Abstract
The extended superfamily of chlorophyll a/b binding proteins comprises the Light-Harvesting Complex Proteins (LHCs), the Early Light-Induced Proteins (ELIPs) and the Photosystem II Subunit S (PSBS). The proteins of the ELIP family were proposed to function in photoprotection or assembly of thylakoid pigment-protein complexes and are further divided into subgroups with one to three transmembrane helices. Two small One-Helix Proteins (OHPs) are expressed constitutively in green plant tissues and their levels increase in response to light stress. In this study, we show that OHP1 and OHP2 are highly conserved in photosynthetic eukaryotes, but have probably evolved independently and have distinct functions in Arabidopsis. Mutations in OHP1 or OHP2 caused severe growth deficits, reduced pigmentation and disturbed thylakoid architecture. Surprisingly, the expression of OHP2 was severely reduced in ohp1 T-DNA insertion mutants and vice versa. In both ohp1 and ohp2 mutants, the levels of numerous photosystem components were strongly reduced and photosynthetic electron transport was almost undetectable. Accordingly, ohp1 and ohp2 mutants were dependent on external organic carbon sources for growth and did not produce seeds. Interestingly, the induction of ELIP1 expression and Cu/Zn superoxide dismutase activity in low light conditions indicated that ohp1 mutants constantly suffer from photo-oxidative stress. Based on these data, we propose that OHP1 and OHP2 play an essential role in the assembly or stabilization of photosynthetic pigment-protein complexes, especially photosystem reaction centers, in the thylakoid membrane.
Keywords: Early light-induced protein family, phylogeny, pigment-protein complexes, photosynthesis, photoprotection
Introduction
Photosynthetic energy conversion in plants and algae forms the basis for almost all life on earth. Oxygenic photosynthesis most likely evolved in ancestral cyanobacteria around 2.7–3.5 billon years ago (Nowicka and Kruk, 2016). More than 1.5 billion years ago, engulfment of a cyanobacterium by a eukaryotic host cell gave rise to chloroplasts and thereby photo-autotrophic eukaryotes (Archibald, 2015). During the co-evolution of plastids and their host cells, the major part of the organelle genome was transferred to the host nucleus, generating the need for protein import to maintain organelle structure and function (Zimorski et al., 2014). Early on, the photosynthetic eukaryotes split into several lineages, giving rise to the now living Glaucophytes, Chloroplastida, and Rhodophytes. In both the “green” and the “red” lineage, further endocytobiosis events gave rise to secondary or even tertiary plastids (Zimorski et al., 2014).
A common feature of all photosynthetic organisms is that the conversion of light energy into chemical energy is performed by large pigment-protein complexes in specialized membrane systems, the thylakoids (Nelson and Ben-Shem, 2004). In plants, the thylakoids are structurally and functionally heterogeneous: Grana stacks predominantly contain the water-oxidizing photosystem II (PSII) while stroma lamellae harbor PS I and the ATP-synthase complex (Pribil et al., 2014). Assembling the photosynthetic complexes in the thylakoid membrane requires a tight coordination of protein synthesis and folding with pigment synthesis and delivery. Failures of this coordination can lead to protein mis-folding or accumulation of uncoupled pigments that initiate deleterious processes rather than funneling the absorbed energy to the photosynthetic reaction centers (Rochaix, 2011; Komenda et al., 2012; Wang and Grimm, 2015).
Recent studies on cyanobacteria demonstrated that the family of High Light Induced Proteins (HLIPS) plays important functions as carriers of newly synthesized pigments during the assembly of photosystem II and potentially also photosystem I (Chidgey et al., 2014; Knoppová et al., 2014; Akulinkina et al., 2015). As their name implies, HLIPs were identified by their up-regulation during high light stress, when photo-damaged photosystems have to be degraded and replaced. In this process, HLIPs may additionally function in the recycling of pigments from damaged photosystems or antennae (Dolganov et al., 1995; Havaux et al., 2003).
HLIP-like proteins are also present in all photosynthetic eukaryotes and are probably the evolutionary ancestors of the Light Harvesting Complex (LHC) superfamily of proteins (Engelken et al., 2012). The common feature of all LHCs and LHC-like proteins are conserved transmembrane helices that mediate the binding of chlorophyll (Chl) and carotenoids. In higher plants, the LHC superfamily contains members with one to four transmembrane helices. Parallel to the structural diversification, also the function of the proteins has changed. The three-helix LHCs form the membrane-integral inner and outer antennae of both photosystems and thus contribute to the efficiency of light harvesting (Büchel, 2015). In contrast, the four-helix protein PSBS is an important factor in the induction of excess energy dissipation within the LHC antenna (Li et al., 2000; Niyogi and Truong, 2013). Another class of three-helix proteins, the Early Light Induced Proteins (ELIPs) are almost exclusively expressed during light stress and de-etiolation of proplastids. They do not contribute to light harvesting but may act as pigment carriers during the assembly of photosynthetic complexes (Montané and Kloppstech, 2000; Adamska, 2001). Mutant analyses revealed only a minor function of ELIPs in light stress tolerance, whereas they contributed to the regulation of germination (Casazza et al., 2005; Rossini et al., 2006; Rojas-Stütz, 2008; Rizza et al., 2011). The two-helix LHC-like (LIL) or Stress Enhanced Proteins (SEPs) were proposed to function in photoprotection (Heddad et al., 2012). For two recently diverged LIL3 isoforms in Arabidopsis, an important function in stabilizing or localizing protein complexes mediating late steps in Chl biosynthesis has been shown (Tanaka et al., 2010; Takahashi et al., 2014; Lohscheider et al., 2015).
The One-helix proteins (OHPs) resemble most closely the ancestral cyanobacterial HLIPs and almost all photosynthetic eukaryotes contain at least one member of each of the two sub-classes OHP1 and OHP2 (Engelken et al., 2012). Based on the gene expression patterns of Arabidopsis OHP1 (At5g02120) and OHP2 (At1g34000) and the localization of both OHP1 and OHP2 in thylakoid membranes, functions in light stress protection and de-etiolation of proplastids have been proposed (Jansson et al., 2000; Andersson et al., 2003; Stawski et al., 2014). In this study, we analyzed the evolutionary relationship between OHP1 and OHP2 and directly compared their gene expression patterns under various conditions. To unravel their functions in planta, we characterized ohp1 and ohp2 T-DNA insertion mutants. Surprisingly, we found that ohp1 and ohp2 mutants of Arabidopsis showed nearly identical phenotypes with chlorotic appearance, high Chl fluorescence, strongly decreased levels of photosystems resulting in the prevention of photoautotrophic growth and altered thylakoid ultrastructure. Complementation analyses confirmed the specificity of the mutant phenotypes and demonstrated that OHP1 and OHP2 have essential, non-redundant functions in Arabidopsis.
Materials and methods
Phylogenetic analysis
Sequence data were collected from NCBI databases (http://www.ncbi.nlm.nih.gov), protein sequences were aligned using the ClustalW algorithm (Larkin et al., 2007) and the alignment was manually refined in Bioedit (Hall, 1999). Phylogenetic analysis was performed with PHYML (http://www.atgc-montpellier.fr/phyml) with 100 bootstrap replicates (Guindon et al., 2010).
Plant material and growth conditions
Arabidopsis thaliana (L.) Heynh., ecotype Col-0 was obtained from the NASC (Stock-Nr. N60000) and T-DNA insertion mutants ohp1-1 (GABI_362D02, N434694), ohp1-2 (GABI_631G03, N460555), and ohp2-1 (GABI_071E10, N406778) were obtained from the GABI-KAT project (Kleinboelting et al., 2012). Surface-sterilized seeds were grown on Murashige and Skoog plant medium (Duchefa Biochemie, Harleem, The Netherlands) solidified with 0.8% (w/v) agar and supplemented with 3% (w/v) sucrose (unless stated otherwise), with or without 10 mg L−1 sulfadiazine. Plants were grown in a growth chamber under either continuous illumination or a 8 h light/16 h dark cycle with light intensities of 10 to 100 μmol photons m−2 s−1 at 23°C (±2°C). Homozygous and heterozygous mutants were identified by PCR on genomic DNA using gene-specific primers in combination with a T-DNA specific primer (Supplemental Table 1). For transcript and protein analyses, wildtype (WT) plants were grown in individual pots in a phytochamber with an 8 h day/16 h night cycle at 21/17°C and a light intensity of 120 μmol photons m−2 s−1 at a relative humidity of 60%. Heterozygous mutants for seed production were grown on soil in a greenhouse with at least 16 h of light per day.
Isolation of genomic DNA and northern blotting
Genomic DNA was extracted from leaves using the High Pure GMO Sample Preparation Kit (Roche, Basel, Switzerland) according to the manufacturer's instructions. Total RNA was isolated from frozen leaf material (−80°C) using Trizol (Thermo Fisher Scientific, Carlsbad, CA, USA) and the RNeasy Kit (Qiagen, Hilden, Germany). Northern blots were carried out using the DIG labeling and detection system (Roche, Basel, Switzerland). Labeled probes comprising the entire CDS of OHP1 or OHP2 or approximately 300 bp of ELIP1 or ACT2 were generated by PCR, column-purified and diluted in high-SDS hybridization buffer (see Supplemental Table 1 for primer sequences). RNA separation, transfer to a nylon membrane (Pall Corp., Port Washington, NY, USA), hybridization and detection were performed as described by Woitsch and Römer (2003).
Mutant complementation
The CDSs of OHP1 or OHP2 were inserted by InFusion cloning (Takara Bio Europe, Saint-Germain-en-Laye, France) into a pEG100-derived plant transformation vector containing a synthetic riboswitch in the 3′-UTR of the 35S-driven expression cassette (Earley et al., 2006; Ausländer et al., 2010). Agrobacterium tumefaciens strain GV3101 was used to introduce the constructs into heterozygous ohp1-1 or ohp2-1 mutants by floral dip (Clough and Bent, 1998).
Protein isolation and analysis
Plant material was frozen in liquid nitrogen and crushed using either mortar and pestle or a Tissue Lyzer (Qiagen, Hilden, Germany). Samples were suspended in 50 mM Tris-HCl pH 8, 5 mM MgCl2 and centrifuged at 16,000 g for 10 min at 4°C. Soluble and membrane-associated proteins were removed by washing the pellet with 50 mM Tris-HCl, pH 7.5 supplemented with increasing NaCl concentrations of 0, 250, and 500 mM. Chl was removed by washing the pellet with 80% (v/v) acetone. For denaturing gel electrophoresis, membrane proteins were solubilized in sample buffer containing 50 mM Tris-HCl, pH 7.5, 2% (w/v) LDS, 50 mM DTT, 0.01% (w/v) bromphenol blue and 10% (v/v) glycerol for 30 min at 45°C (modified from Leto and Young, 1984). Protein concentrations were determined using the RC/DC Protein Determination Kit (Bio-Rad, Hercules, CA, USA). For efficient separation of proteins bigger than 15 kDa, SDS-PAGE was performed according to Laemmli (1970) using Biorad Minigel Systems (Bio-Rad, Hercules, CA, USA). For the analysis of smaller proteins, Tris-Tricine buffered SDS-PAGE was performed as described by Schägger and von Jagow (1987). Ten or twenty microgram protein was loaded per lane.
Immunoblotting was carried out according to Towbin et al. (1979) using polyvinylidene difluoride (PVDF) membranes with 0.45 μm pore size (GE-Healthcare, Little Chalfont, UK) at a current of 0.5 mA cm−2 of membrane for 40 min for small proteins (below 15 kDa) and 1 mA cm−2 of membrane for 60 min for proteins above 15 kDa. Membranes were blocked in 5% (w/v) non-fat dry milk dissolved in PBS containing 0.1% (v/v) Tween 20. Horseradish peroxidase-coupled secondary antibodies and enhanced chemiluminescence reagents (ECL Plus, GE Healthcare, Little Chalfont, UK) were used as a detection system. Antibody sources: anti-OHP1 (see below), anti-OHP2 (Andersson et al., 2003), anti-ELIP1 (Heddad et al., 2006), anti-33 kDa protein of the oxygen-evolving complex (PSBO; Lundin et al., 2008), all other antibodies were obtained from Agrisera AB (Vännäs, Sweden).
Green-Native PAGE of total protein extracts from entire rosettes was performed according to Allen and Staehelin (1991).
Recombinant expression of OHP1 and production of polyclonal antibodies
The OHP1 coding sequence was amplified with specific primers (Supplemental Table 1) from cDNA and inserted into the E. coli expression vector pBAD/Thio-TOPO (Thermo Fisher Scientific, Carlsbad, CA, USA). Overexpression was performed according to the manufacturer's protocol. The fusion protein accumulated in inclusion bodies that were purified according to Chen et al. (1991), resuspended in 10 mM NaHPO4 pH 7.2, 0.9% (w/v) NaCl and used to raise a polyclonal OHP1-antiserum in rabbit (TFA, University of Konstanz, Konstanz, Germany). The antiserum reacted strongly with recombinant OHP1 but showed cross-reactivity with another thylakoid protein that migrated to the same position in SDS gels.
Pigment isolation and analysis
Plant material was processed as for membrane protein extraction, except that pigments were extracted by vortexing and sonication in the presence of 80% acetone. Cell debris was removed by centrifugation for 10 min at 16,000 g and 4°C. The absorbance of extracts was determined at 662 nm, 645 nm and 470 nm and the pigment content was calculated as described by Lichtenthaler (1987). To analyze pigment composition, extracts normalized on fresh weight were analyzed by thin layer chromatography. The stationary phase consisted of TLC aluminum sheets coated with silica gel 60. Pigments were separated in a mobile phase consisting of petroleum ether:diethyl ether:chloroform:methanol:acetone at the ratio 8:2:2:1:1.
Electron microscopy
Rosette leaves of 6-week-old Arabidopsis plants were cut into small pieces of 2 × 3 mm and pre-fixed immediately with 2% (v/v) glutaraldehyde in 0.1 M phosphate buffer, pH 7.2, for 2 h at room temperature. Samples were then rinsed five times and post-fixed with 1% (w/v) osmium tetroxide for 2 h in the same buffer at room temperature. After dehydration in a graded ethanol series, samples were infiltrated and embedded in a mixture of Spurr's and Epon/Araldite resins. After polymerization, ultrathin sections of about 50 nm thickness were cut with a diamond knife and mounted on copper grids. Sections were stained with uranyl acetate first and post-stained with aqueous lead citrate (0.1 M, pH 13). Micrographs were taken with a Hitachi H 7000 electron microscope at 75 kV (Hitachi, Tokyo, Japan).
Pulse amplitude modulated (PAM) fluorimetry
Chl fluorescence was monitored using an Imaging PAM Fluorometer (Walz GmbH, Effeltrich, Germany) equipped with a standard measuring head using the ImagingWin software provided by the supplier. Program settings used in kinetics experiments were: measuring light intensity 1, measuring light frequency 1, actinic light intensity 1 or 11 (corresponding to 8 or 150 μmol photons m−2 s−1, respectively), actinic light width 0, damping 2, gain 5, saturating pulse intensity 10, yield filter 3 and Fm-factor 1.024. Before the measurements, single leaves or whole plants were dark-adapted for 5 min and exposed to a light regime consisting of one saturating flash in the dark-adapted state followed by a period of actinic illumination with further saturating flashes until the photochemical quantum efficiency (ΦPSII) reached a steady state. After each measurement, absorptivity of the leaves or seedlings was estimated as suggested by the ImagingWin software and photosynthetic parameters were calculated by the software.
Detection of reactive oxygen species and SOD activity
H2O2 and were detected in situ by infiltration of excised mature leaves with 3,3-diaminobenzidine or NBT solution, respectively, according to Thordal-Christensen et al. (1997) and Bournonville and Díaz-Ricci (2011) except that 0.02% (v/v) Silwet were included in the staining solutions. Superoxide dismutase (SOD) and catalase activities were estimated by in-gel activity staining according to Beauchamp and Fridovich (1971) and Clare et al. (1984). Entire rosettes were ground in 10 ml/g ice-cold extraction buffer [100 mM KHPO4 pH 7.5, 1 mM DTT, 3 mM EDTA, 0.4% (v/v) Triton X-100]. Soluble proteins were obtained after centrifugation at 25,000 g for 10 min at 4°C and quantified with Bradford's reagent. Per lane, 30 μg (for SODs) or 10 μg (for catalase) of total protein was separated on native polyacrylamide gels. After staining, digital images of the gels were processed with the ImageJ software (Schneider et al., 2012).
Results
Two distinct types of OHPs are present in plants
Phylogenetic analyses based on the signature element of ELIP sequences, the conserved transmembrane helices with a predicted chlorophyll (Chl) binding motive, had very little power to resolve the evolutionary origin of the sub-families of OHP and SEP/LIL proteins (Engelken et al., 2012). We used a manually refined alignment of OHP and SEP/LIL sequences around the conserved transmembrane helix corresponding to the entire length of mature OHP1 to calculate a maximum likelihood tree (Figure 1 and Supplemental Figure 1). In this tree, OHP1 and OHP2 sequences are clearly assigned to separate branches, while the bootstrap values for the individual branches between the OHP and SEP/LIL proteins are too low to deduce an exact genealogy. Within each branch, the topology roughly follows the species tree of Viridiplantae (Rodríguez-Ezpeleta et al., 2005) with a separate branch of OHP2 sequences in organisms of the red algae lineage. In the red lineage, no nuclear-encoded OHP1 was identified, but they contain a putatively ancestral plastid-encoded High Light Induced Protein (HLIP) that is not present in the green lineage (Engelken et al., 2010). Among higher plants, OHP1 sequences and likewise OHP2 sequences are highly conserved and typically have more than 75% identical amino acids in the analyzed region within the monocot and dicot clades, whereas OHP1 sequences share only 10–20% identical amino acids with OHP2 (Supplemental Figure 1).
Figure 1.
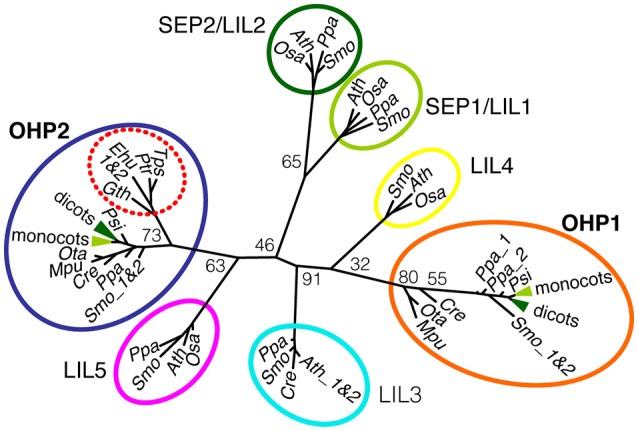
Phylogenetic analysis of OHP1 and OHP2. OHP and SEP/LIL sequences from selected plants and algae were aligned and used to calculate an unrooted maximum likelihood tree (See Supplemental Figure 1 for an excerpt of the alignment). Numbers at branching points are bootstrap values from 100 replicates. OHP1 and OHP2 proteins belong to two evolutionary distinct groups that most likely evolved independently in photosynthetic eukaryotes. Notably, secondary algae in the red lineage only contain OHP2 sequences (red dotted circle) but no nuclear-encoded OHP1. For a better overview, the highly conserved dicot (dark green) and monocot (light green) sequences were collapsed in the OHP1 and OHP2 branches of the tree and SEP/LIL sequences were only included from a small group of green algae and land plants. In addition to the species mentioned, OHP1 and OHP2 sequences from the dicots apple (Malus domestica), barrel clover (Medicago truncatula) and poplar (Populus trichocarpa) were included for the calculation of the tree. Seed plants: Ath, Arabidopsis thaliana; Pop, Populus trichocarpa; Zma, Zea mays, Osa, Oryza sativa; Psi, Picea sitchensis. Lower land plants: Ppa, Physcomitrella patens; Smo, Selaginella moellendorffii. Green algae: Cre, Chlamydonomas reinhardtii; Ota, Ostreococcus tauri; Mpu, Micromonas pusilla. Diatoms: Ptr, Phaeodactylum tricornutum; Tps, Thalassiosira pseudonana. Haptophyte: Ehu, Emiliana huxleyi. Cryptophyte: Gth, Guillardia theta.
Arabidopsis OHP genes are expressed constitutively and are upregulated upon light stress
As expected for thylakoid membrane proteins, transcripts of both OHP genes showed the highest abundance in leaves and were present in all green plant tissues, whereas both OHP1 and OHP2 transcripts were undetectable in roots (Figure 2A). Diurnal regulation of OHP1 expression had been described previously (Jansson et al., 2000). We observed that transcript levels of OHP1 and OHP2 showed very similar fluctuations during an 8 h light/16 h night cycle with the highest transcript levels before the onset and during the first half of the photoperiod being 25% higher than during the second half of the light phase and the beginning of the night (Figure 2B). Like other members of the ELIP family, OHP1 and OHP2 have been implicated in light stress responses (Jansson et al., 2000; Andersson et al., 2003). We confirmed that mRNA levels of both OHP1 and OHP2 were increased by approximately 40 and 60%, respectively, after 3 h of exposure to excess light (Figure 2C). In comparison to ELIP1, the increase in transcript levels was less pronounced for OHP1 and OHP2, especially since both OHP genes showed much higher basal expression levels. ELIP1 transcript levels were shown to increase during de-etiolation similarly to the response to light stress (Casazza et al., 2005). Also transcript levels of both OHP1 and OHP2 increased transiently by more than 100% during de-etiolation of dark-grown seedlings, peaking together with ELIP1 after 2–4 h of exposure to light (Figure 2D).
Figure 2.
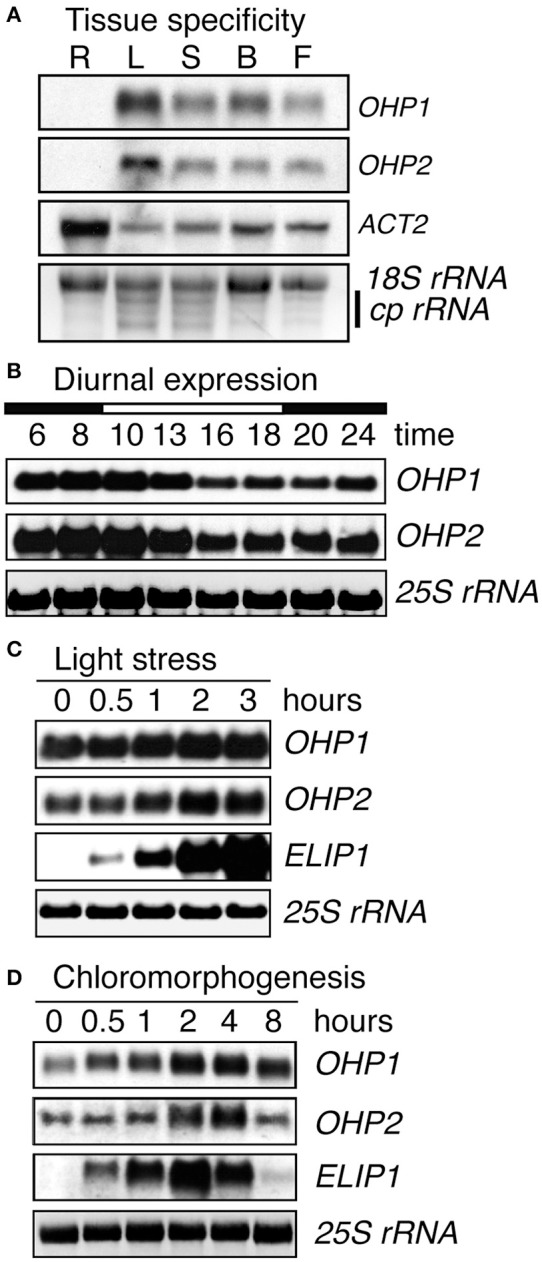
Light regulation of OHP1 and OHP2 expression. Transcript levels of OHP1, OHP2, ELIP1, and ACT2a in Col-0 (WT) plants were analyzed by northern blotting. (A) Different organs from mature plants in the reproductive phase were analyzed separately. R, Roots; L, Leaves; S, Stems; B, Buds; F, Flowers. Detection of ACT2 demonstrates the successful isolation of mRNA from roots. EtBR-stained 18S rRNA and chloroplast rRNAs (cp rRNA) are shown as loading control. (B) Diurnal regulation of OHP1 and OHP2 transcript levels in mature leaves of 6-week-old WT plants cultivated in an 8 h light/16 h dark cycle at a light intensity of 120 μmol photons m−2 s−1. The light period (10 a.m. to 6 p.m.) is indicated as a white bar on top of the time scale. (C) Light stress was induced by exposing detached leaves of plants cultivated at 120 μmol photons m−2 s−1 to a light intensity of 1000 μmol photons m−2 s−1. Induction of ELIP1 expression served as a control to demonstrate the presence of light stress. (D) De-etiolation was induced by exposing 2-week-old, dark-grown WT seedlings to white light at an intensity of 100 μmol photons m−2 s−1. Again, expression of ELIP1 served as a control to monitor the progression of de-etiolation. In (B–D), EtBR-stained 25S rRNA serves as loading control.
Co-expression analysis of OHP1 and OHP2 using AttedII revealed that OHP1 and OHP2 expression is similarly regulated: both fall within the top 60 of co-expressed genes for each other (Obayashi et al., 2014). The top 100 lists of co-expressed genes for OHP1 and OHP2 share 22 common entries. Analysis of the complete ranked list of co-expressed genes for enriched GO terms for processes with GOrilla suggested a strong connection of OHP1 and OHP2 with glyceraldehyde-3-phosphate metabolism, isopentenyl pyrophosphate (IPP) metabolism and thylakoid membrane organization, along with other important chloroplast-associated processes (Eden et al., 2009).
T-DNA insertion mutants of OHP1 or OHP2 show pale green phenotypes and are infertile
To unravel the function of OHP1 and OHP2 in thylakoids, we identified and characterized Arabidopsis T-DNA insertion mutants (Figure 3 and Supplemental Figure 2). The segregation of sulfadiazine resistance in the progeny of heterozygous plants of the T-DNA insertion line GABI_362D02 (ohp1-1) was consistent with a single T-DNA insertion (data not shown). As reported in the Chloroplast Function Database (Myouga et al., 2013), roughly one fourth of the progeny from plants heterozygous for the T-DNA insertion in OHP1 stayed very small and were of pale green color (Figure 3A). PCR analysis of the pale green plants confirmed that they were homozygous for the T-DNA in the OHP1 gene (Supplemental Figure 2). No OHP1 transcripts could be detected in these plants (Figure 3B), demonstrating that ohp1-1 is a complete loss-of-function mutant. A polyclonal antiserum raised against recombinant OHP1 detected residual amounts of a protein with the same molecular weight as OHP1 in protein extracts of homozygous ohp1-1 mutants, indicating that the serum is not monospecific (Figure 3C). So far, we did not identify the cross-reacting protein. In MS-based protein detection of gel regions corresponding to the expected position of OHP1, three tryptic peptides of OHP1 were detected a total of 18 times (corresponding to 3.5% of all peptides identified unambiguously) in a wildtype (WT) sample (Supplemental Figure 2). In a corresponding gel slice with proteins from ohp1-1 mutants, no OHP1-specific peptides were detected while peptides derived from other thylakoid proteins were detected with similar counts as in the WT sample, strongly suggesting the actual lack of the OHP1 protein in the ohp1-1 mutant. Heterozygous ohp1-1 mutants were indistinguishable from WT plants in size and coloration but they displayed reduced levels of OHP1 mRNA (Figures 3A,B). In a second T-DNA insertion line, GABI_631G03 (ohp1-2), the T-DNA is located 72 bp upstream of the start codon of OHP1. Homozygous ohp1-2 mutants showed low levels of residual OHP1 expression and a similar but less severe mutant phenotype compared to ohp1-1 mutants (Supplemental Figure 2D).
Figure 3.
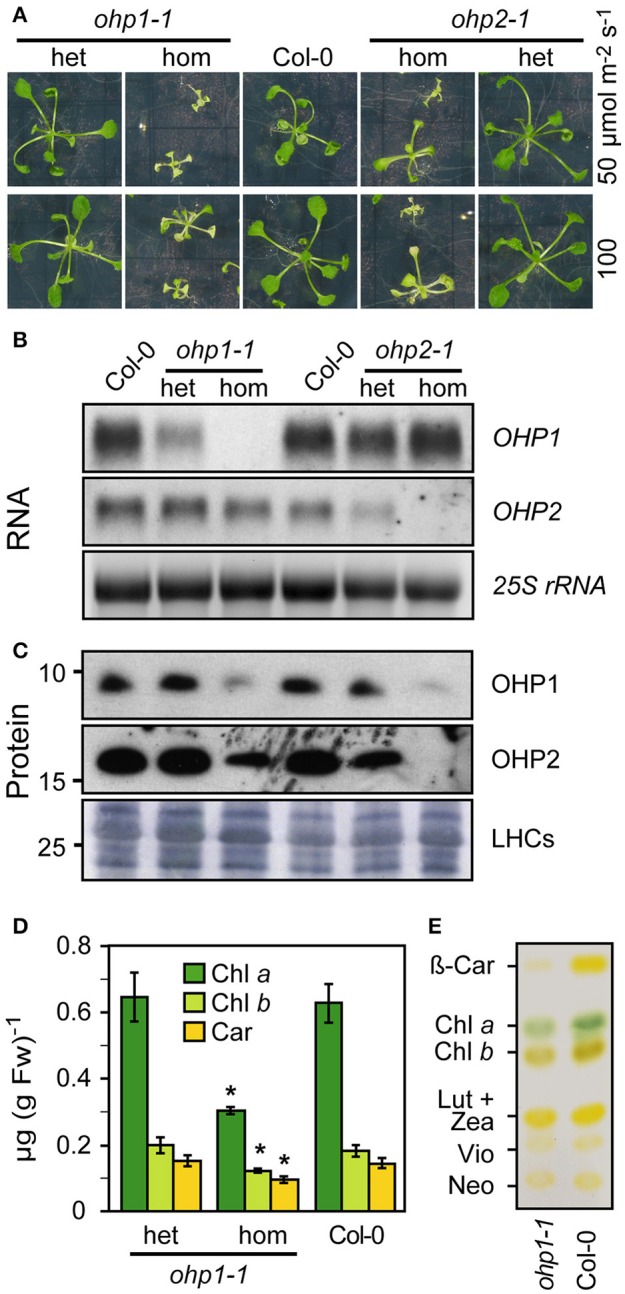
Characterization of ohp1 and ohp2 mutants. (A) Representative images of 3-week-old homozygous (hom) or heterozygous (het) ohp1-1 and ohp2-1 mutants in comparison to Col-0 WT plants of the same age. The plants were grown in axenic culture with continuous light. (B) Northern blot analysis of OHP1 and OHP2 transcript levels in the respective homozygous or heterozygous mutants in comparison to WT Col-0 plants. Fifteen microgram of total RNA were loaded per lane and EtBR-stained 25S rRNA is shown as loading control. (C) Western blot analysis of OHP1 and OHP2 protein levels in corresponding samples. Ten microgram of LDS-solubilized insoluble proteins were loaded per lane and Coomassie-stained LHC proteins are shown as loading control. (D) Pigments from leaves of Col-0 WT plants or heterozygous and homozygous ohp1-1 mutants were extracted with 80% acetone and quantified photometrically. Values represent the mean±SD (N = 4), Asterisks indicate significant differences from the respective WT value (p ≤ 0.01 by Student's t-test). (E) Pigments of homozygous ohp1-1 mutants and WT seedlings separated by thin layer chromatography.
The line GABI_071E10 (ohp2-1) initially carried at least two T-DNA insertions. After backcrossing to Col-0 plants, a line with a single insertion in OHP2 was identified. After self-fertilization, this line showed approximately 3:1 segregation of the sulfadiazine resistance marker and, surprisingly, homozygous ohp2-1 mutants phenocopied ohp1-1 mutants (Figure 3A). OHP2 transcripts and protein were undetectable in homozygous ohp2-1 seedlings (Figures 3B,C). The similar mutant phenotypes of ohp1-1 and ohp2-1 mutants indicated that OHP1 and OHP2 might act together in the same process despite their different evolutionary origin. Strikingly, hardly any OHP2 protein was detected in ohp1-1 mutants and similarly, OHP1 protein levels were strongly reduced in ohp2-1 mutants. In contrast, the transcript level of OHP1 was not affected in ohp2-1 mutants and likewise, ohp1 mutants had unchanged transcript levels of OHP2.
The reduced growth and the pale color of ohp1 and ohp2 mutants indicated severe deficits in photosynthesis, which might cause problems during photosynthetically active phases of embryo development. Indeed, the occurrence of homozygous ohp1-1 mutants was 34% less frequent than expected (Supplemental Figure 2G). Also among the progeny of heterozygous ohp2-1 mutant plants, 28% less homozygous individuals than expected were observed, explaining the slightly lower than expected proportion of sulfadiazine-resistant plants. The defects of ohp1-1 and ohp2-1 seedlings in post-embryonic development could be a symptom of starvation as a result of impaired photosynthesis. In the absence of external sucrose, seedlings of both mutant lines were not able to develop beyond cotyledon stage, while increasing the sucrose concentration from 2 to 3% (w/v) improved the growth of the mutants (data not shown). Even in the presence of high sucrose concentrations, a developmental arrest and rapid bleaching of most ohp1-1 and ohp2-1 seedlings was observed in a day/night cycle with a light intensity of 100 μmol photons m−2 s−1. Illumination with very low light (10 to 15 μmol photons m−2 s−1), lowering the temperature from 23° to 19°C and/or supplementing the growth medium with Gamborg's vitamin mix allowed homozygous ohp1-1 and ohp2-1 mutants to develop further (data not shown). However, even under improved cultivation conditions photosynthetic performance remained very low. Not all homozygous mutants survived until flowering and none of them produced seeds (data not shown).
The modified growth regime allowed the generation of sufficient biomass of ohp1-1 seedlings to investigate the pigmentation defect in more detail. Chl and carotenoids were extracted and quantified photometrically (Figure 3D). Confirming the visual impression, the pigment concentrations were very similar in WT and heterozygous ohp1-1 mutants. In contrast, homozygous ohp1-1 mutants had less than 50% of the Chl a concentration of WT seedlings and also Chl b and carotenoids were lower in ohp1-1 mutants, although to a lesser extent than Chl a (Chl b: 68%, carotenoids 65%). The differential reduction in pigment concentrations was also reflected in altered pigment ratios. The Chl a to Chl b ratio and the total Chl to carotenoids ratio were reduced by 30 or 20%, respectively, in ohp1-1 mutants as compared to WT plants. Thin-layer chromatography of isolated pigments demonstrated that mainly a decrease in β-carotene was the cause for the reduced carotenoid concentration in ohp1-1 mutants, while xanthophyll levels were very similar compared to WT plants (Figure 3E).
Chloroplast ultrastructure is altered in ohp1 mutants
In a previous study the pale green appearance of homozygous ohp1 mutants in the Ler background had been associated with altered chloroplast ultrastructure (Chloroplast Function Database, Myouga et al., 2013). To verify the requirement of OHP1 expression for normal thylakoid architecture, we analyzed the Col-0-derived mutant line ohp1-1 by transmission electron microscopy (Figure 4). Micrographs of sections of true leaves showed identical pictures for WT plants and heterozygous ohp1-1 mutants with well-differentiated grana stacks and stroma lamellae with interspersed plastoglobules. In contrast, stroma lamellae were virtually absent in leaves of homozygous ohp1-1 mutants and plastoglobules appeared fragmented. Grana stacks were present in chloroplasts of ohp1-1 mutants, but in non-appressed regions at grana stack margins the thylakoid lumen was strongly inflated. In comparison to the stroma, the content of the marginal thylakoids had a very low electron density, indicating that it contained only a low concentration of macromolecules. Additionally, starch grains were virtually absent in leaves of homozygous ohp1-1 mutants, while they occurred with similar frequencies in plastids of WT plants and heterozygous ohp1-1 mutants (data not shown). The structural aberrations observed in our specimens were different from the ones in the pictures contained in the Chloroplast Function Database, indicating ecotype- or tissue-specific effects of OHP1 depletion (Myouga et al., 2013).
Figure 4.
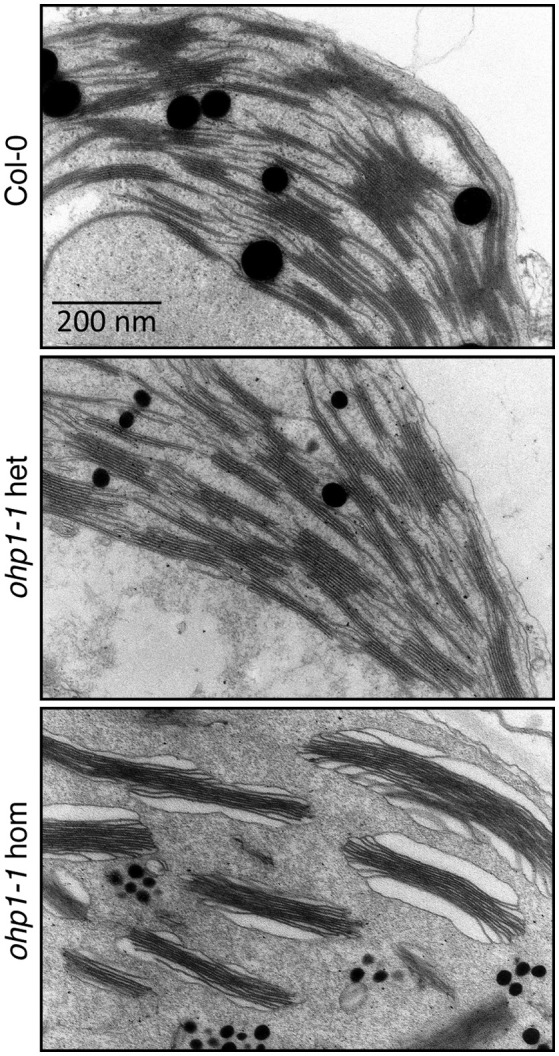
Chloroplast ultrastructure is altered in ohp1-1 mutants. Mature leaves of 6-week-old plants cultivated at 15 μmol photons m−2 s−1 in an 8 h light/16 h dark cycle were analyzed by transmission electron microscopy. The micrographs show representative sectors of chloroplasts from WT plants in comparison to homozygous and heterozygous ohp1-1 mutants. While heterozygous mutants have plastids with a similar appearance as the WT, homozygous ohp1-1 mutants display strongly disturbed thylakoid ultrastructure.
ohp1 and ohp2 mutants are impaired in photosynthesis
To examine whether reduced pigment content of ohp1-1 and ohp2-1 mutants impaired photosynthesis, we monitored Chl fluorescence using pulse amplitude-modulated (PAM) fluorimetry. During de-etiolation all seedlings showed similar levels of ground state fluorescence (F0). In WT and heterozygous seedlings F0 declined during further development, whereas homozygous ohp1-1 and ohp2-1 maintained a high F0 that only declined when the leaves bleached and died (Figure 5A and data not shown). Induction kinetics showed typical fluorescence traces in WT plants and heterozygous ohp mutants. In contrast, fluorescence emission in homozygous ohp1-1 and ohp2-1 mutants was nearly independent of the light intensity (Figure 5B and Supplemental Figure 3). In mature true leaves of homozygous mutants, F0 values were six to nine times as high as in heterozygous or WT plants (Figure 5C). Maximal fluorescence values (Fm) during a saturating light flash were only slightly higher than F0 in homozygous ohp1-1 and ohp2-1 mutants. Fm values were approximately 1.5- or 1.9-fold higher in homozygous ohp1-1 or ohp2-1 mutants, respectively, compared to heterozygous mutants and WT plants. Maximum quantum efficiency of PSII in the dark-adapted state (Fv/Fm) was close to 0 in homozygous mutants, whereas heterozygous mutants and WT plants showed values close to 0.8 typical for photosynthetically fully competent leaves (Björkman and Demmig, 1987; Johnson et al., 1993). The Fv/Fm values in homozygous ohp1-1 mutants were highest (up to 0.13) in very young leaves but rapidly declined to 0 with increasing leaf age (Supplemental Figure 3). The quantum efficiency of PSII in the light acclimated state (ΦPSII) was typically below 0.1 in leaves of homozygous ohp1-1 and ohp2-1 mutants, compared to expected values around 0.7 in heterozygous mutants and WT plants. Upon exposure to high light (150 μmol photons m−2 s−1), the fluorescence dropped rapidly in photosynthetically competent young leaves of homozygous ohp1-1 mutants concomitant with a transient increase in NPQ values. In contrast, fluorescence stayed at levels similar to F0 in older leaves and NPQ remained low (Supplemental Figure 3).
Figure 5.
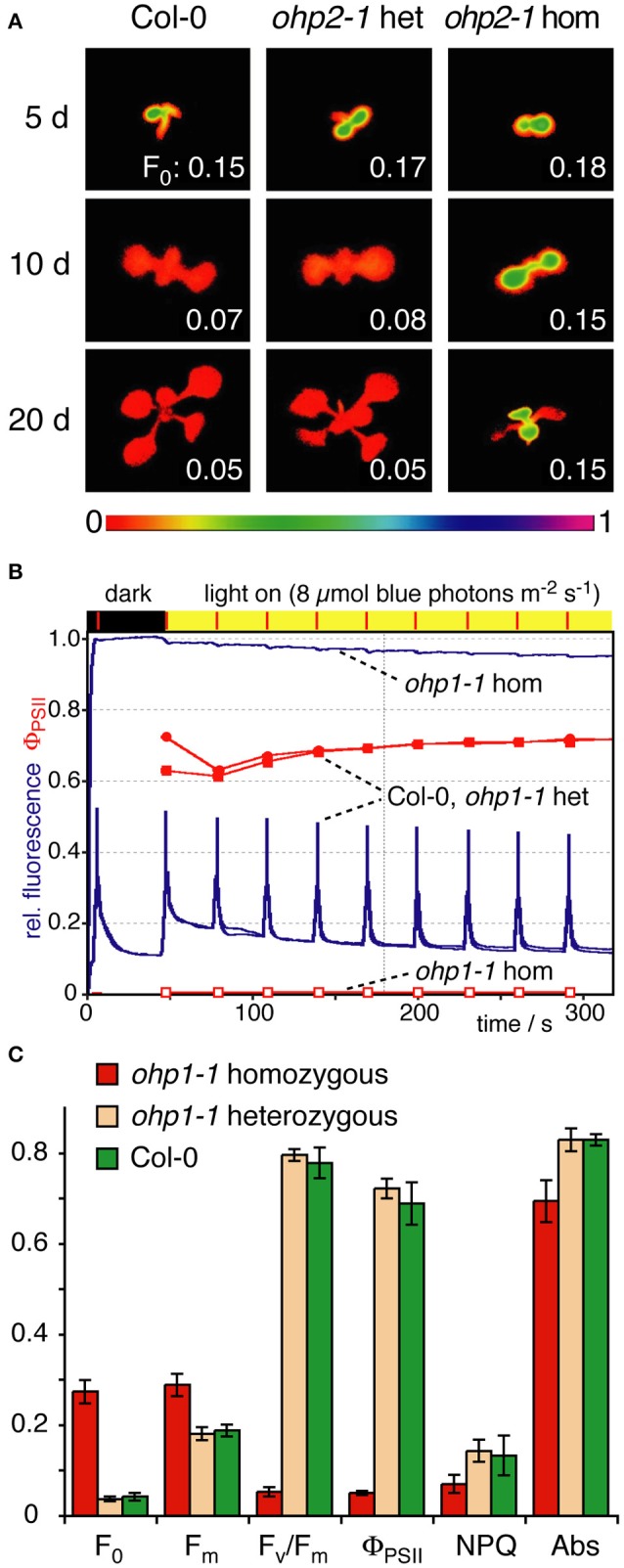
Photosynthetic performance of ohp1 and ohp2 mutants. (A) F0 images of WT plants, heterozygous and homozygous ohp2 mutants were recorded 5, 10, and 20 d after germination. Plants were grown in a 12 h light/12 h dark cycle at a light intensity of 10 μmol photons m−2 s−1 at 18°C. The bar below represents the color to numeric value conversion. The F0 values indicated in the panels are the average over the area of one cotyledon (5 and 10 d after germination) or a true leaf (at 20 d). (B) Fluorescence traces (blue lines) of an induction kinetic using mature true leaves of 3-week-old homozygous and heterozygous ohp1 mutants as well as WT plants. The plants were exposed to blue actinic illumination of 8 μmol photons m−2 s−1, which is corresponding to the cultivation conditions with 15 μmol photons m−2 s−1 white light. The operating efficiency of PSII (ΦPSII) is shown in red. (C) Fluorescence-derived photosynthetic parameters of mature leaves analyzed as described above. F0, Minimal fluorescence of dark-adapted leaves; Fm, Maximal fluorescence of dark-adapted leaves; Fv/Fm, Maximal photochemical efficiency of dark-adapted leaves; NPQ, Non-photochemical quenching; Abs, Relative absorptivity of the leaves. Values are mean±SD (N = 5). Homozygous mutants differed significantly from WT and heterozygous plants in all analyzed parameters (Student's t-test, p < 0.05).
PSI and PSII reaction centers are affected by depletion of OHP proteins
The similar regulation of OHP1 and OHP2 expression and the nearly identical mutant phenotypes indicated that OHP1 and OHP2 are required for PSII function and that both proteins might be functionally linked. Our western blot analysis revealed that only low levels of OHP2 protein were detected in homozygous ohp1-1 mutants, and likewise, ohp2-1 mutants lacked OHP1 protein (Figure 3). Comparative analysis of photosynthetic complexes by native gel electrophoresis showed that homozygous ohp1-1 mutants lack several components of the photosynthetic machinery (Figure 6). To further investigate these defects in both ohp1-1 and ohp2-1 mutants, we analyzed the levels of photosynthetic proteins in 10-week-old plants cultivated at 15 μmol photons m−2 s−1 with a short-day light regime. The abundance of several antenna proteins of PSII and PSI was reduced in ohp1-1 and ohp2-1 mutants compared to WT plants (Figure 6 and Supplemental Figure 4). Strikingly, levels of the reaction center (RC) proteins of both PSII (PSBA) and PSI (PSAB) were near or below the detection limit. Furthermore, the 33 kDa-subunit of the oxygen-evolving complex (PSBO) and PSAK, another subunit of PSI, were strongly reduced in ohp1-1 mutants. These differences became more pronounced when comparing older plants or when plants were grown at an illumination intensity of 50 μmol m−2 s−1 (data not shown). Interestingly, homozygous ohp1-1 and ohp2-1 mutants but not heterozygous mutants or WT plants showed expression of ELIP1, a phenomenon generally associated with light stress (Adamska et al., 1992).
Figure 6.
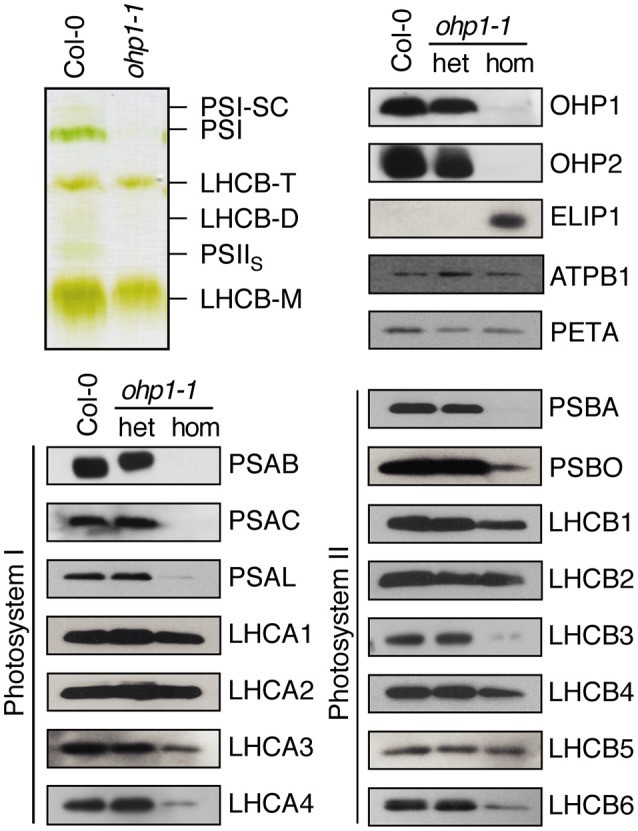
Deletion of OHP1 affects specific pigment-protein complexes in the thylakoid membrane. Total membrane protein extracts of entire rosettes of 10-week-old homozygous and heterozygous ohp1-1 mutants and WT plants cultivated at 15 μmol photons m−2 s−1 in an 8 h light/16 h dark cycle were analyzed by green native PAGE or Western blotting after denaturing SDS-PAGE. Equal amounts of protein were loaded in each lane.
Loss of OHP1 causes induction of the antioxidant defense
The high Chl fluorescence yield and almost complete absence of reaction centers observed in homozygous ohp1-1 and ohp2-1 mutants indicated poor usage of excitation energy, which is very likely to cause increased formation of reactive oxygen species (ROS). Staining of leaves with trypan blue revealed that ohp1-1 mutants did not contain elevated levels of dead cells (data not shown). Staining of leaves for and H2O2 levels produced very similar pictures for homozygous ohp1-1 mutants and WT plants (Figure 7). Therefore, we analyzed the activity of superoxide dismutase (SOD) and catalase in leaf extracts. The activities of catalase and Mn-SOD were very similar in WT plants and ohp1-1 mutants. In contrast, the activities of Fe-SOD and Cu/Zn-SOD were increased in homozygous ohp1-1 mutants, especially Cu/Zn SOD, for which the activity was increased approximately 2-fold.
Figure 7.
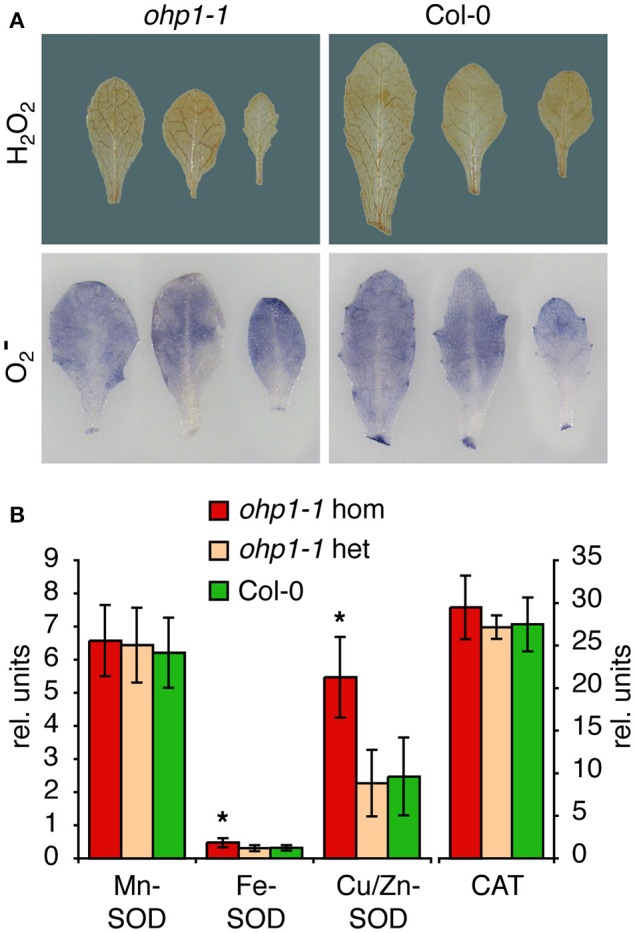
ROS staining and ROS-scavenging enzyme activities in ohp1-1 mutants. (A) Leaves of 4-week-old ohp1-1 mutants and WT plants cultivated under continuous illumination at 15 μmol photons m−2 s−1 were stained for H2O2 content with 3,3-diaminobenzidine or for content with NBT. Leaves were vacuum-infiltrated with staining solution and incubated for 2 h or 20 min, respectively, at 15 μmol photons m−2 s−1. Three representative leaves of each genotype are displayed. (B) Relative activities of the three superoxide dismutase (SOD) isoforms and catalase were quantified in leaf extracts by in-gel staining after native gel electrophoresis. SOD activities were normalized by the total SOD activity of WT leaves for each gel. Values represent the mean ± SD in relative units (N = 8 for SODs, N = 3 for catalase), asterisks indicate significant difference from the respective activity in WT leaves (p ≤ 0.02 in a Student's t-test).
OHP1 and OHP2 are not functionally redundant
The highly similar phenotypes of ohp1-1 and ohp2-1 mutants despite the distinct evolutionary origin of both proteins suggested that they might either have a redundant, dose-dependent function or act together in the assembly or maintenance of photosynthetic complexes. To test for functional redundancy and the specificity of the mutant phenotypes, we transformed heterozygous mutants with constructs for overexpression of OHP1 or OHP2. Already in the T1 generation, homozygous ohp1-1 plants with WT-like appearance were identified in lines carrying a 35S-OHP1 construct but never with a 35S-OHP2 construct. In the T2 generation, also the offspring of heterozygous ohp1-1 mutants carrying the 35S-OHP1 construct contained homozygous ohp1-1 mutants that showed normal growth and pigmentation and had a photosynthetic capacity (Fv/Fm) similar to WT plants (Figure 8A). Among the offspring of heterozygous ohp1-1 mutants carrying the 35S-OHP2 construct, all homozygous ohp1-1 mutants were pale and showed high Chl fluorescence independent of the presence of the 35S-OHP2 construct (Figure 8B). Likewise, 35S-OHP2 but not 35S-OHP1 complemented the phenotype of homozygous ohp2-1 mutants. Transcript analysis confirmed that the complementation constructs, which produce longer transcripts due to the insertion of a riboswitch in the 3′-UTR, were expressed even in the lines that failed to show cross-complementation (Figure 8C). These findings demonstrate that the loss of OHP expression is the definite cause for the mutant phenotypes and that OHP1 and OHP2 have clearly distinct functions.
Figure 8.
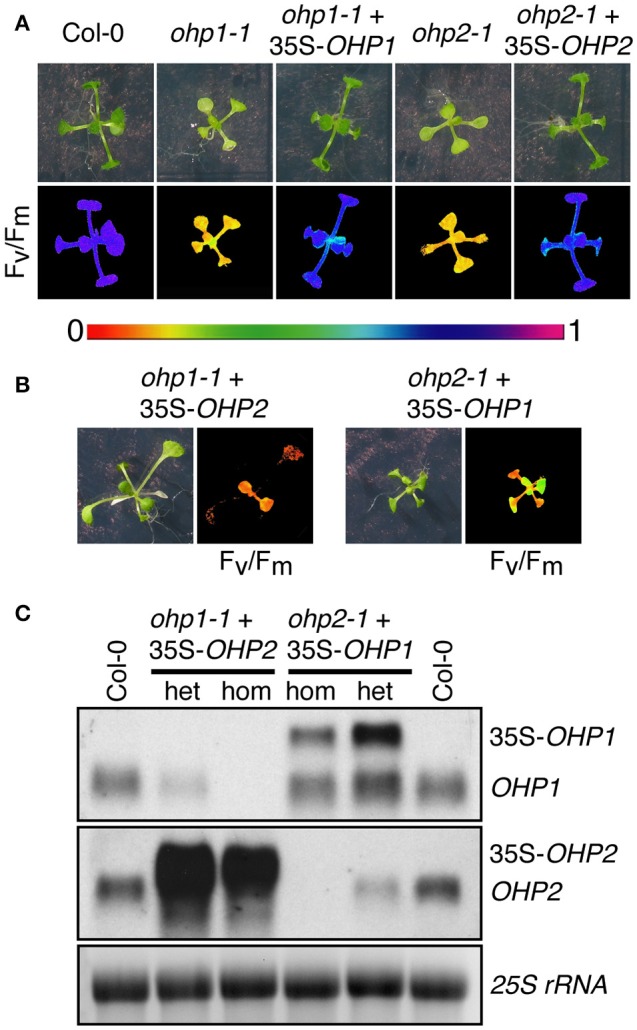
Complementation of ohp mutants. (A,B) Photos and Fv/Fm images of 2-week-old WT plants, homozygous ohp1-1 or ohp2-1 mutants and complemented mutants (T2 generation). Color scale for Fv/Fm is shown between (A,B). In the T1 generation after transformation of heterozygous ohp mutants with OHP overexpression constructs, plants homozygous or heterozygous for the ohp1-1 or ohp2-1 T-DNA insertions were identified by PCR. T2 plants were selected for BASTA resistance conferred by the overexpression constructs, sulfadiazine resistance conferred by the KO-constructs and transferred to plates without herbicide 5 days after germination. (C) Transcript analysis of plants from the cross-complementation attempt. Transcripts of the rescue constructs (35S-OHP1 and 35S-OHP2) can be distinguished from endogenous transcripts by their bigger size due to artificial UTRs. Fifteen microgram of total RNA were loaded per lane and EtBR-stained 25S rRNA is shown as loading control.
Discussion
OHP1 and OHP2 proteins are highly conserved
It is striking that all well annotated genomes of phototrophic eukaryotes except dinoflagellates and chlorarachniophytes contain at least one HLIP-encoding (glaucophytes and red algae) or OHP1-encoding (green algae and plants) gene and one gene encoding an OHP2 protein (Engelken et al., 2012). Despite the similar predicted secondary structure with a single transmembrane helix, OHPs are most likely not a monophyletic group of proteins (Figure 1; Engelken, 2010). The phylogenetic tree derived from the alignment comprising the entire sequence of the mature OHP1 protein roughly displays the expected species topology in the green lineage with a gradual evolution via green algal, bryophyte and pteridophyte OHP1 sequences to higher land plants (Rodríguez-Ezpeleta et al., 2005). While OHP1-type proteins were probably directly derived from cyanobacterial HLIP ancestors, OHP2-type proteins are more closely related to the eukaryotic Light Harvesting-Like proteins (LILs) than to OHP1 and HLIPs and have probably evolved by the loss of the second transmembrane helix present in LILs (Figure 1; Engelken et al., 2012).
The broad abundance and strong conservation can be taken as an indication that both OHP1 and OHP2 fulfill essential functions in thylakoids. In contrast to the high diversity of HLIPs and their variable number per organism, OHP1 and OHP2 sequences typically occur as one copy per organism and they are highly conserved, especially in land plants including mosses (Figure 1 and Supplemental Figure 1). Synechocystis mutants missing all four HLIPs were not viable while growing under high light conditions and it could be shown that HLIPs in cyanobacteria stabilize PSI trimers under exposure to high irradiance (He et al., 2001; Wang et al., 2008). More recently, cyanobacterial HLIPs were associated with delivery of newly synthesized or recycled pigments during assembly of photosystem II and potentially also Photosystem I (Komenda and Sobotka, 2016). They were found to interact in pairs or as dimers with photosystems and other proteins involved in pigment synthesis and pigment-protein assembly factors (Chidgey et al., 2014; Knoppová et al., 2014; Akulinkina et al., 2015; Staleva et al., 2015). The strong conservation suggests that OHPs in plants fulfill similar functions while some of the functions of HLIPs might be taken over by LILs or ELIPs in plants, thus allowing the reduction to one copy each of OHP1 and OHP2.
Co-regulation of OHP1 and OHP2 expression suggests a common function
Expression of both OHP1 and OHP2 was found to be regulated at the transcript level in response to different light conditions (Jansson et al., 2000; Andersson et al., 2003; Stawski et al., 2014). Our detailed expression analysis revealed that OHP1 and OHP2 transcript levels are co-regulated during the diurnal light cycle, during de-etiolation and in response to excess light (Figure 2). In contrast to ELIP1 transcripts, OHP1 and OHP2 mRNAs were detected under all conditions in photosynthetically active tissues, indicating that the function of OHPs is not limited to light stress protection. Transcript levels of OHP1 and OHP2 were increased by approximately 25, 50, and 100% at the beginning of the light phase, in response to high light stress and during de-etiolation of dark-grown seedlings, respectively, indicating that OHP proteins may be especially important when the synthesis or repair rates of photosynthetic protein complexes in the thylakoid membrane are high. A high degree of co-expression across the total range of transcriptome analyses summarized in the AttedII database indicates that co-regulation of OHP1 and OHP2 expression may not be limited to light responses (Obayashi et al., 2014). OHP1 and OHP2 expression was additionally co-regulated with genes of the IPP metabolism, which is the starting point for isoprenoid biosynthesis (Vranová et al., 2013). Among the isoprenoids, especially the terpenoids like phytols, tocopherols and carotenoids are essential for assembly and functionality of photosynthetic pigment-protein complexes as well as for photoprotection (Domonkos et al., 2013; Mokrosnop, 2014).
OHP1 and OHP2 are essential for functional photosystems in arabidopsis
Based on expression patterns during light stress, photoprotective functions were proposed for ELIP family members, both in cyanobacteria and higher plants (Heddad et al., 2012). However, detailed information about the molecular and physiological functions of proteins of the ELIP family is still scarce. Most of the functional studies were performed on three-helix ELIPs or two-helix LILs/SEPs in higher plants and on HLIPs in cyanobacteria. Besides light stress protection, interaction studies and mutant analyses indicated functions in pigment synthesis, pigment transfer and recycling as well as pigment protein complex assembly (Heddad et al., 2012; Sinha et al., 2012; Yao et al., 2012; Stawski et al., 2014; Takahashi et al., 2014; Lohscheider et al., 2015; Komenda and Sobotka, 2016).
In this study, we demonstrate that OHP1 and OHP2 play a crucial role in assembly or maintenance of functional photosystems in Arabidopsis thylakoid membranes. The stepwise assembly of photosystem II in higher plants and the repair cycle that enables selective replacement of damaged D1 proteins in photosystem II reaction centers has been analyzed in great detail and many structural and accessory proteins have been identified (Järvi et al., 2015; Lu, 2016). However, none of the studies on these topics have identified a contribution of OHP1 or OHP2. As described in the present study, the depletion of OHP1 and OHP2 led to drastically reduced levels of both photosystem II and photosystem I reaction center proteins (Figure 6) concomitant with impairment or complete loss of photosynthesis in ohp mutants (Figure 5 and Supplemental Figure 3). Induction kinetics of Chl fluorescence quenching demonstrated that leaves of ohp mutants rapidly lost any detectable photochemical quenching which is in accordance with the loss of reaction centers. Also inducible non-photochemical fluorescence quenching was barely detectable in ohp mutant leaves although a fraction of the LHC antenna was expressed at almost the same level as in WT plants. Pigment concentrations in ohp mutant leaves were strongly reduced, especially of Chl and β-carotene. Xanthophyll levels were very similar in leaves of WT plants and ohp1 mutants, indicating that xanthophyll-mediated light harvesting or energy dissipation in the antenna are not the primary targets of OHP action. Despite the low Chl concentration, ground state fluorescence (F0) was much higher in ohp mutants than in WT plants, indicating that either the antenna had insufficient possibilities to dissipate the excitation energy or that additionally free pigments accumulated in the mutants. Probably the early seedling lethal phenotype has prevented the identification of ohp mutants in previous screens for altered Chl fluorescence (Meurer et al., 1996; Shikanai et al., 1999).
The loss of reaction centers and photosystems also resulted in drastic changes of thylakoid ultrastructure in ohp1-1 mutants (Figure 4). The presence of LHCII antenna probably stabilized the remaining grana stacks, while the absence of stroma lamella and the swelling of the marginal thylakoids clearly indicate that OHP proteins are essential to build or maintain the correct architecture of the thylakoids. Interestingly, the changes observed in an ohp1 mutant in the Ler background were different from what we observed in the Col-0-derived mutant (Myouga et al., 2013). It remains to be determined if these difference reflect an ecotype-specific effect or are due to differences in cultivation conditions or the age of the analyzed leaves.
By complementation of the ohp1-1 and ohp2-1 mutants we demonstrated that the mutant phenotypes are specific and exclusively caused by the loss of expression of individual OHPs (Figure 8). Cross-complementation was not observed, indicating that OHP1 and OHP2 have separate functions, although they may act together in the same process. The downregulation of OHP1 protein levels in ohp2 mutants and vice versa complicates the assignment of a specific function to either of the OHP proteins in Arabidopsis. To get more detailed information about the molecular function of OHPs in higher plants, the identification of interaction partners and potential pigment binding capacities need to be determined in the future. We aim to use riboswitch-mediated de-stabilization of OHP transcripts in our complemented lines to investigate the consequences of OHP depletion in fully developed chloroplasts.
For cyanobacterial HLIPs, homodimer or heterodimer formation has been postulated as a prerequisite of pigment binding (Knoppová et al., 2014; Staleva et al., 2015; Komenda and Sobotka, 2016). Binding of HLIPs to photosystem II reaction centers has been clearly demonstrated, while also an association with photosystem I complexes is still debated (Akulinkina et al., 2015; Komenda and Sobotka, 2016). Mutant analyses and characterization of isolated protein complexes indicate a function of HLIPs in pigment delivery and energy dissipation during early steps of reaction center protein assembly (Komenda and Sobotka, 2016).
In a similar manner a complex comprising both OHP1 and OHP2 may be formed in higher plants, depending on either protein for functionality. Such a complex might not interact strongly with the structural photosystem components but might be essential for pigment delivery to both reaction centers during biogenesis or repair, which would explain the severe mutant phenotype. Loss of one OHP might not only abolish the function of such a complex, but additionally cause de-stabilization of the other OHP protein, which would explain the nearly identical mutant phenotypes. Similar stabilizing interactions have been observed previously for other thylakoid proteins, e.g., Geranylgeranyl Reductase was found to be unstable in the absence of LIL3 proteins in Arabidopsis and point mutations in Synechocystis Cytochrome b559 de-stabilized the entire PS II complex (Hung et al., 2007; Tanaka et al., 2010).
Oxidative stress in ohp mutants
High Chl fluorescence is typically caused by an increased lifetime of excited Chl molecules, which at the same time increases the risk that excitation energy or excited electrons are transferred to molecular oxygen, leading to the formation of singlet oxygen or superoxide radicals, respectively (Pospíšil, 2012). Superoxide is rapidly converted to the more stable hydrogen peroxide either by chemical dismutation or by the action of superoxide dismutases (Alscher et al., 2002). We did not detect elevated levels of superoxide or hydrogen peroxide in ohp1-1 mutants, indicating that oxidative stress is not the major reason for the seedling lethality of the mutants. Nevertheless, upregulated activity of chloroplastic FeSOD and Cu/ZnSOD activities indicated that ROS levels or ROS signaling were disturbed in ohp1-1 mutants (Alscher et al., 2002; Pilon et al., 2011). The absence of strong oxidative stress in ohp mutants indicates that OHP proteins do not primarily function in protection against excess light stress.
Based on the improved growth of ohp mutants by external supply of sugar and vitamins together with the selective absence of reaction centers, we propose that OHP proteins fulfill a crucial function in either assembly or maintenance of photosynthetic reaction center complexes, similar to their evolutionary ancestors, the cyanobacterial HLIPs.
Author contributions
IA, JB, MR, SA, JL, and DF conceived the study. MR, JB, SA, and DF screened and identified ohp1 and ohp2 mutant lines. MR and DF analyzed transcript expression patterns. UA generated OHP-specific antisera. JB and SA carried out immunoblot analysis of thylakoid membrane proteins. JB analyzed pigment-protein complexes by green native gel electrophoresis. JL performed phylogenetic and in silico co-expression analysis. MR, JB, and JL performed analysis of pigment content and composition. DF, MR, SA, and JB measured Chl fluorescence parameters in WT and ohp mutant plants. DF generated and analyzed complemented mutants. KM provided and analyzed EM micrographs. JB, JL, and DF wrote the manuscript with critical input by all other authors.
Funding
This work was supported by the German Research Foundation (DFG, grant Nrs. AD92/7-3 to IA and LO2018/2-1 to JL) and the University of Konstanz. The publication of this article was supported by a DFG grant for open access publishing to the library of the University of Konstanz.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors are grateful for continuous support by the team of the botanical garden of the University of Konstanz as well as technical support from Roswitha Miller, Silvia Kuhn and Regina Grimm. We thank Heinz Vahlenkamp for assistance in obtaining the EM micrographs. Prof. Bernhard Grimm (Humboldt-Universität zu Berlin) is acknowledged for critical review of the manuscript.
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.00007/full#supplementary-material
References
- Adamska I. (2001). The Elip familiy of stress proteins in the thylakoid membrane of pro- and eukaryota. Adv. Photosyn. Respir. 11, 487–505. 10.1007/0-306-48148-0_28 [DOI] [Google Scholar]
- Adamska I., Ohad I., Kloppstech K. (1992). Synthesis of the early light-inducible protein is controlled by blue light and related to light stress. Proc. Natl. Acad. Sci. U.S.A. 89, 2610–2613. 10.1073/pnas.89.7.2610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akulinkina D. V., Bolychevtseva Y. V., Elanskaya I. V., Karapetyan N. V., Yurina N. P. (2015). Association of high light-inducible HliA/HliB stress proteins with photosystem 1 trimers and monomers of the Cyanobacterium Synechocystis PCC 6803. Biochem. Biokhim. 80, 1254–1261. 10.1134/S0006297915100053 [DOI] [PubMed] [Google Scholar]
- Allen K. D., Staehelin L. A. (1991). Resolution of 16 to 20 chlorophyll-protein complexes using a low ionic strength native green gel system. Anal. Biochem. 194, 214–222. 10.1016/0003-2697(91)90170-X [DOI] [PubMed] [Google Scholar]
- Alscher R. G., Erturk N., Heath L. S. (2002). Role of superoxide dismutases (SODs) in controlling oxidative stress in plants. J. Exp. Bot. 53, 1331–1341. 10.1093/jexbot/53.372.1331 [DOI] [PubMed] [Google Scholar]
- Andersson U., Heddad M., Adamska I. (2003). Light stress-induced one-helix protein of the chlorophyll a/b-binding family associated with photosystem I. Plant Physiol. 132, 811–820. 10.1104/pp.102.019281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archibald J. M. (2015). Endosymbiosis and eukaryotic cell evolution. Curr. Biol. CB 25, R911–R921. 10.1016/j.cub.2015.07.055 [DOI] [PubMed] [Google Scholar]
- Ausländer S., Ketzer P., Hartig J. S. (2010). A ligand-dependent hammerhead ribozyme switch for controlling mammalian gene expression. Mol. Biosyst. 6, 807–814. 10.1039/b923076a [DOI] [PubMed] [Google Scholar]
- Beauchamp C., Fridovich I. (1971). Superoxide dismutase: improved assays and an assay applicable to acrylamide gels. Anal. Biochem. 44, 276–287. 10.1016/0003-2697(71)90370-8 [DOI] [PubMed] [Google Scholar]
- Björkman O., Demmig B. (1987). Photon yield of O2 evolution and chlorophyll fluorescence characteristics at 77 K among vascular plants of diverse origins. Planta 170, 489–504. 10.1007/BF00402983 [DOI] [PubMed] [Google Scholar]
- Bournonville C. F., Díaz-Ricci J. C. (2011). Quantitative determination of superoxide in plant leaves using a modified NBT staining method. Phytochem. Anal. 22, 268–271. 10.1002/pca.1275 [DOI] [PubMed] [Google Scholar]
- Büchel C. (2015). Evolution and function of light harvesting proteins. J. Plant Physiol. 172, 62–75. 10.1016/j.jplph.2014.04.018 [DOI] [PubMed] [Google Scholar]
- Casazza A. P., Rossini S., Rosso M. G., Soave C. (2005). Mutational and expression analysis of ELIP1 and ELIP2 in Arabidopsis thaliana. Plant Mol. Biol. 58, 41–51. 10.1007/s11103-005-4090-1 [DOI] [PubMed] [Google Scholar]
- Chen Z., Koelsch G., Han H. P., Wang X. J., Lin X. L., Hartsuck J. A., et al. (1991). Recombinant rhizopuspepsinogen. Expression, purification, and activation properties of recombinant rhizopuspepsinogens. J. Biol. Chem. 266, 11718–11725. [PubMed] [Google Scholar]
- Chidgey J. W., Linhartová M., Komenda J., Jackson P. J., Dickman M. J., Canniffe D. P., et al. (2014). A cyanobacterial chlorophyll synthase-HliD complex associates with the Ycf39 protein and the YidC/Alb3 insertase. Plant Cell 26, 1267–1279. 10.1105/tpc.114.124495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clare D. A., Duong M. N., Darr D., Archibald F., Fridovich I. (1984). Effects of molecular oxygen on detection of superoxide radical with nitroblue tetrazolium and on activity stains for catalase. Anal. Biochem. 140, 532–537. 10.1016/0003-2697(84)90204-5 [DOI] [PubMed] [Google Scholar]
- Clough S. J., Bent A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. 10.1046/j.1365-313x.1998.00343.x [DOI] [PubMed] [Google Scholar]
- Dolganov N. A., Bhaya D., Grossman A. R. (1995). Cyanobacterial protein with similarity to the chlorophyll a/b binding proteins of higher plants: evolution and regulation. Proc. Natl. Acad. Sci. U.S.A. 92, 636–640. 10.1073/pnas.92.2.636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domonkos I., Kis M., Gombos Z., Ughy B. (2013). Carotenoids, versatile components of oxygenic photosynthesis. Prog. Lipid Res. 52, 539–561. 10.1016/j.plipres.2013.07.001 [DOI] [PubMed] [Google Scholar]
- Earley K. W., Haag J. R., Pontes O., Opper K., Juehne T., Song K., et al. (2006). Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 45, 616–629. 10.1111/j.1365-313X.2005.02617.x [DOI] [PubMed] [Google Scholar]
- Eden E., Navon R., Steinfeld I., Lipson D., Yakhini Z. (2009). GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics 10:48. 10.1186/1471-2105-10-48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelken J. (2010). Evolution of the Extended LHC Protein Superfamily in Photosynthesis. PhD. thesis, University of Konstanz. [Google Scholar]
- Engelken J., Brinkmann H., Adamska I. (2010). Taxonomic distribution and origins of the extended LHC (light-harvesting complex) antenna protein superfamily. BMC Evol. Biol. 10:233. 10.1186/1471-2148-10-233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelken J., Funk C., Adamska I. (2012). The extended Light-Harvesting Complex (LHC) protein superfamily: classification and evolutionary dynamics, in Functional Genomics and Evolution of Photosynthetic Systems, eds Burnap R. L., Vermaas W. F. J. (Dordrecht: Springer; ), 265–284. [Google Scholar]
- Guindon S., Dufayard J. F., Lefort V., Anisimova M., Hordijk W., Gascuel O. (2010). New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321. 10.1093/sysbio/syq010 [DOI] [PubMed] [Google Scholar]
- Hall T. A. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl. Acids Symp. Ser. 41, 95–98. [Google Scholar]
- Havaux M., Guedeney G., He Q., Grossman A. R. (2003). Elimination of high-light-inducible polypeptides related to eukaryotic chlorophyll a/b-binding proteins results in aberrant photoacclimation in Synechocystis PCC6803. Biochim. Biophys. Acta 1557, 21–33. 10.1016/S0005-2728(02)00391-2 [DOI] [PubMed] [Google Scholar]
- He Q., Dolganov N., Bjorkman O., Grossman A. R. (2001). The high light-inducible polypeptides in Synechocystis PCC6803. Expression and function in high light. J. Biol. Chem. 276, 306–314. 10.1074/jbc.M008686200 [DOI] [PubMed] [Google Scholar]
- Heddad M., Engelken J., Adamska I. (2012). Light stress proteins in viruses, cyanobacteria and Photosynthetic Eukaryota, in Photosynthesis: Plastid Biology, Energy Conversion and Carbon Assimilation, eds Eaton-Rye J. J., Tripathy B. C., Sharkey T. D. (Dordrecht: Springer; ), 299–317. [Google Scholar]
- Heddad M., Norén H., Reiser V., Dunaeva M., Andersson B., Adamska I. (2006). Differential expression and localization of early light-induced proteins in Arabidopsis. Plant Physiol. 142, 75–87. 10.1104/pp.106.081489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung C. H., Huang J. Y., Chiu Y. F., Chu H. A. (2007). Site-directed mutagenesis on the heme axial-ligands of cytochrome b559 in photosystem II by using cyanobacteria Synechocystis PCC 6803. Biochim. Biophys. Acta 1767, 686–693. 10.1016/j.bbabio.2007.02.016 [DOI] [PubMed] [Google Scholar]
- Jansson S., Andersson J., Kim S. J., Jackowski G. (2000). An Arabidopsis thaliana protein homologous to cyanobacterial high-light-inducible proteins. Plant Mol. Biol. 42, 345–351. 10.1023/A:1006365213954 [DOI] [PubMed] [Google Scholar]
- Järvi S., Suorsa M., Aro E. M. (2015). Photosystem II repair in plant chloroplasts–Regulation, assisting proteins and shared components with photosystem II biogenesis. Biochim. Biophys. Acta 1847, 900–909. 10.1016/j.bbabio.2015.01.006 [DOI] [PubMed] [Google Scholar]
- Johnson G., Young A., Scholes J., Horton P. (1993). The dissipation of excess excitation energy in British plant species. Plant Cell Environ. 16, 673–679. 10.1111/j.1365-3040.1993.tb00485.x [DOI] [Google Scholar]
- Kleinboelting N., Huep G., Kloetgen A., Viehoever P., Weisshaar B. (2012). GABI-Kat SimpleSearch: new features of the Arabidopsis thaliana T-DNA mutant database. Nucleic Acids Res. 40, D1211–D1215. 10.1093/nar/gkr1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoppová J., Sobotka R., Tichy M., Yu J., Konik P., Halada P., et al. (2014). Discovery of a chlorophyll binding protein complex involved in the early steps of photosystem II assembly in Synechocystis. Plant Cell 26, 1200–1212. 10.1105/tpc.114.123919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komenda J., Sobotka R. (2016). Cyanobacterial high-light-inducible proteins–Protectors of chlorophyll-protein synthesis and assembly. Biochim. Biophys. Acta 1857, 288–295. 10.1016/j.bbabio.2015.08.011 [DOI] [PubMed] [Google Scholar]
- Komenda J., Sobotka R., Nixon P. J. (2012). Assembling and maintaining the Photosystem II complex in chloroplasts and cyanobacteria. Curr. Opin. Plant Biol. 15, 245–251. 10.1016/j.pbi.2012.01.017 [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685. 10.1038/227680a0 [DOI] [PubMed] [Google Scholar]
- Larkin M. A., Blackshields G., Brown N. P., Chenna R., Mcgettigan P. A., Mcwilliam H., et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948. 10.1093/bioinformatics/btm404 [DOI] [PubMed] [Google Scholar]
- Leto K. J., Young R. (1984). Improved sample buffer for LDS PAGE of thylakoid membrane proteins. MNL 58, 205–207. [Google Scholar]
- Li X. P., Björkman O., Shih C., Grossman A. R., Rosenquist M., Jansson S., et al. (2000). A pigment-binding protein essential for regulation of photosynthetic light harvesting. Nature 403, 391–395. 10.1038/35000131 [DOI] [PubMed] [Google Scholar]
- Lichtenthaler H. K. (1987). Chlorophylls and carotenoids: pigments of photosynthetic biomembranes, in Methods in Enzymology, eds Packer L., Douce R. (San Diego; London: Academic Press; ), 350–382. 10.1016/0076-6879(87)48036-1 [DOI] [Google Scholar]
- Lohscheider J. N., Rojas-Stütz M. C., Rothbart M., Andersson U., Funck D., Mendgen K., et al. (2015). Altered levels of LIL3 isoforms in Arabidopsis lead to disturbed pigment-protein assembly and chlorophyll synthesis, chlorotic phenotype and impaired photosynthetic performance. Plant Cell Environ. 38, 2115–2127. 10.1111/pce.12540 [DOI] [PubMed] [Google Scholar]
- Lu Y. (2016). Identification and roles of photosystem II Assembly, stability, and repair factors in Arabidopsis. Front. Plant Sci. 7:168. 10.3389/fpls.2016.00168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundin B., Nurmi M., Rojas-Stuetz M., Aro E. M., Adamska I., Spetea C. (2008). Towards understanding the functional difference between the two PsbO isoforms in Arabidopsis thaliana–insights from phenotypic analyses of psbo knockout mutants. Photosyn. Res. 98, 405–414. 10.1007/s11120-008-9325-y [DOI] [PubMed] [Google Scholar]
- Meurer J., Meierhoff K., Westhoff P. (1996). Isolation of high-chlorophyll-fluorescence mutants of Arabidopsis thaliana and their characterisation by spectroscopy, immunoblotting and northern hybridisation. Planta 198, 385–396. 10.1007/BF00620055 [DOI] [PubMed] [Google Scholar]
- Mokrosnop V. M. (2014). Functions of tocopherols in the cells of plants and other photosynthetic organisms. Ukr. Biochem. J. 86, 26–36. 10.15407/ubj86.05.026 [DOI] [PubMed] [Google Scholar]
- Montané M.-H., Kloppstech K. (2000). The family of light-harvesting-related proteins (LHCs, ELIPs, HLIPs): was the harvesting of light their primary function? Gene 258, 1–8. 10.1016/S0378-1119(00)00413-3 [DOI] [PubMed] [Google Scholar]
- Myouga F., Akiyama K., Tomonaga Y., Kato A., Sato Y., Kobayashi M., et al. (2013). The Chloroplast Function Database II: a comprehensive collection of homozygous mutants and their phenotypic/genotypic traits for nuclear-encoded chloroplast proteins. Plant Cell Physiol. 54, e2. 10.1093/pcp/pcs171 [DOI] [PubMed] [Google Scholar]
- Nelson N., Ben-Shem A. (2004). The complex architecture of oxygenic photosynthesis. Nat. Rev. Mol. Cell Biol. 5, 971–982. 10.1038/nrm1525 [DOI] [PubMed] [Google Scholar]
- Niyogi K. K., Truong T. B. (2013). Evolution of flexible non-photochemical quenching mechanisms that regulate light harvesting in oxygenic photosynthesis. Curr. Opin. Plant Biol. 16, 307–314. 10.1016/j.pbi.2013.03.011 [DOI] [PubMed] [Google Scholar]
- Nowicka B., Kruk J. (2016). Powered by light: phototrophy and photosynthesis in prokaryotes and its evolution. Microbiol. Res. 186–187, 99–118. 10.1016/j.micres.2016.04.001 [DOI] [PubMed] [Google Scholar]
- Obayashi T., Okamura Y., Ito S., Tadaka S., Aoki Y., Shirota M., et al. (2014). ATTED-II in 2014: evaluation of gene coexpression in agriculturally important plants. Plant Cell Physiol. 55, e6. 10.1093/pcp/pct178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilon M., Ravet K., Tapken W. (2011). The biogenesis and physiological function of chloroplast superoxide dismutases. Biochim. Biophys. Acta 1807, 989–998. 10.1016/j.bbabio.2010.11.002 [DOI] [PubMed] [Google Scholar]
- Pospíšil P. (2012). Molecular mechanisms of production and scavenging of reactive oxygen species by photosystem II. Biochim. Biophys. Acta 1817, 218–231. 10.1016/j.bbabio.2011.05.017 [DOI] [PubMed] [Google Scholar]
- Pribil M., Labs M., Leister D. (2014). Structure and dynamics of thylakoids in land plants. J. Exp. Bot. 65, 1955–1972. 10.1093/jxb/eru090 [DOI] [PubMed] [Google Scholar]
- Rizza A., Boccaccini A., Lopez-Vidriero I., Costantino P., Vittorioso P. (2011). Inactivation of the ELIP1 and ELIP2 genes affects Arabidopsis seed germination. New Phytol. 190, 896–905. 10.1111/j.1469-8137.2010.03637.x [DOI] [PubMed] [Google Scholar]
- Rochaix J. D. (2011). Assembly of the photosynthetic apparatus. Plant Physiol. 155, 1493–1500. 10.1104/pp.110.169839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas-Stütz M. (2008). Molecular Mechanisms of Light Stress Protection in Higher Plants. PhD. University of Konstanz.
- Rodríguez-Ezpeleta N., Brinkmann H., Burey S. C., Roure B., Burger G., Löffelhardt W., et al. (2005). Monophyly of primary photosynthetic eukaryotes: green plants, red algae, and glaucophytes. Curr. Biol. 15, 1325–1330. 10.1016/j.cub.2005.06.040 [DOI] [PubMed] [Google Scholar]
- Rossini S., Casazza A. P., Engelmann E. C., Havaux M., Jennings R. C., Soave C. (2006). Suppression of both ELIP1 and ELIP2 in Arabidopsis does not affect tolerance to photoinhibition and photooxidative stress. Plant Physiol. 141, 1264–1273. 10.1104/pp.106.083055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schägger H., von Jagow G. (1987). Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166, 368–379. 10.1016/0003-2697(87)90587-2 [DOI] [PubMed] [Google Scholar]
- Schneider C. A., Rasband W. S., Eliceiri K. W. (2012). NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675. 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shikanai T., Munekage Y., Shimizu K., Endo T., Hashimoto T. (1999). Identification and characterization of Arabidopsis mutants with reduced quenching of chlorophyll fluorescence. Plant Cell Physiol. 40, 1134–1142. 10.1093/oxfordjournals.pcp.a029498 [DOI] [PubMed] [Google Scholar]
- Sinha R. K., Komenda J., Knoppová J., Sedlárová M., Pospíšil P. (2012). Small CAB-like proteins prevent formation of singlet oxygen in the damaged photosystem II complex of the cyanobacterium Synechocystis sp. PCC 6803. Plant Cell Environ. 35, 806–818. 10.1111/j.1365-3040.2011.02454.x [DOI] [PubMed] [Google Scholar]
- Staleva H., Komenda J., Shukla M. K., Šlouf V., Kana R., Polívka T., et al. (2015). Mechanism of photoprotection in the cyanobacterial ancestor of plant antenna proteins. Nat. Chem. Biol. 11, 287–291. 10.1038/nchembio.1755 [DOI] [PubMed] [Google Scholar]
- Stawski K., Banach M., Goc A. (2014). Expression patterns of convergently overlapping Arabidopsis thaliana gene pairs Ohp-Ndp1 and Ohp2-Mes14. Acta Biol. Cracov. Ser. Bot. 56, 80–89. 10.2478/abcsb-2014-0006 [DOI] [Google Scholar]
- Takahashi K., Takabayashi A., Tanaka A., Tanaka R. (2014). Functional analysis of light-harvesting-like protein 3 (LIL3) and its light-harvesting chlorophyll-binding motif in Arabidopsis. J. Biol. Chem. 289, 987–999. 10.1074/jbc.M113.525428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka R., Rothbart M., Oka S., Takabayashi A., Takahashi K., Shibata M., et al. (2010). LIL3, a light-harvesting-like protein, plays an essential role in chlorophyll and tocopherol biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 107, 16721–16725. 10.1073/pnas.1004699107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thordal-Christensen H., Zhang Z. G., Wei Y. D., Collinge D. B. (1997). Subcellular localization of H2O2 in plants. H2O2 accumulation in papillae and hypersensitive response during the barley-powdery mildew interaction. Plant J. 11, 1187–1194. 10.1046/j.1365-313X.1997.11061187.x [DOI] [Google Scholar]
- Towbin H., Staehelin T., Gordon J. (1979). Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. U.S.A. 76, 4350–4354. 10.1073/pnas.76.9.4350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vranová E., Coman D., Gruissem W. (2013). Network analysis of the MVA and MEP pathways for isoprenoid synthesis. Annu. Rev. Plant Biol. 64, 665–700. 10.1146/annurev-arplant-050312-120116 [DOI] [PubMed] [Google Scholar]
- Wang P., Grimm B. (2015). Organization of chlorophyll biosynthesis and insertion of chlorophyll into the chlorophyll-binding proteins in chloroplasts. Photosyn. Res. 126, 189–202. 10.1007/s11120-015-0154-5 [DOI] [PubMed] [Google Scholar]
- Wang Q., Jantaro S., Lu B., Majeed W., Bailey M., He Q. (2008). The high light-inducible polypeptides stabilize trimeric photosystem I complex under high light conditions in Synechocystis PCC 6803. Plant Physiol. 147, 1239–1250. 10.1104/pp.108.121087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woitsch S., Römer S. (2003). Expression of xanthophyll biosynthetic genes during light-dependent chloroplast differentiation. Plant Physiol. 132, 1508–1517. 10.1104/pp.102.019364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao D. C., Brune D. C., Vavilin D., Vermaas W. F. (2012). Photosystem II component lifetimes in the cyanobacterium Synechocystis sp. strain PCC 6803: small Cab-like proteins stabilize biosynthesis intermediates and affect early steps in chlorophyll synthesis. J. Biol. Chem. 287, 682–692. 10.1074/jbc.M111.320994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimorski V., Ku C., Martin W. F., Gould S. B. (2014). Endosymbiotic theory for organelle origins. Curr. Opin. Microbiol. 22, 38–48. 10.1016/j.mib.2014.09.008 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


