Abstract
Background
Health care workers (HCWs) use their mobile phones during working hours or medical care. There is evidence that the instruments are colonized with pathogenic microorganisms. Here, we describe levels of Enterobacteriaceae contamination (EC) in cell phones and the risk factors associated with EC in Peruvian intensive care units (ICUs).
Methods
This was a 5-month cohort study among 114 HCWs of 3 pediatric and 2 neonatology ICUs from 3 Peruvian hospitals. A baseline survey collected data on risk factors associated with EC. Swabs were collected from HCWs’ phones every other week.
Results
Three-quarters of HCWs never decontaminated their phones, and 47% reported using the phones in the ICU >5 times while working. EC was frequent across samplings and sites and was substantially higher in subjects with longer follow-up. Potential risk factors identified did not have strong associations with positive samples (relative risk, 0.7–1.5), regardless of significance. Half of the phones were colonized with an Enterobacteriaceae at least once during the 4 samplings attained on average during the study period. Half of the isolates were multidrug resistant (MDR), and 33% were extended-spectrum β-lactamase producers.
Conclusions
EC on HCWs’ phones was frequent and apparently randomly distributed through the hospitals without clear clustering or strongly associated risk factors for having a positive sample. Based on the level of EC, phones may be considered as potential bacterial reservoirs of MDR and ESBL bacteria.
Keywords: Infectious disease transmission, professional-to-patient; Cell phones; Enterobacteriaceae; Health personnel
Intensive care units (ICUs) require a high level of sanitation and infection control because of the critical condition of their patients and the high risk for complications and nosocomial infections. Outbreaks of healthcare–associated infections with multidrug-resistant bacteria have been widely described in pediatric and neonatology ICUs, causing significant morbidity and mortality and increasing health care costs and length of stay.1–3
The most common cause of outbreaks in ICUs is Enterobacteriaceae.3,4 These pathogens are coresistant to different classes of drugs and often contain antibiotic resistance genes such as extended-spectrum β-lactamase (ESBL). ESBL infections are associated with increased morbidity, mortality, and the need for carbapenem therapy, for example, leaving few therapeutic options available for patient treatment.5,6
In the last decade, cell phone use has penetrated clinical practice, providing rapid access to medical information and allowing efficient communication with colleagues worldwide.7 However, cell phone use in sensitive settings and lack of disinfection, coupled with their portability, makes them a potential source of infection.8,9 Evidence of cell phone contamination at hospitals has been observed, with up to 94% of phones testing positive for a wide range of bacteria; these may be implicated in outbreaks at ICUs.10,11 Therefore, cell phones probably represent a constant infection risk for patients,12 and developing countries are likely at greater risk.
The objective of this study was to describe levels of Enterobacteriaceae contamination in cell phones in Peruvian ICUs. This study also investigated potential risk factors associated with cell phone contamination in these settings.
MATERIALS AND METHODS
Study design
From February–June 2012, we conducted a 5-month cohort study among health care workers (HCWs) of ICUs from 3 national hospitals in Lima, Peru. HCWs completed a baseline questionnaire, and swabs were collected from their cell phones (referred to as phones from here onward) every other week (10 samplings total) for Enterobacteriaceae culture.
The Ethics Committees of Universidad Peruana Cayetano Heredia (SIDISI 58415) and the hospitals approved the study protocol and all of its procedures. All participants underwent oral informed consent and were provided with an information sheet for their understanding of study procedures.
Population and sample
The study was conducted in 3 pediatric and 2 neonatology ICUs of 2 pediatric and 1 general hospitals. All physicians, residents in training, and professional and technical nursing personnel were invited to participate. Personnel without phones or with appointments in other wards at the same hospitals were excluded. Most HCWs were enrolled at the first visit, and others were assessed in subsequent dates. Consenting participants were asked to enroll their phones, choosing one device if they had multiple eligible devices. Tablets or other type of devices were not included in the study.
Enrollment and baseline assessments
Ten visits were scheduled every other week at each ICU. All eligible subjects present were asked to participate by providing their phones. A baseline survey was applied for collecting demographics, phone use at work, hygiene-related practices, and knowledge about contamination and transmission of pathogens through phones. Phone characteristics were recorded for identification during the follow-up. In every visit, enrolled HCWs were asked to provide their enrolled phones, and a swab sample was collected without asking any additional information or providing prior microbiologic results. Study personnel verified that the phone presented was the device initially enrolled. Some HCWs were not found in each sampling visit because of their variable work schedule, resulting in a variable number of samples collected from the phone of each HCW.
Phone swab collection and testing for Enterobacteriaceae
All samples were collected using a sterile technique for each phone. A sterile cotton swab moistened with trypticase soy broth was rotated on the phone covering the entire surface (back and keyboard and screen or touchscreen, depending of the type of phone). The swab was then submerged into a 3 mL trypticase soy broth tube and incubated aerobically for 18–24 hours at 35°C. After incubation, the swab was plated onto MacConkey agar and incubated under the same conditions. Bacterial isolates were characterized at the species level by standard microbiologic procedures.
Antimicrobial susceptibility testing
Antibiotic susceptibility was determined by the disk diffusion method according to the Clinical and Laboratory Standards Institute.13 The Enterobacteriaceae antibiogram included aztreonam 30 μg, cefepime 30 μg, cefotaxime 30 μg, ceftazidime 30 μg, amoxicillin–clavulanic acid 30 μg, imipenem 10 μg, meropenem 10 μg, ertapenem 10 μg, cefoxitin 30 μg, sulfamethoxazole-trimethoprim (SXT) 25 μg, amikacin 30 μg, gentamicin 10 μg, tobramycin 10 μg, and ciprofloxacin 5 μg.
The Clinical and Laboratory Standards Institute’s ESBL confirmatory test was used to confirm ESBL production in all strains with a reduced inhibition zone to aztreonam, cefepime, cefotaxime, or ceftazidime.13 Metallo-β-lactamases, carbapenemase, and AmpC β-lactamases were screened using 3 combined disk procedures as previously described.14–16
Molecular identification of bla genes
The genotyping characterization of the bla genes associated with ESBL-producing bacteria was done by polymerase chain reaction. DNA was extracted by the thermal shock method, and blaTEM, blaSHV, and blaCTX-M genes were amplified as previously described.17–19 Polymerase chain reaction products were separated by electrophoresis on 1% agarose gel and revealed by RunSafe (Cleaver Scientific, Rugby, UK).
Statistical analysis
The frequency of positive Enterobacteriaceae isolates was estimated during the entire study. We used a binomial family generalized linear model with a logarithmic link and estimated risk ratios (RRs) for different characteristics of the HCWs to determine the association with a positive swab conducted in a visit or sampling. We calculated the proportion of positive swabs (with at least 1 positive culture) collected from each subject out of the total samples taken from of the 10 sampling times. We also calculated the proportion of positive phones (one or more of the attained samples positive) using a binomial distribution. For data analysis purposes, physicians were grouped with residents and nurses, including both technical and professional personnel. The calculation of the percent swab positivity was analyzed including the number of samples collected and may be affected by the rejection sampling or the lack of follow-up.
Differences in antibiotic resistance to each drug were compared between ESBL- and non–ESBL-producing isolates for the most frequent Enterobacteriaceae using χ2 and Fisher exact tests as needed. Levels of intermediate antibiotic resistance based on the susceptibility testing were considered resistant to simplify the data analysis. Also, we defined multidrug resistance as being not susceptible to ≥3 antibiotic families. Analyses were conducted using Stata version 13.0 (StataCorp, College Station, TX), and significant associations were considered if P < .05.
RESULTS
A total of 114 HCWs provided 1 phone each and were enrolled in the study. HCWs were similarly distributed through hospitals, were primarily nurses (66.7%), and mainly were from pediatric ICUs (61.4%). Seventy-six percent (86/113) of HCWs reported never using anything to decontaminate their phones, and 47.4% reported using the phone >5 times while working at the ICU (Table 1).
Table 1.
Characteristics of the 114 health care workers enrolled, frequency of Enterobacteriaceae isolation, and associations
| Characteristic | n (%)* | Positive samples, n/N (%) | Risk ratio | P value | Adjusted risk ratio† | 95% CI | P value |
|---|---|---|---|---|---|---|---|
| Hospital | |||||||
| A | 39 (34.2) | 39/179 (21.7) | Reference | — | Reference | — | — |
| B | 35 (30.7) | 29/137 (21.1) | 1.02 | .906 | 1.19 | 0.70–2.01 | .523 |
| C | 40 (35.1) | 36/175 (20.5) | 0.99 | .969 | 1.36 | 0.86–2.15 | .188 |
| Occupational group | |||||||
| Physicians | 38 (33.3) | 35/175 (20.0) | Reference | — | Reference | — | — |
| Nurses | 76 (66.7) | 69/316 (21.8) | 1.09 | .652 | 1.13 | 0.78–1.65 | .515 |
| ICU | |||||||
| Neonatology | 44 (38.6) | 36/201 (17.9) | Reference | — | Reference | — | — |
| Pediatrics | 70 (61.4) | 68/290 (23.4) | 1.31 | .185 | 1.39 | 0.95–2.02 | .089 |
| Time working at ICU | |||||||
| <4 wk | 15 (13.2) | 13/75 (17.3) | Reference | — | Reference | — | — |
| 4 wk–6 y | 41 (35.9) | 38/182 (20.9) | 1.20 | .483 | 1.21 | 0.71–2.06 | .492 |
| >6 y | 58 (50.9) | 53/234 (22.6) | 1.30 | .271 | 1.46 | 0.88–2.43 | .143 |
| Type of cell phone | |||||||
| With keyboard (no touchscreen) | 70 (61.4) | 74/319 (23.2) | Reference | — | Reference | — | — |
| Touchscreen | 44 (38.6) | 30/172 (17.4) | 0.75 | .176 | 0.87 | 0.59–1.29 | .491 |
| Do you use a disinfectant on your cell phone? | |||||||
| No | 86 (76.1) | 70/371 (18.9) | Reference | — | Reference | — | — |
| Yes | 27 (23.9) | 33/115 (28.7) | 1.52 | .043 | 1.47 | 1.01–2.16 | .047 |
| No. of times you use your cell phone while working | |||||||
| 0–5 times | 60 (52.6) | 55/262 (21.0) | Reference | — | Reference | — | — |
| 6–10 times | 26 (22.8) | 28/115 (24.3) | 1.16 | .506 | 1.32 | 0.87–2.01 | .197 |
| >10 times | 28 (24.6) | 21/114 (18.4) | 0.88 | .612 | 0.87 | 0.57–1.35 | .545 |
| Answer for: Can bacteria be transmitted through your hands? | |||||||
| No | 11 (9.7) | 9/37 (24.3) | Reference | — | Reference | — | — |
| Yes | 103 (90.3) | 95/454 (20.9) | 0.86 | .597 | 0.59 | 0.30–1.13 | .109 |
| Answer for: Can cell phones be contaminated with bacteria? | |||||||
| No | 20 (17.5) | 24/87 (27.6) | Reference | — | Reference | — | — |
| I do not know | 9 (7.9) | 14/47 (29.8) | 1.08 | .785 | 0.88 | 0.48–1.60 | .673 |
| Yes | 85 (74.6) | 66/357 (18.5) | 0.67 | .053 | 0.64 | 0.43–0.97 | .034 |
| Answer for: Can bacteria be transmitted through cell phones? | |||||||
| No | 22 (19.3) | 25/99 (25.3) | Reference | — | Reference | — | — |
| I do not know | 13 (11.4) | 18/68 (26.5) | 1.04 | .874 | 1.01 | 0.60–1.71 | .961 |
| Yes | 79 (69.3) | 61/324 (18.8) | 0.75 | .249 | 0.81 | 0.52–1.28 | .37 |
| No. of samplings | |||||||
| 1–3 | 43 (37.7) | 12/103 (11.7) | Reference | — | Reference | — | — |
| 4–6 | 59 (51.8) | 64/296 (21.6) | 1.86 | .050 | 1.77 | 0.96–3.29 | .069 |
| 7–10 | 12 (10.5) | 28/92 (30.4) | 2.61 | .005 | 2.54 | 1.35–4.82 | .004 |
CI, confidence interval; ICU, intensive care unit.
n represents the total number of phones included in the study.
Adjusted by number of samplings and the use of a disinfectant.
In the 10 sampling dates, phones were sampled an average of 4 times (interquartile range, 3–6), for a total of 491 phone samples; of these, 104 (21.1%) were positive for Enterobacteriaceae. There was a strong correlation between the number of times a phone was swabbed and the percent positivity of all of the swab samples collected: 11.7% positivity among samples of phones swabbed 1–3 times, 21.6% in phones swabbed 4–6 times, and 30.6% in phones swabbed 7–9 times (P = .005). No other significant associations were found in bivariate or multiple regression analyses between the frequency of positive visits (samplings) and demographics, type of phone, hygiene-related practices, and knowledge variables, except for reporting use of disinfectant on their phones (P < .05). After adjustment for the number of visits, the RRs were in a very narrow range (0.59–1.47), showing poor discriminatory ability (Table 1).
Half of the phones (61/114, 53.5%) were colonized with at least 1 Enterobacteriaceae during the study period (4 samplings only on average), and out of 12 subjects with 6–10 samplings, 83% were positive at least once. Of all of the 61 colonized phones, 54.0% (n = 33) had only non–ESBL-producing bacteria isolates, 30.0% (n = 18) had at least 1 ESBL-producing bacteria, and 16.0% (n = 10) had isolates exclusively with bacteria that harbor ESBL.
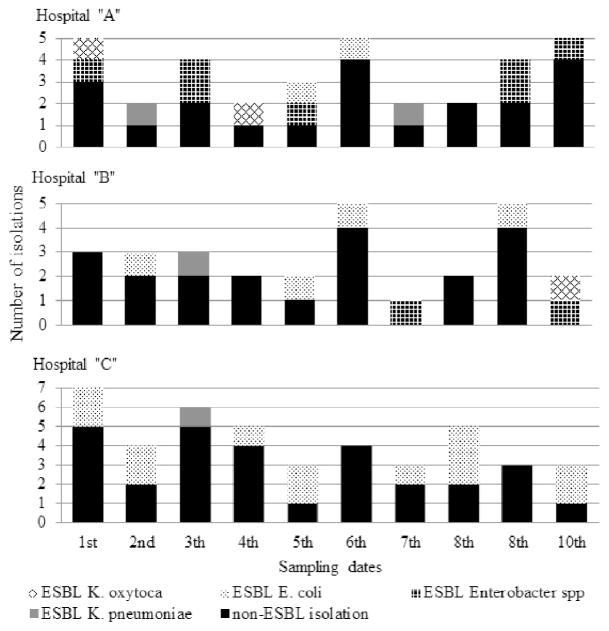
Enterobacteriaceae were frequently isolated in HCWs’ phones throughout the study period and across the 3 sites. In general, ESBL-producing bacteria were isolated frequently (range, 1–5) in every sampling across the study period (Fig 1). However, Enterobacter spp and Escherichia coli were isolated more frequently at hospital A and C, respectively (P = .049) (Table 2).
Fig. 1.
Frequency of Enterobacteriaceae isolated in 5 Peruvian neonatology and pediatric intensive care units of 3 hospitals. Extended-spectrum β-lactamase (ESBL)–producing Enterobacteriaceae contamination on health care workers’ phones was frequent and present across the sampling sites.
Table 2.
Characteristics associated with pathogenic bacteria isolates in health care workers’ cell phones
| Characteristic | Enterobacter spp (n = 48) | Escherichia coli (n = 34) | Klebsiella pneumoniae (n = 13) | Klebsiella oxytoca (n = 10) | P value* |
|---|---|---|---|---|---|
| Hospital | .049 | ||||
| A | 20 (41.7) | 5 (14.7) | 4 (30.7) | 4 (40.0) | |
| B | 15 (31.2) | 8 (23.5) | 4 (30.7) | 1 (10.0) | |
| C | 13 (27.1) | 21 (61.8) | 5 (38.6) | 5 (50.0) | |
| Occupational group | .833 | ||||
| Physician | 16 (33.3) | 12 (35.3) | 4 (30.8) | 2 (20.0) | |
| Nurse | 32 (66.7) | 22 (64.7) | 9 (69.2) | 8 (80.0) | |
| Antibiotic resistance | |||||
| T | 8 (16.7) | 16 (47.1) | 4 (30.7) | 1 (10.0) | .012 |
| G | 8 (16.7) | 11 (32.4) | 4 (30.7) | 1 (10.0) | .243 |
| A | 3 (6.3) | 2 (5.9) | 1 (7.69) | 0 (0.0) | .999 |
| F | 15 (31.3) | 3 (8.8) | 3 (23.1) | 1 (10.0) | .075 |
| C | 8 (16.7) | 21 (61.8) | 2 (15.4) | 2 (20.0) | <.001 |
| S | 9 (18.8) | 15 (44.1) | 4 (30.7) | 10 (10.0) | .043 |
| MDR | .099 | ||||
| No | 23 (47.9) | 12 (35.3) | 6 (46.2) | 8 (80.0) | |
| Yes | 25 (52.1) | 22 (64.7) | 7 (53.8) | 2 (20.0) | |
| ESBL | .005 | ||||
| No | 39 (81.2) | 15 (44.1) | 9 (69.2) | 7 (70.0) | |
| Yes | 9 (18.8) | 19 (55.9) | 4 (30.8) | 3 (30.0) | |
NOTE. Values are n (%) or as otherwise indicated.
A, amikacin; C, ciprofloxacin; ESBL, extended-spectrum β-lactamase; F, cefoxitin; G, gentamicin; MDR, multidrug resistance, nonsusceptible to ≥3 antibiotic families; S, sulfamethoxazole-trimethoprim; T, tobramycin.
Fisher exact or χ2 test as appropriate.
A total of 105 Enterobacteriaceae were isolated; the most common isolates were 48 (45.7%) Enterobacter spp, 34 (32.4%) E coli, 13 (12.4%) Klebsiella pneumoniae, and 10 (9.5%) Klebsiella oxytoca (Table 2). A third of the isolates were ESBL producers (35/105, 33.3%), and ESBL production was found in all types of Enterobacteriaceae (P = .004), but it was predominately higher in E coli (55.9%) and K pneumoniae (30.8%).
Nearly half of all bacteria isolated were multidrug resistant (MDR; 56/105, 53.3%). K oxytoca strains were marginally less MDR than the other Enterobacteriaceae isolated (20.0% vs 52.1%–64.7%, respectively; P = .099). Also, Enterobacter spp and K oxytoca presented lower levels of ESBL (18.8%–30.0%) than K pneumoniae and E coli (30.8%–55.9%, P < .005).
Tobramycin, gentamicin, SXT, and ciprofloxacin resistance were significantly associated with ESBL production in all isolates (Table 3). Additionally, nearly all of the isolates (99/105, 94.3%) were susceptible to amikacin, and 71.4% (50/70) of non-ESBL isolates were susceptible to all drugs tested in this study (Table 4). Two ESBL-producing strains, 1 E coli and 1 K pneumoniae, were resistant to all of the drugs tested. Four other ESBL-producing strains, 3 Enterobacter spp and 1 E coli, were resistant to all but 1 of the drug families tested (carbapenem). No resistance to carbapenem group A, metallo-β-lactamase, or AmpC β-lactamase production were found by phenotypic methods.
Table 3.
Antibiotic resistance pattern of Enterobacteriaceae isolated from health care workers’ cell phones
| Organism and drug resistance | Phenotype: ESBL negative | Phenotype: ESBL positive | P value* |
|---|---|---|---|
| Enterobacter spp (n = 48; ESBL negative: n = 39; ESBL positive: n = 9) | |||
| Tobramycin | 0.0 (0/39) | 88.9 (8/9) | <.001 |
| Gentamicin | 0.0 (0/39) | 88.9 (8/9) | <.001 |
| Amikacin | 0.0 (0/39) | 33.3 (3/9) | .005 |
| Cefoxitin | 38.5 (15/39) | 0.0 (0/9) | .042 |
| Ciprofloxacin | 0.0 (0/39) | 88.9 (8/9) | <.001 |
| Sulfamethoxazole-trimethoprim | 0.0 (0/39) | 100.0 (9/9) | <.001 |
| Escherichia coli (n = 34; ESBL negative: n = 15; ESBL positive: n = 19) | |||
| Tobramycin | 6.7 (1/15) | 78.9 (15/19) | <.001 |
| Gentamicin | 6.7 (1/15) | 52.6 (10/19) | .005 |
| Amikacin | 0.0 (0/15) | 10.5 (2/19) | .492 |
| Cefoxitin | 6.7 (1/15) | 10.5 (2/19) | .441 |
| Ciprofloxacin | 13.3 (2/15) | 100.0 (19/19) | <.001 |
| Sulfamethoxazole-trimethoprim | 20.0 (3/15) | 63.2 (12/19) | .017 |
| Klebsiella pneumoniae (n = 13; ESBL negative: n = 9; ESBL positive: n = 4) | |||
| Tobramycin | 0.0 (0/9) | 100.0 (4/4) | <.001 |
| Gentamicin | 0.0 (0/9) | 100.0 (4/4) | <.001 |
| Amikacin | 0.0 (0/9) | 25.0 (1/4) | .308 |
| Cefoxitin | 11.1 (1/9) | 50.0 (2/4) | .203 |
| Ciprofloxacin | 0.0 (0/9) | 75.0 (3/4) | .014 |
| Sulfamethoxazole-trimethoprim | 11.1 (1/9) | 75.0 (3/4) | .052 |
| Klebsiella oxytoca (n = 10; ESBL negative: n = 7; ESBL positive: n = 3) | |||
| Tobramycin | 0.0 (0/7) | 33.3 (1/3) | .300 |
| Gentamicin | 0.0 (0/7) | 33.3 (1/3) | .300 |
| Amikacin | 0.0 (0/7) | 0.0 (0/3) | 1.000 |
| Cefoxitin | 0.0 (0/7) | 33.3 (1/3) | .300 |
| Ciprofloxacin | 0.0 (0/7) | 66.7 (2/3) | .067 |
| Sulfamethoxazole-trimethoprim | 0.0 (0/7) | 33.3 (1/3) | .300 |
NOTE. Values are % (n/N) or as otherwise indicated.
ESBL, extended-spectrum β-lactamase.
Fisher exact or χ2 test as appropriate.
Table 4.
Phenotypes of Enterobacteriaceae isolated for 6 different antibiotics
| Resistance phenotypes | Enterobacter spp (n = 48) | Escherichia coli (n = 34) | Klebsiella pneumoniae (n = 13) | Klebsiella oxytoca (n = 10) | Total (N = 105) | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Non-ESBL isolates (n = 70) | ||||||||||
| . | . | . | . | . | . | 24 | 12 | 7 | 7 | 50 |
| F | . | . | . | . | . | 15 | 0 | 1 | 0 | 16 |
| F | S | . | . | . | . | 0 | 1 | 0 | 0 | 1 |
| . | S | . | G | T | C | 0 | 1 | 0 | 0 | 1 |
| . | S | . | G | . | C | 0 | 1 | 0 | 0 | 1 |
| . | S | . | . | . | . | 0 | 0 | 1 | 0 | 1 |
| ESBL isolates (n = 35) | ||||||||||
| . | S | . | G | T | C | 5 | 7 | 0 | 0 | 12 |
| . | S | A | G | T | C | 3 | 0 | 0 | 0 | 3 |
| . | . | . | . | T | C | 0 | 3 | 0 | 0 | 3 |
| . | . | . | . | . | C | 0 | 3 | 0 | 0 | 3 |
| F | S | A | G | T | C | 0 | 1 | 1 | 0 | 2 |
| . | S | . | G | T | . | 0 | 0 | 2 | 0 | 2 |
| . | S | . | . | T | C | 0 | 2 | 0 | 0 | 2 |
| . | S | . | . | . | . | 1 | 0 | 0 | 1 | 2 |
| F | S | . | G | T | C | 0 | 1 | 0 | 0 | 1 |
| F | . | . | G | T | C | 0 | 0 | 1 | 0 | 1 |
| F | . | . | . | . | C | 0 | 0 | 0 | 1 | 1 |
| . | S | A | . | T | C | 0 | 1 | 0 | 0 | 1 |
| . | . | . | G | T | C | 0 | 0 | 0 | 1 | 1 |
| . | . | . | G | . | C | 0 | 1 | 0 | 0 | 1 |
A, amikacin resistant; C, ciprofloxacin resistant; ESBL, extended-spectrum β-lactamase; F, cefoxitin resistant; G, gentamicin resistant; S, sulfamethoxazole-trimethoprim resistant; T, tobramycin resistant;., susceptible.
Among ESBL producers, the blaTEM gene was the most frequent (31/35, 88.6%), followed by blaCTX-M (29/35, 82.9%), whereas the blaSHV gene was only present in 4 isolates (11.4%). Of the 35 ESBL-positive bacteria, 2 (5.7%) harbored the 3 bla genes tested, 24 (68.6%) harbored blaCTX-M and blaTEM, and 1 (2.9%) harbored blaTEM and blaSHV. Four bacteria (11.4%) only carried the blaTEM gene, 3 bacteria (8.6%) only carried the blaCTX-M gene, and 1 bacterium (2.9%) only carried the blaSHV gene. All E coli and five Enterobacter spp carried the blaCTX-M gene and blaTEM gene simultaneously. Additionally, 2 Enterobacter spp were blaCTX-M only and other 2 were blaTEM positive only. Two K oxytoca were blaTEM-positive carriers and another one was a blaCTX-M-positive carrier. All of the K pneumoniae were blaSHV positive, and 2 of them harbored the blaCTX-M and blaTEM genes at the same time. Another one just harbored the blaTEM gene. Only 1 K pneumoniae was negative for the detection of the blaCTX-M and blaTEM genes.
DISCUSSION
We observed that Enterobacteriaceae contamination on phones of ICU health care workers was frequent and distributed fairly uniformly across the studied hospitals during the study period. Hospitals, occupational group, time working at the ICU, and frequency of phone use were not statistically associated with having bacterial isolates from phones. A significant but weak association between increased phone contamination and disinfectant use (RR, 1.47; P = .043) suggests that disinfection is used in response to concerns of contamination but may not reduce the risk. Type of ICU and knowledge of pathogen transmission and contamination had borderline significant associations with presence of bacteria but without important risk discrimination (RRs, 0.58–1.46). Despite having nearly 500 samples, we failed to observe the increased risk of having a positive sample associated with a touchscreen phone, as shown by Lee et al.20 The frequent contamination observed apparently rose from phone use inside the ICUs: 47.4% of HCWs used their phones >5 times while working, and 76.1% did not disinfect their phones in general. We did not study other hygiene-related practices like whether hand-washing and decontamination of shared equipment affect bacterial contamination in phones; this deserves further exploration.
Previous studies have described bacterial contamination in phones,8–12,20 but so far there is no clear understanding of whether contaminated phones introduce bacteria into hospitals, take pathogens from hospitals to the community, or both. Our study design did not allow us to answer such mechanistic questions, partially because of the absence of a nonhospital comparison group. Further studies are needed to better understand the directionality of the relationship between phone and ICU bacterial contamination. The MDR bacteria identified on the phones suggest that phone contamination is a marker of the nosocomial pathogens that circulate in the ICU because our study participants only work in their respective hospitals or ICUs because of our inclusion criteria, and the patterns of resistance observed are rare outside hospitals. The potential physical sources of phone contamination inside the ICU also remain unclear; however, it is probably safe to assume that most phones were exposed to bacteria by the contaminated hands of ICU personnel. However, how contamination reached the hands of HCWs is also important and still unknown: other HCWs, patients, shared devices, the environment, or other sources. The overall conclusion is that phones can help to spread contamination from ICUs to the community, other hospitals, or wards, and neither bans to their use in ICUs nor a policy of mandatory periodic disinfection in resource-limited settings could be issued. Disinfecting phones efficiently may be a partial solution, but this does not address the greater issue: phones probably should not be brought into ICUs.
We isolated ESBL MDR Enterobacteriaceae constantly across the whole study period and in the 3 hospitals. We found a high prevalence of ESBL-producing Enterobacteriaceae, predominantly E coli and K pneumoniae, which are the most common bacteria described and related to outbreaks in ICUs.1,2,4,21 We observed several different antimicrobial ESBL patterns. Such diversity may suggest multiple, separate introductions of different bacteria instead of either a few introductions of pathogens with specific drug resistance patterns or the persistence of some particular bacteria throughout the study period. Also, the weak or lack of association between potential risk factors and bacterial positivity might be the result of multiple introductions of contamination lacking a single, main pattern. Therefore, based on our data, it can be hypothesized that phones may not maintain the circulation of bacteria over long periods but instead allow multiple and even continuous introductions of both susceptible and resistant bacteria.
We characterized the genes associated with ESBL-producing bacteria and found at least 1 in all 35 ESBL-positive isolates. The coexistence of ≥2 bla genes and its role in the production of ESBL should be interpreted with caution because only the blaCTX-M gene encodes ESBL enzymes, whereas the blaTEM and blaSHV genes are not necessarily ESBL enzymes.22 It is possible that the ESBL-producer status is attributable to the presence of the blaCTX-M gene regardless of simultaneously carrying the blaTEM or blaSHV gene. Also, 1 K pneumoniae isolate (chromosomal blaSHV carrier) was negative for the blaTEM and the blaCTX-M genes but was ESBL positive by phenotypic tests. It is possible that a particular type of ESBL blaSSHV gene or another variety of ESBL enzyme is associated with ESBL production in this case.
Bacterial contamination in phones of ICU staff was frequent and diverse, leading to a large number of isolates. This suggests that phones may be sensitive indicators to monitor bacterial contamination in settings where phones are frequently used, not regularly disinfected, and exposed to the environment. However, we observed no clusters of isolates of exactly the same bacteria in a hospital and period. This absence of detected outbreaks could suggest that the bacteria isolated on phones may not be capable of causing human infections because of the lack of a point source. Alternatively, bacteria may have caused either small or self-contained outbreaks that did not spread or were not identified because of our spaced weekly sampling. Our data clearly indicate that phones have the potential to act as bacterial reservoirs but may lack the specificity necessary to detect outbreaks and serve as early warning systems. Further study is needed to accurately estimate their contribution to the overall burden of nosocomial infections and their value as a possible surveillance tool.
Compliance with all planned study visits was partial because of changes in personnel’s schedules, lack of interest among participants, and potentially other factors. Only 4 of 10 potential visits were conducted on average, and revisits could not be scheduled because of the narrow time frame of the study. The positive correlation between the number of phone swabs and the positivity rate may be the result of surveillance biases resulting from greater interest in participants who may have suspected that their phones were contaminated. However, adjustment by the number of samples did not alter importantly the associations (or lack thereof) observed between positivity rate and potential risk factors, suggesting that any biases may have only a limited and partially correctable effect in the results and conclusions. Additionally, incomplete sampling also limited our ability to determine bacterial persistence over time and clearly identify bacterial clusters or similar antimicrobial patterns that suggest transmission in the ICU per sampling date. Finally, the associations between self-reported knowledge and behaviors and positive bacteria isolation were not significant, probably because of the relatively small sample size and potentially small effect sizes. Stronger associations such as those between drug resistance and ESBL-producing bacteria were highly significant even with the much smaller sample size of the number of E coli and K pneumoniae infections.
In summary, our data suggest that phones represent an important source of ESBL bacteria in ICUs in the developing world. Based on our findings, the portability of phones and poor hygiene may facilitate the transmission of ESBL-producing and MDR bacteria within and across wards. Also, phones may serve in keeping a myriad of different pathogens circulating in the ICU for a prolonged time and even could be carried outside the source hospital. HCWs use their phones excessively in the ICU and do not disinfect their phones regularly. Therefore, strict adherence to phone bans in ICUs and compliance to reduce the use of phones in sensitive settings could be encouraged to minimize the threat of nosocomial infections in vulnerable patients.
Acknowledgments
Funding/Support: Supported by a competitive student award received from the “Fondo de Apoyo a la Investigación 2011” program offered by the School of Medicine “Alberto Hurtado,” Universidad Peruana Cayetano Heredia, Lima, Peru and the “Peru Infectious Diseases Epidemiology Research Training Consortium,” sponsored by the Fogarty International Center of the US National Institutes of Health (grant 2D43 TW007393) and awarded to A.G.L.
Footnotes
Conflicts of Interest: None to report.
Disclaimer: The views expressed in this article are those of the authors and do not necessarily reflect the official policy or position of the Universidad Peruana Cayetano Heredia, Department of the Navy, Department of Defense, or the U.S. Government.
Additional Information: Several authors of this article are military service members or employees of the U.S. Government. This work was prepared as part of their official duties. Title 17 U.S.C. §105 provides that “Copyright protection under this title is not available for any work of the United States Government.” Title 17 U.S.C. §101 defines a U.S. Government work as a work prepared by a military service member or employee of the U.S. Government as part of that person’s official duties.
References
- 1.Martinez-Aguilar G, Alpuche-Aranda C, Anaya C, Alcantar-Curiel D, Gayosso C, Daza C, et al. Outbreak of nosocomial sepsis and pneumonia in a newborn intensive care unit by multiresistant extended-spectrum beta-lactamase-producing Klebsiella pneumoniae: high impact on mortality. Infect Control Hosp Epidemiol. 2001;22:725–8. doi: 10.1086/501855. [DOI] [PubMed] [Google Scholar]
- 2.Stone P, Gupta A, Loughrey M, Della-Latta P, Cimiotti J, Larson E, et al. Attributable costs and lenght of stay of an extended-spectrum beta-lactamase-producing Klebsiella pneumoniae outbreak in a neonatal intensive care unit. Infect Control Hosp Epidemiol. 2003;24:601–6. doi: 10.1086/502253. [DOI] [PubMed] [Google Scholar]
- 3.Benner KW, Prabhakaran P, Lowros AS. Epidemiology of infections due to extended-spectrum Beta-lactamase-producing bacteria in a pediatric intensive care unit. J Pediatr Pharmacol Ther. 2014;19:83–90. doi: 10.5863/1551-6776-19.2.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morgan DJ, Lomotan LL, Agnes K, McGrail L, Roghmann MC. Characteristics of healthcare-associated infections contributing to unexpected in-hospital deaths. Infect Control Hosp Epidemiol. 2010;31:864–6. doi: 10.1086/655018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee CC, Lee NY, Yan JJ, Lee HC, Chen PL, Chang CM, et al. Bacteremia due to extended-spectrum-beta-lactamase-producing Enterobacter cloacae: role of carbapenem therapy. Antimicrob Agents Chemother. 2010;54:3551–6. doi: 10.1128/AAC.00055-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chopra T, Marchaim D, Veltman J, Johnson P, Zhao JJ, Tansek R, et al. Impact of cefepime therapy on mortality among patients with bloodstream infections caused by extended-spectrum-β-lactamase-producing Klebsiella pneumoniae and Escherichia coli. Antimicrob Agents Chemother. 2012;56:3936–42. doi: 10.1128/AAC.05419-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burdette SD, Herchline TE, Oehler R. Surfing the web: practicing medicine in a technological age: using smartphones in clinical practice. Clin Infect Dis. 2008;47:117–22. doi: 10.1086/588788. [DOI] [PubMed] [Google Scholar]
- 8.Ramesh J, Carter AO, Campbell MH, Gibbons N, Powlett C, Moseley H, et al. Use of mobile phones by medical staff at Queen Elizabeth Hospital, Barbados: evidence for both benefit and harm. J Hosp Infect. 2008;70:160–5. doi: 10.1016/j.jhin.2008.06.007. [DOI] [PubMed] [Google Scholar]
- 9.Nwankwo EO, Ekwunife N, Mofolorunsho KC. Nosocomial pathogens associated with the mobile phones of healthcare workers in a hospital in Anyigba, Kogi state, Nigeria. J Epidemiol Glob Health. 2014;4:135–40. doi: 10.1016/j.jegh.2013.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brady RR, Wasson A, Stirling I, McAllister C, Damani NN. Is your phone bugged? The incidence of bacteria known to cause nosocomial infection on healthcare workers’ mobile phones. J Hosp Infect. 2006;62:123–5. doi: 10.1016/j.jhin.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 11.Ulger F, Esen S, Dilek A, Yanik K, Gunaydin M, Leblebicioglu H. Are we aware how contaminated our mobile phones with nosocomial pathogens? Ann Clin Microbiol Antimicrob. 2009;8:1–4. doi: 10.1186/1476-0711-8-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ustun C, Cihangiroglu M. Health care workers’ mobile phones: a potential cause of microbial cross-contamination between hospitals and community. J Occup Environ Hyg. 2012;9:538–42. doi: 10.1080/15459624.2012.697419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing: twenty-third informational supplement M100-S23. Wayne (PA): Clinical Laboratory Standards Institute; 2013. [Google Scholar]
- 14.Yong D, Lee K, Yum JH, Shin HB, Rossolini GM, Chong Y. Imipenem-EDTA disk method for differentiation of metallo-beta-lactamase-producing clinical isolates of Pseudomonas spp. and Acinetobacter spp. J Clin Microbiol. 2002;40:3798–801. doi: 10.1128/JCM.40.10.3798-3801.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pasteran F, Mendez T, Guerriero L, Rapoport M, Corso A. Sensitive screening tests for suspected class A carbapenemase production in species of Enterobacteriaceae. J Clin Microbiol. 2009;47:1631–9. doi: 10.1128/JCM.00130-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tan TY, Ng LS, He J, Koh TH, Hsu LY. Evaluation of screening methods to detect plasmid-mediated AmpC in Escherichia coli, Klebsiella pneumoniae, and Proteus mirabilis. Antimicrob Agents Chemother. 2009;53:146–9. doi: 10.1128/AAC.00862-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arlet G, Philippon A. Construction by polymerase chain reaction and use of intragenic DNA probes for three main types of transferable beta-lactamases (TEM, SHV, CARB) [corrected] FEMS Microbiol Lett. 1991;66:19–25. doi: 10.1016/0378-1097(91)90414-6. [DOI] [PubMed] [Google Scholar]
- 18.Babini GS, Livermore DM. Are SHV beta-lactamases universal in Klebsiella pneumoniae? Antimicrob Agents Chemother. 2000;44:2230. doi: 10.1128/aac.44.8.2230-2230.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Edelstein M, Pimkin M, Palagin I, Edelstein I, Stratchounski L. Prevalence and molecular epidemiology of CTX-M extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae in Russian hospitals. Antimicrob Agents Chemother. 2003;47:3724–32. doi: 10.1128/AAC.47.12.3724-3732.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee YJ, Yoo CG, Lee CT, Chung HS, Kim YW, Han SK, et al. Contamination rates between smart cell phones and non-smart cell phones of healthcare workers. J Hosp Med. 2013;8:144–7. doi: 10.1002/jhm.2011. [DOI] [PubMed] [Google Scholar]
- 21.Mammina C, Di Carlo P, Cipolla D, Giuffrè M, Casuccio A, Di Gaetano V, et al. Surveillance of multidrug-resistant gram-negative bacilli in a neonatal intensive care unit: prominent role of cross transmission. Am J Infect Control. 2007;35:222–30. doi: 10.1016/j.ajic.2006.04.210. [DOI] [PubMed] [Google Scholar]
- 22.Bradford PA. Extended-spectrum beta-lactamases in the 21st century: characterization, epidemiology, and detection of this important resistance threat. Clin Microbiol Rev. 2001;14:933–51. doi: 10.1128/CMR.14.4.933-951.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]