Abstract
The rapid advancement of next generation sequencing protocols in recent years has led to the diversification in the methods used to study microbial communities; however, how comparable the data generated from these different methods are, remains unclear. In this study we compared the taxonomic composition and seasonal dynamics of the bacterial community determined by two distinct 16s amplicon sequencing protocols: sequencing of the V6 region of the 16s rRNA gene using 454 pyrosequencing vs the V4 region of the 16s rRNA gene using the Illumina Hiseq 2500 platform. Significant differences between relative abundances at all taxonomic levels were observed; however, their seasonal dynamics between phyla were largely consistent between methods. This study highlights that care must be taken when comparing datasets generated from different methods.
Keywords: Marine bacterial community, 16S, Pyrosequencing, Illumina
1. Introduction
In recent years, studies based on large next generation sequencing datasets have unveiled the extensive diversity and complex structure of microbial communities in the oceans, and the potential consequences on the functioning of the ecosystem such as impact on biogeochemical cycles (Lima-Mendez et al., 2015, Sunagawa et al., 2015). Such studies provide unprecedented insights into the key players and the biological processes in the open ocean.
Underpinning this progress is the rapid advancement in sequencing technologies, leading to the rapid shift in the methodology used to study the structure of microbial communities. Early studies are dominated by the use of the 454 pyrosequencing technology, but in recent years, the Illumina platform have been preferred over 454 pyrosequencing due to its lower cost, lower error rate, and higher throughput (Glenn, 2011). In addition, new primer sets have been developed to cover distinct regions of the 16s rRNA gene allowing better taxonomic assignments and better coverage of the taxonomic diversity (Apprill et al., 2015, Caporaso et al., 2011, Parada et al., 2016). Several 16s rRNA regions (V4, V6, V7, or V9 for instance) have been targeted in studies investigating the composition of bacterial communities. Recent studies have shown that the sequencing of the V4 region was the most reliable to describe the diversity and composition of the bacterial communities (Ghyselinck et al., 2013, Tremblay et al., 2015). Thus protocols have evolved rapidly, however, little is known about our ability to compare microbial communities analyses across distinct sequencing protocols.
A few studies have investigated the impact of samples processing (primers and sequencing technology) on the microbial community structure (Caporaso et al., 2012, Claesson et al., 2010, Nelson et al., 2014). However, these studies usually focus on the analyses of a single sample or very different samples, and have not investigated the impact of the methods on the observed temporal dynamics in microbial communities.
In this study we compared the bacterial community structure and dynamic over a year at one sampling location using two 16s rRNA sequencing protocols to evaluate the impact of the sequencing method on the relative abundances and seasonal dynamics.
2. Methods
2.1. Sampling
Water samples were collected from the surface at the L4 sampling site (50°15.00′N, 4°13.02′W) of the Western Channel Observatory (http://www.westernchannelobservatory.org.uk, accessed on March 31st 2016) between January 2008 and December 2008. The sampling was performed on 17 sampling dates during this period (Table 1). For each sampling occasion, 5 l of water were filtered through a 0.22 μm Sterivex cartridge (Millipore), which was then stored at − 80 °C until processing.
Table 1.
Sequencing method used for each sampling date (x indicates the samples used for the sequencing).
| Sampling date | V6–454 | V4–Illumina |
|---|---|---|
| 28/01/2008 | x | |
| 20/02/2008 | x | x |
| 05/03/2008 | x | |
| 17/03/2008 | x | x |
| 21/04/2008 | x | |
| 06/05/2008 | x | |
| 28/05/2008 | x | |
| 02/06/2008 | x | |
| 23/06/2008 | x | x |
| 21/07/2008 | x | x |
| 20/08/2008 | x | x |
| 22/09/2008 | x | x |
| 21/10/2008 | x | |
| 27/10/2008 | x | |
| 17/11/2008 | x | |
| 08/12/2008 | x | |
| 22/12/2008 | x |
2.2. DNA extraction and sequencing
DNA was extracted for each sample from the filters according to Neufeld et al. (2007) except that the filter was first removed from the Sterivex casing and transferred to a sterile 2 ml container. In order to compare two sequencing protocols, 12 samples were processed for sequencing of the V6 region of the 16s rRNA gene using 454 sequencing pyrosequencing (hereafter referred to as V6-454 samples) according to Gilbert et al. (2009) and Huber et al. (2007), and 11 samples (Table 1) were processed and barcoded according to Caporaso et al. (2011) (hereafter referred to as V4–Illumina samples). Within these samples, 6 were sequenced using both methods independently.
For the V6-454 samples, the V6 region was amplified using a pool of 5 forward primers (967F-PP 5′-gcctccctcgcgccatcagCNACGCGAAGAACCTTANC-3′; 967F-UC1 5′-gcctccctcgcgccatcagCAACGCGAAAAACCTTACC-3′; 967F-UC2 5′-gcctccctcgcgccatcagCAACGCGCAGAACCTTACC-3′; 967F-UC3 5′-gcctccctcgcgccatcagATACGCGARGAACCTTACC-3′; 967F-AQ 5′-gcctccctcgcgccatcagCTAACCGANGAACCTYACC-3′) and 4 reverse primers (1046R 5′-gccttgccagcccgctcagCGACAGCCATGCANCACCT-3′; 1046R-PP 5′-gccttgccagcccgctcagCGACAACCATGCANCACCT-3′; 1046R-AQ1 5′-gccttgccagcccgctcagCGACGGCCATGCANCACCT-3′; 1046R-AQ2 5′-gccttgccagcccgctcagCGACGACCATGCANCACCT-3′) according to Huber et al. (2007). The samples were sequenced using the 454 GS-flx platform and the LR70 kit.
For the V4–Illumina samples, the V4 region of the 16s rRNA gene was amplified using the forward primer 515F (5′-GTGCCAGCMGCCGCGGTAA-3′) and the reverse primer 806R (5′-GGACTACHVGGGTWTCTAAT-3′) according to Caporaso et al. (2011). Replica real-time PCRs were run alongside the samples destined for Illumina sequences to ensure that the Illumina PCR samples were removed during the log amplification phase of the PCR. The multiplex sequencing of the V4 region of the 16s rRNA gene was performed using Illumina HiSeq 2500 at the University of Exeter sequencing service facility.
For the V4–Illumina samples, the raw sequences have been deposited and are available at the European Nucleotide Archive (ENA) under the accession number PRJEB14618. For the V6–454 samples, the raw sequences are available at ENA under the accession number ERP000118.
2.3. Sequence processing and data analyses
The sequences obtained from the 454 sequencing were processed as described in (Gilbert et al., 2010, Gilbert et al., 2012). For the present study a subsampled OTU table (to 4101 sequences per samples) was used for further analyses.
For the V4-Illumina sequences, the quality of the HiSeq 2500 pair-end sequences was checked using Fastqc. Due to low quality of Read 2, only Read 1 was processed further. The primer and adaptor sequences were removed from the reads, and the sequences were trimmed to the same length (80 bases). The sequences were then processed using Qiime 1.8 (Caporaso et al., 2010), OTUs cluster with 97% identity, and the Silva reference database (release 119) was used to perform the taxonomic annotation. OTUs assigned to Archaea, mitochondria or chloroplast were removed for downstream analyses. To avoid bias due to differences in sequencing depth among samples, the OTU table was subsampled to the lowest number of sequences of a sample (79,302 sequences). To characterize the total diversity at the order level of the V4-Illumina dataset, the 200 most abundant OTUs (representing approximately 93% of the sequences) were extracted, the number of reads for each order were summed among samples to obtain the total yearly abundances and the diversity at order level was visualized using Krona (Ondov et al., 2011).
To compare the community composition and dynamics over the year 2008 of both datasets, the relative abundances of main bacterial groups (phylum) were calculated on the datasets containing all the bacterial OTUs. For each bacterial group a polynomial-fitting curve was applied to the data represented over time; this was done to highlight the seasonal dynamics of each group.
3. Results and discussion
The Illumina Hiseq method allowed for a greater sequencing depth than 454 pyrosequencing (increased by approximately 1830%), resulting in the number of OTUs identified to increase by 100% (Table 2). The increase sequencing depth has highlighted the presence of a hidden diversity in the 2008 samples sequenced using the 454 pyrosequencing method. In addition, the increased sequencing depth has allowed for the reduction in the percentage of singletons in the data set; a reduction of nearly half the singletons (Table 2). Increased sequencing depth allowed not only a reduction in the number of spurious OTUs (Pinto and Raskin, 2012) but it permits a better identification of true rare OTUs over sequencing errors.
Table 2.
Summary of the OTU dataset obtained with the V6–454 and V4–Illumina methods.
| Method | Level of sub-sampling (number of sequences) | Number of OTUs | % singletons (in the subsampled dataset) |
|---|---|---|---|
| V6–454a | 4101 | 1459 | 48.5% |
| V4–Illumina | 79,302 | 2919 | 25.6% |
Taken from (Gilbert et al., 2010, Gilbert et al., 2012).
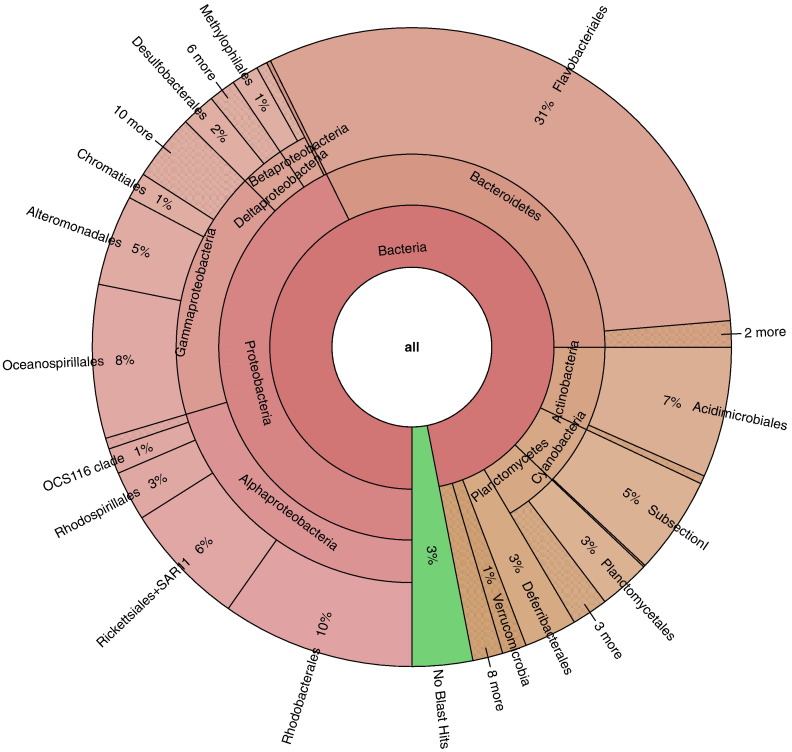
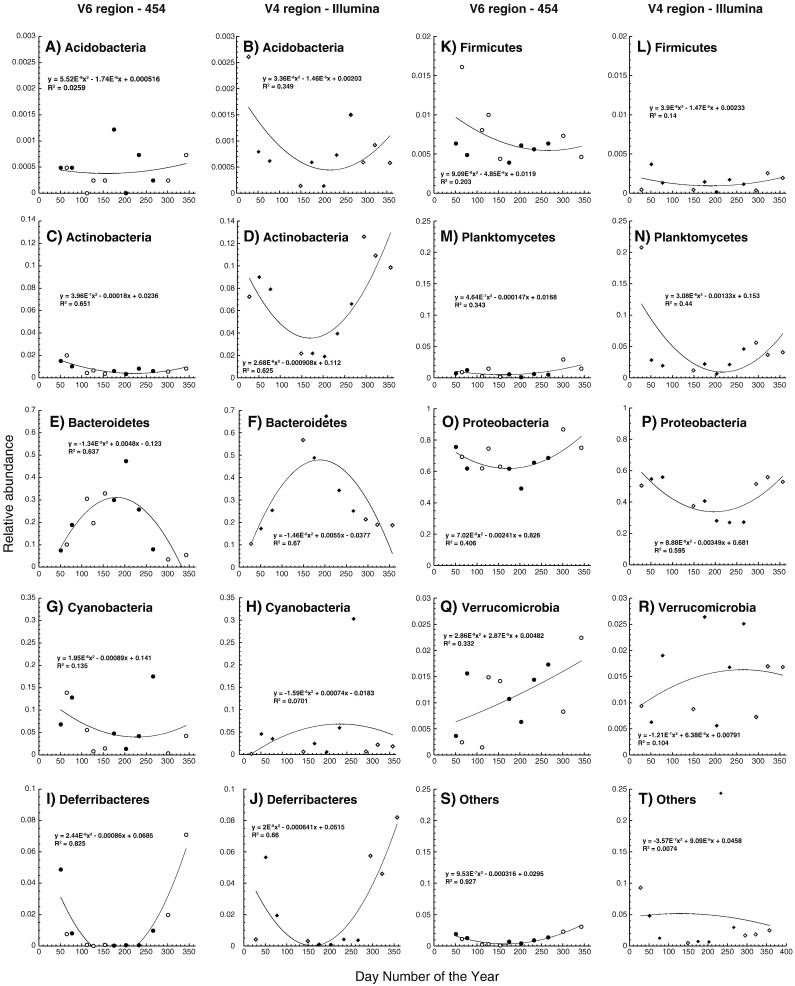
For the V4-Illumina method, the 200 most abundant OTUs were dominated by the class of the Bacteroidetes (with the order of Flavobacteriales), followed by the Alphaproteobacteria (with the orders of Rhodobacterales and Rickettsiales with SAR11) and the Gamma Proteobacteria (Fig. 1). These taxa were also the taxa that dominated the V6-454 bacterial community as reported by (Gilbert et al., 2012), although some differences in the relative abundances were observed in some of these taxa (Fig. 2). For both sequencing strategies, the relative abundances of Verrucomicrobia and Cyanobacteria were similar. However, the Proteobacteria and the Deferribacteres had lower relative abundances with the V4-Illumina methods than with the V6-454 methods (Fig. 1O, P, I, and J). In a previous study, Caporaso et al. (2012) showed differences in community composition of a single sample which have been sequenced by the 454 and the Illumina platforms. This results might partially explain the differences in relative abundances observed in the current study, but others factors can have important roles. For instance, the lower abundances of the Proteobacteria can be partly explained by the differences in primers selectivity. Hence, the primer combination used for the Illumina data set has been shown to underestimate the SAR11 clade belonging to the Alpha Proteobacteria (Apprill et al., 2015, Parada et al., 2016). The lower relative abundance of the Proteobacteria with the V4-Illumina method most likely explained the observed higher relative abundances of the other bacteria groups in comparison with the V6-454 method (Fig. 2).
Fig. 1.
Krona chart showing the relative abundance and diversity at the order level of the 200 most abundant OTUs for the V4–Illumina dataset.
Fig. 2.
Seasonal dynamics of the relative abundances of bacteria phyla determined using the V6-454 and V4-Illumina protocols (the circles and the diamonds represent the V6-454 and the V4-Ilummina protocols respectively, the full symbols shows the samples that were sequenced with both methods, the empty symbols represent the sampling dates that were only sequenced with one of the two protocols). A polynomial fitting curve was added to highlight seasonal changes in relative abundances.
For the seasonal dynamics, the abundant bacterial groups such as the Bacteroidetes and the Proteobacteria showed similar seasonal trends for both methods (Fig. 2E, F, O, and P). Also, the seasonal pattern of the Deferibacteres was very similar for both methods (Fig. 2I and J). For the Planktomycete, the seasonal dynamics were comparable for the two sequencing methods, particularly when excluding the first sampling date sample sequenced with the Illumina technology which was not used for the 454 pyrosequencing (Fig. 2M and N). For both the Acidobacteria and the Actinobacteria, the seasonal patterns were more pronounced with the Illumina method, showing clearly that both bacterial groups are more abundant in the winter months (Fig. 2A, B, C, and D). Larger sequencing depth of the V4-Illumina protocol has likely increased the resolution in the non-dominant taxa. For the Cyanobacteria and the Verrucomicrobia, the seasonal patterns identified with the V6-454 and the V4-Illumina protocols were less comparable (Fig. 2G, H, Q, and R).
4. Conclusions
In conclusion, the datasets obtained from different sequencing strategies need to be interpreted carefully, but the seasonal dynamics of several bacterial groups were similar and could be directly compared. However, extreme care must be taking for direct comparison of relative abundance of bacterial phyla.
Acknowledgments
This study was funded by FP7-OCEAN-2011 call, MicroB3 (grant number 287589) to D.S. and a post-doctoral research grant from the Swedish Research Council (Vetenskapsrådet; grant number 637-2014-6821) to K.L. We would also like to thank the University of Exeter sequencing service for their support and advice.
References
- Apprill A., McNally S., Parsons R., Weber L. Minor revision to V4 region SSU rRNA 806R gene primer greatly increases detection of SAR11 bacterioplankton. Aquat. Microb. Ecol. 2015;75:129–137. [Google Scholar]
- Caporaso J.G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F.D., Costello E.K. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso J.G., Lauber C.L., Walters W.A., Berg-Lyons D., Lozupone C.A., Turnbaugh P.J. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. PNAS. 2011;108:4516–4522. doi: 10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso J.G., Paszkiewicz K., Field D., Knight R., Gilbert J.A. The Western English Channel contains a persistent microbial seed bank. ISME J. 2012;6:1089–1093. doi: 10.1038/ismej.2011.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claesson M.J., Wang Q., O'Sullivan O., Greene-Diniz R., Cole J.R., Ross R.P. Comparison of two next-generation sequencing technologies for resolving highly complex microbiota composition using tandem variable 16S rRNA gene regions. Nucleic Acids Res. 2010;38 doi: 10.1093/nar/gkq873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghyselinck J., Pfeiffer S., Heylen K., Sessitsch A., De Vos P. The effect of primer choice and short read sequences on the outcome of 16S rRNA gene based diversity studies. Plos One. 2013;8 doi: 10.1371/journal.pone.0071360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert J.A., Field D., Swift P., Newbold L., Oliver A., Smyth T. The seasonal structure of microbial communities in the Western English Channel. Environ. Microbiol. 2009;11:3132–3139. doi: 10.1111/j.1462-2920.2009.02017.x. [DOI] [PubMed] [Google Scholar]
- Gilbert J.A., Field D., Swift P., Thomas S., Cummings D., Temperton B. The taxonomic and functional diversity of microbes at a temperate coastal site: a ‘multi-omic’ study of seasonal and diel temporal variation. PLoS One. 2010;5 doi: 10.1371/journal.pone.0015545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert J.A., Steele J.A., Caporaso J.G., Steinbrueck L., Reeder J., Temperton B. Defining seasonal marine microbial community dynamics. ISME J. 2012;6:298–308. doi: 10.1038/ismej.2011.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glenn T.C. Field guide to next-generation DNA sequencers. Mol. Ecol. Resour. 2011;11:759–769. doi: 10.1111/j.1755-0998.2011.03024.x. [DOI] [PubMed] [Google Scholar]
- Huber J.A., Mark Welch D.B., Morrison H.G., Huse S.M., Neal P.R., Butterfield D.A. Microbial population structures in the deep marine biosphere. Science. 2007;318:97–100. doi: 10.1126/science.1146689. [DOI] [PubMed] [Google Scholar]
- Lima-Mendez G., Faust K., Henry N., Decelle J., Colin S., Carcillo F. Determinants of community structure in the global plankton interactome. Science. 2015;348 doi: 10.1126/science.1262073. [DOI] [PubMed] [Google Scholar]
- Nelson M.C., Morrison H.G., Benjamino J., Grim S.L., Graf J. Analysis, optimization and verification of illumina-generated 16s rRNA gene amplicon surveys. PLoS One. 2014;9 doi: 10.1371/journal.pone.0094249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neufeld J.D., Schäfer H., Cox M.J., Boden R., McDonald I.R., Murrell J.C. Stable-isotope probing implicates Methylophaga spp and novel Gammaproteobacteria in marine methanol and methylamine metabolism. ISME J. 2007;1:480–491. doi: 10.1038/ismej.2007.65. [DOI] [PubMed] [Google Scholar]
- Ondov B.D., Bergman N.H., Phillippy A.M. Interactive metagenomic visualization in a Web browser. BMC Bioinforma. 2011;12 doi: 10.1186/1471-2105-12-385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parada A., Needham D.M., Fuhrman J.A. Every base matters: assessing small subunit rRNA primers for marine microbiomes with mock communities, time-series and global field samples. Environ. Microbiol. 2016;18:1403–1414. doi: 10.1111/1462-2920.13023. [DOI] [PubMed] [Google Scholar]
- Pinto A.J., Raskin L. PCR biases distort bacterial and archaeal community structure in pyrosequencing datasets. PLoS One. 2012;7 doi: 10.1371/journal.pone.0043093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunagawa S., Coelho L.P., Chaffron S., Kultima J.R., Labadie K., Salazar G. Structure and function of the global ocean microbiome. Science. 2015;348 doi: 10.1126/science.1261359. [DOI] [PubMed] [Google Scholar]
- Tremblay J., Singh K., Fern A., Kirton E.S., He S., Woyke T. Primer and platform effects on 16S rRNA tag sequencing. Front. Microbiol. 2015;6 doi: 10.3389/fmicb.2015.00771. [DOI] [PMC free article] [PubMed] [Google Scholar]