Abstract
Intraspecific variation is a major component of biodiversity, yet it has received relatively little attention from governmental and nongovernmental organizations, especially with regard to conservation plans and the management of wild species. This omission is ill‐advised because phenotypic and genetic variations within and among populations can have dramatic effects on ecological and evolutionary processes, including responses to environmental change, the maintenance of species diversity, and ecological stability and resilience. At the same time, environmental changes associated with many human activities, such as land use and climate change, have dramatic and often negative impacts on intraspecific variation. We argue for the need for local, regional, and global programs to monitor intraspecific genetic variation. We suggest that such monitoring should include two main strategies: (i) intensive monitoring of multiple types of genetic variation in selected species and (ii) broad‐brush modeling for representative species for predicting changes in variation as a function of changes in population size and range extent. Overall, we call for collaborative efforts to initiate the urgently needed monitoring of intraspecific variation.
Keywords: ecosystem function and services, functional variation, genetic variation, neutral variation, non‐neutral variation
1. Introduction
The future of the Earth system will be heavily influenced by human‐induced environmental changes that have detrimental effects on biodiversity. The consequent loss of diversity impacts not only local ecosystems and the services they provide (Cardinale et al., 2012; Díaz, Fargione, Chapin, & Tilman, 2006), but also biodiversity on regional and global scales (Faith et al., 2010). Under the Convention on Biological Diversity (CBD), biodiversity is considered to encompass variation at all levels, such as within and among ecosystems, communities, species, and populations. However, to date, most discussions and efforts that considered the impacts of human‐induced environmental change focused on ecosystems and communities. Although some programs, such as the US Endangered Species Act (ESA) and Canadian Species at Risk Act (SARA), have long cited the importance of intraspecific variation for protecting endangered species, intraspecific variation is typically overlooked (Hoban et al., 2013; Laikre, 2010), especially for nonendangered but possibly ecologically important species. Here, we argue that ignoring intraspecific variation in management decisions can lead to irreversible consequences for biodiversity and the services and benefits it provides. In particular, we will argue that human activities are dramatically changing the structure of neutral and functional variations in natural populations that are critical for species persistence, community structure, and ecosystem services and hence the integration of strategies to monitor this variation is urgently required.
In this review, “intraspecific variation” refers to all forms of variation within a species, both within and among populations, including variations in phenotypes and genomes. “Genetic variation” refers to all forms of genetic variation within a species, including neutral and functional sequence variations and variation in gene expression. The term “molecular genetic diversity’” is used when the variation is measured by molecular tools (e.g., microsatellites or single‐nucleotide polymorphisms). Some of this variation influences morphological, physiological, and other types of functional genetic variation that affect the performance of individuals and populations. Therefore, this “functional (non‐neutral) genetic variation” has important consequences for population dynamics, species interactions, and ecosystem function. Importantly, this functional variation might or might not be “adaptive,” that is, improving fitness (survival and reproduction). By contrast, “neutral (nonfunctional) genetic variation” has no such direct consequences and is more commonly considered as a proxy indicator for important population parameters, such as effective population size, gene flow, genetic integrity, or evolutionary potential (e.g., Parker, Snow, Schug, Booton, & Fuerst, 1998; Sunnucks, 2000).
Our goal was to explain the need for, and outline a strategy for, incorporating intraspecific variation into monitoring programs. We first illustrate how this variation promotes and maintains relevant levels of biodiversity, community integrity, and ecosystem function. We then outline how human‐induced environmental changes impact intraspecific variation, imposing environmental, economic, and social costs. Finally, we outline some potential monitoring strategies to observe and predict regional and global changes in intraspecific variation. Work along these lines would greatly increase our ability to predict, prevent, and mitigate detrimental ecosystem changes.
2. Intraspecific Variation Is Critical for Population Dynamics, Community Structure, and Ecosystem Function
Intraspecific variation is both the product of, and the foundation of, evolutionary and ecological processes (summarized in Table 1). Therefore, understanding the origins, architecture, and maintenance of genetic variation is critical for predicting the short‐ and long‐term responses of populations, communities, and ecosystems to novel and changing environments (Hendry et al., 2011; Lankau, Jørgensen, Harris, & Sih, 2011).
Table 1.
Roles of intraspecific variation in ecological and evolutionary processes with representative open access articles
| Levels | Processes | Summary | Examples of open access articles |
|---|---|---|---|
| Population | Portfolio effects | Genetic variation (and biodiversity) reduces risks and buffers negative impacts of changing environments. Individuals with various genotypes may produce a wide range of responses to the environment, thus contributing to population stability | Schindler et al. (2015) reviewed existing papers to illustrate the importance of diversity, both inter‐ and intraspecific variations for population persistence and evolutionary potentials |
| Connectivity, effective population size, and mating success | Genetic variation increases effective population size and reduces risks of inbreeding depression, thus ensuring offspring survival | Hoffman et al. (2014) suggested that higher neutral genetic variation reduces the impact of inbreeding depression and the negative impact on population health | |
| Adaptability/evolvability | Genetic variation provides genotypes for new selections in a changing environment and contributes to populations fitting into the new environment | Merilä and Hendry (2014) reviewed evolutionary responses to climate changes. Additional examples of environmental changes are listed in the text | |
| Community and ecosystems | Species diversityAbundancePrimary productivityPlant–soil interaction | Increasing genetic and phenotypic variations within species typically increases its primary productivity, species diversity, and abundance of mutualistic and antagonistic species (e.g., herbivores), and influences in plant–soil interactions | Crutsinger (2016) reviewed a number of examples illustrating how genetic variation influences the diversity and abundance of surrounding species, productivity, and plant–soil interactions |
| Stability of ecosystem processes | Due to the above effects, genetic variation contributes to the stability of ecological processes and functions | Genung et al. (2010) found that the genetic variation of flowering species increases the floral abundance and number of visiting pollinators, thus ensuring the reproduction of the species and a sustainable food supply for pollinators |
2.1. Effects on population dynamics
Environmental change will often harm populations that are poorly suited to the new conditions, which can lead to population declines, extirpation, and extinction (e.g., Green, Cornell, Scharlemann, & Balmford, 2005; Pörtner & Knust, 2007; Thomas et al., 2004). These negative effects can be offset, in part, when diversity within and among populations can help to buffer these problems through the so‐called portfolio effect (Leimu, Vergeer, Angeloni, & Ouborg, 2010; Moore, Yeakel, Peard, Lough, & Beere, 2014; Schindler, Armstrong, & Reed, 2015; Schindler et al., 2010). This effect predicts that higher biodiversity minimizes the overall risk in stability of ecosystem functions. In addition, populations can respond to detrimental environmental changes through migration to more optimal locations, adaptive plasticity, or evolutionary (genetic) adaptation. However, the first two options are often constrained to the point that evolutionary adaptation becomes a critical component of a species’ persistence in the face of environmental change (Phillimore, Had, Jones, & Smithers, 2012; Visser, 2008). Therefore, careful attention needs to be paid to how organisms evolve in response to environmental change.
Evidence for evolutionary adaptation to environmental change is widespread in a spatial context (i.e., populations in different environments show local adaptation to those environments), which reflects the action of past selection in shaping biodiversity (Schluter, 2000). However, these spatial patterns typically arise over long timescales, raising the question as to whether or not a similar adaptation can occur over much shorter timescales that typify human‐induced environmental change (Merilä & Hendry, 2014). The short answer appears to be “yes,” at least in some cases, in that a large number of studies have demonstrated adaptive evolution over time frames ranging from years to decades (reviewed in Reznick & Ghalambor, 2001; Hendry, Farrugia, & Kinnison, 2008). Such contemporary evolutionary responses have been observed in response to hunting/harvesting (Coltman et al., 2003; Pigeon, Festa‐Bianchet, Coltman, & Pelletier, 2016), pollution (Antonovics, Bradshaw, & Turner, 1971; Levinton et al., 2003), introduced species (Strauss, Lau, & Carroll, 2006), novel and changing climates (Bradshaw & Holzapfel, 2001; Colautti & Barrett, 2013; Merilä & Hendry, 2014), and novel environments (Prentis, Wilson, Dormontt, Richardson, & Lowe, 2008). At the same time, however, many other populations that have faced environmental change clearly did not evolve rapidly enough, as evidenced by frequent extirpations and extinctions (Barnosky et al., 2011; Hughes, Daily, & Ehrlich, 1997). Thus, the critical question becomes: What factors determine the potential for evolution to avert the extirpation of populations and the extinction of species threatened by human‐induced environmental change?
A critical determinant of the potential for adaptive evolution is the amount of genetic variation in fitness and, thus, in fitness‐related traits: a potential often assessed as additive genetic variance (V A) or heritability (h 2 = V A/V P), where V P is phenotypic variance (Hoffmann & Merilä, 1999; Visscher, Hill, & Wray, 2008) as the concepts were initially introduced by Fisher (1930) and Wright (1920). In theory, the adaptive evolutionary rate is directly proportional to V A (Fisher, 1930). However, given that V A is not easily measured in natural populations (Kruuk, 2004), many studies have instead used molecular genetic diversity as a proxy for the overall functional and neutral genetic variations. Although molecular genetic diversity is not always correlated with functional variation in natural populations (Reed & Frankham, 2001), recent experimental studies have shown that molecular genetic diversity and genotypic diversity, the variation in genotypes among individuals, can predict population responses to environmental change (see Figure 1a; Vázquez‐Domínguez, Piñero, & Ceballos, 1999; Reusch, Ehlers, Hämmerli, & Worm, 2005; Hoffman et al., 2014). Moreover, recent innovations in molecular biology enable the direct assessment of functional genetic variants responsible for adaptation (discussed later in “What types of variation should be monitored?”). This ability augurs a new era for biodiversity monitoring with a more direct measurement of functional genetic variation, which is obviously most relevant to predicting evolutionary responses in many species facing the environmental changes.
Figure 1.
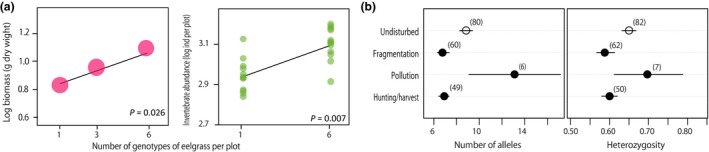
Roles of genetic diversity in ecosystems but dramatically influenced by human‐induced environmental changes. (a) Genetic diversity of eelgrass (Zostera marina) affected by ecosystem functioning and resilience (replotted from Reusch et al., 2005, Copyright (2005) National Academy of Sciences, USA). Experimental plots were designed with one, three, and six eelgrass genotypes and the mean biomass of eelgrass (left panel) and the number of invertebrates within each plot at the end of a 4‐month experiment were measured (right panel). (b) Shown are the results of a meta‐analysis by DiBattista et al. (2008) comparing variation in microsatellite markers between undisturbed populations and populations subject to various types of human disturbance (redraw from the original). Fragmentation and hunting/harvesting tend to decrease genetic diversity (left: number of alleles, right: heterozygosity), whereas pollution has less predictable effects. The number of studies is indicated in parenthesis
2.2. Effects on communities and ecosystems
The theoretical and empirical studies have indicated that ecological system functionality and species interaction, which provides fundamental services for humanity, are affected by biodiversity (Hooper et al., 2005; May, 1973). Most studies addressing this topic focus on interspecific diversity (e.g., the number of species or functional groups); however, intraspecific variation can also be substantial and important in communities and ecosystems (Siefert et al., 2015). For example, increasing genetic and phenotypic variations within a species typically increases species diversity and abundance and primary productivity, promotes positive plant–soil interactions (reviewed in Crutsinger, 2016), and stabilizes ecosystem functions (Genung et al., 2010; Prieto et al., 2015). Although species and individual interactions due to variance in individuals are less predictable, these effects appear to be strongest when the species in question plays an important role in the ecosystem (Hendry, 2016; Hughes, Inouye, Johnson, Underwood, & Vellend, 2008); that is, it is a “keystone species,” “foundation species,” “niche constructor,” “strong interactor,” and so on.
Two primary mechanisms can explain the positive relationship between diversity (both among and within species) and various community/ecosystem processes (Loreau & Hector, 2001). First, diverse communities use a wider range of resources due to resource partitioning or positive species interactions (i.e., “complementarity effects”), which can increase productivity and nutrient cycling. For example, genetic variation in foundation tree species helps to maintain the diversity of associated plants, animals, and fungi because different tree genotypes are advantageous to different species interactions (Barbour et al., 2009; Zytynska, Fay, Penney, & Preziosi, 2011). Second, communities with greater diversity have a better chance of including a few key species that have large effects on ecosystem processes (i.e., “selection effects”). At the same time, it is important to recognize that greater diversity is not always “better,” such as when it compromises local adaptations (Hansen, Carter, & Pélabon, 2006) or allows one species to dominate and have detrimental effects on other species. The many ways that intraspecific variation influences community/ecosystem function and stability are context‐specific and are still being discovered (Hendry, 2016). Overall, then, diversity, both within and among species, is best thought of as providing not just “ecosystem services” but, more generally, “evosystem services” (Faith et al., 2010).
3. Human Activities Dramatically Influence Crucial Aspects of Intraspecific Variation
The many potential influences of intraspecific variation, as described above, motivate a need to consider how this variation is influenced by human activities, such as habitat loss and degradation, harvesting and hunting, pollution, species introductions, climate change, and so on. One major consequence is decline in populations and extirpations, which are estimated at three to eight times higher than baseline (Hughes et al., 1997). Another major consequence is that humans can directly (e.g., by hunting and harvesting) or indirectly (e.g., climate change and pollution) impose novel selective pressures that lead to evolutionary responses with potential negative effects on future stability, productivity, and persistence (Coltman et al., 2003; Law & Salick, 2005; Pespeni et al., 2013; Stockwell, Hendry, & Kinnison, 2003; Swain, Sinclair, & Mark Hanson, 2007). A third consequence is that human activities can increase the gene flow among populations (the opposite of reduced connectivity) and hybridization among species, which can cause biotic homogenization (McKinney & Lockwood, 1999), outbreeding depression (Dudash & Fenster, 2001), and speciation reversal (De León et al., 2011; Seehausen, Takimoto, Roy, & Jokela, 2008; Vonlanthen et al., 2012). These various effects indicate ways in which management decisions can have a direct influence on genetic variation and evolutionary potential (Santamaría & Mendez, 2012). Below, we illustrate some significant human‐induced environmental changes that affect genetic variation.
3.1. Habitat modification
Land use by humans (e.g., agriculture and settlements) is one of the most important drivers of global biodiversity loss (Ellis, Antill, & Kreft, 2012), partly through the above effects; decreased population size, novel selection, and increased gene flow. First, land use often leads to habitat loss for many species, which can reduce the size and connectivity of the affected populations. Typical outcomes include increased inbreeding and reduced intraspecific variation (Figure 1b; Aguilar, Quesada, Ashworth, Herrerias‐Diego, & Lobo, 2008; Allendorf, Luikart, & Aitken, 2012; DiBattista, 2008; Frankham, Ballou, & Briscoe, 2002), which can impact survival and reproduction by reducing portfolio effects (Bello‐Bedoy & Núñez‐Farfán, 2011; Hoffman et al., 2014; Keller & Waller, 2002; Núñez‐Farfán, Fornoni, & Valverde, 2007), increasing the mutation load (Agrawal & Whitlock, 2012), and hampering evolutionary responses to environmental change (Bijlsma & Loeschcke, 2012). Second, environments altered by land use often impose novel selective pressures, leading to occasionally large phenotypic and genetic responses that alter intraspecific variation. Examples include adaptation to industrial pollution by plants (Antonovics et al., 1971; Bratteler, Lexer, & Widmer, 2006), terrestrial insects (Clarke & Sheppard, 1966; Cook & Saccheri, 2012), and various aquatic organisms (Levinton et al., 2003). Third, habitat loss can lead to altered species interactions, which can increase hybridization. For example, habitat conversion has been followed by an increased chance of hybridization among gray wolves, Canis lupus, and coyotes, C. latrans (Koblmüller, Nord, Wayne, & Leonard, 2009; Lehman et al., 1991).
The above examples follow naturally from the expectation that land use has a strong negative effect on species. However, land‐use changes can have seemingly positive or at least unanticipated effects on some species that nevertheless negatively impact diversity. For example, as illustrated in Figure 2, increased provisioning of human foods for Darwin's finches, Geospiza fortis, reduced the disruptive selection that had historically maintained distinct intraspecific beak size morphs (De León et al., 2011; Hendry et al., 2006). As another example, eutrophication caused by intensive agriculture (Tilman et al., 2001) could increase productivity but it also promotes hybridization, causing the collapse of “species flocks” into “hybrid swarms” (Seehausen, 1997; Vonlanthen et al., 2012). The consequences of land use are, therefore, complex and require careful monitoring of both natural selection (abiotic and biotic environmental changes) and the structure of intraspecific variation.
Figure 2.
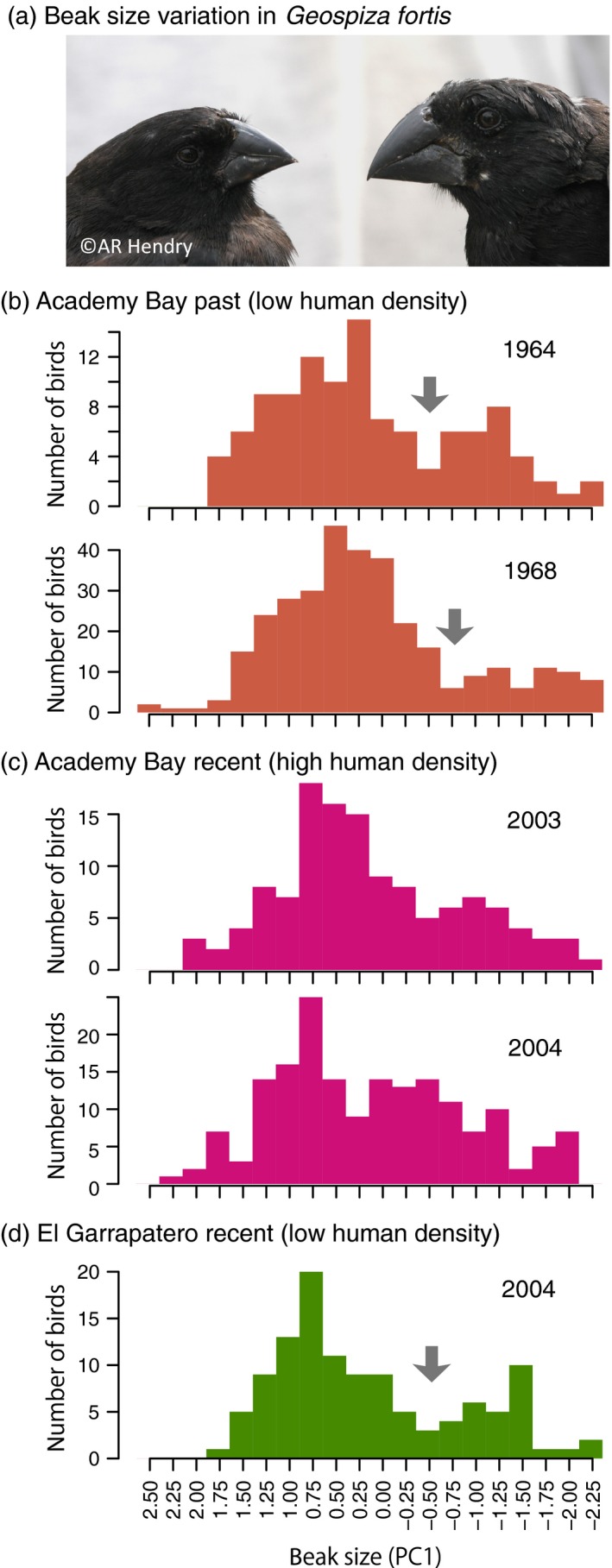
Human‐induced environmental changes affect selection. An example of human activities altering existing selection in (a) the ground finches, Geospiza fortis. (b) The degrees of bimodality in beak sizes within the medium ground finch were stronger in the absence of human influences in 1964 and 1968, (c) than in the presence of human influences in 2003 and 2004 at Academy Bay. (d) The strong bimodality persisted in 2004 at El Garrapatero when the human densities were still low. Gray arrows show discontinuities in beak size variation in the populations statistically confirmed to have the strong bimodality. Beak size variation was calculated as the first principal component (PC1) of the multiple size measurements. These data were replotted from Hendry et al. (2006)
Further complexity comes from the many ways in which connectivity is invoked, considered, and managed. First, land‐use (and other anthropogenic) changes can increase connectivity in some cases and decrease it in others, often dramatically either way. Second, the resulting increasing or decreasing gene flow (and hybridization) can have both positive and negative influences on populations and species (reviewed in Garant, Forde, & Hendry, 2007). Gene flow can increase an effective population size, reduce inbreeding, aid adaptive responses to environmental change, and spread adaptive variants among populations, while it reduces genetic differences among populations which can limit portfolio effects and hamper local adaptation by introducing maladapted genes from other environments (García‐Ramos & Kirkpatrick, 1997; Lenormand, 2002; Slatkin, 1987). Given that changes in connectivity and a release of translocated or captive raised individuals are one of the more easily implemented management actions (e.g., “genetic restoration” and “assisted migration”), these various effects are often debated both theoretically and practically (Aitken & Whitlock, 2013; Laikre, Schwartz, Waples, & Ryman, 2010; Santamaría & Mendez, 2012).
3.2. Climate change
Human‐induced climate change is altering patterns of temperature, precipitation, erosion, and ocean acidification (IPCC 2013). As with other human influences, these consequences of climate change can result in decreased population size, novel selection, and increased gene flow, which can then alter interspecific variation. Changes in population size are a common outcome of climate change, including declines in many species (Both, Bouwhuis, Lessells, & Visser, 2006), but also in increases for other species (Massimino, Johnston, & Pearce‐Higgins, 2015; Rochlin, Ninivaggi, Hutchinson, & Farajollahi, 2013). These changes in abundance can then have various effects on intraspecific variation, as already discussed above. Changes in selection are also common, most obviously in relation to the timing of key life history events (Merilä & Hendry, 2014). For instance, evolutionary responses to climate‐induced selection have been documented for the flowering time in mustards, Boechera stricta (Anderson, Inouye, Mckinney, Colautti, & Mitchell‐olds, 2012), life history timing in pitcher plant mosquitoes, Wyeomyia smithii (Bradshaw & Holzapfel, 2001), and reproductive timing in red squirrels, Tamiasciurus hudsonicus (Réale, McAdam, Boutin, & Berteaux, 2003). Finally, connectivity is often influenced by climate change through the nearly ubiquitous and sometimes dramatic changes in species’ distributions (Bálint et al., 2011; Parmesan, 2006; Pauls, Nowak, Bálint, & Pfenninger, 2013), including range contractions (Habel, Rödder, Schmitt, & Nève, 2011) and range expansions (Hewitt, 1996). These shifting distributions lead to many alterations in species interactions that can also instigate hybridization (Garroway et al., 2010; Mimura, Mishima, Lascoux, & Yahara, 2014). Thus, as for habitat loss, altered (and actively altering) connectivity is frequently discussed with regard to potential management actions in response to climate change (Aitken & Whitlock, 2013; McLachlan, Hellmann, & Schwartz, 2007).
3.3. Harvesting and domestication
Harvesting and domestication can reduce the size of wild populations and thereby alter genetic variation in many of the ways described above (e.g., Allendorf, England, Luikart, Ritchie, & Ryman, 2008; Harris, Wall, Allendorf, Harris, & Wall, 2002). However, many harvested and domesticated species are abundant enough that the problems associated with a small population size are often negligible. Instead, the most obvious effect of harvesting and domestication is typically altered selection. For instance, hunting and fishing practices can inadvertently result in selection for a smaller body size and earlier maturation (Coltman et al., 2003; Law & Salick, 2005; Swain et al., 2007), which can negatively influence survival, resilience, and recovery (Fenberg & Roy, 2008). Like harvesting, domestication (e.g., crop maturity and fish hatcheries) can alter selection and lead to evolutionary changes that alter productivity (Denison, 2012). Domestication can also lead to sustained genetic bottlenecks that dramatically decrease genetic variation (Doebley, Gaut, & Smith, 2006), which can then negatively impact the remaining wild populations through competition or gene flow. For instance, genetic change in captive‐reared fish populations that are then released in the wild can reduce the reproductive potential of natural populations (Araki, Berejikian, Ford, & Blouin, 2008). Importantly, the above effects can persist long after the human activity ceases. For example, harvested silverside fish (Menidia menidia) populations can have evolutionary reductions in growth rate and body size that persist for decades after harvesting is halted (Conover, Munch, & Arnott, 2009).
3.4. Species introductions
Species introductions into new geographic areas sometimes lead to species “invasions” that can reduce the abundance of native species through competition, predation, hybridization, and infection (Pyšek & Richardson, 2010). Again, the above‐described effects of declines in populations can happen to native species. In addition, phenotypic and genetic changes have been observed in many introduced species and the native species with which they interact (Hendry et al., 2008; Mooney & Cleland, 2001). Although the extent to which these changes are adaptive is not always certain (Colautti & Lau, 2015), evolution in introduced species is predicted to influence the rate, extent, and impact of invasions (García‐Ramos & Rodríguez, 2002; Vázquez‐Domínguez, Suárez‐Atilano, Booth, González‐Baca, & Cuarón, 2012). Thus, novel selective pressures can lead to evolutionary changes in both native and invasive species that then influence the abundance of those species, with expected further consequences for intraspecific variation. Although these effects are typically assumed to be negative for native species, the opposite can sometimes also occur, such as when a native species benefits from the introduction of new food resources (Carroll et al., 2005). The result can be a decrease in intraspecific variation owing to increased gene flow (as in the aforementioned Darwin's finch example, Figure 2), or an increase in intraspecific variation owing to the formation of new insect host races (Drès & Mallet, 2002).
4. Toward a Monitoring System for Intraspecific Variation
We have highlighted two basic points: (i) Intraspecific variation has important consequences for population dynamics, community structure, and ecosystem function; and (ii) intraspecific variation is strongly influenced by human‐induced environmental change. From the intersection of these two points comes the need for a monitoring program that can track and assess ongoing changes in intraspecific variation and thus provide important baseline data for assessing the consequences for populations, communities, ecosystems, and human well‐being. Our goal in the rest of this study was to highlight some elements that an appropriate monitoring program might include. We suggest two main strategies: (i) explicit empirical monitoring of variation for selected species and (ii) modeling variation for a larger set of species (illustrated in Figure 3). The first strategy requires measurements of intraspecific variation, ideally including functional variation, in combination with environmental parameters. As the intensive effort required might not be feasible for many species, the second strategy, which is less data‐intensive, uses models to predict how variation will be lost based on environmental changes.
Figure 3.
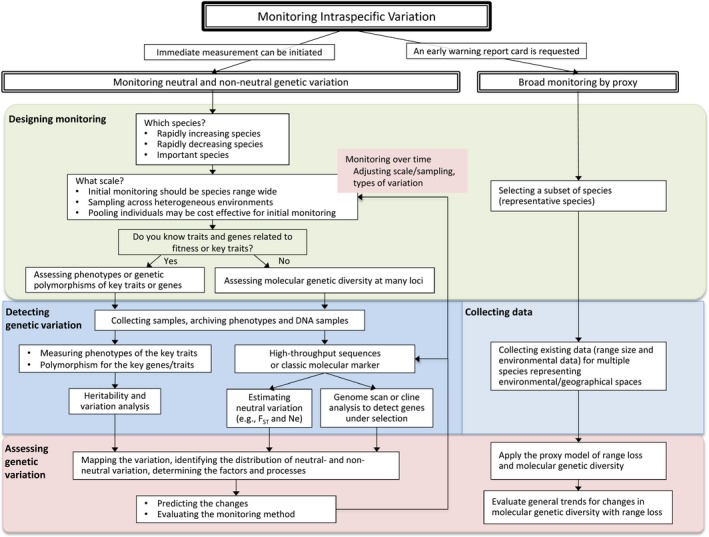
A suggested flowchart for monitoring intraspecific variation
4.1. Monitoring variation for specific target species
4.1.1. Which species should be monitored?
As it is obviously impossible to monitor all species, we need to select specific species for monitoring, presumably either those of a direct conservation or management interest or those that can act as “indicator species.” For instance, suitable indicator species have been suggested to be those that respond rapidly to environmental change over short timescales and that have strong effects on ecosystem function (Pereira & David Cooper, 2006). Along these lines, the Group on Earth Observations Biodiversity Observation Network (GEO BON 2010) defined three main categories of species that might be appropriate to monitor, which we summarize as (i) rapidly declining species, (ii) rapidly increasing species, and (iii) important species. Here, we focus on these categories and describe some successful examples of their monitoring during human‐induced environmental change. In cases where causal inferences are desired, it could also be valuable to track genetic changes in “control” species that are not rapidly increasing, rapidly decreasing, or “important.”
Rapidly declining species are typically listed as critically endangered on the IUCN Red List or by the Evolutionarily Distinct Globally Endangered Program (EDGE, http://www.edgeofexistence.org). The loss of variation in such species could reflect—and contribute to—a decline leading to extinction. Such species are typically of intense interest and are therefore frequently monitored for various aspects of intraspecific variation. As one example, a comparative genomic study across avian species discovered that both the loss of genetic variation and the accumulation of deleterious mutations of protein‐coding genes contributed to major genetic defects in endangered species (Li et al., 2014). They also implied that some sets of genes could contribute to averting extinction or enhancing recovery in endangered crested ibis (Nipponia nippon) populations. All endangered and rapidly declining species would benefit from similar assessments and monitoring.
Typical examples of rapidly increasing species include many invasive alien species, novel pests, and emerging infectious diseases (EIDs). These species are important to monitor, not only because of their negative impacts on biodiversity and human well‐being, but also because their rapid increases often have clear genomic signatures. For example, introduced yellow monkey flower (Mimulus guttatus) populations show a reduced neutral genetic variation but also signatures of positive selection at genomic regions linked to flowering time and stress responses (Puzey & Vallejo‐Marín, 2014). Similarly, genetic elements that enhance invasiveness have been tracked by comparing molecular genetic diversity between introduced and native populations of an ant species, Cardiocondyla obscurior (Schrader et al., 2014), and a wetland grass species, Phalaris arundinacea (Lavergne & Molofsky, 2007). Importantly, the genomic signatures that point toward invasions or EIDs can sometimes be detected before the expansion begins in earnest, and so, monitoring of introduced species can provide an early‐warning system for upcoming challenges. Given that rapidly increasing species can be widespread, international collaborations will often be required for such monitoring. A good example of this is the “Global Garlic Mustard Field Survey” (http://www.GarlicMustard.org/) that collects field data and seeds of this invasive species.
Important species could be commercially important (fisheries, forestry, or agriculture), ecologically important (keystone, foundation, or ecosystem engineer), or culturally important (including “flagship” species that attract considerable public attention). These species are also considered in CBD Aichi Biodiversity Targets (i.e., Target 13). Monitoring such species is critical not only due to their importance but also because that importance can have consequences for intraspecific variation. For instance, species under cultivation can suffer from bottlenecks, strong selection, and the propagation of specific strains that reduce intraspecific variation and thereby have negative consequences for safeguarding food security and for “option values” (Jump, Marchant, & Peñuelas, 2009). An existing application of this thinking is the ongoing effort by agronomists to collect and evaluate landraces and wild relatives of crop species (Hyten et al., 2006; Plucknett, Smith, Williams, & Anishetty, 1987), and to find a source of new genetic markers for future improvements. Another example is the monitoring of changes in life history traits in the commercially harvested fish species, Atlantic cod, Gadus morhua (Olsen et al., 2005), which can signal disastrous collapses of fisheries and thereby perhaps motivate mitigating management actions.
4.1.2. What types of variation should be monitored?
Neutral genetic variation
Human‐induced factors that are likely to affect genetic variation include decreased population size, novel selection, and increased gene flow (as previously described). Neutral molecular markers can be used to detect changes in population size and gene flow (Table 2) through the estimation of key parameters of genetic variation, such as the number of alleles (A), heterozygosity (He), effective population size (N e), inbreeding (F), and population divergence (F ST). As theoretical and empirical studies suggest (Leberg, 2002; Nei, Maruyama, & Chakraborty, 1975; Spencer, Neigel, & Leberg, 2000), the number of alleles (A) can be a more sensitive indicator of population decline than heterozygosity (He) in a variety of scenarios (Hoban et al., 2014). Effective population size (N e) is also an important indicator for monitoring changes in population size, and several statistical methods to estimate N e have been developed and evaluated for their efficacy in early detection of population declines (e.g., Antao, Pérez‐Figueroa, & Luikart, 2011). Although translating the population divergence parameter (F ST) as an indirect measurement of gene flow is unrealistic (Whitlock & Mccauley, 1999), several statistical approaches using various molecular markers allow a relatively accurate estimation of migration rate, many of which use coalescence‐based Bayesian approaches (e.g., Beaumont, 2010; Beerli & Palczewski, 2010; Hey, 2010; Wilson & Rannala, 2003). These measures are considered to be relatively affordable and rapid surrogate measures of key population genetic parameters. Such markers certainly can be a key component of any monitoring program, and appropriate methods have been frequently reviewed elsewhere (e.g., Luikart, Sherwin, Steele, & Allendorf, 1998; Schwartz, Luikart, & Waples, 2007). Given this existing literature, here we focus more directly on monitoring functional (non‐neutral) variation, which may have significant impacts on population persistence and therefore management options for species exposed to novel selective pressures.
Table 2.
Examples of techniques currently most adequate for monitoring genetic variation and the impacts of human‐induced environmental changes. The cost for these techniques is constantly changing; thus, it is not included
| Examples of suitable techniques for monitoring | Types of variation | Advantages | Disadvantages | Detection of human‐induced changes | |
|---|---|---|---|---|---|
| Population size/gene flow (neutral variation) | Selection (non‐neutral variation) | ||||
| Genetic monitoring | |||||
| Traditional molecular markers | |||||
| Codominant markers (e.g., microsatellites) | Mostly neutral | Cheap and reproducible. Microsatellites are usually hypervariable. Codominant markers. Multiplex PCR (up to 10‐12 loci) is cost‐effective | Often requires species‐ or genera‐specific primer information. Location of the markers on the genome is usually unknown. Often limited number of loci (10‐100 loci). Only polymorphic loci is used, which may cause biased estimations. Poor‐levels of interlaboratory calibration | Yes, but using only polymorphic loci can lead to biased estimation | Limited, depending on the number of loci |
| Dominant markers (e.g., AFLP—amplified fragment length polymorphism) | Mostly neutral | Genome information not required. Relatively cheap. Highly polymorphic at hundreds of loci | Less reproducible. Location of the markers on the genome is usually unknown. Dominant markers. Only polymorphic loci is used, which may cause biased estimations. Poor‐levels of interlaboratory calibration | Yes, but using only polymorphic loci can lead to biased estimation | Limited, depending on the number of loci |
| SNPs and sequences | |||||
| Sanger DNA sequencing | Neutral and functional | Useful for DNA barcoding. Suitable if genes responsible for key traits are known prior to monitoring. Low error rate and high data transferability between laboratories | Lower polymorphism than traditional molecular markers. Mostly species‐ or genera‐specific for functional genes; thus, genome or primer information is required | Limited, due to low polymorphism if few loci are available | Yes, with prior information of the target loci associated with key traits |
| SNP chips | Neutral and functional; mostly functional for qPCR‐based SNP chips | Array of genes allows screening of candidate loci. Can deal with hundreds to 105 loci. Low error rate and high data transferability between laboratories | Require prior genome information for constructing arrays. Only polymorphic loci is used, which may cause biased estimations | Limited, due to potentially nonneutral polymorphisms | Yes, effective screening of the candidate loci from a set of genes |
| Reduced representative sequencing (RRS) (e.g., genotyping‐by‐sequencing and RADseq) | Neutral and functional | 104 to 105 loci detected from genome‐wide in nonmodel organisms. Multiple individuals can be processed at once. A reference sequence is not required, but preferable for annotating DNA and some analyses. With enough read depth, low error rate and high data transferability between laboratories | Representing roughly 1% of genome sequences. Putative functions of loci are often unknown if reference sequences are not available | Yes, with increased accuracy | Yes, allows the genome‐wide screening to identify candidate loci from anonymous or annotated DNA |
| Whole‐genome resequencing | Neutral and functional | (Almost) complete genome polymorphism. Unbiased estimation of population genetic parameters. With enough read depth, low error rate and high data transferability between laboratories | Data often overkill for standard population estimations (e.g., F ST and N e). Relatively expensive. Requires genome information (e.g., reference sequences) | Yes, data is often more than needed but best accuracy is obtained without subsetting a set of loci | Most appropriate. Allows scanning the whole genome to identify the candidate loci |
| Phenotype monitoring | |||||
| Trait measurement | |||||
| Trait variance, heritability and additive genetic (co)variance of traits | Phenotypic, neutral, and functional | Provide direct evidence for how the traits might respond to environmental changes. Provide crucial information for interpretation of genomic data | Labor‐ and cost‐intensive to measure phenotypes in wild populations. Subject to genotype–environmental interaction. Often require common garden experiments to identify genetic‐based variation | No, not accurate estimation | Yes, direct evidence in association with survival, growth and reproduction (fitness) |
| Gene expression | |||||
| Quantitative PCR (qPCR), RNAseq, qPCR‐based DNA chips | Mostly functional (expressing genes) | Useful to detect changes in gene expression of target genes or a whole genome | Require prior information for genes already known to have important influences on (fitness‐related) traits | No | Allow measurements of changes in expression levels of fitness‐related genes among environments |
Functional variation
Monitoring variation in ecologically important and heritable phenotypes (e.g., phenology, growth, and physiology) provides direct evidence for how those traits might respond to environmental change. For instance, many long‐term studies have tracked changes in mean phenotypes to infer the effects of disturbances such as in fisheries (Kuparinen & Merilä, 2007), hunting (Coltman et al., 2003; Pigeon et al., 2016), or with climate change (Parmesan, 2006). Some studies have also shown changes in trait variance, such as declining variance in body size over nine decades in fished cod populations (Olsen, Carlson, Gjøsaeter, & Stenseth, 2009). Of additional interest are the quantitative genetic parameters underlying these changes, such as heritability and additive genetic (co)variances estimated from controlled breeding experiments or “animal model” analyses of intensively monitored and pedigree populations (Bock et al., 2014; Charmantier & Garant, 2005; Charmantier, Perrins, McCleery, & Sheldon, 2006; DiBattista, Feldheim, Garant, Gruber, & Hendry, 2011; Merilä & Hendry, 2014; Wilson et al., 2010). These approaches can often reveal temporal changes in quantitative genetic parameters and the contribution of genetic and plastic effects to temporal trait changes. At the same time, they can be extremely labor‐intensive and subject to strong complications from genotype‐by‐environmental interactions. For example, Kellermann, van Heerwaarden, Sgrò, and Hoffmann (2009) showed that tropical rainforest Drosophila species had a very low ability to evolve desiccation resistance in response to a reduced humidity (10% relative humidity) expected under environmental change, whereas van Heerwaarden and Sgrò (2014) showed that the same species had a high evolvability of desiccation resistance when tested under a more realistic reduction in humidity (35% relative humidity).
An alternative to working with phenotypes is to quantify variation in functional molecular loci (or other physically linked markers) expected to be under strong selection in the case of environmental change. For instance, in some cases, particular alleles in particular genes are known to be differentially sensitive to different temperatures (e.g., Feder & Hofmann, 1999) or salinities (e.g., Munns & Tester, 2008), and so on. In particular, key phenotypes can be controlled by complex nonadditive genetic mechanisms. Thus, understanding the effects of those genes on phenotypes is often essential for understanding the effects of genetic variation on population performance and its consequential effect on ecosystems. Therefore, quantifying the variation in these genes can reveal the evolutionary potential. In addition to monitoring sequence variation in functional genes, one can also monitor gene expression, such as through real‐time PCR (Satake et al., 2013). Of course, these methods are limited to genes already known to have important influences, and the amount of fitness variance explained by such genes is often very low.
An alternative to focusing on specific genes expected a priori to be under strong selection is that one can employ next‐generation sequencing (NGS) to genotype tens to hundreds of thousands of loci in hundreds of individuals at reasonable cost. The usefulness of genomic approaches has received increased attention for its value in conservation biology (reviewed in Allendorf, Hohenlohe, & Luikart, 2010; Shafer et al., 2015). Some common NGS approaches include whole genome resequencing, reduced‐representation sequencing (RRS), and pooled DNA sequencing (Pool‐seq). RRS methods include genotyping‐by‐sequencing (GBS; Davey et al., 2011), restriction site‐associated DNA sequencing (RADseq; Baird, Etter, Atwood, & Currey, 2008; Peterson, Weber, Kay, Fisher, & Hoekstra, 2012), and multiplexed ISSR genotyping‐by‐sequencing (MIGseq) (Suyama & Matsuki, 2015). By restricting sequencing to a fraction of the genome (e.g., restriction enzyme sites ~1% of the genome), GBS and RADseq can typically generate tens of thousands of polymorphisms for monitoring variation. These approaches do not require a pre‐existing reference genome, although availability of such a genome allows for more accurate calls for SNPs and a better ability to infer selection (discussed below). Pool‐seq (Futschik & Schlotterer, 2010) combines DNA from multiple individuals into a single sequencing run, which greatly reduces the cost of obtaining allele frequency data but sacrifices information about the linkage among genetic polymorphisms. Finally, RNA‐seq can be used to measure expression differences across thousands of genes without relying on a priori assumptions about which genes are important.
With enough loci generated by GBS, one can statistically partition loci into genomic regions or—when lacking a reference genome—”markers” that are neutral and regions that are under selection (Vitti, Grossman, & Sabeti, 2013). The former loci can be used to infer population genetic parameters such as those described at the beginning of this section, whereas the latter loci can be used to monitor functional markers. When a reference genome is available, one set of statistical approaches uses the linkage disequilibrium to infer selective sweeps on the genome (e.g., Sabeti et al., 2002). Another set of statistical approaches uses the distribution across loci as measures of population differentiation (e.g., F ST) to infer candidate loci under selection in a Bayesian framework (Beaumont & Balding, 2004; Foll & Gaggiotti, 2008). Moreover, a set of approaches looks for spatial or temporal associations among alleles at particular loci and environmental variables (e.g., temperature, pollution, fishing, hunting, and land use) thought to impose selection on organisms (Joost et al., 2007; de Villemereuil & Gaggiotti, 2015). As each of these approaches has its own strengths and weaknesses, it is commonly suggested that multiple methods be used for optimal inference (Hansen, Olivieri, Waller, & Nielsen, 2012).
In summary, it is clear that no single parameter is a sufficient metric of intraspecific variation. Instead, different parameters yield different insights and their combination is necessary for robust inferences (Table 2). For instance, by combining genotypic and phenotypic data of Atlantic salmon (Salmo salar), Barson et al. (2015) found a sex‐dependent dominance of the gene affecting age maturity; a heterozygote induces late maturity in females and early maturity in males. This finding in the genetic mechanism controlling the key traits may have significant impact on the population managements where the harvesting consequentially selected for early maturation (e.g., Olsen et al., 2005). Initial monitoring of genetic variation in broader contexts with multiple parameters is crucial for understanding the underlying mechanisms that determine the population performance. Hence, monitoring of both neutral and functional genetic variations and phenotypic changes may be necessary to effectively detect and interpret the impact of environmental/management changes on genetic variation. However, linking functional variation in changing environments to population viability and persistence is a challenging task. To aid such integration, we encourage field sampling protocols that archive phenotypes and DNA in ways that allow both current and future analyses depending on changes in resources and technologies. Obvious examples of such protocols are photographs, archiving well‐documented, complete, or partial specimens (such as fish scales or wood samples) in museums and herbaria (assuming that collecting specimens for preservation do not compromise the viability of the population), and long‐term preservation of DNA and RNA.
4.1.3. What spatiotemporal scales should be monitored?
The simplest approach to predicting responses to environmental change would be a single‐time spatial survey relating intraspecific variation to environmental variation. This “space‐for‐time” substitution approach can be informative but many factors, most obviously different timescales, can dictate that spatial patterns will not always accurately predict temporal responses to environmental change (Fukami & Wardle, 2005; Merilä & Hendry, 2014). In short, temporal monitoring of populations is an essential component of any attempt to understand and predict how intraspecific variation will change with ongoing environmental change (Hoban et al., 2014; Schwartz et al., 2007).
4.1.4. What timescale of monitoring is necessary?
The answer here will be species‐specific, taking into account information on life histories and generation times, and also environment‐specific, taking into account information on the timescale and “color” (autocorrelation) of environmental variation (e.g., de Barba et al., 2010; Dowling et al., 2014; Gotanda & Hendry, 2014; Hansen et al., 2012; Schwartz et al., 2007). At the very least, a few generations are necessary to reliably infer trends; however, many studies have found that considerably longer time frames are needed for reliable inference. For example, evolutionary responses of bighorn trophy rams to harvesting (Coltman et al., 2003), and red squirrels to climate change (Réale et al., 2003) that were initially inferred from at least a decade of data, were found to be quite different in the following decade (Pigeon et al., 2016). Simulation studies suggested that even in a 90% decline in population size it might be difficult to detect a signature in sensitive parameters (i.e., number of alleles) within a few generations (Hoban et al., 2014). Long‐term monitoring is therefore optimal but is also not feasible for many organisms. Fortunately, retrospective sampling (e.g., herbaria, museums, sediments, fish scales, or otoliths, and seed banks) can sometimes greatly extend the monitoring timescale by providing insights into past genotypes and phenotypes (Morinaga, Iwasaki, & Suyama, 2014; Wandeler, Hoeck, & Keller, 2007). In some cases, seeds and eggs are still viable over decades, allowing for the “resurrection” of past genotypes for direct comparison of current genotypes (Angeler, 2007; Franks et al., 2008) Finally, in long‐lived organisms, comparative evaluations of genetic variation patterns in adults and juveniles can provide an “early warning” of potential genetic changes (Kettle, Hollingsworth, Jaffré, Moran, & Ennos, 2007; Lowe, Cavers, Boshier, Breed, & Hollingsworth, 2015).
4.1.5. What spatial scale of monitoring is necessary?
If all populations across a species range experienced the same environmental change and responded similarly to that environmental change, then monitoring a single population would be sufficient. However, environmental changes vary dramatically across species’ ranges and different populations respond differently even to the same environmental change (Both & Visser, 2001; Hampe & Petit, 2005). Thus, the optimal monitoring strategy would take into account the spatial grain of environmental change and the spatial grain of population responses to a given environmental change.
For most species, a range‐wide assessment first needs to be conducted so as to understand broad‐scale intraspecific variation and how it is structured within and among populations in relation to spatial (distance) and environmental variations. These assessments serve multiple purposes as they point to key factors, such as connectivity and gene flow (and the potential for portfolio effects), associations between environments and genotypes/phenotypes (the value of which was noted above), and which specific populations contain particularly high or low amounts of within‐population diversity or provide particularly important contributions to among‐population diversity. For instance, populations in the core part of a species’ range often, but not always, contain the most within‐population genetic variation, whereas those from the range peripheries are often, but not always, the most distinct from other populations and also the most susceptible to population size changes (Hampe & Petit, 2005; Smith & Steyn, 2005).
Information from range‐wide assessments can then be used to select particular populations for temporal monitoring. This selection can be aided by systematic methodologies to adjust (scale‐down) geographic observation scales for population sampling to those that best describe spatial trends in genetic variation within a species (Pauls et al., 2013; Pfenninger, Bálint, & Pauls, 2012). Decisions of which populations to monitor can also depend on specific policy and management concerns, locations where environmental change is greatest, particularly unique populations, populations expected to play a major role in range expansions or contractions, and many other considerations. For an invasive species, sampling that covers both the native and the introduced range can trace the invasion and capture evolutionary changes from the ancestral state.
4.1.6. How intensive should sampling be within populations?
For severely endangered species that have only a few individuals remaining, an optimal strategy could be “complete genotyping (or ubiquitous genotyping),” that is, genotyping all individuals. An ongoing and successful example of this approach is the program in Japan where all individuals of more than 20 critically endangered plant species have been recorded and genotyped with microsatellite markers (Isagi & Kaneko, 2014). For larger populations, the minimum strategy should be to obtain good estimates of allele frequencies, for which sample sizes of 30–50 are typically sufficient (Dale & Fortin, 2014; Nei, 1978) and individual‐level data are unnecessary, such as in Pool‐seq (Futschik & Schlotterer, 2010). However, many questions benefit greatly from phenotyping and genotyping large numbers of individuals on fine spatial scales. For instance, individual‐level data allow the estimation of linkage disequilibrium, which facilitates estimates of effective population size as a sensitive indicator of population declines (Antao et al., 2011). In addition, Anderson et al. (2010) suggested that sampling resolution should be smaller than dispersal distances and home ranges so as to best evaluate population connectivity. Finally, high‐resolution individual‐level sampling better covers heterogeneous environments within the selected geographic area (Anderson et al., 2010; Oyler‐McCance, Fedy, & Landguth, 2012; Prunier et al., 2013).
4.1.7. Assessing, evaluating, and improving the monitoring
The scope of empirical sampling will inevitably be incomplete for all but a few species. Fortunately, spatial simulation approaches in landscape genetics can help to cope with heterogeneity and incompleteness and can identify factors affecting genetic variation (Epperson et al., 2010; Landguth, Cushman, & Balkenhol, 2015). For instance, simulations have successfully identified agents that restricted gene flow in heterogeneous environments for American pikas, Ochotona princeps (Castillo, Epps, Davis, & Cushman, 2014), and American marten, Martes americana (Wasserman, Cushman, Schwartz, & Wallin, 2010). Simulations also identified the spread of adaptive genetic variation with range expansion (White, Perkins, Heckel, & Searle, 2013) and projected genetic consequences of future climate scenarios (Wasserman, Cushman, Littell, Shirk, & Landguth, 2013). It should be noted that evaluating monitoring methods (e.g., number of samples and sampling locations) is urgently needed to develop better monitoring methods, especially with new molecular techniques and complex ecological contexts. Simulation modeling with empirical data collected by initial monitoring helps to evaluate monitoring methods (Balkenhol & Fortin, 2015). Such efforts are greatly aided by the recent development of software to simulate genetic consequences in complex ecological contexts (reviewed in Hoban, 2014; Landguth et al., 2015). For instance, spatially explicit simulation software such as SPLATCHE2 (Ray, Currat, Foll, & Excoffier, 2010) and CDPOP (Landguth & Cushman, 2010) can use genetic data on heterogeneous landscapes to evaluate landscape resistance and range expansion in the context of environmental change. The utility of such approaches will escalate with the increasing availability of environmental data, including climate variables (e.g., Worldclim; http://www.worldclim.org) (Hijmans, Cameron, Parra, Jones, & Jarvis, 2005) and digital elevation model (DEM) variables, with free programs for geographic information systems (e.g., Quantum GIS, http://qgis.org).
4.2. Broad monitoring of representative species by proxy
Direct empirical monitoring, including simulations based on incomplete empirical data, will not be feasible in some cases, such as species that are very rare or hard to catch or genetically uncharacterized species. Moreover, detailed monitoring may not be the first choice if it detracts from the immediate needs for action to prevent extinction (Lindenmayer, Piggott, & Wintle, 2013). In such cases, simple predictive models that are broadly applicable across many species can provide some rapid broad‐brush insight. Indeed, global monitoring programs such as GEO BON wish to report regularly on such a global report card. One classic model‐based approach relates changes in population size (or habitat area as a proxy) to changes in genetic variation (Allendorf, 1986; Boecklen, 1986; Boyce, 1992). Such a model seems reasonable based on the established observation that species with larger population sizes or ranges have greater variation (Ellstrand & Elam, 1993; Frankham, 2012). We will now illustrate one way in which such a model might be implemented, but the specific model is intended to be only that—an illustration—and more sophisticated models should be developed, with some ideas introduced below.
One good initial candidate for general “proxy” model is a power law relationship, (G 0/G 1) = (R 0/R 1)z, where G is the genetic variation and R is the range extent; the subscript 0 indicates the original value and the subscript 1 indicates the new value (Neto, de Oliveira, Rosas, & Campos, 2011; also see Rauch & Bar‐Yam, 2004). Box 1 presents this approach schematically by showing how the power law can be used to predict the loss of genetic variation as a function of the loss of range area, while making a number of simplifying assumptions (e.g., random loss of area from a species’ range). Likely violations of these assumptions indicate that real species in real landscapes can deviate considerably from the simple prediction by having either a greater or lesser than expected loss of genetic variation for a given loss of range area. Thus, accurate predictions for specific species would require additional information and more sophisticated models that avoid some of the simplifying assumptions. However, the simple power curve might adequately capture the average outcome when calculated across many species and so provide a useful early‐warning report card of the general expected losses of within‐species genetic variation.
Box 1. Proxies for within‐species genetic variation.
1.
There is some evidence in support of a power law model linking within‐species genetic diversity loss to range loss (e.g., Neto et al., 2011). A power law model linking range and genetic variation is also supported, at least indirectly, by Morlon et al. (2011), who found support for a power relationship between the amount of phylogenetic diversity (PD) sampled and the sampled area. As an example, we have explored such a model through an analysis of genetic variation and range data from Alsos et al. (2012). They examined genetic data for 27 plant species over many populations covering the range of each species. They linked scenarios of range‐area loss to genetic variation loss through a random sampling approach. For a given species, they randomly removed an increasing number of grid cells (from the distribution of the species) and recorded the loss of alleles (loss of genetic variation). They repeated this random sampling 1000 times to find the median number of alleles (i.e., markers) lost for any given total number of grid cells removed (total range‐area lost). Based on these data, we explored the power curve relationship model relating the range extent loss to the genetic variation loss for each of the 27 species. While a power curve model consistently provided good fit, the power curve z values (see formula in main text) varied among these models, suggesting that there were difficulties applying one general model to predict genetic variation loss, given only fractional range‐loss information, over any set of representative species. However, the z values varied somewhat predictably (Figure a) according to the estimated population differentiation value (F ST) of the species (F ST values provided in Alsos et al., 2012).
This suggests that a report card on the loss of genetic variation based on loss of geographic range extent for a given representative set of species is possible, if we have some estimates of genetic differentiation (e.g., estimates of F ST). A practical strategy may be the use of two pooled models for species broadly categorized as having large versus small differentiation values. Analysis of the Alsos et al. (2012) data suggests that this may be an effective, simple option (Figure b). Such a broad categorization also allows for expert opinion and other sources of information (such as dispersal or life history information) to be used to categorize species. Future work could explore whether a power curve with an intermediate z value (e.g., of 0.25) provides a robust proxy approach applicable to the tens of thousands of species in the Map of Life.
An alternative approach is possible when we have information, for a given species, about the pattern of losses of its distribution of sites in an environmental space (Figure c).
Figure Analysis of the genetic variation and range data from Alsos et al. (2013). (a) Z values from significant power curve models (24 species) linking range loss to genetic variation loss are predicted well by species F ST values. However, middle range values are predicted less well. (b) Single power curve models for the high F ST and for the low F ST species are well supported, suggesting that the report card might simply use course categories for species that reflect a magnitude of F ST. Note that, as expected, high F ST species will have a more rapid loss of genetic variation as their range extent is lost. (c) The ED method (see Faith et al., 2004) indicates loss of genetic variation for a given species as sites are lost from an environmental space. When losses from environmental space are random, the genetic diversity loss indicated by ED again approximates a power curve (linear when the axes are log‐transformed; shown by the black line). However, ED indicates a range of possible degrees of loss of genetic diversity (shown as the shaded area) depending on the actual pattern of site losses in environmental space.
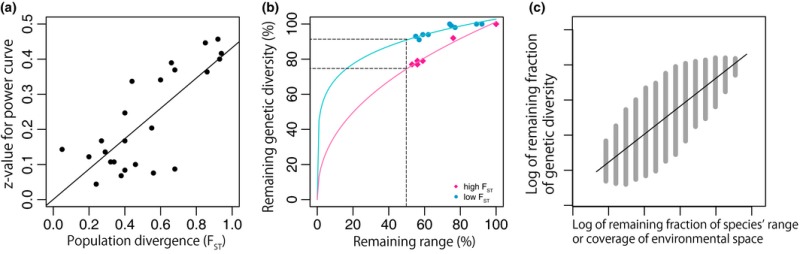
This strategy, based on range loss, considers consequences for neutral variation, but might not be effective at estimating changes in non‐neutral variation, such as that reflecting adaptation to environmental variation across the range of a species. More relevant predictions could be generated by employing information (for many species) on actual species distributions and estimated habitat/environmental loss (from Geographical Information System records or the “Map of Life”) to estimate how much of a species’ “environmental space” is lost. The “environmental diversity” or “ED” method (Faith, Ferrier, & Walker, 2004; Faith & Walker, 1996) can be applied to estimate fractional genetic variation loss for a given pattern of loss of sites in a species’ environmental space. Depending on whether losses in species’ environmental space are spread out or “clumped,” the same fractional loss of sites in the species’ environmental space can mean a high or low loss of functional genetic variation (Box 1). Similar approaches could be used for neutral genetic variation, providing an alternative to the simplifying assumption of random loss of range.
As with direct empirical monitoring, the large subset of specific species for which broad monitoring by proxy would be conducted would need to be selected. If a subset of species is selected that is representative of environmental space, then the estimated loss of genetic diversity for that subset of species may be an indicator of the more general losses. Here, we provide one possible process to select such a set of representative species:
For any two species, calculate their “dissimilarity” based on the difference in their locations in environmental (or geographic) space. For example, dissimilarity may be defined as the average distance in environmental space between the locations of the two species.
For any nominated target number (call it k) of representative species, use the dissimilarities to derive k clusters of a species. For example, k‐means clustering algorithms can directly use dissimilarities to derive k clusters. Choose one member from each cluster to form the subset of k representative species.
For the k species, apply the proxy model to infer loss of genetic variation based on the loss of geographic (or environmental) range extent.
4.3. The way forward
A recent study revealed our limited knowledge of global distribution of genetic diversity (Miraldo et al 2016). Thus, we need a global monitoring strategy for intraspecific variation. Such a strategy will require integration and cooperation across researchers, stakeholders, countries, and all levels of government. Many monitoring and database efforts are already underway (e.g., international and national LTER: Long‐Term Ecological Research network; GBIF: Global Biodiversity Information Facility; TRY: plant trait database). For instance, LTER programs collect and archive individual data (e.g., presence/absence, density, and growth data) from multiple plots with repeated observations. Genotyping such individuals contributes to developing global databases of spatiotemporal trends in intraspecific variation. Together with these monitoring efforts, genetic variation can be mapped with other demographic data, including trends in species distribution and population growth. This can be used to account for changes in these trends, to understand how genetic mechanisms are altering population performances, to interpret the effects of the consequences of human activities on ecosystem function via changes in genetic variation, and to illustrate the relative importance of genetic variation among other factors in an ecological and evolutionary context. The discussion that attended such efforts would also serve to predict the changes, identify key gaps in monitoring, assess current indicators and improve them, seek appropriate funding for expanding and better coordinating monitoring efforts, and aid the design and promotion of standardized protocols that could be implemented across taxa and contexts. Settling on these protocols will involve discussion and optimization of answers to the above questions, such as which species to monitor, what types of variation to monitor, and what spatiotemporal scales to monitor. The integrated development and implementation of broad‐brush modeling approaches for situations where direct empirical monitoring is not feasible will also be valuable. To achieve all of these goals, collaboration with other ecological and environmental monitoring networks will be essential.
5. Conclusion
The status of, and trends in, intraspecific (genetic and phenotypic) variation through time and space will determine the fate of a population and species, the biodiversity and structure of communities, and the state of ecosystem functions and services. Human activities are having profound effects on variation within and among many populations and species through increased population size, novel selection, and increased gene flow (connections between species/populations), and can thereby shape all of those fates. For these reasons, it is critical to (i) establish monitoring programs for genetic variation, (ii) link observed changes in variation to specific environmental changes and management decisions, and (iii) develop predictive frameworks for changes in variation and its consequences. We call for the development of a globally‐coordinated observation network for monitoring intraspecific variation and its potential consequences for human well‐being.
Acknowledgements
We thank C. Primmer and anonymous reviewers for helpful comments on the manuscript. We acknowledge the support of the bioGENESIS and the funding provided by JSPS Global COE program, JSPS KAKENHI Grant Numbers JP15H02640, JP16H02553, JP25840161, and the Environment Research and Technology Development Funds (S9 and 4‐1601) of the Ministry of the Environment, Japan.
Literature cited
- Agrawal, A. F. , & Whitlock, M. C. (2012). Mutation load: The fitness of individuals in populations where deleterious alleles are abundant. Annual Review of Ecology, Evolution, and Systematics, 43, 115–135. [Google Scholar]
- Aguilar, R. , Quesada, M. , Ashworth, L. , Herrerias‐Diego, Y. , & Lobo, J. (2008). Genetic consequences of habitat fragmentation in plant populations: Susceptible signals in plant traits and methodological approaches. Molecular Ecology, 17, 5177–5188. [DOI] [PubMed] [Google Scholar]
- Aitken, S. N. , & Whitlock, M. C. (2013). Assisted gene flow to facilitate local adaptation to climate change. Annual Review of Ecology, Evolution, and Systematics, 44, 367–388. [Google Scholar]
- Allendorf, F. W. (1986). Genetic drift and the loss of alleles versus heterozygosity. Zoo Biology, 5, 181–190. [Google Scholar]
- Allendorf, F. W. , England, P. R. , Luikart, G. , Ritchie, P. A. , & Ryman, N. (2008). Genetic effects of harvest on wild animal populations. Trends in Ecology & Evolution, 23, 327–337. [DOI] [PubMed] [Google Scholar]
- Allendorf, F. W. , Hohenlohe, P. A. , & Luikart, G. (2010). Genomics and the future of conservation genetics. Nature Genetics, 11, 697–709. [DOI] [PubMed] [Google Scholar]
- Allendorf, F. W. , Luikart, G. H. , & Aitken, S. N. (2012). Conservation and the genetics of populations, 2nd ed Oxford: Blackwell Publishing Ltd. [Google Scholar]
- Alsos, I. G. , Ehrich, D. , Thuiller, W. , Eidesen, P. B. , Tribsch, A. , Schönswetter, P. , … Brochmann, C. (2012). Genetic consequences of climate change for northern plants. Proceedings of the Royal Society B‐Biological Sciences, 279, 2042–2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, C. D. , Epperson, B. K. , Fortin, M. J. , Holderegger, R. , James, P. M. A. , Rosenberg, M. S. , … Spear, S. (2010). Considering spatial and temporal scale in landscape‐genetic studies of gene flow. Molecular Ecology, 19, 3565–3575. [DOI] [PubMed] [Google Scholar]
- Anderson, J. T. , Inouye, D. W. , Mckinney, A. M. , Colautti, R. I. , & Mitchell‐olds, T. (2012). Phenotypic plasticity and adaptive evolution contribute to advancing flowering phenology in response to climate change. Proceedings of the Royal Society B‐Biological Sciences, 279, 3843–3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angeler, D. G. (2007). Resurrection ecology and global climate change research in freshwater ecosystems resurrection ecology and global climate change research in freshwater ecosystems. Journal of the North American Benthological Society, 26, 12–22. [Google Scholar]
- Antao, T. , Pérez‐Figueroa, A. , & Luikart, G. (2011). Early detection of population declines: High power of genetic monitoring using effective population size estimators. Evolutionary Applications, 4, 144–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonovics, J. , Bradshaw, A. D. , & Turner, R. G. (1971). Heavy metal tolerance in plants. Advances in Ecological Research, 7, 1–85. [Google Scholar]
- Araki, H. , Berejikian, B. A. , Ford, M. J. , & Blouin, M. S. (2008). Fitness of hatchery‐reared salmonids in the wild. Evolutionary Applications, 1, 342–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird, N. , Etter, P. , Atwood, T. , & Currey, M. (2008). Rapid SNP discovery and genetic mapping using sequenced RAD markers. PLoS ONE, 3, e3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bálint, M. , Domisch, S. , Engelhardt, C. H. M. , Haase, P. , Lehrian, S. , Sauer, J. , … Nowak, C. (2011). Cryptic biodiversity loss linked to global climate change. Nature Climate Change, 1, 313–318. [Google Scholar]
- Balkenhol, N. , & Fortin, M.‐J. (2015). Basics of study design: Sampling landscape heterogeneity and genetic variation for landscape genetic studies In Balkenhol N., Cushman S., Storfer A., & Waits L. (Eds.), Landscape genetics: Concepts, methods, applications. Hoboken, NJ: Wiley Blackwell. [Google Scholar]
- de Barba, M. , Waits, L. P. , Garton, E. O. , Genovesi, P. , Randi, E. , Mustoni, A. , & Groff, C. (2010). The power of genetic monitoring for studying demography, ecology and genetics of a reintroduced brown bear population. Molecular Ecology, 19, 3938–3951. [DOI] [PubMed] [Google Scholar]
- Barbour, R. C. , O'Reilly‐Wapstra, J. M. , De Little, D. W. , Jordan, G. J. , Steane, D. A. , Humphreys, J. R. , … Potts, B. M. (2009). A geographic mosaic of genetic variation within a foundation tree species and its community‐level consequences. Ecology, 90, 1762–1772. [DOI] [PubMed] [Google Scholar]
- Barnosky, A. D. , Matzke, N. , Tomiya, S. , Wogan, G. O. U. , Swartz, B. , Quental, T. B. , … Ferrer, E. A. (2011). Has the Earth's sixth mass extinction already arrived? Nature, 471, 51–57. [DOI] [PubMed] [Google Scholar]
- Barson, N. J. , Aykanat, T. , Hindar, K. , Baranski, M. , Bolstad, G. H. , Fiske, P. , … Primmer, C. R. (2015). Sex‐dependent dominance at a single locus maintains variation in age at maturity in salmon. Nature, 528, 405–408. [DOI] [PubMed] [Google Scholar]
- Beaumont, M. A. (2010). Approximate Bayesian computation in evolution and ecology. Annual Review of Ecology, Evolution, and Systematics, 41, 379–406. [Google Scholar]
- Beaumont, M. A. , & Balding, D. J. (2004). Identifying adaptive genetic divergence among populations from genome scans. Molecular Ecology, 13, 969–980. [DOI] [PubMed] [Google Scholar]
- Beerli, P. , & Palczewski, M. (2010). Unified framework to evaluate panmixia and migration direction among multiple sampling locations. Genetics, 185, 313–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bello‐Bedoy, R. , & Núñez‐Farfán, J. (2011). The effect of inbreeding on defence against multiple enemies in Datura stramonium . Journal of Evolutionary Biology, 24, 518–530. [DOI] [PubMed] [Google Scholar]
- Bijlsma, R. , & Loeschcke, V. (2012). Genetic erosion impedes adaptive responses to stressful environments. Evolutionary Applications, 5, 117–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock, A. , Sparks, T. H. , Estrella, N. , Jee, N. , Casebow, A. , Schunk, C. , … Menzel, A. (2014). Changes in first flowering dates and flowering duration of 232 plant species on the island of Guernsey. Global Change Biology, 20, 3508–3519. [DOI] [PubMed] [Google Scholar]
- Boecklen, W. J. (1986). Optimal design of nature reserves: Consequences of genetic drift. Biological Conservation, 38, 323–338. [Google Scholar]
- Both, C. , Bouwhuis, S. , Lessells, C. M. , & Visser, M. E. (2006). Climate change and population declines in a long‐distance migratory bird. Nature, 441, 81–83. [DOI] [PubMed] [Google Scholar]
- Both, C. , & Visser, M. E. (2001). Adjustment to climate change is constrained by arrival date in a long‐distance migrant bird. Nature, 411, 296–298. [DOI] [PubMed] [Google Scholar]
- Boyce, M. S. (1992). Population viability analysis. Annual Review of Ecology and Systematics, 23, 481–506. [Google Scholar]
- Bradshaw, W. E. , & Holzapfel, C. M. (2001). Genetic shift in photoperiodic response correlated with global warming. Proceedings of the National Academy of Sciences, 98, 14509–14511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bratteler, M. , Lexer, C. , & Widmer, A. (2006). Genetic architecture of traits associated with serpentine adaptation of Silene vulgaris . Journal of Evolutionary Biology, 19, 1149–1156. [DOI] [PubMed] [Google Scholar]
- Cardinale, B. J. , Duffy, J. E. , Gonzalez, A. , Hooper, D. U. , Perrings, C. , Venail, P. , … Naeem, S. (2012). Biodiversity loss and its impact on humanity. Nature, 486, 59–67. [DOI] [PubMed] [Google Scholar]
- Carroll, S. P. , Loye, J. E. , Dingle, H. , Mathieson, M. , Famula, T. R. , & Zalucki, M. P. (2005). And the beak shall inherit—evolution in response to invasion. Ecology Letters, 8, 944–951. [DOI] [PubMed] [Google Scholar]
- Castillo, J. A. , Epps, C. W. , Davis, A. R. , & Cushman, S. A. (2014). Landscape effects on gene flow for a climate‐sensitive montane species, the American pika. Molecular Ecology, 23, 843–856. [DOI] [PubMed] [Google Scholar]
- Charmantier, A. , & Garant, D. (2005). Environmental quality and evolutionary potential: Lessons from wild populations. Proceedings of the Royal Society B‐Biological Sciences, 272, 1415–1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charmantier, A. , Perrins, C. , McCleery, R. H. , & Sheldon, B. C. (2006). Evolutionary response to selection on clutch size in a long‐term study of the mute swan. American Naturalist, 167, 453–465. [DOI] [PubMed] [Google Scholar]
- Clarke, C. A. , & Sheppard, P. M. (1966). A local survey of the distribution of industrial melanic forms in the moth Biston betularia and estimates of the selective values of these in an industrial environment. Proceedings of the Royal Society B‐Biological Sciences, 165, 424–439. [Google Scholar]
- Colautti, R. I. , & Barrett, S. C. H. (2013). Rapid adaptation to climate facilitates range expansion of an invasive plant. Science, 342, 364–366. [DOI] [PubMed] [Google Scholar]
- Colautti, R. I. , & Lau, J. A. (2015). Contemporary evolution during invasion: Evidence for differentiation, natural selection, and local adaptation. Molecular Ecology, 24, 1999–2017. [DOI] [PubMed] [Google Scholar]
- Coltman, D. W. , O'Donoghue, P. , Jorgenson, J. T. , Hogg, J. T. , Strobeck, C. , & Festa‐Bianchet, M. (2003). Undesirable evolutionary consequences of trophy hunting. Nature, 426, 655–658. [DOI] [PubMed] [Google Scholar]
- Conover, D. O. , Munch, S. B. , & Arnott, S. A. (2009). Reversal of evolutionary downsizing caused by selective harvest of large fish. Proceedings of the Royal Society B‐Biological Sciences, 276, 2015–2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook, L. M. , & Saccheri, I. J. (2012). The peppered moth and industrial melanism: Evolution of a natural selection case study. Heredity, 110, 207–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crutsinger, G. M. (2016). A community genetics perspective: Opportunities for the coming decade. New Phytologist, 210, 65–70. [DOI] [PubMed] [Google Scholar]
- Dale, M. R. T. , & Fortin, M.‐J. (2014). Spatial analysis: A guide for ecologists. Cambridge: Cambridge University Press. [Google Scholar]
- Davey, J. W. , Hohenlohe, P. A. , Etter, P. D. , Boone, J. Q. , Catchen, J. M. , & Blaxter, M. L. (2011). Genome‐wide genetic marker discovery and genotyping using next‐generation sequencing. Nature Reviews Genetics, 12, 499–510. [DOI] [PubMed] [Google Scholar]
- De León, L. F. , Raeymaekers, J. A. M. , Bermingham, E. , Podos, J. , Herrel, A. , & Hendry, A. P. (2011). Exploring possible human influences on the evolution of Darwin's finches. Evolution, 65, 2258–2272. [DOI] [PubMed] [Google Scholar]
- Denison, F. R. (2012). Darwinian agriculture: How understanding evolution can improve agriculture. Princeton, NJ: Princeton University Press. [Google Scholar]
- Díaz, S. , Fargione, J. , Chapin, F. S. , & Tilman, D. (2006). Biodiversity loss threatens human well‐being. PLoS Biology, 4, e277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiBattista, J. D. (2008). Patterns of genetic variation in anthropogenically impacted populations. Conservation Genetics, 9, 141–156. [Google Scholar]
- DiBattista, J. D. , Feldheim, K. A. , Garant, D. , Gruber, S. H. , & Hendry, A. P. (2011). Anthropogenic disturbance and evolutionary parameters: A lemon shark population experiencing habitat loss. Evolutionary Applications, 4, 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebley, J. F. , Gaut, B. S. , & Smith, B. D. (2006). The molecular genetics of crop domestication. Cell, 127, 1309–1321. [DOI] [PubMed] [Google Scholar]
- Dowling, T. E. , Turner, T. F. , Carson, E. W. , Saltzgiver, M. J. , Adams, D. , Kesner, B. , & Marsh, P. C. (2014). Time‐series analysis reveals genetic responses to intensive management of razorback sucker (Xyrauchen texanus). Evolutionary Applications, 7, 339–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drès, M. , & Mallet, J. (2002). Host races in plant‐feeding insects and their importance in sympatric speciation. Philosophical Transactions of the Royal Society B‐Biological Sciences, 357, 471–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudash, M. , & Fenster, C. B. (2001). Inbreeding and outbreeding depression in fragmented populations In Young A. G., & Clarke G. M. (Eds.), Genetics, demography, and viability of fragmented populations (pp. 33–35). Cambridge: Cambridge University Press. [Google Scholar]
- Ellis, E. C. , Antill, E. C. , & Kreft, H. (2012). All is not loss: Plant biodiversity in the anthropocene. PLoS ONE, 7, e30535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellstrand, N. C. , & Elam, D. R. (1993). Consequences of small population size: Implications for plant conservation. Conservation Genetics, 24, 217–242. [Google Scholar]
- Epperson, B. B. K. , McRae, B. B. H. , Scribner, K. , Cushman, S. A. , Rosenberg, M. S. M. , Fortin, M. M.‐J. , … Dale, M. R. (2010). Utility of computer simulations in landscape genetics. Molecular Ecology, 19, 3549–3564. [DOI] [PubMed] [Google Scholar]
- Faith, D. P. , Ferrier, S. , & Walker, P. A. (2004). The ED strategy: How species‐level surrogates indicate general biodiversity patterns through an ‘environmental diversity’ perspective. Journal of Biogeography, 31, 1207–1217. [Google Scholar]
- Faith, D. P. , Magallón, S. , Hendry, A. P. , Conti, E. , Yahara, T. , & Donoghue, M. J. (2010). Evosystem services: An evolutionary perspective on the links between biodiversity and human well‐being. Current Opinion in Environmental Sustainability, 2, 66–74. [Google Scholar]
- Faith, D. P. , & Walker, P. A. (1996). Environmental diversity: On the best‐possible use of surrogate data for assessing the relative biodiversity of sets of areas. Biodiversity and Conservation, 5, 399–415. [Google Scholar]
- Feder, M. E. , & Hofmann, G. E. (1999). Heat‐shock proteins, molecular chaperones, and the stress response: Evolutionary and ecological physiology. Annual Review of Physiology, 61, 243–282. [DOI] [PubMed] [Google Scholar]
- Fenberg, P. B. , & Roy, K. (2008). Ecological and evolutionary consequences of size‐selective harvesting: How much do we know? Molecular Ecology, 17, 209–220. [DOI] [PubMed] [Google Scholar]
- Fisher, R. A. (1930). The genetical theory of natural selection. Oxford: Clarendon Press. [Google Scholar]
- Foll, M. , & Gaggiotti, O. (2008). A genome‐scan method to identify selected loci appropriate for both dominant and codominant markers: A Bayesian perspective. Genetics, 180, 977–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankham, R. (2012). How closely does genetic diversity in finite populations conform to predictions of neutral theory? Large deficits in regions of low recombination. Heredity, 108, 167–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankham, R. , Ballou, J. D. , & Briscoe, D. A. (2002). Introduction of conservation genetics. Cambridge: Cambridge University Press. [Google Scholar]
- Franks, S. J. , Avise, J. C. , Bradshaw, W. E. , Conner, J. K. , Julie, R. , Mazer, S. J. , … Etterson, J. R. (2008). The resurrection initiative: Storing ancestral genotypes to capture evolution in action. BioScience, 58, 870–873. [Google Scholar]
- Fukami, T. , & Wardle, D. A. (2005). Long‐term ecological dynamics: Reciprocal insights from natural and anthropogenic gradients. Proceedings of the Royal Society B‐Biological Sciences, 272, 2105–2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futschik, A. , & Schlotterer, C. (2010). The next generation of molecular markers from massively parallel sequencing of pooled DNA samples. Genetics, 186, 207–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garant, D. , Forde, S. E. , & Hendry, A. P. (2007). The multifarious effects of dispersal and gene flow on contemporary adaptation. Functional Ecology, 21, 434–443. [Google Scholar]
- García‐Ramos, G. , & Kirkpatrick, M. (1997). Genetic models of adaptation and gene flow in peripheral populations. Evolution, 51, 21–28. [DOI] [PubMed] [Google Scholar]
- García‐Ramos, G. , & Rodríguez, D. (2002). Evolutionary speed of species invasions. Evolution, 56, 661–668. [DOI] [PubMed] [Google Scholar]
- Garroway, C. J. , Bowman, J. , Cascaden, T. J. , Holloway, G. L. , Mahan, C. G. , Malcolm, J. R. , … Wilson, P. J. (2010). Climate change induced hybridization in flying squirrels. Global Change Biology, 16, 113–121. [Google Scholar]
- Genung, M. A. , Lessard, J.‐P. , Brown, C. B. , Bunn, W. A. , Cregger, M. A. , Reynolds, W. M. N. , … Bailey, J. K. (2010). Non‐additive effects of genotypic diversity increase floral abundance and abundance of floral visitors. PLoS ONE, 5, e8711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GEO BON . (2010). Group on earth observations biodiversity observation network (GEO BON) Detailed Implementation Plan, version 1.0. Group on Earth Observations Biodiversity Network.
- Gotanda, K. M. , & Hendry, A. P. (2014). Using adaptive traits to consider potential consequences of temporal variation in selection: Male guppy colour through time and space. Biological Journal of the Linnean Society, 112, 108–122. [Google Scholar]
- Green, R. E. , Cornell, S. J. , Scharlemann, J. P. W. , & Balmford, A. (2005). Farming and the fate of wild nature. Science, 307, 550–555. [DOI] [PubMed] [Google Scholar]
- Habel, J. C. , Rödder, D. , Schmitt, T. , & Nève, G. (2011). Global warming will affect the genetic diversity and uniqueness of Lycaena helle populations. Global Change Biology, 17, 194–205. [Google Scholar]
- Hampe, A. , & Petit, R. J. (2005). Conserving biodiversity under climate change: The rear edge matters. Ecology Letters, 8, 461–467. [DOI] [PubMed] [Google Scholar]
- Hansen, T. F. , Carter, A. J. R. , & Pélabon, C. (2006). On adaptive accuracy and precision in natural populations. American Naturalist, 168, 168–181. [DOI] [PubMed] [Google Scholar]
- Hansen, M. M. , Olivieri, I. , Waller, D. M. , & Nielsen, E. E. (2012). Monitoring adaptive genetic responses to environmental change. Molecular Ecology, 21, 1311–1329. [DOI] [PubMed] [Google Scholar]
- Harris, R. B. , Wall, W. A. , Allendorf, F. W. , Harris, B. , & Wall, A. (2002). Genetic consequences of hunting: What do we know and what should we do? Wildlife Society Bulletin, 30, 634–643. [Google Scholar]
- van Heerwaarden, B. , & Sgrò, C. M. (2014). Is adaptation to climate change really constrained in niche specialists? Proceedings of the Royal Society B‐Biological Sciences, 281, 20140396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendry, A. P. (2016). Eco‐evolutionary dynamics. Princeton, NJ: Princeton University Press. [Google Scholar]
- Hendry, A. P. , Farrugia, T. J. , & Kinnison, M. T. (2008). Human influences on rates of phenotypic change in wild animal populations. Molecular Ecology, 17, 20–29. [DOI] [PubMed] [Google Scholar]
- Hendry, A. P. , Grant, P. R. , Rosemary Grant, B. , Ford, H. A. , Brewer, M. J. , & Podos, J. (2006). Possible human impacts on adaptive radiation: Beak size bimodality in Darwin's finches. Proceedings of the Royal Society B‐Biological Sciences, 273, 1887–1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendry, A. P. , Kinnison, M. T. , Heino, M. , Day, T. , Smith, T. B. , Fitt, G. , … Carroll, S. P. (2011). Evolutionary principles and their practical application. Evolutionary Applications, 4, 159–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewitt, G. M. (1996). Some genetic consequences of ice ages, and their role in divergence and speciation. Biological Journal of the Linnean Society, 58, 247–276. [Google Scholar]
- Hey, J. (2010). Isolation with migration models for more than two populations. Molecular Biology and Evolution, 27, 905–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hijmans, R. J. , Cameron, S. E. , Parra, J. L. , Jones, P. G. , & Jarvis, A. (2005). Very high resolution interpolated climate surfaces for global land areas. International Journal of Climatology, 25, 1965–1978. [Google Scholar]
- Hoban, S. (2014). An overview of the utility of population simulation software in molecular ecology. Molecular Ecology, 23, 2383–2401. [DOI] [PubMed] [Google Scholar]
- Hoban, S. , Arntzen, J. A. , Bruford, M. W. , Godoy, J. A. , Hoelzel, R. , Segelbacher, A. G. , … Bertorelle, G. (2014). Comparative evaluation of potential indicators and temporal sampling protocols for monitoring genetic erosion. Evolutionary Applications, 7, 984–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoban, S. M. , Hauffe, H. C. , Pérez‐Espona, S. , Arntzen, J. W. , Bertorelle, G. , Bryja, J. , … Bruford, M. W. (2013). Bringing genetic diversity to the forefront of conservation policy and management. Conservation Genetic Resources, 5, 593–598. [Google Scholar]
- Hoffman, J. I. , Simpson, F. , David, P. , Rijks, J. M. , Kuiken, T. , Thorne, M. A. S. , … Dasmahapatra, K. K. (2014). High‐throughput sequencing reveals inbreeding depression in a natural population. Proceedings of the National Academy of Sciences, 111, 3775–3780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann, A. A. , & Merilä, J. (1999). Heritable variation and evolution under favourable and unfavourable conditions. Trends in Ecology and Evolution, 14, 96–101. [DOI] [PubMed] [Google Scholar]
- Hooper, D. U. , Chapin, F. S. III , Ewel, J. J. , Hector, A. , Inchausti, P. , Lavorel, S. , … Wardle, D. A. (2005). Effects of biodiversity on ecosystem functioning: A consensus of current known knowledge. Ecological Monographs, 75, 3–35. [Google Scholar]
- Hughes, J. B. , Daily, G. C. , & Ehrlich, P. R. (1997). Population diversity: Its extent and extinction. Science, 278, 689–692. [DOI] [PubMed] [Google Scholar]
- Hughes, A. R. , Inouye, B. D. , Johnson, M. T. J. , Underwood, N. , & Vellend, M. (2008). Ecological consequences of genetic diversity. Ecology Letters, 11, 609–623. [DOI] [PubMed] [Google Scholar]
- Hyten, D. L. , Song, Q. , Zhu, Y. , Choi, I.‐Y. , Nelson, R. L. , Costa, J. M. , … Cregan, P. B. (2006). Impacts of genetic bottlenecks on soybean genome diversity. Proceedings of the National Academy of Sciences of the United States of America, 103, 16666–16671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IPCC . (2013). Summary for policymakers In Stocker T. F., Qin D., Plattner G.‐K., Tignor M., Allen S. K., Boschung J., Nauels A., Xia Y., And V. B., & Midgley P. M. (Eds.), Climate Change 2013: The physical science basis. Contribution of working group I to the fifth assessment report of the intergovernmental panel on climate change. Cambridge: Cambridge University Press. [Google Scholar]
- Isagi, Y. , & Kaneko, S. (2014). Ubiquitous genotyping for conservation of endangered plant species In Nakano S., Yahara T., & Nakashizuka T. (Eds.), Asia‐Pacific biodiversity observation network – integrative observation and assessment. NY: Springer. [Google Scholar]
- Joost, S. , Bonin, A. , Bruford, M. W. , Després, L. , Coord, C. , Erhard, G. , & Taberlet, P. (2007). A spatial analysis method (SAM) to detect candidate loci for selection: Towards a landscape genomics approach to adaptation. Molecular Ecology, 16, 3955–3969. [DOI] [PubMed] [Google Scholar]
- Jump, A. S. , Marchant, R. , & Peñuelas, J. (2009). Environmental change and the option value of genetic diversity. Trends in Plant Science, 14, 51–58. [DOI] [PubMed] [Google Scholar]
- Keller, L. F. , & Waller, D. M. (2002). Inbreeding effects in wild populations. Trends in Ecology & Evolution, 17, 230–241. [Google Scholar]
- Kellermann, V. , van Heerwaarden, B. , Sgrò, C. M. , & Hoffmann, A. A. (2009). Fundamental evolutionary limits in ecological traits drive drosophila species distributions. Science, 325, 1244–1246. [DOI] [PubMed] [Google Scholar]
- Kettle, C. J. , Hollingsworth, P. M. , Jaffré, T. , Moran, B. , & Ennos, R. A. (2007). Identifying the early genetic consequences of habitat degradation in a highly threatened tropical conifer, Araucaria nemorosa Laubenfels. Molecular Ecology, 16, 3581–3591. [DOI] [PubMed] [Google Scholar]
- Koblmüller, S. , Nord, M. , Wayne, R. K. , & Leonard, J. A. (2009). Origin and status of the Great Lakes wolf. Molecular Ecology, 18, 2313–2326. [DOI] [PubMed] [Google Scholar]
- Kruuk, L. E. B. (2004). Estimating genetic parameters in natural populations using the “animal model”. Philosophical Transactions of the Royal Society B‐Biological Sciences, 359, 873–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuparinen, A. , & Merilä, J. (2007). Detecting and managing fisheries‐induced evolution. Trends in Ecology and Evolution, 22, 652–659. [DOI] [PubMed] [Google Scholar]
- Laikre, L. (2010). Genetic diversity is overlooked in international conservation policy implementation. Conservation Genetics, 11, 349–354. [Google Scholar]
- Laikre, L. , Schwartz, M. K. , Waples, R. S. , & Ryman, N. (2010). Compromising genetic diversity in the wild: Unmonitored large‐scale release of plants and animals. Trends in Ecology and Evolution, 25, 520–529. [DOI] [PubMed] [Google Scholar]
- Landguth, E. L. , & Cushman, S. A. (2010). cdpop, a spatially explicit cost distance population genetics program. Molecular Ecology Resources, 10, 156–161. [DOI] [PubMed] [Google Scholar]
- Landguth, E. , Cushman, S. A. , & Balkenhol, N. (2015). Simulation modeling in landscape genetics In Balkenhol N., Cushman S., Storfer A. & Waits L. (Eds.), Landscape genetics: Concepts, methods, applications (pp. 101–113). West Sussex: Wiley Blackwell. [Google Scholar]
- Lankau, R. , Jørgensen, P. S. , Harris, D. J. , & Sih, A. (2011). Incorporating evolutionary principles into environmental management and policy. Evolutionary Applications, 4, 315–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavergne, S. , & Molofsky, J. (2007). Increased genetic variation and evolutionary potential drive the success of an invasive grass. Proceedings of the National Academy of Sciences, 104, 3883–3888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law, W. , & Salick, J. (2005). Human‐induced dwarfing of Himalayan snow lotus, Saussurea laniceps (Asteraceae). Proceedings of the National Academy of Sciences, 102, 10218–10220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leberg, P. L. (2002). Estimating allelic richness: Effects of sample size and bottleneck. Molecular Ecology, 11, 2445–2449. [DOI] [PubMed] [Google Scholar]
- Lehman, N. , Eisenhawer, A. , Hansen, K. , Mech, L. D. , Peterson, R. O. , Gogan, P. J. P. , & Wayne, R. K. (1991). Introgression of coyote mitochondrial‐DNA into sympatric North‐American gray wolf populations. Evolution, 45, 104–119. [DOI] [PubMed] [Google Scholar]
- Leimu, R. , Vergeer, P. , Angeloni, F. , & Ouborg, N. J. (2010). Habitat fragmentation, climate change, and inbreeding in plants. Annals of the New York Academy of Sciences, 1195, 84–98. [DOI] [PubMed] [Google Scholar]
- Lenormand, T. (2002). Gene flow and the limits to natural selection. Trends in Ecology and Evolution, 17, 183–189. [Google Scholar]
- Levinton, J. S. , Suatoni, E. , Wallace, W. , Junkins, R. , Kelaher, B. , & Allen, B. J. (2003). Rapid loss of genetically based resistance to metals after the cleanup of a Superfund site. Proceedings of the National Academy of Sciences, 100, 3595–3600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, S. , Li, B. , Cheng, C. , Xiong, Z. , Liu, Q. , Lai, J. , … Yan, J. (2014). Genomic signatures of near‐extinction and rebirth of the crested ibis and other endangered bird species. Genome Biology, 15, 557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindenmayer, D. B. , Piggott, M. P. , & Wintle, B. A. (2013). Counting the books while the library burns: Why conservation monitoring programs need a plan for action. Frontiers in Ecology and the Environment, 11, 549–555. [Google Scholar]
- Loreau, M. , & Hector, A. (2001). Partitioning selection and complementarity in biodiversity experiments. Nature, 412, 72–76. [DOI] [PubMed] [Google Scholar]
- Lowe, A. J. , Cavers, S. , Boshier, D. , Breed, M. F. , & Hollingsworth, P. M. (2015). The resilience of forest fragmentation genetics—no longer a paradox—we were just looking in the wrong place. Heredity, 115, 97–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luikart, G. , Sherwin, W. B. , Steele, B. M. , & Allendorf, F. W. (1998). Usefulness of molecular markers for detecting population bottlenecks via monitoring genetic change. Molecular Ecology, 7, 963–974. [DOI] [PubMed] [Google Scholar]
- Massimino, D. , Johnston, A. , & Pearce‐Higgins, J. W. (2015). The geographical range of British birds expands during 15 years of warming. Bird Study, 62, 523–534. [Google Scholar]
- May, R. M. (1973). Stability and complexity in model exosystems. Princeton, NJ: Princeton University Press. [Google Scholar]
- McKinney, M. , & Lockwood, J. (1999). Biotic homogenization: A few winners replacing many losers in the next mass extinction. Trends in Ecology & Evolution, 14, 450–453. [DOI] [PubMed] [Google Scholar]
- McLachlan, J. S. , Hellmann, J. J. , & Schwartz, M. W. (2007). A framework for debate of assisted migration in an era of climate change. Conservation Biology, 21, 297–302. [DOI] [PubMed] [Google Scholar]
- Merilä, J. , & Hendry, A. P. (2014). Climate change, adaptation, and phenotypic plasticity: The problem and the evidence. Evolutionary Applications, 7, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimura, M. , Mishima, M. , Lascoux, M. , & Yahara, T. (2014). Range shift and introgression of the rear and leading populations in two ecologically distinct Rubus species. BMC Evolutionary Biology, 14, 209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miraldo, A. , Li, S. , Borregaard, M. K. , Flórez‐Rodríguez, A. , Gopalakrishnan, S. , Rizvanovic, M. , Wang, Z. , ... Nogués‐Bravo, D . (2016) An Anthropocene map of genetic diversity. Science, 353, 1532–1535. [DOI] [PubMed] [Google Scholar]
- Mooney, H. A. , & Cleland, E. E. (2001). The evolutionary impact of invasive species. Proceedings of the National Academy of Sciences of the United States of America, 98, 5446–5451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore, J. W. , Yeakel, J. D. , Peard, D. , Lough, J. , & Beere, M. (2014). Life‐history diversity and its importance to population stability and persistence of a migratory fish: Steelhead in two large North American watersheds. Journal of Animal Ecology, 83, 1035–1046. [DOI] [PubMed] [Google Scholar]
- Morinaga, S.‐I. , Iwasaki, T. , & Suyama, Y. (2014). Eco‐evolutionary genomic observation for local and global environmental changes In Nakano S., Yahara T., & Nakashizuka T. (Eds.), Asia‐Pacific biodiversity observation network – integrative observation and assessment. NY: Springer. [Google Scholar]
- Morlon, H. , Schwilk, D. W. , Bryant, J. A. , Marquet, P. A. , Rebelo, A. G. , Tauss, C. , … Green, J. L. (2011). Spatial patterns of phylogenetic diversity. Ecology Letters, 14, 141–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munns, R. , & Tester, M. (2008). Mechanisms of salinity tolerance. Annual Review of Plant Biology, 59, 651–681. [DOI] [PubMed] [Google Scholar]
- Nei, M. (1978). Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics, 89, 583–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei, M. , Maruyama, T. , & Chakraborty, R. (1975). The Bottleneck effect and genetic variability in natural populations. Evolution, 29, 1–10. [DOI] [PubMed] [Google Scholar]
- Neto, E. D. C. , de Oliveira, V. M. , Rosas, A. , & Campos, P. R. A. (2011). The effect of spatially correlated environments on genetic diversity‐area relationships. Journal of Theoretical Biology, 288, 57–65. [DOI] [PubMed] [Google Scholar]
- Núñez‐Farfán, J. , Fornoni, J. , & Valverde, P. L. (2007). The evolution of resistance and tolerance to herbivores. Annual Review of Ecology, Evolution, and Systematics, 38, 541–566. [Google Scholar]
- Olsen, E. M. , Carlson, S. M. , Gjøsaeter, J. , & Stenseth, N. C. (2009). Nine decades of decreasing phenotypic variability in Atlantic cod. Ecology Letters, 12, 622–631. [DOI] [PubMed] [Google Scholar]
- Olsen, E. M. , Lilly, G. R. , Heino, M. , Morgan, M. J. , Brattey, J. , & Dieckmann, U. (2005). Assessing changes in age and size at maturation in collapsing populations of Atlantic cod (Gadus morhua). Canadian Journal of Fisheries and Aquatic Sciences, 823, 811–823. [Google Scholar]
- Oyler‐McCance, S. J. , Fedy, B. C. , & Landguth, E. L. (2012). Sample design effects in landscape genetics. Conservation Genetics, 14, 275–285. [Google Scholar]
- Parker, P. G. , Snow, A. A. , Schug, M. D. , Booton, G. C. , & Fuerst, P. A. (1998). What molecules can tell us about populations: Choosing and using a molecular marker. Ecology, 79, 361–382. [Google Scholar]
- Parmesan, C. (2006). Ecological and evolutionary responses to recent climate change. Annual Review of Ecology, Evolution, and Systematics, 37, 637–669. [Google Scholar]
- Pauls, S. U. , Nowak, C. , Bálint, M. , & Pfenninger, M. (2013). The impact of global climate change on genetic diversity within populations and species. Molecular Ecology, 22, 925–946. [DOI] [PubMed] [Google Scholar]
- Pereira, H. M. , & David Cooper, H. (2006). Towards the global monitoring of biodiversity change. Trends in Ecology and Evolution, 21, 123–129. [DOI] [PubMed] [Google Scholar]
- Pespeni, M. H. , Sanford, E. , Gaylord, B. , Hill, T. M. , Hosfelt, J. D. , Jaris, H. K. , … Palumbi, S. R. (2013). Evolutionary change during experimental ocean acidification. Proceedings of the National Academy of Sciences, 110, 6937–6942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson, B. K. , Weber, J. N. , Kay, E. H. , Fisher, H. S. , & Hoekstra, H. E. (2012). Double digest RADseq: An inexpensive method for de novo SNP discovery and genotyping in model and non‐model species. PLoS ONE, 7, e37135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfenninger, M. , Bálint, M. , & Pauls, S. U. (2012). Methodological framework for projecting the potential loss of intraspecific genetic diversity due to global climate change. BMC Evolutionary Biology, 12, 224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillimore, A. B. , Had, J. D. , Jones, O. R. , & Smithers, R. J. (2012). Differences in spawning date between populations of common frog reveal local adaptation. Proceedings of the National Academy of Sciences, 109, 5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pigeon, G. , Festa‐Bianchet, M. , Coltman, D. W. , & Pelletier, F. (2016). Intense selective hunting leads to artificial evolution in horn size. Evolutionary Applications, 9, 521–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plucknett, D. L. , Smith, N. J. H. , Williams, J. T. , & Anishetty, N. M. (1987). Gene Banks and the world's food. Princeton, NJ: Princeton University Press. [Google Scholar]
- Pörtner, H. O. , & Knust, R. (2007). Climate change affects marine fishes through the oxygen limitation of thermal tolerance. Science, 315, 95–97. [DOI] [PubMed] [Google Scholar]
- Prentis, P. J. , Wilson, J. R. U. , Dormontt, E. E. , Richardson, D. M. , & Lowe, A. J. (2008). Adaptive evolution in invasive species. Trends in Plant Science, 13, 288–294. [DOI] [PubMed] [Google Scholar]
- Prieto, I. , Violle, C. , Barre, P. , Durand, J. , Ghesquiere, M. , & Litrico, I. (2015). Complementary effects of species and genetic diversity on productivity and stability of sown grasslands. Nature Plants, 1, 15033. [DOI] [PubMed] [Google Scholar]
- Prunier, J. G. , Kaufmann, B. , Fenet, S. , Picard, D. , Pompanon, F. , Joly, P. , & Lena, J. P. (2013). Optimizing the trade‐off between spatial and genetic sampling efforts in patchy populations: Towards a better assessment of functional connectivity using an individual‐based sampling scheme. Molecular Ecology, 22, 5516–5530. [DOI] [PubMed] [Google Scholar]
- Puzey, J. , & Vallejo‐Marín, M. (2014). Genomics of invasion: Diversity and selection in introduced populations of monkeyflowers (Mimulus guttatus). Molecular Ecology, 23, 4472–4485. [DOI] [PubMed] [Google Scholar]
- Pyšek, P. , & Richardson, D. M. (2010). Invasive species, environmental change and management, and health. Annual Review of Environment and Resources, 35, 25–55. [Google Scholar]
- Rauch, E. M. , & Bar‐Yam, Y. (2004). Theory predicts the uneven distribution of genetic diversity within species. Nature, 431, 449–452. [DOI] [PubMed] [Google Scholar]
- Ray, N. , Currat, M. , Foll, M. , & Excoffier, L. (2010). SPLATCHE2: A spatially explicit simulation framework for complex demography, genetic admixture and recombination. Bioinformatics, 26, 2993–2994. [DOI] [PubMed] [Google Scholar]
- Réale, D. , McAdam, A. G. , Boutin, S. , & Berteaux, D. (2003). Genetic and plastic responses of a northern mammal to climate change. Proceedings of the Royal Society B‐Biological Sciences, 270, 591–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed, D. H. , & Frankham, R. (2001). How closely correlated are molecular and quantitative measures of genetic variation? A meta‐analysis. Evolution, 55, 1095–1103. [DOI] [PubMed] [Google Scholar]
- Reusch, T. B. H. , Ehlers, A. , Hämmerli, A. , & Worm, B. (2005). Ecosystem recovery after climatic extremes enhanced by genotypic diversity. Proceedings of the National Academy of Sciences, 102, 2826–2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reznick, D. N. , & Ghalambor, C. K. (2001). The population ecology of contemporary adaptations: What empirical studies reveal about the conditions that promote adaptive evolution. Genetica, 112–113, 183–198. [PubMed] [Google Scholar]
- Rochlin, I. , Ninivaggi, D. V. , Hutchinson, M. L. , & Farajollahi, A. (2013). Climate change and range expansion of the Asian Tiger Mosquito (Aedes albopictus) in Northeastern USA: Implications for public health practitioners. PLoS ONE, 8, 60874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabeti, P. C. , Reich, D. E. , Higgins, J. M. , Levine, H. Z. P. , Richter, D. J. , Schaffner, S. F. , … Lander, E. S. (2002). Detecting recent positive selection in the human genome from haplotype structure. Nature, 419, 832–837. [DOI] [PubMed] [Google Scholar]
- Santamaría, L. , & Mendez, P. (2012). Evolution in biodiversity policy–current gaps and future needs. Evolutionary Applications, 5, 202–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satake, A. , Kawagoe, T. , Saburi, Y. , Chiba, Y. , Sakurai, G. , & Kudoh, H. (2013). Forecasting flowering phenology under climate warming by modelling the regulatory dynamics of flowering‐time genes. Nature Communications, 4, 2303. [DOI] [PubMed] [Google Scholar]
- Schindler, D. E. , Armstrong, J. B. , & Reed, T. E. (2015). The portfolio concept in ecology and evolution. Frontiers in Ecology and the Environment, 13, 257–263. [Google Scholar]
- Schindler, D. E. , Hilborn, R. , Chasco, B. , Boatright, C. P. , Quinn, T. P. , Rogers, L. A. , & Webster, M. S. (2010). Population diversity and the portfolio effect in an exploited species. Nature, 465, 609–612. [DOI] [PubMed] [Google Scholar]
- Schluter, D. (2000). The ecology of adaptative radiation. Oxford: Oxford University Press. [Google Scholar]
- Schrader, L. , Kim, J. W. , Ence, D. , Zimin, A. , Klein, A. , Wyschetzki, K. , … Oettler, J. (2014). Transposable element islands facilitate adaptation to novel environments in an invasive species. Nature Communications, 5, 5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz, M. K. , Luikart, G. , & Waples, R. S. (2007). Genetic monitoring as a promising tool for conservation and management. Trends in Ecology & Evolution, 22, 25–33. [DOI] [PubMed] [Google Scholar]
- Seehausen, O. (1997). Cichlid fish diversity threatened by eutrophication that curbs sexual selection. Science, 277, 1808–1811. [Google Scholar]
- Seehausen, O. , Takimoto, G. , Roy, D. , & Jokela, J. (2008). Speciation reversal and biodiversity dynamics with hybridization in changing environments. Molecular Ecology, 17, 30–44. [DOI] [PubMed] [Google Scholar]
- Shafer, A. B. A. , Wolf, J. B. W. , Alves, P. C. , Bergström, L. , Bruford, M. W. , Brännström, I. , … Zieliński, P. (2015). Genomics and the challenging translation into conservation practice. Trends in Ecology and Evolution, 30, 78–87. [DOI] [PubMed] [Google Scholar]
- Siefert, A. , Violle, C. , Chalmandrier, L. , Albert, C. H. , Taudiere, A. , Fajardo, A. , … Wardle, D. A. (2015). A global meta‐analysis of the relative extent of intraspecific trait variation in plant communities. Ecology Letters, 18, 1406–1419. [DOI] [PubMed] [Google Scholar]
- Slatkin, M. (1987). Gene flow and the geographic structure of natural populations. Science, 236, 787–792. [DOI] [PubMed] [Google Scholar]
- Smith, G. F. , & Steyn, E. M. A. (2005). Notes on the phenology, natural geographical distribution range and taxonomy of Aloe dichotoma (Aloaceae). Bradleya, 23, 17–22. [Google Scholar]
- Spencer, C. C. , Neigel, J. E. , & Leberg, P. L. (2000). Experimental evaluation of the usefulness of microsatellite DNA for detecting demographic bottlenecks. Molecular Ecology, 9, 1517–1528. [DOI] [PubMed] [Google Scholar]
- Stockwell, C. A. , Hendry, A. P. , & Kinnison, M. T. (2003). Contemporary evolution meets conservation biology. Trends in Ecology & Evolution, 18, 94–101. [Google Scholar]
- Strauss, S. Y. , Lau, J. A. , & Carroll, S. P. (2006). Evolutionary responses of natives to introduced species: What do introductions tell us about natural communities? Ecology Letters, 9, 357–374. [DOI] [PubMed] [Google Scholar]
- Sunnucks, P. (2000). Efficient genetic markers for population biology. Trends in Ecology & Evolution, 15, 199–203. [DOI] [PubMed] [Google Scholar]
- Suyama, Y. , & Matsuki, Y. (2015). MIG‐seq: An effective PCR‐based method for genome‐wide single‐nucleotide polymorphism genotyping using the next‐generation sequencing platform. Scientific Reports, 5, 16963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swain, D. P. , Sinclair, A. F. , & Mark Hanson, J. (2007). Evolutionary response to size‐selective mortality in an exploited fish population. Proceedings of the Royal Society B‐Biological Sciences, 274, 1015–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, C. D. , Cameron, A. , Green, R. E. , Bakkenes, M. , Beaumont, L. J. , Collingham, Y. C. , … Williams, S. E. (2004). Extinction risk from climate change. Nature, 427, 145–148. [DOI] [PubMed] [Google Scholar]
- Tilman, D. , Fargione, J. , Wolff, B. , D'Antonio, C. , Dobson, A. , Howarth, R. , … Swackhamer, D. (2001). Forecasting agriculturally driven global environmental change. Science, 292, 281–284. [DOI] [PubMed] [Google Scholar]
- Vázquez‐Domínguez, E. , Piñero, D. , & Ceballos, G. (1999). Linking heterozygosity, demography, and fitness of tropical populations of Liomys pictus . Journal of Mammalogy, 80, 810–822. [Google Scholar]
- Vázquez‐Domínguez, E. , Suárez‐Atilano, M. , Booth, W. , González‐Baca, C. , & Cuarón, A. D. (2012). Genetic evidence of a recent successful colonization of introduced species on islands: Boa constrictorimperator on Cozumel Island. Biological Invasions, 14, 2101–2116. [Google Scholar]
- de Villemereuil, P. , & Gaggiotti, O. E. (2015). A new F ST‐based method to uncover local adaptation using environmental variables. Methods in Ecology and Evolution, 6, 1248–1258. [Google Scholar]
- Visscher, P. M. , Hill, W. G. , & Wray, N. R. (2008). Heritability in the genomics era‐concepts and misconceptions. Nature Reviews Genetics, 9, 255–266. [DOI] [PubMed] [Google Scholar]
- Visser, M. E. (2008). Keeping up with a warming world; assessing the rate of adaptation to climate change. Proceedings of the Royal Society B‐Biological Sciences, 275, 649–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitti, J. J. , Grossman, S. R. , & Sabeti, P. C. (2013). Detecting natural selection in genomic data. Annual Review of Genetics, 47, 97–120. [DOI] [PubMed] [Google Scholar]
- Vonlanthen, P. , Bittner, D. , Hudson, A. G. , Young, K. A. , Müller, R. , Lundsgaard‐Hansen, B. , … Seehausen, O. (2012). Eutrophication causes speciation reversal in whitefish adaptive radiations. Nature, 482, 357–362. [DOI] [PubMed] [Google Scholar]
- Wandeler, P. , Hoeck, P. E. A. , & Keller, L. F. (2007). Back to the future: Museum specimens in population genetics. Trends in Ecology & Evolution, 22, 634–642. [DOI] [PubMed] [Google Scholar]
- Wasserman, T. N. , Cushman, S. A. , Littell, J. S. , Shirk, A. J. , & Landguth, E. L. (2013). Population connectivity and genetic diversity of American marten (Martes americana) in the United States northern Rocky Mountains in a climate change context. Conservation Genetics, 14, 529–541. [Google Scholar]
- Wasserman, T. N. , Cushman, S. A. , Schwartz, M. K. , & Wallin, D. O. (2010). Spatial scaling and multi‐model inference in landscape genetics: Martes americana in northern Idaho. Landscape Ecology, 25, 1601–1612. [Google Scholar]
- White, T. A. , Perkins, S. E. , Heckel, G. , & Searle, J. B. (2013). Adaptive evolution during an ongoing range expansion: The invasive bank vole (Myodes glareolus) in Ireland. Molecular Ecology, 22, 2971–2985. [DOI] [PubMed] [Google Scholar]
- Whitlock, M. C. , & Mccauley, D. E. (1999). Indirect measures of gene flow and migration: FST≠1/(4Nm+1). Heredity, 82, 117–125. [DOI] [PubMed] [Google Scholar]
- Wilson, G. A. , & Rannala, B. (2003). Bayesian inference of recent migration rates using multilocus genotypes. Genetics, 163, 1177–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, A. J. , Réale, D. , Clements, M. N. , Morrissey, M. M. , Postma, E. , Walling, C. A. , … Nussey, D. H. (2010). An ecologist's guide to the animal model. The Journal of Animal Ecology, 79, 13–26. [DOI] [PubMed] [Google Scholar]
- Wright, S. (1920). The relative importance of heredity and environment in determining the piebald pattern of guinea‐pigs. Proceedings of the National Academy of Sciences of the United States of America, 6, 320–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zytynska, S. E. , Fay, M. F. , Penney, D. , & Preziosi, R. F. (2011). Genetic variation in a tropical tree species influences the associated epiphytic plant and invertebrate communities in a complex forest ecosystem. Philosophical Transactions of the Royal Society B‐Biological Sciences, 366, 1329–1336. [DOI] [PMC free article] [PubMed] [Google Scholar]


