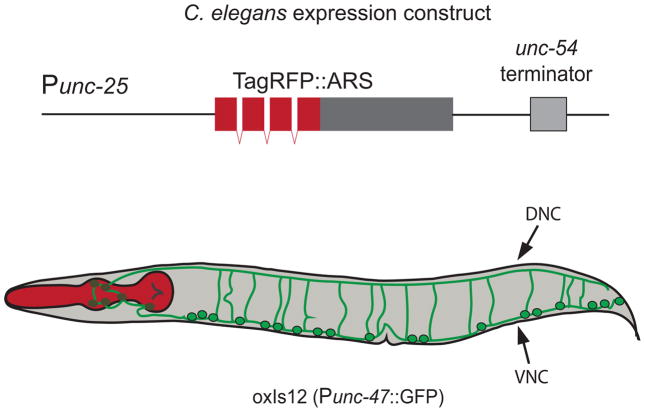
Figure 5. Studying ARS toxicity in C. elegans.
GABA-neuron-specific expression of a fluorescently tagged ARS protein (TagRFP::ARS) is achieved via a GABA-specific promoter (Punc-25). The construct is injected into the worm strain oxls12, which stably expresses GFP in GABAergic neurons via the unc-47 promoter (Punc-47). Axons (green) extend ventrally from the cell bodies (green, black outline). The dorsal (DNC) and ventral (VNC) nerve cords are indicated. Fluorescent microscopy is used to assess for abnormalities in axonal morphology (e.g., Figure 1F) and thrash assays are used to quantify motor neuron function.
Models to study the mechanism of ARS mutations in human disease
The functional strategies outlined above are useful for predicting the pathogenicity of newly identified ARS variants. Interestingly, the majority of previous studies employing these approaches revealed loss-of-function effects for ARS mutations implicated in recessive and dominant disease phenotypes. A loss-of-function effect is clearly the predominant hypothesis for the molecular pathology of ARS-associated recessive diseases, which is supported by functional analyses and the types of alleles identified (e.g., many patients are compound heterozygous for one frameshift or nonsense allele and one hypomorphic missense allele). However, even though many (if not all) ARS mutations implicated in dominant peripheral neuropathy (i.e., CMT disease) cause impaired enzyme function (Table 2), the precise mechanism of disease has not been defined; multiple gain- and loss-of-function hypotheses have been presented [2,36,73], and none of these mechanisms are mutually exclusive. For example, the loss-of-function characteristics of missense mutations identified in patients with dominant ARS-associated neuropathy in addition to the lack of complete null alleles (nonsense, frameshift, etc.) raises the possibility of a dominant-negative effect. Here, the mutant protein may interfere with, and reduce the function of, the remaining wild-type protein. As such, it is important to briefly consider the models that have advanced our understanding of the pathogenic mechanism of ARS-related disease toward developing accurate assays to test newly identified variants.
In addition to directly showing that ARS mutations impair enzyme kinetics, molecular and cellular biology techniques have been useful in teasing out the mechanism of ARS-associated disease. For example, mutations in mitochondrial ARS enzymes that cause recessive diseases can directly affect protein solubility [74] or impair mitochondrial protein translation [4,5,46,50,56–59,63,75–79], consistent with a loss-of-function effect. With respect to ARS mutations that cause dominant peripheral neuropathy, structural analyses have revealed that the majority of GARS mutations affect residues within the dimer interface [80]. Subsequent molecular studies have shown that these mutations alter the dimer interface, which may mediate a gain-of-function effect via the acquisition of protein partners (see below) [81,82]; one caveat of these latter studies is that this effect was also observed for S581L GARS [81], which is a non-pathogenic variant [34]. Finally, studies in cultured neurons have shown that certain GARS and YARS mutations alter the sub-cellular localization of the protein suggesting a localized loss-of-function effect [11,34].
Animal models have also been useful in determining how ARS mutations cause recessive and dominant human disease phenotypes. For example, gene knock-down studies showed that reduced glutaminyl-(qars) and isoleucyl-tRNA synthetase (iars) function in zebrafish cause, respectively: (i) reduced eye size and pigmentation, smaller brains, and lack of coordination; and (ii) growth retardation and brain deformity [47,54]. These data support a loss-of-function effect of QARS and IARS mutations in the associated autosomal recessive disease phenotypes (Table 1). Furthermore, the zebrafish models provide an in vivo system to test the pathogenicity of newly identified QARS and IARS alleles through determining if they are able to rescue the fish phenotype. The pathogenic role of ARS mutations in dominant CMT disease has also been studied in animal models including fly, zebrafish, and mouse. For example, analysis of three disease-associated GARS mutations and three disease-associated YARS mutations in flies revealed that each resulted in reduced protein translation compared to the respective wild-type proteins; however, this effect was independent of impaired tRNA charging and a gain-of-function mechanism was therefore proposed [83]. In contrast, studies in zebrafish have shown that loss-of-function gars alleles impair neuromuscular junction morphology (consistent with an axonal CMT disease phenotype) and that dimerization is required for missense mutations to have this effect [84]. These data support the dominant-negative mechanism associated with YARS mutations [11]; however, data directly implicating a dominant-negative effect of GARS mutations have not been reported. Finally, three mouse models of Gars mutations support a gain-of-function mechanism. These studies revealed that: (1) missense mutations in Gars cause dominant neurodegeneration in mouse [85,86]; (2) over-expression of wild-type human GARS does not rescue the mouse phenotypes [87]; (3) GARS mutations act via inappropriate binding to neuropilin-1 [81]; (4) impaired neuromuscular junction development followed by pre-synaptic defects are part of the neuropathy phenotype [88,89]; and (5) carnitine is decreased in the spinal cord of mutant Gars mice, and treatment with carnitine improves motor performance but had no effect on axon number, axon size, or neuromuscular junction innervation, deeming the relevance of these findings unclear [90]. Combined, extensive studies on neuropathy-associated ARS mutations suggest that both gain- and loss-of-function mechanisms may be at play. Interestingly, an additional mouse model revealed that homozygosity for a mutation in Aars that impairs editing of misacylated tRNAs causes recessive cerebellar Purkinje cell loss and ataxia, but no peripheral neuropathy [91,92]). While such mutations have not been identified in human populations, it may eventually be important to assess ARS editing activity when evaluating newly identified variants [13]. For all of the studies presented in this section, a more complete set of pathogenic and non-pathogenic alleles must be tested to validate the findings and to determine if the results are broadly applicable to ARS mutations and the respective patient phenotypes.
Moving forward: New disease-associated ARS loci and alleles
It is important to emphasize that building a strong genetic argument is essential for implicating any newly identified ARS locus or allele in human Mendelian disease. Specifically, the mode of inheritance (autosomal recessive or autosomal dominant; all ARS loci are encoded on autosomes) must be considered in the context of factors such as the disease prevalence, the frequency of the implicated variant in control populations, and the genotypes of individuals in control populations that carry the variant. A major issue facing human geneticists is the ease at which a newly identified variant in a known disease-causing gene can be erroneously reported as pathogenic; conclusive genetic data should also be required in these cases and we feel that S581L GARS serves as a cautionary tale [34].
In addition to genetic evidence for pathogenicity, we argue that our current understanding of disease-causing ARS mutations warrants the careful employment of functional studies to assist in predicting the pathogenicity of newly identified ARS alleles. For variants identified in patients with recessive phenotypes, testing for impaired ARS function using in vitro and in vivo model systems is rather straightforward; a loss-of-function mechanism is the leading hypothesis in these diseases. However, while the downstream effect on patient phenotypes is not clear, reduced gene function is also useful for predicting the pathogenicity of ARS missense variants identified in patients with dominant CMT disease. This argument is supported by studies showing that most, if not all, CMT-associated ARS missense mutations impair enzyme function (Table 2) and that loss-of-function missense mutations in GARS, YARS, AARS, and HARS have not, to date, been reported in unaffected individuals or in the general population. Multiple animal models have also been developed to test loss-of-function missense mutations for dominant toxicity and these models should be used to complement assays that test for the impaired function of ARS variants identified in patients with dominant disease.
Careful design and interpretation of functional studies is required to assess the consequences of ARS variants identified in patients with inherited diseases. First and foremost, positive and negative results should be interpreted in the context of the molecular, cellular, and tissue environment of the respective assay. For example, reduced enzyme activity should be tested in the appropriate tissue when feasible and substrate-specific effects (e.g., amino acid, tRNA, ATP) on enzyme activity should be considered before ruling out a loss-of-function mechanism. When performing yeast complementation assays, the human gene should be utilized if possible to rule out any species-specific effects of certain ARS mutations, and correlations between yeast data and in vitro tRNA charging data should be evaluated. Yeast assays also provide a rapid, tractable system to screen for molecules that can improve ARS function. Indeed, supplementation with methionine improved the in vivo function of mutant methionyl-tRNA synthetase (MARS) alleles that cause pulmonary alveolar proteinosis [52]. Finally, current worm and fly models rely on over-expression of the mutant allele to test for pathogenicity. Models should be developed to express ARS variants from the endogenous locus—similar to the mouse models of Gars mutations [85,86]—and should include the analysis of non-pathogenic control alleles.
ARS variants are being identified in patients with inherited disease at a rapid rate and databases such as gnomAD (i.e., ExAC) are expanding the number of known ARS variants in the general population. As such, it is becoming impractical to test the functional consequences of individual variants as they are identified. Screens are needed to assess the functional consequences of all possible ARS variants toward developing a dataset to allow rapid predictions of pathogenicity. For example, massively parallel mutagenesis [93] of the human GARS open-reading frame followed by growth assays in allelically heterogeneous yeast cultures would provide a quantitative assessment of allele function. If all possible GARS missense mutations were tested in such a system, the resulting data could be used by clinical geneticists to build an argument for or against the pathogenicity of a newly identified GARS allele; however, we caution that these data alone would not be sufficient to determine the role of a variant in disease. Tools are also available to test the pathogenic potential of mutations in any ARS locus. This is particularly important because, while it is clear that mutations in any ARS gene could lead to a severe recessive phenotype, it is not clear if mutations in any ARS could lead to dominant CMT disease or if there is something unique about the four loci implicated to date—GARS, YARS, AARS, and HARS. For example, potentially pathogenic missense variants in an ARS locus not yet associated with CMT disease could be predicted via conservation analysis. Subsequently, the missense variants could be tested for a loss-of-function effect in enzyme kinetic and yeast assays. Loss-of-function missense mutations could then be evaluated for dominant toxicity in fly or worm, with any toxic alleles modeled in mouse to test for a peripheral neuropathy phenotype. These experiments would inform the allelic and locus heterogeneity of ARS-associated CMT disease, provide additional models to study disease pathogenesis, and direct future patient screening. Importantly, non-pathogenic control alleles need to be included in the above experiments to confirm the ability to distinguish between pathogenic and benign variants, and the suggestions presented here will likely be modified as we gain more information on the pathological mechanisms of ARS-associated disease.
Highlights.
Aminoacyl-tRNA synthetases (ARSs) are ubiquitously expressed, essential enzymes that ligate tRNA molecules to cognate amino acids
Genes encoding ARSs are associated with a spectrum of human inherited diseases
Implicating an ARS locus or allele in genetic disease requires strong genetic evidence
The majority of ARS mutations implicated in human disease cause impaired enzyme function
Functional studies should be carefully employed to predict the pathogenicity of newly identified ARS alleles
Acknowledgments
We would like to thank all of the patients and their families for agreeing to participate in the studies reviewed here; each of our colleagues for contributing to our current knowledge of ARS-associated disease; and Rebecca Meyer for critical evaluation of the manuscript.
FUNDING
L.B.G. was supported by the NIH Cellular and Molecular Biology Training Grant (GM007315), the NIH Medical Scientist Training Grant (GM07863), and an NIH F30 NRSA (NS092238). A.A.B is supported by grants from the Muscular Dystrophy Association (MDA382300) and the National Institute of Neurological Disease and Stroke (NS094678). A.A. is supported by grants from the Muscular Dystrophy Association (MDA294479) and the National Institute of General Medical Sciences (GM118647).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Antonellis A, Green ED. The role of aminoacyl-tRNA synthetases in genetic diseases. Annu Rev Genomics Hum Genet. 2008;9:87–107. doi: 10.1146/annurev.genom.9.081307.164204. [DOI] [PubMed] [Google Scholar]
- 2.Li L, Francklyn C, Carter CW. Aminoacylating urzymes challenge the RNA world hypothesis. Journal of Biological Chemistry. 2013;288:26856–26863. doi: 10.1074/jbc.M113.496125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Steenweg ME, Ghezzi D, Haack T, Abbink TEM, Martinelli D, van Berkel CGM, et al. Leukoencephalopathy with thalamus and brainstem involvement and high lactate “LTBL” caused by EARS2 mutations. Brain. 2012;135:1387–1394. doi: 10.1093/brain/aws070. [DOI] [PubMed] [Google Scholar]
- 4.Scheper GC, van der Klok T, van Andel RJ, van Berkel CGM, Sissler M, Smet J, et al. Mitochondrial aspartyl-tRNA synthetase deficiency causes leukoencephalopathy with brain stem and spinal cord involvement and lactate elevation. Nat Genet. 2007;39:534–539. doi: 10.1038/ng2013. [DOI] [PubMed] [Google Scholar]
- 5.Elo JM, Yadavalli SS, Euro L, Isohanni P, Gotz A, Carroll CJ, et al. Mitochondrial phenylalanyl-tRNA synthetase mutations underlie fatal infantile Alpers encephalopathy. Hum Mol Genet. 2012;21:4521–4529. doi: 10.1093/hmg/dds294. [DOI] [PubMed] [Google Scholar]
- 6.Pierce SB, Chisholm KM, Lynch ED, Lee MK, Walsh T, Opitz JM, et al. Mutations in mitochondrial histidyl tRNA synthetase HARS2 cause ovarian dysgenesis and sensorineural hearing loss of Perrault syndrome. Proc Natl Acad Sci USa. 2011;108:6543–6548. doi: 10.1073/pnas.1103471108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wolf NI, Salomons GS, Rodenburg RJ, Pouwels PJW, Schieving JH, Derks TGJ, et al. Mutations in RARS cause hypomyelination. Ann Neurol. 2014;76:134–139. doi: 10.1002/ana.24167. [DOI] [PubMed] [Google Scholar]
- 8.Taft RJ, Vanderver A, Leventer RJ, Damiani SA, Simons C, Grimmond SM, et al. REPOR TMutations in DARS Cause Hypomyelination with Brain Stem and Spinal Cord Involvement and Leg Spasticity. The American Journal of Human Genetics. 2013;92:774–780. doi: 10.1016/j.ajhg.2013.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Safka Brozkova D, Deconinck T, Beth Griffin L, Ferbert A, Haberlova J, Mazanec R, et al. Loss of function mutations in HARScause a spectrum of inherited peripheral neuropathies. Brain. 2015;138:2161–2172. doi: 10.1093/brain/awv158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Antonellis A, Ellsworth RE, Sambuughin N, Puls I, Abel A, Lee-Lin S-Q, et al. Glycyl tRNA synthetase mutations in Charcot-Marie-Tooth disease type 2D and distal spinal muscular atrophy type V. The American Journal of Human Genetics. 2003;72:1293–1299. doi: 10.1086/375039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jordanova A, Irobi J, Thomas FP, Van Dijck P, Meerschaert K, Dewil M, et al. Disrupted function and axonal distribution of mutant tyrosyl-tRNA synthetase in dominant intermediate Charcot-Marie-Tooth neuropathy. Nat Genet. 2006;38:197–202. doi: 10.1038/ng1727. [DOI] [PubMed] [Google Scholar]
- 12.Latour P, Thauvin-Robinet C, Baudelet-MEry C, Soichot P, Cusin V, Faivre L, et al. REPOR TA Major Determinant for Binding and Aminoacylationof tRNA. The American Journal of Human Genetics. 2010;86:77–82. doi: 10.1016/j.ajhg.2009.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McLaughlin HM, Sakaguchi R, Giblin W, Wilson TE, Biesecker L, et al. Intramural Sequencing Center NIH. A Recurrent loss-of-function alanyl-tRNA synthetase (AARS) mutation in patients with charcot-marie-tooth disease type 2N (CMT2N) Hum Mutat. 2011;33:244–253. doi: 10.1002/humu.21635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vester A, Velez-Ruiz G, McLaughlin HM, Lupski JR, Talbot K, et al. Comparative Sequencing Program NISC. A Loss-of-Function Variant in the Human Histidyl-tRNA Synthetase (HARS) Gene is Neurotoxic In Vivo. Hum Mutat. 2012;34:191–199. doi: 10.1002/humu.22210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Skre H. Genetic and clinical aspects of Charcot-Marie-Tooth’s disease. Clin Genet. 1974;6:98–118. doi: 10.1111/j.1399-0004.1974.tb00638.x. [DOI] [PubMed] [Google Scholar]
- 16.Dyck PJ, Lambert EH. Lower motor and primary sensory neuron diseases with peroneal muscular atrophy. II. Neurologic, genetic, and electrophysiologic findings in various neuronal degenerations. Arch Neurol. 1968;18:619–625. doi: 10.1001/archneur.1968.00470360041003. [DOI] [PubMed] [Google Scholar]
- 17.Burton PR, Tobin MD, Hopper JL. Key concepts in genetic epidemiology. Lancet. 2005;366:941–951. doi: 10.1016/S0140-6736(05)67322-9. [DOI] [PubMed] [Google Scholar]
- 18.Jordanova A, Thomas FP, Guergueltcheva V, Tournev I, Gondim FAA, Ishpekova B, et al. Dominant intermediate Charcot-Marie-Tooth type C maps to chromosome 1p34-p35. The American Journal of Human Genetics. 2003;73:1423–1430. doi: 10.1086/379792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Christodoulou K, Kyriakides T, Hristova AH, Georgiou DM, Kalaydjieva L, Yshpekova B, et al. Mapping of a distal form of spinal muscular atrophy with upper limb predominance to chromosome 7p. Hum Mol Genet. 1995;4:1629–1632. doi: 10.1093/hmg/4.9.1629. [DOI] [PubMed] [Google Scholar]
- 20.Ionasescu V, Searby C, Sheffield VC, Roklina T, Nishimura D, Ionasescu R. Autosomal dominant Charcot-Marie-Tooth axonal neuropathy mapped on chromosome 7p (CMT2D) Hum Mol Genet. 1996;5:1373–1375. doi: 10.1093/hmg/5.9.1373. [DOI] [PubMed] [Google Scholar]
- 21.Pericak-Vance MA, Speer MC, Lennon F, West SG, Menold MM, Stajich JM, et al. Confirmation of a second locus for CMT2 and evidence for additional genetic heterogeneity. Neurogenetics. 1997;1:89–93. doi: 10.1007/s100480050013. [DOI] [PubMed] [Google Scholar]
- 22.Sambuughin N, Sivakumar K, Selenge B, Lee HS, Friedlich D, Baasanjav D, et al. Autosomal dominant distal spinal muscular atrophy type V (dSMA-V) and Charcot-Marie-Tooth disease type 2D (CMT2D) segregate within a single large kindred and map to a refined region on chromosome 7p15. Journal of the Neurological Sciences. 1998;161:23–28. doi: 10.1016/s0022-510x(98)00264-0. [DOI] [PubMed] [Google Scholar]
- 23.Ellsworth RE, Ionasescu V, Searby C, Sheffield VC, Braden VV, Kucaba TA, et al. The CMT2D locus: refined genetic position and construction of a bacterial clone-based physical map. Genome Res. 1999;9:568–574. [PMC free article] [PubMed] [Google Scholar]
- 24.Dawn Teare M, Barrett JH. Genetic linkage studies. Lancet. 2005;366:1036–1044. doi: 10.1016/S0140-6736(05)67382-5. [DOI] [PubMed] [Google Scholar]
- 25.MORTON NE. Sequential tests for the detection of linkage. The American Journal of Human Genetics. 1955;7:277–318. [PMC free article] [PubMed] [Google Scholar]
- 26.Kruglyak L, Lander ES. A nonparametric approach for mapping quantitative trait loci. Genetics. 1995;139:1421–1428. doi: 10.1093/genetics/139.3.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ott J, Wang J, Leal SM. Genetic linkage analysis in the age of whole-genome sequencing. Nat Rev Genet. 2015;16:275–284. doi: 10.1038/nrg3908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ng SB, Turner EH, Robertson PD, Flygare SD, Bigham AW, Lee C, et al. Targeted capture and massively parallel sequencing of 12 human exomes. Nature. 2009;461:272–276. doi: 10.1038/nature08250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Turner EH, Lee C, Ng SB, Nickerson DA, Shendure J. Massively parallel exon capture and library-free resequencing across 16 genomes. Nat Methods. 2009;6:315–316. doi: 10.1038/nmeth.f.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ng PC, Kirkness EF. Whole genome sequencing. Methods Mol Biol. 2010;628:215–226. doi: 10.1007/978-1-60327-367-1_12. [DOI] [PubMed] [Google Scholar]
- 31.Bamshad MJ, Ng SB, Bigham AW, Tabor HK, Emond MJ, Nickerson DA, et al. Exome sequencing as a tool for Mendelian disease gene discovery. Nat Rev Genet. 2011;12:745–755. doi: 10.1038/nrg3031. [DOI] [PubMed] [Google Scholar]
- 32.Wang Z, Sadovnick AD, Traboulsee AL, Ross JP, Bernales CQ, Encarnacion M, et al. Nuclear Receptor NR1H3 in Familial Multiple Sclerosis. Neuron. 2016;90:948–954. doi: 10.1016/j.neuron.2016.04.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.James PA, Cader MZ, Muntoni F, Childs A-M, Crow YJ, Talbot K. Severe childhood SMA and axonal CMT due to anticodon binding domain mutations in the GARS gene. Neurology. 2006;67:1710–1712. doi: 10.1212/01.wnl.0000242619.52335.bc. [DOI] [PubMed] [Google Scholar]
- 34.Griffin LB, Sakaguchi R, McGuigan D, Gonzalez MA, Searby C, Züchner S, et al. Impaired Function is a Common Feature of Neuropathy-Associated Glycyl-tRNA Synthetase Mutations. Hum Mutat. 2014:n/a–n/a. doi: 10.1002/humu.22681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jordan DM, Frangakis SG, Golzio C, Cassa CA, Kurtzberg J, et al. Task Force for Neonatal Genomics. Identification of cis-suppression of human disease mutations by comparative genomics. Nature. 2015;524:225–229. doi: 10.1038/nature14497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Motley WW, Talbot K, Fischbeck KH. GARS axonopathy: not every neuron’s cup of tRNA. Trends Neurosci. 2010;33:59–66. doi: 10.1016/j.tins.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yao P, Fox PL. Aminoacyl-tRNA synthetases in medicine and disease. EMBO Mol Med. 2013;5:332–343. doi: 10.1002/emmm.201100626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Delarue M. Aminoacyl-tRNA synthetases. Curr Opin Struct Biol. 1995;5:48–55. doi: 10.1016/0959-440x(95)80008-o. [DOI] [PubMed] [Google Scholar]
- 39.Francklyn CS, First EA, Perona JJ, Hou Y-M. Methods for kinetic and thermodynamic analysis of aminoacyl-tRNA synthetases. Methods. 2008;44:100–118. doi: 10.1016/j.ymeth.2007.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moldave K, Castelfranco P, Meister A. The Synthesis and Some Properties of Amino Acyl Adenylates. Journal of Biological Chemistry. 1959;234:1–9. [PubMed] [Google Scholar]
- 41.Baldwin AN, Berg P. Transfer Ribonucleic Acid-induced Hydrolysis of Valyladenylate Bound to Isoleucyl Ribonucleic Acid Synthetase. Journal of Biological Chemistry. 1966;241:1–8. [PubMed] [Google Scholar]
- 42.Froelich CA, First EA. Dominant Intermediate Charcot-Marie-Tooth disorder is not due to a catalytic defect in tyrosyl-tRNA synthetase. Biochemistry. 2011;50:7132–7145. doi: 10.1021/bi200989h. [DOI] [PubMed] [Google Scholar]
- 43.Schreier AA, Schimmel PR. Transfer ribonucleic acid synthetase catalyzed deacylation of aminoacyl transfer ribonucleic acid in the absence of adenosine monophosphate and pyrophosphate. Biochemistry. 1972;11:1582–1589. doi: 10.1021/bi00759a006. [DOI] [PubMed] [Google Scholar]
- 44.Simons C, Griffin LB, Helman G, Golas G, Pizzino A, Bloom M, et al. Loss-of-Function Alanyl-tRNA Synthetase Mutations Cause an Autosomal-Recessive Early-Onset Epileptic Encephalopathy with Persistent Myelination Defect. The American Journal of Human Genetics. 2015;96:675–681. doi: 10.1016/j.ajhg.2015.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Coughlin CR, II, Scharer GH, Friederich MW, Yu H-C, Geiger EA, Creadon-Swindell G, et al. Mutations in the mitochondrial cysteinyl-tRNA synthase gene, CARS2,lead to a severe epileptic encephalopathy and complex movement disorder. J Med Genet. 2015;52:532–540. doi: 10.1136/jmedgenet-2015-103049. [DOI] [PubMed] [Google Scholar]
- 46.Almalki A, Alston CL, Parker A, Simonic I, Mehta SG, He L, et al. Biochimica et Biophysica Acta, BBA - Molecular Basis of Disease. 2014;1842:56–64. doi: 10.1016/j.bbadis.2013.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kopajtich R, Murayama K, Janecke AR, Haack TB, Breuer M, Knisely AS, et al. REPOR TBiallelic IARS Mutations Cause Growth Retardation with Prenatal Onset, Intellectual Disability, Muscular Hypotonia, and Infantile Hepatopathy. The American Journal of Human Genetics. 2016:1–9. doi: 10.1016/j.ajhg.2016.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McLaughlin HM, Sakaguchi R, Liu C, Igarashi T, Pehlivan D, Chu K, et al. REPOR TCompound Heterozygosity for Loss-of-Function Lysyl-tRNA Synthetase Mutationsin a Patient with Peripheral Neuropathy. The American Journal of Human Genetics. 2010;87:560–566. doi: 10.1016/j.ajhg.2010.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li R, Guan MX. Human Mitochondrial Leucyl-tRNA Synthetase Corrects Mitochondrial Dysfunctions Due to the tRNALeu(UUR) A3243G Mutation, Associated with Mitochondrial Encephalomyopathy, Lactic Acidosis, and Stroke-Like Symptoms and Diabetes. Molecular and Cellular Biology. 2010;30:2147–2154. doi: 10.1128/MCB.01614-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Riley LG, Rudinger-Thirion J, Schmitz-Abe K, Thorburn DR, Davis RL, Teo J, et al. JIMD Reports. Springer Berlin Heidelberg; Berlin, Heidelberg: 2015. LARS2 Variants Associated with Hydrops, Lactic Acidosis, Sideroblastic Anemia, and Multisystem Failure; pp. 49–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.van Meel E, Wegner DJ, Cliften P, Willing MC, White FV, Kornfield S, et al. Rare recessive loss-of-function methionyl-tRNA synthetase mutations presenting as a multi-organ phenotype. 2013:1–10. doi: 10.1186/1471-2350-14-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hadchouel A, Wieland T, Griese M, Baruffini E, Lorenz-Depiereux B, Enaud L, et al. REPOR TBiallelic Mutations of Methionyl-tRNA Synthetase Cause a Specific Type of Pulmonary Alveolar Proteinosis Prevalent on Réunion Island. The American Journal of Human Genetics. 2015;96:826–831. doi: 10.1016/j.ajhg.2015.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bayat V, Thiffault I, Jaiswal M, Tétreault M, Donti T, Sasarman F, et al. Mutations in the Mitochondrial Methionyl-tRNA Synthetase Cause a Neurodegenerative Phenotype in Flies and a Recessive Ataxia (ARSAL) in Humans. PLoS Biol. 2012;10:e1001288. doi: 10.1371/journal.pbio.1001288.s014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang X, Ling J, Barcia G, Jing L, Wu J, Barry BJ, et al. Mutations in QARS, Encoding Glutaminyl-tRNA Synthetase, Cause Progressive Microcephaly, Cerebral-Cerebellar Atrophy, and Intractable Seizures. The American Journal of Human Genetics. 2014;94:547–558. doi: 10.1016/j.ajhg.2014.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Edvardson S, Shaag A, Kolesnikova O, Gomori JM, Tarassov I, Einbinder T, et al. Deleterious Mutation in the Mitochondrial Arginyl–Transfer RNA Synthetase Gene Is Associated with Pontocerebellar Hypoplasia. The American Journal of Human Genetics. 2007;81:857–862. doi: 10.1086/521227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Belostotsky R, Ben-Shalom E, Rinat C, Becker-Cohen R, Feinstein S, Zeligson S, et al. REPOR TMutations in the Mitochondrial Seryl-tRNA Synthetase Cause Hyperuricemia, Pulmonary Hypertension, Renal Failure in Infancy and Alkalosis, HUPRA Syndrome. The American Journal of Human Genetics. 2011;88:193–200. doi: 10.1016/j.ajhg.2010.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Diodato D, Melchionda L, Haack TB, Dallabona C, Baruffini E, Donnini C, et al. VARS2and TARS2Mutations in Patients with Mitochondrial Encephalomyopathies. Hum Mutat. 2014;35:983–989. doi: 10.1002/humu.22590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sasarman F, Nishimura T, Thiffault I, Shoubridge EA. A novel mutation in YARS2 causes myopathy with lactic acidosis and sideroblastic anemia. Hum Mutat. 2012;33:1201–1206. doi: 10.1002/humu.22098. [DOI] [PubMed] [Google Scholar]
- 59.Riley LG, Cooper S, Hickey P, Rudinger-Thirion J, McKenzie M, Compton A, et al. AR TICLEMutation of the Mitochondrial Tyrosyl-tRNA Synthetase Gene, YARS2, Causes Myopathy, Lactic Acidosis, and Sideroblastic Anemia—MLASA Syndrome. The American Journal of Human Genetics. 2010;87:52–59. doi: 10.1016/j.ajhg.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gonzaga-Jauregui C, Harel T, Gambin T, Kousi M, Griffin LB, Francescatto L, et al. Exome Sequence Analysis Suggests that Genetic Burden Contributes to Phenotypic Variability and Complex Neuropathy. Cell Rep. 2015;12:1169–1183. doi: 10.1016/j.celrep.2015.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Curan BP, Bugeja V. Basic investigations in Saccharomyces cerevisiae. Methods in Molecular Biology. 2006;313:1–13. doi: 10.1385/1-59259-958-3:001. [DOI] [PubMed] [Google Scholar]
- 62.Boeke JD, Trueheart J, Natsoulis G, Fink GR. 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Meth Enzymol. 1987;154:164–175. doi: 10.1016/0076-6879(87)54076-9. [DOI] [PubMed] [Google Scholar]
- 63.Dallabona C, Diodato D, Kevelam SH, Haack TB, Wong L-J, Salomons GS, et al. Novel (ovario) leukodystrophy related to AARS2 mutations. Neurology. 2014;82:2063–2071. doi: 10.1212/WNL.0000000000000497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pierce SB, Gersak K, Michaelson-Cohen R, Walsh T, Lee MK, Malach D, et al. REPOR TMutations in LARS2, Encoding Mitochondrial Leucyl-tRNA Synthetase, Lead to PrematureOvarian Failure and Hearing Loss in Perrault Syndrome. The American Journal of Human Genetics. 2013;92:614–620. doi: 10.1016/j.ajhg.2013.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Antonellis A, Lee-Lin SQ, Wasterlain A, Leo P, Quezado M, Goldfarb LG, et al. Functional Analyses of Glycyl-tRNA Synthetase Mutations Suggest a Key Role for tRNA-Charging Enzymes in Peripheral Axons. Journal of Neuroscience. 2006;26:10397–10406. doi: 10.1523/JNEUROSCI.1671-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chihara T, Luginbuhl D, Luo L. Cytoplasmic and mitochondrial protein translation in axonal and dendritic terminal arborization. Nat Neurosci. 2007;10:828–837. doi: 10.1038/nn1910. [DOI] [PubMed] [Google Scholar]
- 67.Motley WW, Griffin LB, Mademan I, Baets J, De Vriendt E, De Jonghe P, et al. A novel AARS mutation in a family with dominant myeloneuropathy. Neurology. 2015;84:2040–2047. doi: 10.1212/WNL.0000000000001583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Storkebaum E, Leitão-Gonçalves R, Godenschwege T, Nangle L, Mejia M, Bosmans I, et al. Dominant mutations in the tyrosyl-tRNA synthetase gene recapitulate in Drosophila features of human Charcot-Marie-Tooth neuropathy. Proc Natl Acad Sci USa. 2009;106:11782–11787. doi: 10.1073/pnas.0905339106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Leitão-Gonçalves R, Ermanoska B, Jacobs A, De Vriendt E, Timmerman V, Lupski JR, et al. Drosophila as a platform to predict the pathogenicity of novel aminoacyl-tRNA synthetase mutations in CMT. Amino Acids. 2011;42:1661–1668. doi: 10.1007/s00726-011-0868-4. [DOI] [PubMed] [Google Scholar]
- 70.Yook K. Complementation. WormBook; 2005. pp. 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schuske K, Beg AA, Jorgensen EM. The GABA nervous system in C. elegans. Trends Neurosci. 2004;27:407–414. doi: 10.1016/j.tins.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 72.Buckingham SD, Sattelle DB. Fast, automated measurement of nematode swimming (thrashing) without morphometry. BMC Neurosci. 2009;10:84. doi: 10.1186/1471-2202-10-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wallen RC, Antonellis A. To charge or not to charge: mechanistic insights into neuropathy-associated tRNA synthetase mutations. Curr Opin Genet Dev. 2013;23:302–309. doi: 10.1016/j.gde.2013.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sauter C, Lorber B, Gaudry A, Karim L, Schwenzer H, Wien F, et al. Neurodegenerative disease-associated mutants of a human mitochondrial aminoacyl-tRNA synthetase present individual molecular signatures. Nature Publishing Group. 2015;5:17332. doi: 10.1038/srep17332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Götz A, Tyynismaa H, Euro L, Ellonen P, Hyötyläinen T, Ojala T, et al. REPOR TExome Sequencing Identifies Mitochondrial Alanyl-tRNA Synthetase Mutationsin Infantile Mitochondrial Cardiomyopathy. The American Journal of Human Genetics. 2011;88:635–642. doi: 10.1016/j.ajhg.2011.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Talim B, Pyle A, Griffin H, Topaloglu H, Tokatli A, Keogh MJ, et al. Multisystem fatal infantile disease caused by a novel homozygous EARS2 mutation. Brain. 2013;136:e228–e228. doi: 10.1093/brain/aws197. [DOI] [PubMed] [Google Scholar]
- 77.Schwartzentruber J, Buhas D, Majewski J, Sasarman F, Papillon-Cavanagh S, Thiffaut I, et al. Mutation in The Nuclear-Encoded Mitochondrial Isoleucyl-tRNA Synthetase IARS2in Patients with Cataracts, Growth Hormone Deficiency with Short Stature, Partial Sensorineural Deafness, and Peripheral Neuropathy or with Leigh Syndrome. Hum Mutat. 2014:n/a–n/a. doi: 10.1002/humu.22629. [DOI] [PubMed] [Google Scholar]
- 78.Sofou K, Kollberg G, Holmström M, Dávila M, Darin N, Gustafsson CM, et al. Whole exome sequencing reveals mutations in NARS2and PARS2, encoding the mitochondrial asparaginyl-tRNA synthetase and prolyl-tRNA synthetase, in patients with Alpers syndrome. Mol Genet Genomic Med. 2014;3:59–68. doi: 10.1002/mgg3.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Vanlander AV, Menten B, Smet J, De Meirleir L, Sante T, De Paepe B, et al. Two Siblings with Homozygous Pathogenic Splice-Site Variant in Mitochondrial Asparaginyl-tRNA Synthetase (NARS2) Hum Mutat. 2015;36:222–231. doi: 10.1002/humu.22728. [DOI] [PubMed] [Google Scholar]
- 80.Nangle LA, Zhang W, Xie W, Yang X-L, Schimmel P. Charcot-Marie-Tooth disease-associated mutant tRNA synthetases linked to altered dimer interface and neurite distribution defect. Proc Natl Acad Sci USa. 2007;104:11239–11244. doi: 10.1073/pnas.0705055104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.He W, Bai G, Zhou H, Wei N, White NM, Lauer J, et al. CMT2D neuropathy is linked to the neomorphic binding activity of glycyl-tRNA synthetase. Nature. 2015;526:710–714. doi: 10.1038/nature15510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mo Z, Zhang Q, Liu Z, Lauer J, Shi Y, Sun L, et al. Neddylation requires glycyl-tRNA synthetase to protect activated E2. Nat Struct Mol Biol. 2016;23:730–737. doi: 10.1038/nsmb.3250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Niehues S, Bussmann J, Steffes G, Erdmann I, Köhrer C, Sun L, et al. Impaired protein translation in Drosophila models for Charcot–Marie–Tooth neuropathy caused by mutant tRNA synthetases. Nature Communications. 2015;6:7520. doi: 10.1038/ncomms8520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Malissovas N, Griffin LB, Antonellis A, Beis D. Dimerization is required for GARS-mediated neurotoxicity in dominant CMT disease. Hum Mol Genet. 2016;25:1528–1542. doi: 10.1093/hmg/ddw031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Seburn KL, Nangle LA, Cox GA, Schimmel P, Burgess RW. An active dominant mutation of glycyl-tRNA synthetase causes neuropathy in a Charcot-Marie-Tooth 2D mouse model. Neuron. 2006;51:715–726. doi: 10.1016/j.neuron.2006.08.027. [DOI] [PubMed] [Google Scholar]
- 86.Achilli F, Bros-Facer V, Williams HP, Banks GT, AlQatari M, Chia R, et al. An ENU-induced mutation in mouse glycyl-tRNA synthetase (GARS) causes peripheral sensory and motor phenotypes creating a model of Charcot-Marie-Tooth type 2D peripheral neuropathy. Disease Models & Mechanisms. 2009;2:359–373. doi: 10.1242/dmm.002527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Motley WW, Seburn KL, Nawaz MH, Miers KE, Cheng J, Antonellis A, et al. Charcot-Marie-Tooth-linked mutant GARS is toxic to peripheral neurons independent of wild-type GARS levels. PLoS Genet. 2011;7:e1002399. doi: 10.1371/journal.pgen.1002399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sleigh JN, Grice SJ, Burgess RW, Talbot K, Cader MZ. Neuromuscular junction maturation defects precede impaired lower motor neuron connectivity in Charcot-Marie-Tooth type 2D mice. Hum Mol Genet. 2014;23:2639–2650. doi: 10.1093/hmg/ddt659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Spaulding EL, Sleigh JN, Morelli KH, Pinter MJ, Burgess RW, Seburn KL. Synaptic Deficits at Neuromuscular Junctions in Two Mouse Models of Charcot-Marie-Tooth Type 2d. Journal of Neuroscience. 2016;36:3254–3267. doi: 10.1523/JNEUROSCI.1762-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bais P, Beebe K, Morelli KH, Currie ME, Norberg SN, Evsikov AV, et al. Metabolite profile of a mouse model of Charcot-Marie-Tooth type 2D neuropathy: implications for disease mechanisms and interventions. Biol Open. 2016;5:908–920. doi: 10.1242/bio.019273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lee JW, Beebe K, Nangle LA, Jang J, Longo-Guess CM, Cook SA, et al. Editing-defective tRNA synthetase causes protein misfolding and neurodegeneration. Nature. 2006;443:50–55. doi: 10.1038/nature05096. [DOI] [PubMed] [Google Scholar]
- 92.Stum M, McLaughlin HM, Kleinbrink EL, Miers KE, Ackerman SL, Seburn KL, et al. An assessment of mechanisms underlying peripheral axonal degeneration caused by aminoacyl-tRNA synthetase mutations. Molecular and Cellular Neuroscience. 2011;46:432–443. doi: 10.1016/j.mcn.2010.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kitzman JO, Starita LM, Lo RS, Fields S, Shendure J. Massively parallel single-aminoacid mutagenesis. Nat Methods. 2015;12:203–6. doi: 10.1038/nmeth.3223. 4 p following 206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lynch DS, Zhang WJ, Lakshmanan R, Kinsella JA, Uzun GA, Karbay M, et al. Analysis of Mutations in AARS2 in a Series of CSF1R-Negative Patients With Adult-Onset Leukoencephalopathy With Axonal Spheroids and Pigmented Glia. JAMA Neurol. 2016 doi: 10.1001/jamaneurol.2016.2229. [DOI] [PubMed] [Google Scholar]
- 95.Yang Y, Liu W, Fang Z, Shi J, Che F, He C, et al. A Newly Identified Missense Mutation in FARS2Causes Autosomal-Recessive Spastic Paraplegia. Hum Mutat. 2015;37:165–169. doi: 10.1002/humu.22930. [DOI] [PubMed] [Google Scholar]
- 96.Raviglione F, Conte G, Ghezzi D, Parazzini C, Righini A, Vergaro R, et al. Clinical findings in a patient with FARS2 mutations and early-infantile-encephalopathy with epilepsy. Am J Med Genet. 2016;170:3004–3007. doi: 10.1002/ajmg.a.37836. [DOI] [PubMed] [Google Scholar]
- 97.McMillan HJ, Schwartzentruber J, Smith A, Lee S, Chakraborty P, Bulman DE, et al. Compound heterozygous mutations in glycyl-tRNA synthetase are a proposed cause of systemic mitochondrial disease. 2014:1–6. doi: 10.1186/1471-2350-15-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Puffenberger EG, Jinks RN, Sougnez C, Cibulskis K, Willert RA, Achilly NP, et al. Genetic mapping and exome sequencing identify variants associated with five novel diseases. PLoS ONE. 2012;7:e28936. doi: 10.1371/journal.pone.0028936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.McMillan HJ, Humphreys P, Smith A, Schwartzentruber J, Chakraborty P, Bulman DE, et al. Congenital Visual Impairment and Progressive Microcephaly Due to Lysyl-Transfer Ribonucleic Acid (RNA) Synthetase (KARS) Mutations: The Expanding Phenotype of Aminoacyl-Transfer RNA Synthetase Mutations in Human Disease. J Child Neurol. 2015;30:1037–1043. doi: 10.1177/0883073814553272. [DOI] [PubMed] [Google Scholar]
- 100.Santos-Cortez RLP, Lee K, Azeem Z, Antonellis PJ, Pollock LM, Khan S, et al. REPOR TMutations in KARS, Encoding Lysyl-tRNA Synthetase, Cause Autosomal-Recessive Nonsyndromic Hearing Impairment DFNB89. The American Journal of Human Genetics. 2013;93:132–140. doi: 10.1016/j.ajhg.2013.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Casey JP, McGettigan P, Lynam-Lennon N, McDermott M, Regan R, Conroy J, et al. Molecular Genetics and Metabolism. Molecular Genetics and Metabolism. 2012;106:351– 358. doi: 10.1016/j.ymgme.2012.04.017. [DOI] [PubMed] [Google Scholar]
- 102.Gonzalez M, McLaughlin H, Houlden H, Guo M, Yo-Tsen L, Hadjivassilious M, et al. Exome sequencing identifies a significant variant in methionyl-tRNA synthetase (MARS) in a family with late-onset CMT2. J Neurol Neurosurg Psychiatr. 2013;84:1247–1249. doi: 10.1136/jnnp-2013-305049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Webb BD, Wheeler PG, Hagen JJ, Cohen N, Linderman MD, Diaz GA, et al. Novel, Compound Heterozygous, Single-Nucleotide Variants in MARS2Associated with Developmental Delay, Poor Growth, and Sensorineural Hearing Loss. Hum Mutat. 2015;36:587–592. doi: 10.1002/humu.22781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Karaca E, Harel T, Pehlivan D, Jhangiani SN, Gambin T, Coban Akdemir Z, et al. Genes that Affect Brain Structure and Function Identified by Rare Variant Analyses of Mendelian Neurologic Disease. Neuron. 2015;88:499–513. doi: 10.1016/j.neuron.2015.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Taylor RW, Pyle A, Griffin H, Blakely EL, Duff J, He L, et al. Use of Whole-Exome Sequencing to Determine the Genetic Basis of Multiple Mitochondrial Respiratory Chain Complex Deficiencies. Jama. 2014;312:68. doi: 10.1001/jama.2014.7184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Baertling F, Alhaddad B, Seibt A, Budaeus S, Meitinger T, Strom TM, et al. Neonatal encephalocardiomyopathy caused by mutations in VARS2. Metab Brain Dis. 2016:1–4. doi: 10.1007/s11011-016-9890-2. [DOI] [PubMed] [Google Scholar]
- 107.Nowaczyk MJM, Huang L, Tarnopolsky M, Schwartzentruber J, Majewski J, Bulman DE, et al. A novel multisystem disease associated with recessive mutations in the tyrosyl-tRNA synthetase (YARS) gene. Am J Med Genet. 2016 doi: 10.1002/ajmg.a.37973. [DOI] [PubMed] [Google Scholar]
- 108.Lin K-P, Soong B-W, Yang C-C, Huang L-W, Chang M-H, Lee I-H, et al. The Mutational Spectrum in a Cohort of Charcot-Marie-Tooth Disease Type 2 among the Han Chinese in Taiwan. PLoS ONE. 2011;6:e29393. doi: 10.1371/journal.pone.0029393.s004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zhao Z, Hashiguchi A, Hu J, Sakiyama Y, Okamoto Y, Tokunaga S, et al. Alanyl-tRNA synthetase mutation in a family with dominant distal hereditary motor neuropathy. 2012:1–6. doi: 10.1212/WNL.0b013e3182574f8f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Rohkamm B, Reilly MM, Lochmüller H, Schlotter-Weigel B, Barisic N, Schöls L, et al. Further evidence for genetic heterogeneity of distal HMN type V, CMT2 with predominant hand involvement and Silver syndrome. Journal of the Neurological Sciences. 2007;263:100–106. doi: 10.1016/j.jns.2007.06.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lee JH, Park J, Khriezanou N, Park JM, Hur Y-M, Choi BO, et al. Two novel mutations of GARS in Korean families with distal hereditary motor neuropathy type V. 2012:1–4. doi: 10.1111/j.1529-8027.2012.00442.x. [DOI] [PubMed] [Google Scholar]
- 112.Abe A, Hayasaka K. The GARS gene is rarely mutated in Japanese patients with Charcot–Marie–Tooth neuropathy. 2009;54:310–312. doi: 10.1038/jhg.2009.25. [DOI] [PubMed] [Google Scholar]
- 113.Sivakumar K, Kyriakides T, Puls I, Nicholson GA, Funalot B, Antonellis A, et al. Phenotypic spectrum of disorders associated with glycyl-tRNA synthetase mutations. Brain. 2005;128:2304–2314. doi: 10.1093/brain/awh590. [DOI] [PubMed] [Google Scholar]
- 114.Del Bo R, Locatelli F, Corti S, Scarlato M, Ghezzi S, Prelle A, et al. Coexistence of CMT-2D and distal SMA-V phenotypes in an Italian family with a GARS gene mutation. Neurology. 2006;66:752–754. doi: 10.1212/01.wnl.0000201275.18875.ac. [DOI] [PubMed] [Google Scholar]
- 115.Xie W, Nangle LA, Zhang W, Schimmel P, Yang X-L. Long-range structural effects of a Charcot-Marie-Tooth disease-causing mutation in human glycyl-tRNA synthetase. Proc Natl Acad Sci USa. 2007;104:9976–9981. doi: 10.1073/pnas.0703908104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Hyun YS, Park HJ, Heo SH, Yoon BR, Nam SH, Kim SB, et al. Rare variants in methionyl- and tyrosyl-tRNA synthetasegenes in late-onset autosomal dominant Charcot-Marie-Tooth neuropathy. Clin Genet. 2013;86:592–594. doi: 10.1111/cge.12327. [DOI] [PubMed] [Google Scholar]