Summary
Palmitic acid (C16:0) already makes up approximately 25% of the total fatty acids in the conventional cotton seed oil. However, further enhancements in palmitic acid content at the expense of the predominant unsaturated fatty acids would provide increased oxidative stability of cotton seed oil and also impart the high melting point required for making margarine, shortening and confectionary products free of trans fatty acids. Seed‐specific RNAi‐mediated down‐regulation of β‐ketoacyl‐ACP synthase II (KASII) catalysing the elongation of palmitoyl‐ACP to stearoyl‐ACP has succeeded in dramatically increasing the C16 fatty acid content of cotton seed oil to well beyond its natural limits, reaching up to 65% of total fatty acids. The elevated C16 levels were comprised of predominantly palmitic acid (C16:0, 51%) and to a lesser extent palmitoleic acid (C16:1, 11%) and hexadecadienoic acid (C16:2, 3%), and were stably inherited. Despite of the dramatic alteration of fatty acid composition and a slight yet significant reduction in oil content in these high‐palmitic (HP) lines, seed germination remained unaffected. Regiochemical analysis of triacylglycerols (TAG) showed that the increased levels of palmitic acid mainly occurred at the outer positions, while C16:1 and C16:2 were predominantly found in the sn‐2 position in both TAG and phosphatidylcholine. Crossing the HP line with previously created high‐oleic (HO) and high‐stearic (HS) genotypes demonstrated that HP and HO traits could be achieved simultaneously; however, elevation of stearic acid was hindered in the presence of high level of palmitic acid.
Keywords: palmitic acid, KASII, cotton seed oil, RNAi, fatty acids, TAG
Introduction
Cottonseed oil is a highly polyunsaturated vegetable oil because more than half of its total fatty acid content is linoleic acid (C18:2Δ9,12) that is oxidatively unstable and makes it unsuitable for direct food applications (Jones and King, 1993; O'Brien, 2002). Partial hydrogenation that converts much of linoleic acid into monounsaturated and saturated fatty acids has often been used for whole‐food applications, such as in providing hard stocks with high melting point for margarine and shortening production. However, trans fatty acids are commonly produced as a by‐product of the partial hydrogenation process and have been increasingly recognised to have significant cholesterol‐raising properties and increase the risk of cardiovascular disease based on evidence derived from epidemiologic and clinical studies (Mozaffarian et al., 2006; Oomen et al., 2001; de Souza et al., 2015). A recent systematic review and meta‐analysis of observational studies in the past 10 years, including 41 studies of the association between saturated fat intake and health outcomes, covering more than 300 000 people, and 20 studies of trans fat intake and health outcomes that covered more than 200 000 people, have found positive associations between consumption of trans fat and total coronary heart disease (CHD) and fatal CHD, but not between consumption of saturated fat and CHD, cardiovascular disease (CVD), stroke and type 2 diabetes (de Souza et al., 2015). Although further research on saturated fatty acids is required, there is no doubt that the decades trading saturated fats for trans fat have had an enormous influence on the rising incidences of heart disease and many of the so‐called metabolic syndromes. Alternative cotton seed oils with similar functionality to partially hydrogenated oil could therefore be nutritionally desirable.
Cotton seed oil rich in oxidatively stable fatty acids, such as oleic acid (C18:1Δ9), stearic acid (C18:0) and palmitic acid (C16:0), would meet this purpose. Palmitic acid makes up approximately 25% of the total fatty acids in the conventional cotton seed oil, and its further enhancement is anticipated to not only increase the oxidative stability of cotton seed oil by offsetting the instability of linoleic acid but also impart the high melting point required for making such products as margarine, shortening and confectionary products free of trans fatty acids (Neff and List, 1999; Nzikou et al., 2009).
Tropical oils such as palm oil contain about 50% of palmitic acid, which is increasingly being used as an alternative to partially hydrogenated vegetable oils in baking and processed food applications (Hayes and Pronczuk, 2010; L'Abbe et al., 2009). The mid‐fraction of a palm oil (PMF) rich in symmetrical palmitate triglycerides (POP) has also been most commonly used to formulate cocoa butter substitute by interesterification with oils derived from stearate‐rich tropical oils (L'Abbe et al., 2009; Zaliha et al., 2004). Oil palm plantings have continued to expand in recent years, in response to rapidly increasing demand for food oil, as well as for biofuel. As the conversion of virgin agricultural land to palm plantation in tropical countries has become an ever more sensitive issue, development of cotton seed oil with similar fatty acid composition and functionality to palm oil may offer an attractive alternative because of its continuous availability that is driven by the continuing demand for cotton fibre.
In higher plants, the plastid is the major site of de novo fatty acid biosynthesis pathway, generating fatty acyl precursors for fatty acids of different chain lengths and saturation levels. The de novo fatty acid biosynthesis is performed by a complex of soluble proteins known as fatty acid synthases (FAS), with the β‐ketoacyl‐ACP synthase (KAS) enzyme family catalysing the elongation of malonyl‐acyl carrier protein (malonyl‐ACP) by reiteratively adding C2 units to a growing fatty acyl chain through Claisen condensation (Ohlrogge and Jaworski, 1997). KASIII catalyses the condensation of C2‐CoA to C4, while KASI prefers C4‐ to C14‐ACP substrates leading to the production of palmitoyl‐ACP that is then elongated to stearoyl‐ACP by KASII and subsequently desaturated to form oleoyl‐ACP by Δ9 stearoyl‐ACP desaturase (SAD). The saturated fatty acids, mostly palmitic acid in cotton seed, can be cleaved from palmitoyl‐ACP by the action of the palmitoyl‐ACP thioesterase (FatB), allowing for transportation of free palmitic acid into cytoplasm where it becomes available for further desaturation and triacylglycerol (TAG) assembly.
Because palmitoyl‐ACP is the substrate for two major activities, KASII and FatB, it represents a key branch point in fatty acid biosynthesis (Cahoon and Shanklin, 2000). Most fatty acids in the seed oil pass through a C16 form during biosynthesis, and the content of palmitic acid remaining in the final cotton seed oil is therefore determined, to a large extent, by the competing action of FatB and KASII (Cahoon and Shanklin, 2000; Martz et al., 2006). These enzymes therefore represent possible targets for genetic manipulation of palmitic acid levels in cotton seed oil (Aslan et al., 2015; Pidkowich et al., 2007; Sun et al., 2014).
Genetic engineering of FatB has previously been successful in raising or lowering palmitic acid levels in seeds of other plants. Overexpression of the Arabidopsis FatB1 in seeds resulted in a nearly four fold increase in seed palmitic acid content (Dormann et al., 2000). Transgenic expression of a FatB gene derived from Cuphea hookerinana resulted in raised palmitic acid from 6% to 34% in rapeseed oil (Dehesh et al., 1996). An even more effective approach for raising palmitic acid was demonstrated by RNAi down‐regulation of KAS2 that encodes KASII in Arabidopsis where palmitic acid level was raised to as high as 53% beyond which level the seeds were aborted (Pidkowich et al., 2007). It is assumed that the significantly lowered KASII activity resulting from the attenuated KAS2 transcription reduces the flow of metabolites from palmitoyl‐ACP to stearoyl‐ACP, causing a significant accumulation of the palmitoyl‐ACP and consequently enriched palmitic acid content in TAG.
We have employed a similar approach to raise palmitic acid in cotton seed oil. In this report, we describe the characterisation of two different KAS2 genes from developing cotton embryos and their RNAi down‐regulation that led to substantial enhancement of palmitic acid accumulation. Attempts have also been made to combine the high‐palmitic (HP) trait by crossing with either or both high‐oleic (HO) and high‐stearic (HS) traits that have been previously generated using RNAi‐mediated gene down‐regulation of ghFAD2‐1 or ghSAD‐1, respectively (Liu et al., 2002).
Results
Isolation of two different KAS2 cDNAs from developing cotton seed
From a cDNA library derived from developing embryos of upland cotton (Gossypium hirsutum L.) cv DP‐16, two distinct cDNAs were isolated and designated as ghKAS2‐1 (GenBank accession No. KF611921) and ghKAS2‐2 (GenBank accession No. KF611922). Full length sequences of these cDNAs were completed by 5′ race using RNAs derived from developing embryos as templates. The complete ghKAS2‐1 is 2268 bp long, encoding 577 amino acids. The ghKAS2‐2 cDNA is 2192 bp long, encoding 526 amino acids. The putative polypeptides encoded by the coding region of ghKAS2‐1 and ghKAS2‐2 showed 78.7% identity to each other. The phylogenetic relationship of these two genes and their orthologs in a selected number of plant species is illustrated in Fig. 1. It appears that ghKAS2‐1 and ghKAS2‐2 clustered together with their ortholog derived from Theobroma cacao that is taxonomically closely related to cotton. By searching the G. raimondii genome database, ghKAS2‐1 was mapped onto chromosome 13 and its coding region was interrupted by 12 introns and the 13th intron was found in the 3′ UTR. ghKAS2‐2 was mapped onto chromosome 4, and its coding region was interrupted by 12 introns. The position and length of introns were relatively conserved between ghKAS2‐1 and ghKAS2‐2. It is noteworthy that about half of the ghKAS2‐1 cDNA clones isolated from the cotton developing embryo cDNA library contained intron 12 that was not properly spliced. It is not known whether this constitutes one level of post‐transcriptional regulation and is responsible for the higher level of palmitic acid in cotton seed relative to many other temperate oilseed crops.
Figure 1.
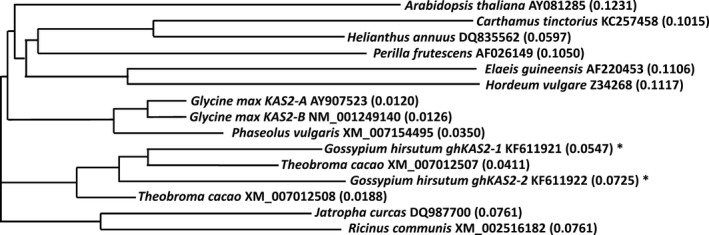
Phylogenetic comparison of cotton ghKAS2 gene family and orthologous KAS2s from some selected plant species. The phylogenetic tree was constructed from multiple sequence alignment of KASII protein from the GenBank generated by the AlignX module of Vector NTI suite using neighbour‐joining clustering algorithm in Clustal X2 program. The GenBank accession numbers and the sources of KAS2 genes and bootstrap values of the clusters are indicated in phylogram. The sequences of ghKASII from this study are marked with an asterisk.
RNAi construct designing and selection of primary transgenic cotton plants
As detailed in the Experimental procedures, a chimeric DNA sequence consisting of an inverted repeat structure of ghKAS2‐1 was used as the trigger DNA sequence in an RNAi cassette. Instead of a typical intron sequence, the upstream adjacent sequence of the 400‐bp sense arm of the inverted repeat was used as a spacer separating the head to head inverted repeat. Although the trigger sequence was designed based on the ghKAS2‐1, it was chosen as the most conserved region of the gene. The trigger sequence has 87.5% homology with ghKAS2‐2, with several stretches of continuous homologous regions longer than 21 bp, enabling it to achieve a level of cross‐silencing of ghKAS2‐2. A seed‐specific promoter derived from soya bean lectin gene LEC1 (Cho et al., 1995) was used to drive the transgene. Six independent cotton lines transformed with the RNAi‐ghKAS2‐1 construct were regenerated and allowed to grow to maturity. Four of the six lines were male sterile, a much higher frequency than seen in other typical cotton transformations in our laboratory, suggesting a possible impact of severe KAS2 down‐regulation rather than a side effect of tissue culture. The other two fertile lines, KIR‐1 and KIR‐10, were carried through for further molecular and biochemical analyses. PCR was first performed to confirm the transgene presence, followed by Southern blot analysis with multiple restriction enzymes on the T1 primary transgenic plants using the promoter region of the transgene as a probe. Single restriction bands were observed in both KIR‐1 and KIR‐10, indicating single transgene insertions (data not shown). Under glasshouse conditions, neither of these two lines exhibited any growth abnormalities nor was there any apparent penalty on plant vigour or seed yield.
Analysis of fatty acid composition was carried out along with the selection and establishment of homozygous transgenic plants. In comparison with wild type (WT) and null segregants, fatty acid profiles of the transgenic seeds derived from KIR‐1 and KIR‐10 have altered significantly, featuring enhanced C16 fatty acids with concomitant reduction in C18 fatty acids. Fig. 2 shows the content of palmitic acid and C16 unsaturated fatty acids in the cotyledons of 65 and 51 randomly selected individual T2 seeds in KIR‐1 and KIR‐10, respectively. Among the T2 seeds, the segregation of null seeds is evident as they have similar fatty acid composition to WT plants, while all other seeds showed raised palmitic acid at various levels, with the majority exceeding 50% of total fatty acids in both transgenic lines. The ratios of transgenic seeds to null segregants for KIR‐1 (54 : 11) and KIR‐10 (49 : 9) fitted a 3 : 1 ratio expected for the segregation of a single dominant gene (x 2 = 0.19 and 0.10, respectively), consistent with the molecular assessment of single copy transgenes as confirmed by Southern blot analysis. In KIR‐1, a single seed with the highest accumulation of total C16 fatty acids reached 85% of total fatty acids. Palmitoleic acid was raised to as high as 11% of total fatty acids. C16:2 that is normally undetectable in cotton seed accounted for up to 3.3% of total fatty acids. Compared to KIR‐1, the extent of increase in palmitic acid and unsaturated C16 fatty acid levels in KIR‐10 seeds is relatively consistent among the transgenic T2 seeds. Most of the transgenic seeds showing palmitic acid level between 45% and 50%. Similar to KIR‐1, the sum of palmitoleic acid and C16:2 was also increased to about 15% of total fatty acids in individual KIR‐10 seeds. As the result, the contents of C18 fatty acids were all reduced, except for cis‐vaccenic acid (C18:1Δ11) that is the elongation product of palmitoleic acid. There is an approximate doubling of cis‐vaccenic acid content in both KIR‐1 and KIR‐10 lines compared to untransformed control.
Figure 2.
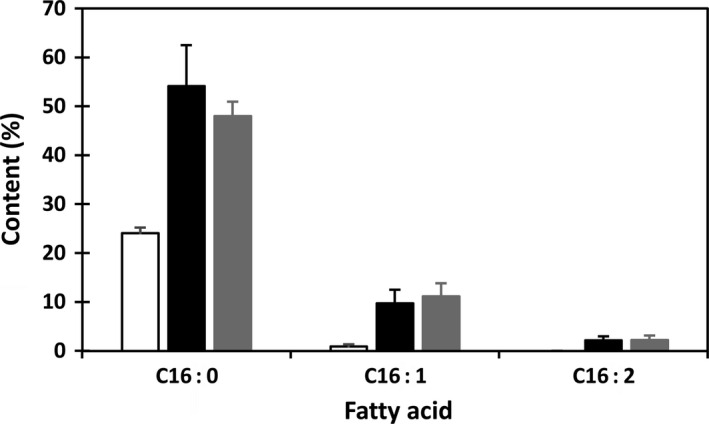
Accumulation of C16 fatty acids in the primary transgenic plants of transgenic lines KIR‐1 and KIR‐10. WT (open box); KIR‐1 (closed box) and KIR‐10 (shaded box).
The T2 seeds with significantly raised palmitic acid were selected and germinated for plant establishment and assessment of the inheritance stability of the gained HP trait in the progenies of both KIR‐1 and KIR‐10. Fifteen individual mature T3 seeds borne on each of the established T2 plants were subjected to analysis of fatty acid composition. There were clear segregation of null seeds with WT‐like fatty acid profile among the T3 seeds, and the homozygous HP lines were able to be established at T4 generation for both KIR‐1 and KIR‐10.
The fatty acid profiles of T2 (null excluded), T3 and T4 seeds were tabulated in Table 1. In all the three generations, the increased contents of palmitic acid and its derivative C16 unsaturated fatty acids were clearly a prominent feature and consistent among generations. The increase of C16 fatty acids was at the expense of all three major C18 fatty acids, including stearic, oleic and linoleic acids. In KIR‐1, distinct from the high variability in primary transgenic seeds, the palmitic acid accumulation in T3–T4 generations was more consistent among individual seeds without significant variation.
Table 1.
Fatty acid composition of transgenic cotton expressing RNAi‐ghKAS2‐1 in three successive progeny generations
| Generation | Genotype | C14:0 | C16:0 | C16:1 | C16:2 | C18:0 | C18:1Δ9 | 18:1Δ11 | C18:2 |
|---|---|---|---|---|---|---|---|---|---|
| T2 | Null | 0.2 ± 0.1 | 24.1 ± 1.1 | 0.9 ± 0.4 | 0.0 ± 0.0 | 2.1 ± 0.6 | 13.3 ± 2.9 | 0.8 ± 0.6 | 58.1 ± 3.1 |
| KIR‐1 | 0.2 ± 0.1 | 54.1 ± 8.4 | 9.7 ± 2.7 | 2.2 ± 0.8 | 1.2 ± 0.5 | 3.5 ± 2.0 | 1.5 ± 0.4 | 26.8 ± 7.2 | |
| KIR‐10 | 0.2 ± 0.1 | 48.0 ± 2.9 | 11.2 ± 2.7 | 2.2 ± 0.9 | 1.0 ± 0.1 | 3.9 ± 1.4 | 1.7 ± 0.2 | 31.3 ± 4.8 | |
| T3 | KIR‐1 | 0.2 ± 0.1 | 51.0 ± 2.0 | 11.2 ± 2.1 | 2.8 ± 0.5 | 1.0 ± 0.0 | 2.3 ± 0.1 | 1.5 ± 0.0 | 29.2 ± 3.4 |
| KIR‐10 | 0.3 ± 0.1 | 49.5 ± 1.2 | 10.7 ± 2.3 | 2.1 ± 0.7 | 1.0 ± 0.1 | 3.7 ± 1.3 | 1.6 ± 0.1 | 30.4 ± 2.1 | |
| T4 | KIR‐1 | 0.2 ± 0.1 | 50.2 ± 1.7 | 10.9 ± 2.0 | 2.2 ± 0.7 | 1.1 ± 0.1 | 4.7 ± 1.8 | 1.8 ± 0.3 | 28.4 ± 2.8 |
| KIR‐10 | 0.2 ± 0.1 | 48.9 ± 1.8 | 11.8 ± 2.2 | 2.61 ± 0.7 | 1.1 ± 0.1 | 4.0 ± 1.6 | 1.7 ± 0.2 | 29.0 ± 2.5 |
Mean ± Std (n = 3).
Oil content analysis and seed germination test
Assessment of oil content was carried out in mature homozygous seeds harvested from the T4 and T5 HP lines and WT plants that were grown alongside each other under controlled greenhouse condition. Across the two generations, there was a slight yet significant decline of oil content in both HP lines (P < 0.05), while there is no significant variation between KIR‐1 and KIR‐10 (Table 2). Two‐way analysis of variance (ANOVA) revealed significant variation in oil content neither between generations nor interaction between genotype and generation (Table S1).
Table 2.
Oil content analysis of T4 and T5 cotton seeds and the germination rate of T4 cotton seeds at two different temperatures (18 °C and 28 °C)
| Genotype | Oil content (%) | Germination rate (%) | ||
|---|---|---|---|---|
| T4 | T5 | 18 °C | 28 °C | |
| Coker 315 | 22.8 ± 0.7a | 23.6 ± 0.5a | 81.7 ± 4.3a | 97.5 ± 3.2b |
| KIR‐1 | 21.3 ± 1.1b | 22.0 ± 0.1b | 80.0 ± 4.7a | 96.7 ± 2.7b |
| KIR‐10 | 21.3 ± 0.4b | 21.5 ± 0.4b | 83.3 ± 6.1a | 95.8 ± 3.2b |
Mean ± Std (oil content analysis, n = 3; germination test, n = 4). Columns with different letters represent significant differences (LSD) at p < 0.05.
Germination response to temperature of homozygous T4 seeds of KIR‐1 and KIR‐10 lines was evaluated at cool (18 °C) and warm (28 °C) temperature regimes. All the seeds derived from both WT and HP lines showed significantly higher germination rate at 28 °C compared to 18 °C, but there was no significant variation among cotton lines (P < 0.05; Table 2; Table S2).
Real‐time RT‐PCR gene expression
To confirm the molecular basis of the alteration in fatty acid composition in KIR‐1 and KIR‐10, real‐time quantitative PCR (RT‐qPCR) was performed to evaluate the transcription level of the target KAS2 gene using template RNAs derived from mid‐maturity developing embryos as previous studies have established that this is the most active period of fatty acid biosynthesis and oil accumulation (Liu et al., 1999). As shown in Fig. 3, the reduction in the expression of both ghKAS2‐1 and ghKAS2‐2 genes in the T4 homozygous transgenic lines was evident compared to WT control. It is also noteworthy that expression of ghKAS2‐1 appeared to be lower than ghKAS2‐2 in developing cotton seeds. This result is consistent with our observation that cDNA library screening yields 3 times more clones of ghKAS2‐2 than ghKAS2‐1. Taken together, we may assume that these two genes do not have equal contributions to the conversion of palmitic acid to stearic acid. Although the ghKAS2‐1 sequence alone was used as the RNAi trigger sequence, the expression of both ghKAS2‐1 and ghKAS2‐2 has been effectively attenuated.
Figure 3.
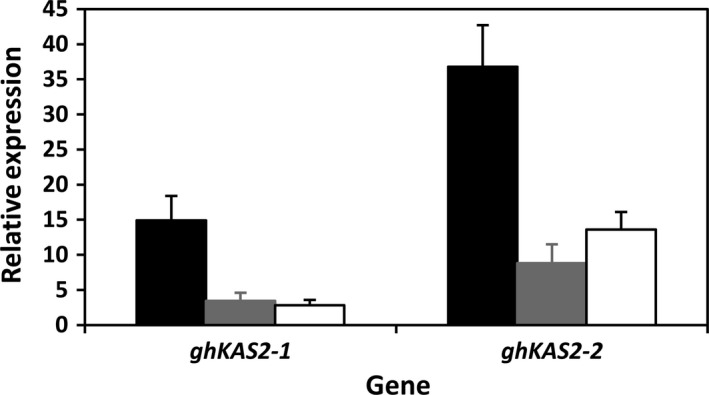
Real‐time RT‐qPCR analysis of ghKAS2‐1 and ghKAS2‐2 in immature cotton embryos at mid‐developmental stage (30 DAA). Coker315 (closed box); KIR‐1 (shaded box) and KIR‐10 (open box).
Lipid analysis of HP cotton seed oil
Palmitic acid content in the total lipids extracted from mature whole cotton seeds of KIR‐1 and KIR‐10 was measured as 52.6% and 50.1%, respectively, compared to 22.2% in WT (Fig. 4). In the TAG fraction, palmitic acid content in KIR‐1 and KIR‐10 seeds was 52.8% and 50.3% of total fatty acids, respectively. Similarly, palmitic acid content in the polar lipid (PL) fraction of KIR‐1 and KIR‐10 seed oil was 34.7% and 35.3%, respectively, compared to 21.6% in WT; palmitic acid content in the phosphatidylcholine (PC) fraction of KIR‐1 and KIR‐10 was 30.5% and 29.7%, respectively, in contrast to 19.5% in WT (Fig. 4). The increase in unsaturated C16 fatty acids was consistent among TAG, total PL and PC. Linoleic acid decreased with a concomitant increase in palmitic acid and other C16 fatty acids, more evident in TAG compared to total PL and PC. The relatively smaller alteration of fatty acid composition in total PL and PC might explain why there was little impact on seed germination and plant growth by RNAi down‐regulation of KAS2 expression in cotton seed.
Figure 4.
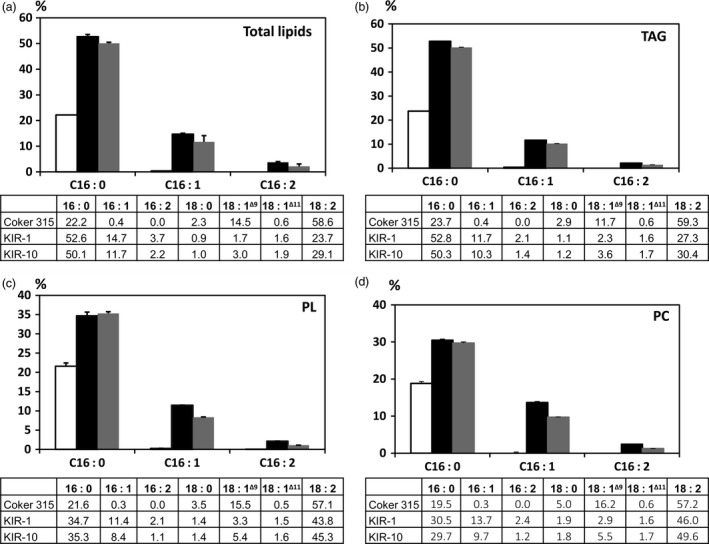
Fatty acid composition (%) of total lipids (a), TAG (b), PL (c) and PC (d). HP cotton seed oil derived from KIR‐1 (closed box) and KIR‐10 (shaded box) were compared to WT (Open box). Lipids were extracted from homozygous T4 seeds and fractionated by TLC to generate neutral and polar lipid fractions. Data are presented as mean ± SD (n = 3). The full fatty acid composition of cotton seed oil is tabulated below the bar diagram.
Positional analysis of palmitic acid
To determine the regiospecificity of palmitic acid and its two C16 derivative fatty acids, palmitoleic acid and C16:2, in KIR‐1 and KIR‐10, the TAG fraction was isolated from the homozygous T4 seeds and digested with appropriate lipase that preferentially cleaves fatty acid from the outer positions (sn‐1 and sn‐3), releasing two free fatty acids and a 2‐monoacylglycerol (2‐MAG) molecule retaining the sn‐2 acyl chain. In KIR‐1 and KIR‐10, consistent with WT, the outer positions of TAG were predominantly occupied by palmitic acid, while the sn‐2 position was mainly occupied by unsaturated fatty acids, mostly linoleic acid. Palmitoleic acid and C16:2 were found in both sn‐2 and outer positions, with some degree of preference for sn‐2 (Fig. 5). This was consistent with the general observation that, in plant seed oil, saturated fatty acids preferentially occupy the outer positions in TAG, whereas unsaturated fatty acids are predominantly found in sn‐2 position.
Figure 5.
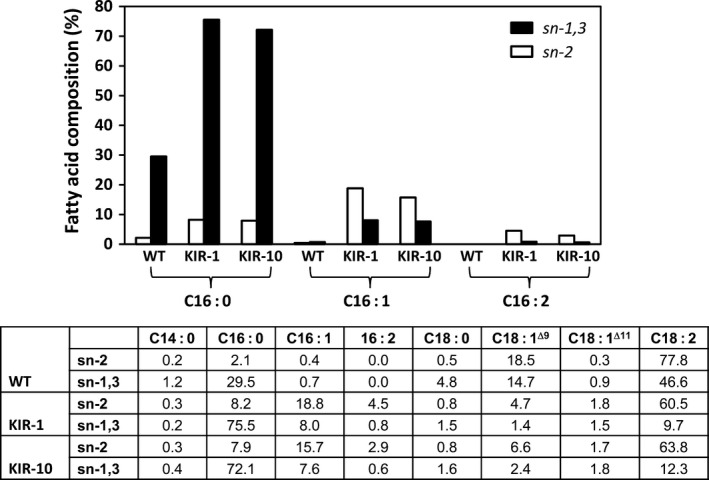
sn Positional distribution of fatty acids in TAG fraction of cotton seed oil derived from HP cotton lines, KIR‐1 and KIR10, compared to the untransformed control Coker 315 (WT). sn‐1,3 (closed box); sn‐2 (open box). The full fatty acid composition of the specified sn position is tabulated below the bar diagram.
For both HP lines, we also analysed the positional distribution of fatty acids on PC, an important intermediate in TAG synthesis and membrane constituents. The isolated PC fraction was digested with phospholipase A2, an enzyme that preferentially cleaves fatty acids from the sn‐2 of PC, releasing a free fatty acid and a lyso‐PC molecule retaining the sn‐1 acyl chain. This positional analysis showed that consistent with WT, the sn‐1 position was predominantly occupied by palmitic acid, while sn‐2 position was mainly occupied by unsaturated fatty acids (Fig. 6). Palmitoleic acid and C16:2 were found on both sn‐1 and sn‐2 positions, with a clear preference for sn‐2. It was also clear that palmitic acid in KIR‐1 and KIR‐10 was mainly found on sn‐1 position, while its increase on sn‐2 position was relatively minor.
Figure 6.
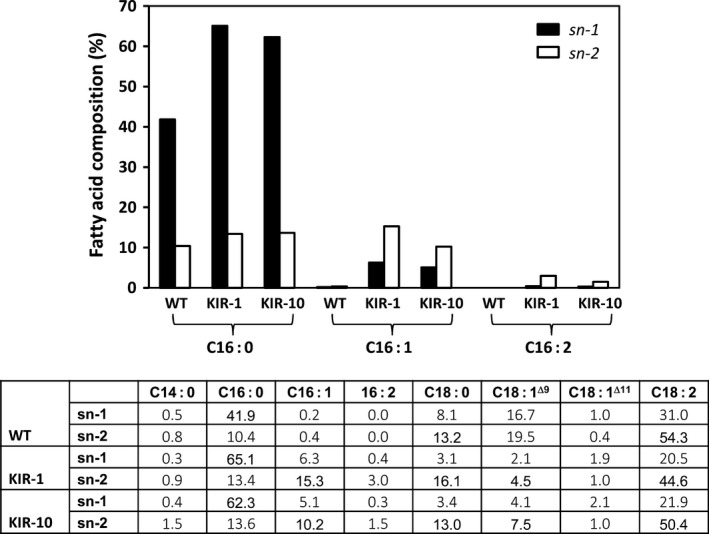
sn Positional distribution of fatty acids in PC fraction of cotton seed oil derived from HP cotton lines, KIR‐1 and KIR10, compared to the untransformed control Coker 315 (WT). sn‐1 (closed box); sn‐2 (open box). The full fatty acid composition of the specified sn position is tabulated below the bar diagram.
Incorporation of HP trait into HS, HO and HSO cotton genotypes
To incorporate the HP trait into some other genotypes with genetically altered fatty acid profiles, KIR‐10 was crossed with previously generated HO, HS lines and their homozygous hybrid (HSO) (Liu et al., 2002). The fatty acid profiles of the total lipids, TAG, PL and PC fractions in the F1 hybrid seeds derived from a cross between KIR‐10 and a HS line, HS‐35, are shown in Fig. 7. Palmitic acid in the F1 seeds was increased to a similar level as in its HP parent in the total lipids and TAG fraction, but its increase in PL and PC fractions was not as prominent. Correspondingly, the reduction in linoleic acid was also significantly more prominent in total lipids and TAG fraction than in polar lipids. In contrast to the high level accumulation of palmitic acid, the stearic acid level in all the four lipid pools in the HP x HS F1 seeds remains low, about 4%, which is significantly higher than WT, but much lower than its HS‐35 parent.
Figure 7.
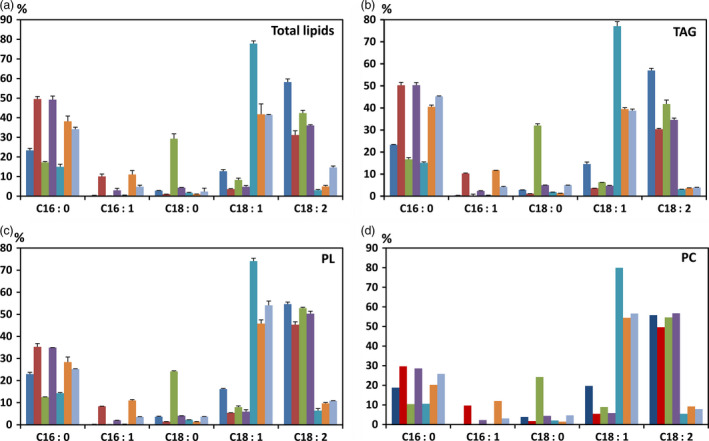
Fatty acid composition of total lipids (a), TAG (b), PL (c) and PC (d) in various cotton seed oil featuring Coker315 (blue), HP (red), HS (green), HP × HS (purple), HO (turquoise), HP × HO (orange), HP × (HS × HO) (grey) genotypes. Total lipids were fractionated by TLC to generate neutral and polar lipids. Data are presented as mean ± SD (n = 3).
The F1 hybrid seeds derived from a cross between KIR‐10 with a HO cotton line, HO‐30, showed the simultaneous increase of both palmitic and oleic acids, mainly at the expense of linoleic acid (Fig. 7). The average level of palmitic acid in the total seed lipids of the HP × HO cross was 41.7%, compared to 50% in the HP parent. The level of oleic acid in the HP × HO cross averaged 38.2% of total seed fatty acids, which in absolute terms is only half of the 78% oleic in the HO parent. However, in the context of the reduced pool of C18 fatty acids in the presence of the HP trait, the 38.2% of oleic acid does represent approx. 85% of the total C18 fatty acids, which is approaching the very high preponderance of oleic acid in the C18 fatty acid pool of the HO parent (94%). Reflecting the effect of FAD2 silencing, the linoleic acid content in the HP × HO hybrid appeared to be at a similar level to that in its HO parent, maintaining below 5%, while its HP parent contained 31.2% linoleic acid. Such a profile was consistent in the TAG, total PL and PC fractions. It is also worthy of noting that the ratio of palmitic/oleic acids was substantially lower in total PL and PC fraction, compared to total lipids and TAG fraction. Palmitoleic acid level remained at a similar level among the four lipid pools examined.
The hybrid (HPSO) was obtained by crossing KIR‐10 with a homozygous HO/HS cotton plant (HO/HS‐9) that was generated through crossing HO‐30 and HS‐35. In its total lipids, similar to what have been observed in the HP × HO and HP × HS, the combination of HP and HO traits was evident, but the stearic acid level was only moderately increased, averaging 3.4%, marginally, yet significantly higher than WT (2.8%). Linoleic acid remained low, about 5%, a similar level to the HO and HP × HO genotypes. Such a trend was consistent in the other three lipid pools, including TAG, PL and PC fractions. This hybrid also showed the common feature with HP and HPO genotypes that in PL and PC the ratio of palmitic acid vs linoleic acid was substantially lower than that in total lipids and TAG fraction.
Regiospecific distributions of fatty acids in the hybrids combining HP and other genotypes with altered fatty acids
The regiospecific distribution of fatty acids on TAG and PC molecules in the hybrids incorporating the HP trait into HS, HO and HSO lines was also analysed and shown in Tables 3 and 4, respectively. In the TAG of high saturate lines, that is HP, HS and HPS, the increases in palmitic acid and stearic acid were mainly concentrated on the sn‐1 and sn‐3 position (Table 3). This was consistent with PC except that stearic acid seemed to prefer sn‐2 in the WT, HP and HO lines where it was present at low levels (Table 4). A general preference of sn‐2 position by unsaturated fatty acids, including palmitoleic acid, C16:2, oleic acid and linoleic acid, was evident in both TAG (Table 3) and PC. However, oleic acid was found on all three sn positions when its accumulation reached very high level in HO‐30.
Table 3.
Positional distribution of fatty acids on TAG in different cotton genotypes containing altered fatty acid composition
| Sample | Position | C14:0 | C16:0 | C16:1 | 16:2 | C18:0 | C18:1Δ9 | C18:1Δ11 | C18:2 |
|---|---|---|---|---|---|---|---|---|---|
| Coker 315 | sn‐2 | 0.2 | 2.5 | 0.4 | 0.0 | 0.7 | 16.9 | 0.3 | 78.8 |
| sn‐1 + 3 | 1.0 | 36.1 | 0.5 | 0.1 | 5.3 | 11.7 | 0.8 | 42.2 | |
| KIR‐10 | sn‐2 | 0.3 | 7.9 | 15.7 | 2.9 | 0.8 | 6.6 | 1.7 | 63.8 |
| sn‐1 + 3 | 0.4 | 72.1 | 7.6 | 0.6 | 1.6 | 2.4 | 1.8 | 12.3 | |
| HS‐35 | sn‐2 | 0.5 | 4.9 | 0.3 | 0.0 | 5.5 | 12.9 | 0.0 | 75.3 |
| sn‐1 + 3 | 0.5 | 26.3 | 0.4 | 0.1 | 41.6 | 6.4 | 0.3 | 21.3 | |
| HPS | sn‐2 | 0.2 | 6.5 | 3.9 | 0.5 | 0.8 | 8.7 | 0.6 | 78.5 |
| sn‐1 + 3 | 0.7 | 72.8 | 2.4 | 0.2 | 7.8 | 3.9 | 0.6 | 9.0 | |
| HO‐30 | sn‐2 | 0.1 | 1.2 | 0.4 | 0.0 | 0.5 | 92.6 | 0.6 | 4.5 |
| sn‐1 + 3 | 1.7 | 19.1 | 1.8 | 0.0 | 4.1 | 65.2 | 1.1 | 5.3 | |
| HPO | sn‐2 | 0.1 | 2.6 | 17.2 | 0.0 | 0.4 | 69.5 | 2.2 | 7.9 |
| sn‐1 + 3 | 0.3 | 58.4 | 8.9 | 0.0 | 2.2 | 24.8 | 1.8 | 1.8 | |
| HPSO | sn‐2 | 0.1 | 3.3 | 6.0 | 0.0 | 0.6 | 80.1 | 1.0 | 8.7 |
| sn‐1 + 3 | 0.5 | 64.6 | 3.5 | 0.0 | 7.5 | 18.9 | 0.5 | 1.4 |
HPS = KIR‐10 × HS‐35; HPO = KIR‐10 × HO‐30; HPSO = KIR‐10 × (HS‐35 × HO‐30).
Table 4.
Positional distribution of fatty acids on PC in different cotton genotypes containing altered fatty acid composition
| Sample | Position | C14:0 | C16:0 | C16:1 | C16:2 | C18:0 | C18:1Δ9 | C18:1Δ11 | C18:2 |
|---|---|---|---|---|---|---|---|---|---|
| Coker 315 | sn‐1 | 0.7 | 48.1 | 0.1 | 0.0 | 10.7 | 13.1 | 1.1 | 25.6 |
| sn‐2 | 2.0 | 18.6 | 0.4 | 0.0 | 21.0 | 14.5 | 0.3 | 41.7 | |
| KIR‐10 | sn‐1 | 0.4 | 62.3 | 5.1 | 0.3 | 3.4 | 4.1 | 2.1 | 21.9 |
| sn‐2 | 1.5 | 13.6 | 10.2 | 1.5 | 13.0 | 7.5 | 1.0 | 50.4 | |
| HS‐35 | sn‐1 | 0.3 | 21.2 | 0.1 | 0.0 | 49.0 | 5.8 | 0.2 | 22.1 |
| sn‐2 | 0.5 | 7.0 | 0.3 | 0.0 | 11.5 | 11.5 | 0.1 | 67.9 | |
| HPS | sn‐1 | 0.3 | 61.2 | 1.0 | 0.0 | 9.4 | 3.7 | 0.7 | 22.8 |
| sn‐2 | 0.7 | 11.0 | 2.7 | 0.3 | 12.6 | 7.6 | 0.3 | 63.3 | |
| HO‐30 | sn‐1 | 0.5 | 23.4 | 0.4 | 0.0 | 4.8 | 65.2 | 1.4 | 3.9 |
| sn‐2 | 0.9 | 9.8 | 0.3 | 0.0 | 15.5 | 68.1 | 0.4 | 4.4 | |
| HPO | sn‐1 | 0.3 | 43.9 | 7.0 | 0.0 | 2.9 | 37.1 | 2.5 | 5.8 |
| sn‐2 | 1.1 | 12.1 | 10.7 | 0.2 | 15.5 | 50.6 | 0.7 | 7.6 | |
| HPSO | sn‐1 | 0.4 | 41.4 | 5.8 | 0.0 | 6.7 | 39.2 | 2.2 | 4.3 |
| sn‐2 | 1.1 | 10.2 | 11.5 | 0.2 | 16.5 | 53.8 | 0.9 | 5.8 |
HPS = KIR‐10 × HS‐35; HPO = KIR‐10 × HO‐30; HPSO = KIR‐10 × (HS‐35 × HO‐30).
Discussion
Palmitic acid is a common saturated fatty acid widely present in our diet system, and reports claiming and disputing a link between ingestion of palmitic acid and heart disease have been well documented (Chowdhury et al., 2014; Martinez‐Ortiz et al., 2006; Narang et al., 2005). Although the overall benefit or risk of HP oils has not been established, it is clear that HP vegetable oil is a better choice than trans fats (de Souza et al., 2015). While emphasis has been placed on reducing saturate fatty acids in the main stream vegetable oils, considerable efforts have also been made in raising palmitic acid in numerous vegetable oils because of its essential functional role in structural food lipids and high oxidative stability in trans‐free fats.
In soya bean, major alleles in at least five loci have been reported to cause an increase in palmitic acid content ranging between 4% and 40%, and the palmitic acid content has been elevated to above 40% of total fatty acids in lines combining HP mutant alleles (Stoltzfus et al., 2000). Some of these HP genotypes have been found to be associated with defective KAS2 genes. For instance, J10 has a deletion of entire gmKASIIA; M22 has a small deletion in an intron of GmKASIIB, which results in mistranslation of approximately 80% of GmKASIIB transcripts (Anai et al., 2012).
Arabidopsis mutant fab1‐1, having approximately double the amount of palmitic acid compared to that of WT plants (James and Dooner, 1990), was identified as a Leu‐337Phe substitution in KASII protein that causes instability due to insufficient space for accommodating the imidazole ring of the mutant Phe‐337 residual (Carlsson et al., 2002; Wu et al., 1994). Enzyme assay on the extracts from fab1‐1 mutant line revealed a 40% reduction of KASII activity in vitro. Seed‐specific down‐regulation of atKAS2 driven by Phaseolin promoter in Arabidopsis illustrated that up to 53% of palmitic acid could be accumulated in seed oil without obvious negative impact on seed development (Pidkowich et al., 2007). In this report, we have demonstrated in cotton that it is possible to raise palmitic acid level substantially and mimic an oil palm‐like profile by genetically altering the expression level of ghKAS2 gene in an otherwise HO genetic background.
KASII enzymes are highly conserved between different plant species, and there is a consensus regarding their role as the general housekeeping fatty acid synthases responsible for the production of most C18 fatty acids in plastids. The characterisation of a T‐DNA knockout mutant of KAS2 in Arabidopsis suggested that a basic level of KASII activity is essential for normal seed viability because the homozygote mutant genotype was not able to be recovered (Pidkowich et al., 2007).
During cotton transformation process, many of the somatic embryos derived from transgenic calli have been aborted at globular stage, which led to substantially fewer number of transgenic plants compared to a standard cotton transformation experiment in our laboratory. In addition to the two fertile transgenic lines we are reporting here, we have also generated a number of male sterile lines at an unusually high rate. When the male sterile lines were pollinated with WT, among a large number of aborted seeds, the recovered viable seeds contained higher level of palmitic acid than those of KIR‐1 and KIR‐10. But the effort has failed to establish a stable transgenic line from their progenies because of the highly variable palmitic acid content and poor seed viability that is likely attributable to the nature of high copy number of ghKAS2‐RNAi transgene. Such a result is consistent with the observation in Arabidopsis that strong suppression atKAS2 resulted in abortion of about one‐fourth of zygotic embryos before torpedo stage, suggesting lethality at homozygosity (Pidkowich et al., 2007).
A slight yet significant reduction in oil content was observed in both KIR‐1 and KIR‐10 seeds. It is well established that the energy requirement for the entire seed germination process including seed imbibition, radical protrusion, elongation and initial seedling development prior to photosynthetic autotrophy is provided by the available nutritive reserves of the cotyledons (Snider et al., 2014; Turley and Chapman, 2010). As seed oil represents the most energy‐dense storage compound of the quiescent cotton seed, it has been suggested that higher seed oil content would enhance seed germination rate and seedling vigour by providing more chemical energy (Bartee and Krieg, 1974; Snider et al., 2014). However, the reduction in seed oil content in both HP lines did not result in compromised seed germination at both cool and warm temperatures. It is particularly interesting that the dramatic increase of palmitic acid that has relatively high melting point compared to unsaturated fatty acids showed a neutral effect on germination rate, which is in sharp contrast to the severe impairment on germination observed in HS cotton seeds generated by RNAi down‐regulation of SAD (Liu et al., 2002). There is no doubt that detailed assessments on a range of seed characteristics need to be carried out, especially in field conditions as early season soil temperatures may strongly influence seed germination and seedling establishment.
Similar to the observation in Arabidopsis, a key feature of the HP cotton lines was a significant increase in levels of the so‐called ω7 fatty acids, including palmitoleic acid, C16:2 and cis‐vaccenic acid when compared with WT cotton. Palmitoleate is found only in trace amounts in most plant tissues, whereas oleate is the main product of plastidial fatty acid biosynthesis. The increased production of C16 unsaturated fatty acids may suggest the presence of a broad substrate specificity of SAD that acts on palmitoyl‐ACP to produce palmitoleic acid that could be either further desaturated by a FAD2 to form C16:2 or catalysed by an elongase to produce cis‐vaccenic acid.
Despite of disputing evidences, palmitoleic acid is gaining attention for its potential to reduce risk factors for coronary heart disease, type 2 diabetes and other obesity‐related diseases (Bernstein et al., 2014; Hiraoka‐Yamamoto et al., 2004). Further, it has not only high oxidative stability, but also lower melting points than oleic acid, which makes it desirable for use as feedstock of biodegradable lubricants or as a valuable precursor for linear low density polyethylene products (Ohlrogge, 1994; Rybak et al., 2008). In sunflower, it has been reported that SAD activity towards stearoyl‐ACP was about 100‐fold higher than for palmitoyl‐ACP (Salas et al., 2004). Therefore, it is likely that the low specificity of cotton SAD towards palmitoyl‐ACP could be the bottleneck for raising palmitoleic acid level in HP cotton seed oil. This might be overcome by co‐expressing a modified SAD with enhanced specificity towards palmitoyl‐ACP, together with the down‐regulation of ghKAS2 expression. In a KAS2‐suppressed Arabidopsis, a modified plastidial desaturase and an extraplastidial palmitate Δ9 desaturase yielded a mean accumulation of ~67% palmitoleic acid and cis‐vaccenic acid, with individual lines showing greater than 71% in the best engineered line (Nguyen et al., 2010). It could be envisaged that as an annual crop amendable to genetic modification, cotton seed oil rich in palmitic acid may become the launching pad for further genetic modification to produce valuable palmitoleic acid in planta.
In cotton seed, a relatively smaller proportion of C16 fatty acids are converted to their dienoic form than are the C18 fatty acids. This might be caused by the cotton FAD2 which had a strong substrate preference to oleic acid in relative to palmitoleic acid on PC. A FAD2 that has high substrate specificity for palmitoleic acid has been reported in safflower (Cao et al., 2013). However, such a palmitate preference has not yet been identified in cotton FAD2 that is comprised of only 4 gene members in contrast to a staggering 11 members as reported in safflower (Cao et al., 2013). Cis‐vaccenic acid is clearly an elongation product of palmitoleic acid, but it remains to be resolved whether the residual KASII in plastids, or the cytoplasmic β‐ketoacyl‐CoA synthase in eukaryotic pathway is the catalysing enzyme (Nguyen et al., 2010).
In addition to chain length and degree of unsaturation of fatty acids, it is now recognised that the positional composition of TAG may be an important determinant of metabolic availability. It has been hypothesised that the saturated fatty acids on the sn‐2 position are hypercholesterolaemic, but neutral on cholesterol metabolism when on the sn‐1 or sn‐3 positions (German and Dillard, 2004; Karupaiah and Sundram, 2007). A plausible mechanism to support this hypothesis is that pancreatic lipase and lipoprotein lipase preferentially hydrolyse the fatty acids in the sn‐1 and sn‐3 position and produce a 2‐MAG that is preferentially transported to liver for LDL cholesterol metabolism (Berry, 2009). Therefore, despite the high level of palmitic acid, the HP cotton seed oil may not be nutritionally undesirable due to its low level of palmitic acid at the sn‐2 position in TAG.
We have previously generated HO and HS cotton genotypes using RNAi down‐regulation of ghFAD2‐1 and ghSAD‐1, respectively (Liu et al., 2002). The HS cotton seed oil, albeit having a significantly raised melting point compared to the conventional cotton seed oil, contained insufficient saturated fatty acids to provide the required texture and spread ability for direct use as a solid fat unless used in combination with another appropriate fat (unpublished data). Therefore, raising palmitic acid content in the HO and HS genotypes has a clear advantage in providing functionality required for the food industry, especially in the area of solid fat applications, such as margarine, shortening and confectionary industries. In addition, simultaneously raised levels of palmitic, stearic and oleic acids could be potentially useful for mimicking the unique TAG profile of cocoa butter that is in high demand by the ever expanding confectionary, cosmetic and pharmaceutical industries. Cocoa butter is one of the most highly valued plant lipids in commerce because of its melting curve properties conferred by symmetrical TAGs such as sn‐1,3 distearoyl sn‐2 oleoyl glycerol (SOS), sn‐1 palmitoyl sn‐2 oleoyl sn‐3 stearoyl glycerol (POS) and sn‐1,3 dipalmitoyl sn‐2 oleoyl glycerol (POP).
We have demonstrated here that both palmitic and oleic acids could be raised simultaneously in cotton seed oil by crossing the HP and HO lines. This is consistent with the HP/HO sunflower mutant CAS‐12 that produces oil with 30% palmitate and 40% oleate (Salas et al., 2004). In contrast, the combination of HP and HS trait appeared to be troublesome as the stearic acid level in the hybrid was only 4% compared to about 30% in its HS parent while palmitic acid was retained at a similar level as its HP parent. In this study, we have shown that such a biased accumulation of saturated fatty acids occurred in both PC and TAG lipids. Although the oil biosynthesis pathway is made of discrete elements, it has been established that there are interactions between such components expediting the dynamic channelling of the metabolic flux through fatty acid elongation, desaturation and TAG assembly (Roughan and Ohlrogge, 1996). Several hypotheses could be proposed to explain why the HP cotton is not able to accumulate stearic acid to a high level. These include the low enzyme specificity of downstream enzymes needed for stearic acid incorporation, and FatB's substrate preference towards palmitic acid. The competition between the two saturated fatty acids for their esterification on TAG molecule might be eased by introducing a FatB enzyme or acyltransferases with substrate preference towards stearic acid.
In summary, we report here that expression of RNAi construct targeting the down‐regulation of KAS2 expression led to substantial increase in palmitic acid and its derivative C16 unsaturated fatty acids at the expense of C18 fatty acids in cotton seed oil. A slight yet significant reduction in oil content has been observed in both HP lines, but seed germination was unaffected. Simultaneous increase of both palmitic acid and oleic acid was also successful. These novel cotton genotypes may provide a temperate vegetable oil, derived from an annual crop, which could potentially replace imported palm oil in many parts of the world. Additional efforts are required for further enhancement of stearic acid in the HP/HO background that may have a clear advantage in providing functionality required for valuable cocoa butter substitute.
Experimental procedures
Plant material and growth conditions
Upland cotton (G. hirsutum) cv Coker315 was used for gene transformation. Homozygous transgenic cotton lines including HO‐30 and HS‐35 were generated as previously described (Liu et al., 2002). Plants were cultivated in greenhouse conditions at 28/18 °C (day/night), with 16‐h photoperiod.
Cloning cotton ghKAS2 genes
The cotton KASII DNA sequences were isolated from a developing cotton seed cDNA library using a DNA fragment corresponding to the first 400 nucleotide of the coding region of KASII gene from Arabidopsis (AF318307) as a probe. Two different KASII cDNAs, ghKASII‐A and ghKASII‐B, were isolated from this cDNA library as previously described (Liu et al., 1999).
Design of RNAi gene silencing construct targeting ghKAS2
We designed an RNAi cassette consisting of inverted repeat structure of partial ghKAS2‐1 DNA sequences driven by a seed‐specific promoter derived from soya bean lec1 gene (Cho et al., 1995). To assemble this RNA cassette, as shown in Fig. 8, we firstly obtained the trigger DNA fragment corresponding to nucleotides from 1001 to 1620 of ghKAS2‐1 by PCR using a pair of primers incorporating the NcoI site (underlined) at both ends: 5′‐CCATGGCTAATCGCGATGGATTTGTCATGG‐3′, and 5′‐CCATGGCTCTTCGGCCAAATGTAAGAAA‐3′. The amplified PCR fragment was then inserted at the NcoI site at nt‐265 of ghKAS2‐1, in an antisense orientation. A DNA fragment consisting of 400 bp of the inverted repeats in a head to head fashion, together with 750 bp adjacent DNA sequence acting as a spacer, was amplified by PCR using the following single primer that is present at the both sides of the inverted repeats and with an additional SmaI restriction site (underlined): 5′‐CCCGGGCGTATTGCCTGTACCGTTGC‐3′. The resulting inverted repeat structure was then inserted at the SmaI site between the lectin promoter and terminator sequences (Cho et al., 1995) in a binary vector using NPTII gene as the selectable marker for gene transformation.
Figure 8.
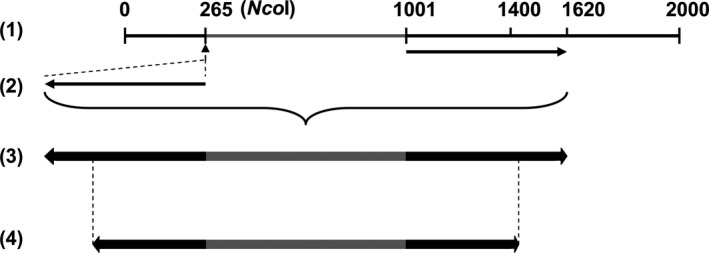
A diagram showing the selection process of trigger DNA sequence for the inverted repeat configuration in RNAi‐ghKAS2 construct. (1) The DNA sequence corresponding to nucleotides from 1001 to 1620 of ghKAS2‐1 was PCR amplified and (2) inserted at the NcoI site at nt 265 of ghKAS2‐1, in an antisense orientation and therefore resulted in the inverted repeat structure as shown in (3). The first 400 bp of the repeated DNA sequences and its adjacent 750‐bp spacer were PCR amplified to create the final RNAi trigger DNA sequence (4).
Transformation of cotton
The above described binary construct was transformed to Agrobacterium tumefaciens strain AGL1 and used to transform cotton variety Coker 315 as described by Liu et al. (2002). Briefly, cotyledons excised from cotton seedlings were used as explants and were infected and co‐cultivated with A. tumefaciens harbouring the RNAi‐ghKAS2‐1 construct. Healthy calli derived from the cotyledon explants were selected by growing on MS medium containing kanamycin until somatic embryo formation. Plantlets developed from the somatic embryos were subsequently transferred to soil and maintained in a glasshouse once leaves and roots were developed, with 28/20 °C (day/night) growth temperature.
Real‐time RT‐qPCR
RNAs from developing embryos of homozygous T4 KIR‐1, KIR‐10 plants were isolated using QIAEX Plant RNA Miniprep kit (Qiagen, Hilden, Germany). The gene expression patterns were studied with RT‐qPCR carried out in triplicate using Platinum SYBR Green qPCR SuperMix (BioRad, Hercules, CA) and run on ABI 7900HT Sequence Detection System. Each 10‐μL PCR contained 20 ng of total RNA template, 800 mm each of the forward and reverse primers, 0.25 μL of reverse transcriptase, 5 μL One‐step RT‐PCR master mix reagents. PCR was carried out at following conditions with an initial cycle at 48 °C for 30 min, and 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 s and 60 °C for 60 s. The following primers were designed such that amplifying a fragment from the ghKAS2‐1 gene (5′‐CCAACAAAGACGTGGGCTTGA‐3′ and 5′‐GGAGGCTTCTTCTTCGTTGTAA‐3′) would give a 290‐bp fragment; and amplifying a ghKAS2‐2 gene (5′‐TCCAAACCCAAGTCCTCAAAACTC‐3′ and 5′‐AGGAGGTTTCTGTTTTGTCATG‐3′) would give rise to a 393‐bp fragment. Cotton unibiquitin‐14 gene (ghUBQ14, GenBank accession number: DW505546) was used as an internal reference gene (Zhang et al., 2013). The primers for ghUBQ14 are sense: 5′‐CAACGCTCCATCTTGTCCTT‐3′ and antisense: 5′‐TGATCGTCTTTCCCGTAAGC‐3′. The calculations were made using the comparative CT method as reported (Livak and Schmittgen, 2001). The data are presented as means ± SD of three reactions performed on independent 96‐well plates.
Oil content analysis, lipid separation and quantification
Seed oil content was quantified by Oxford MQC benchtop nuclear magnetic resonance analyser (NMR) (Oxford Instruments, Oxford, UK). Three‐gram samples of cotton seed were measured and equilibrated to 40 °C for 2 h prior to acquisition of NMR data. The experiment was carried out in triplicate. Two‐way ANOVA was conducted using Microsoft Excel, and LSD test was applied at 5% probability level to compare the differences among treatment means.
Fatty acid profiles of cotton seed oil were determined by GC‐FID analysis of prepared seed oil fatty acid methyl esters (FAME). TAG and polar lipids were fractionated from total lipids on thin layer chromatography (TLC) plates Silica gel 60 (Merck Millipore, Darmstadt, Germany) using hexane/diethyl ether/acetic acid (50/50/1, by volume). Lipid bands were visualised by spraying 0.001% primuline made in 80% acetone in water, and individual lipid classes were identified by running authentic standards on the same TLC plate. Fatty acid profiles of TAG and total polar lipids were determined by incubating corresponding silica bands in 1 N methanolic HCl and by analysing corresponding FAME by GC as above.
Positional distribution of the fatty acids in TAG was analysed by treatment with TAG lipase derived from Rhizopus arrhizus (Fluka, Buchs, Switzerland). Purified TAG (1 mg) in chloroform was transferred to a glass vial, evaporated under nitrogen and mixed strongly using a vortex with 2 U Rhizopus lipase dissolved in 0.4 mL 0.1 m Tris‐HCl (pH 7.7) and 5 mm CaCl2 for 1 min. The reaction was stopped by adding 50 μL 6 m HCl, and lipid was extracted using 1.5 mL CHCl3/MeOH (2/1, by volume). The lipid was fractionated into TAG, DAG, sn‐1/3‐MAG, sn‐2‐MAG and FFA using 15 cm 2.3% boric acid TLC (silica gel‐60, Merck Millipore) and hexane/diethyl ether/acetic acid (50/50/1, by volume) solvent mixture. FAMEs of individual lipid classes were prepared directly by collecting the lipid bands in silica gel isolated from the TLC plate into glass vials and by incubation in 1 N methanolic HCl at 80 °C for 2 h prior to GC analysis.
PC was fractionated from the polar lipid pool using TLC plates Silica gel 60 (Merck Millipore) and chloroform/methanol/acetic acid/water (90/15/10/3, by volume) as the solvent system. Lipid classes were visualised and identified as above, and PC was extracted from silica using chloroform/methanol/water (2/1/1, by volume). Positional distribution of fatty acids in PC was determined by the treatment of 0.1 mg of lipid in 3 U of phospholipase A2 derived from honey bee (Sigma‐Aldrich, St. Louis, MO) as described by Williams et al. (1995). Lipid was further purified using chloroform/methanol/0.1 m KCl (2/1/1, by volume), and it was fractionated into PC, lyso‐PC and free fatty acids by TLC (Silica gel 60, MERCK) in chloroform/methanol/acetic acid/water (68/22/6/4, by volume) as solvent system. FAMEs of individual lipid bands were prepared and analysed as above.
Primary assessment of seed germination rate
To evaluate seed germination response to temperature, 30 seeds from each of the two HP lines, together with the untransformed control Coker 315, were evenly spaced on moist filter paper in a 200 × 100 mm plastic tray with cover and placed in incubators with cool (18 °C) and warm (28 °C) temperatures. The filter paper was kept moist for the duration of the experiment by adding distilled water when required. Seeds were incubated for 5 days at each temperature and were counted as germinated when the radicle protruded beyond the seed coat and elongated to at least 1 cm in length. The experiment was run in quadruplicate (n = 4). Two‐way ANOVA was conducted using Microsoft Excel, and LSD test was applied at 5% probability level to compare the differences among treatment means.
Supporting information
Table S1. Two way ANOVA of oil content in WT, KIR‐1, KIR‐10 across two generations (T4 and T5).
Table S2. Two way ANOVA of germination rate in WT, KIR‐1, KIR‐10 at cool (18 °C) and warm (28 °C) temperatures.
Acknowledgements
The authors wish to Geraldine Lester, Nathalie Niesner and Lijun Tian for excellent technical assistance. This study was supported by Cotton Research & Development Corporation (CRDC) Australia.
References
- Anai, T. , Hoshino, T. , Imai, N. and Takagi, Y. (2012) Molecular characterization of two high‐palmitic‐acid mutant loci induced by X‐ray irradiation in soybean. Breeding Sci. 61, 631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aslan, S. , Hofvander, P. , Dutta, P. , Sitbon, F. and Sun, C. (2015) Transient silencing of the KASII genes is feasible in Nicotiana benthamiana for metabolic engineering of wax ester composition. Sci. Rep. 5, 11213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartee, S.N. and Krieg, D.R. (1974) Cottonseed density: associated physical and chemical properties of 10 cultivars. Agron. J. 66, 433–435. [Google Scholar]
- Bernstein, A.M. , Roizen, M.F. and Martinez, L. (2014) Purified palmitoleic acid for the reduction of high‐sensitivity C‐reactive protein and serum lipids: A double‐blinded, randomized, placebo controlled study. J. Clin. Lipidol. 8, 612–617. [DOI] [PubMed] [Google Scholar]
- Berry, S.E.E. (2009) Triacylglycerol structure and interesterification of palmitic and stearic acid‐rich fats: an overview and implications for cardiovascular disease. Nutr. Res. Rev. 22, 3–17. [DOI] [PubMed] [Google Scholar]
- Cahoon, E.B. and Shanklin, J. (2000) Substrate‐dependent mutant complementation to select fatty acid desaturase variants for metabolic engineering of plant seed oils. Proc. Natl Acad. Sci. USA, 97, 12350–12355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, S. , Zhou, X.‐R. , Wood, C. , Green, A. , Singh, S. , Liu, L. and Liu, Q. (2013) A large and functionally diverse family of Fad2 genes in safflower (Carthamus tinctorius L.). BMC Plant Biol. 13, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlsson, A.S. , LaBrie, S.T. , Kinney, A.J. , Von Wettstein‐Knowles, P. and Browse, J. (2002) A KAS2 cDNA complements the phenotypes of the Arabidopsis fab1 mutant that differs in a single residue bordering the substrate binding pocket. Plant J. 29, 761–770. [DOI] [PubMed] [Google Scholar]
- Cho, M.‐J. , Widholm, J. and Vodkin, L. (1995) Cassettes for seed‐specific expression tested in transformed embryogenic cultures of soybean. Plant Mol. Biol. Rep. 13, 255–269. [Google Scholar]
- Chowdhury, R. , Warnakula, S. , Kunutsor, S. , Crowe, F. , Ward, H.A. , Johnson, L. , Franco, O.H. et al. (2014) Association of dietary, circulating, and supplement fatty acids with coronary risk ‐A systematic review and meta‐analysis. Ann. Intern. Med. 160, 398–406. [DOI] [PubMed] [Google Scholar]
- Dehesh, K. , Jones, A. , Knutzon, D.S. and Voelker, T.A. (1996) Production of high levels of 8:0 and 10:0 fatty acids in transgenic canola by overexpression of Ch FatB2, a thioesterase cDNA from Cuphea hookeriana . Plant J. 9, 167–172. [DOI] [PubMed] [Google Scholar]
- Dormann, P. , Voelker, T.A. and Ohlrogge, J.B. (2000) Accumulation of palmitate in Arabidopsis mediated by the acyl‐acyl carrier protein thioesterase FATB1. Plant Physiol. 123, 637–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- German, J.B. and Dillard, C.J. (2004) Saturated fats: what dietary intake? Am. J. Clin. Nutr. 80, 550–559. [DOI] [PubMed] [Google Scholar]
- Hayes, K.C. and Pronczuk, A. (2010) Replacing trans fat: the argument for palm oil with a cautionary note on interesterification. J. Am. Coll. Nutr. 29, 253s–284s. [DOI] [PubMed] [Google Scholar]
- Hiraoka‐Yamamoto, J. , Ikeda, K. , Negishi, H. , Mori, M. , Hirose, A. , Sawada, S. , Onobayashi, Y. et al. (2004) Serum lipid effects of a monounsaturated (palmitoleic) fatty acid‐rich diet based on macadamia nuts in healthy, young Japanese women. Clin. Exp. Pharmacol. P. 31, S37–S38. [DOI] [PubMed] [Google Scholar]
- James, D.W. Jr and Dooner, H.K. (1990) Novel seed lipid phenotypes in combinations of mutants altered in fatty acid biosynthesis in Arabidopsis. Theor. Appl. Genet. 82, 409–412. [DOI] [PubMed] [Google Scholar]
- Jones, L. and King, C. (1993) Cottonseed Oil. Memphis: National Cottonseed Products Associations, Inc. and the Cotton Foundation. [Google Scholar]
- Karupaiah, T. and Sundram, K. (2007) Effects of stereospecific positioning of fatty acids in triacylglycerol structures in native and randomized fats: a review of their nutritional implications. Nutr. Metab. 4, 16–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- L'Abbe, M.R. , Stender, S. , Skeaff, C.M. , Ghafoorunissa, R. and Tavella, M. (2009) Approaches to removing trans fats from the food supply in industrialized and developing countries. Eur. J. Clin. Nutr. 63, S50–S67.19190645 [Google Scholar]
- Liu, Q. , Singh, S.P. , Brubaker, C.L. , Sharp, P.J. , Green, A.G. and Marshall, D.R. (1999) Molecular cloning and expression of a cDNA encoding a microsomal ω‐6 fatty acid desaturase from cotton (Gossypium hirsutum). Funct. Plant Biol. 26, 101–106. [Google Scholar]
- Liu, Q. , Singh, S.P. and Green, A.G. (2002) High‐stearic and high‐oleic cottonseed oils produced by hairpin RNA‐mediated post‐transcriptional gene silencing. Plant Physiol. 129, 1732–1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak, K.J. and Schmittgen, T.D. (2001) Analysis of relative gene expression data using real‐time quantitative PCR and the (2−ΔΔCt) method. Methods, 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Martinez‐Ortiz, J.A. , Fung, T.T. , Baylin, A. , Hu, F.B. and Campos, H. (2006) Dietary patterns and risk of nonfatal acute myocardial infarction in Costa Rican adults. Eur. J. Clin. Nutr. 60, 770–777. [DOI] [PubMed] [Google Scholar]
- Martz, F. , Kiviniemi, S. , Palva, T.E. and Sutinen, M.L. (2006) Contribution of omega‐3 fatty acid desaturase and 3‐ketoacyl‐ACP synthase II (KASII) genes in the modulation of glycerolipid fatty acid composition during cold acclimation in birch leaves. J. Exp. Bot. 57, 897–909. [DOI] [PubMed] [Google Scholar]
- Mozaffarian, D. , Katan, M.B. , Ascherio, A. , Stampfer, M.J. and Willett, W.C. (2006) Trans fatty acids and cardiovascular disease. New Engl. J. Med. 354, 1601–1613. [DOI] [PubMed] [Google Scholar]
- Narang, D. , Sood, S. , Thomas, M. , Maulik, S.K. and Dinda, A.K. (2005) Dietary palm olein oil augments cardiac antioxidant enzymes and protects against isoproterenol‐induced myocardial necrosis in rats. J. Pharm. Pharmacol. 57, 1445–1451. [DOI] [PubMed] [Google Scholar]
- Neff, W.E. and List, G.R. (1999) Oxidative stability of natural and randomized high‐palmitic and high‐stearic acid oils from genetically modified soybean varieties. JAOCS, 76, 825–831. [Google Scholar]
- Nguyen, H.T. , Mishra, G. , Whittle, E. , Pidkowich, M.S. , Bevan, S.A. , Merlo, A.O. , Walsh, T.A. et al. (2010) Metabolic engineering of seeds can achieve levels of ω‐7 fatty acids comparable with the highest levels found in natural plant sources. Plant Physiol. 154, 1897–1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nzikou, J.M. , Matos, L. , Moussounga, J.E. , Ndangui, C.B. , Pambou‐Tobi, N.P. , Bandzouzi, E.M. , Kimbonguila, A. et al. (2009) Study of oxidative and thermal stability of vegetable oils during frying. Res. J. Appl. Sci. 4, 94–100. [Google Scholar]
- O'Brien, R.D. (2002) Cottonseed oil. In Vegetable Oils in Food Technology: Composition, Properties and Uses ( Gunstone, F.D. , ed), pp. 203–230. Oxford: Blackwell Publishing. [Google Scholar]
- Ohlrogge, J.B. (1994) Design of new plant products: engineering of fatty acid metabolism. Plant Physiol. 104, 821–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlrogge, J.B. and Jaworski, J.G. (1997) Regulation of fatty acid synthesis. Annu. Rev. Plant Phys. 48, 109–136. [DOI] [PubMed] [Google Scholar]
- Oomen, C.M. , Ocke, M.C. , Feskens, E.J. , van Erp‐Baart, M.A. , Kok, F.J. and Kromhout, D. (2001) Association between trans fatty acid intake and 10‐year risk of coronary heart disease in the Zutphen Elderly Study: a prospective population‐based study. Lancet, 357, 746–751. [DOI] [PubMed] [Google Scholar]
- Pidkowich, M.S. , Nguyen, H.T. , Heilmann, I. , Ischebeck, T. and Shanklin, J. (2007) Modulating seed beta‐ketoacyl‐acyl carrier protein synthase II level converts the composition of a temperate seed oil to that of a palm‐like tropical oil. Proc. Natl Acad. Sci. USA, 104, 4742–4747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roughan, P.G. and Ohlrogge, J.B. (1996) Evidence that isolated chloroplasts contain an integrated lipid‐synthesizing assembly that channels acetate into long‐chain fatty acids. Plant Physiol. 110, 1239–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybak, A. , Fokou, P.A. and Meier, M.A.R. (2008) Metathesis as a versatile tool in oleochemistry. Eur. J. Lipid Sci. Tech. 110, 797–804. [Google Scholar]
- Salas, J.J. , Martínez‐Force, E. and Garcés, R. (2004) Biochemical characterization of a high‐palmitoleic acid Helianthus annuus mutant. Plant Physiol. Bioch. 42, 373–381. [DOI] [PubMed] [Google Scholar]
- Snider, J.L. , Collins, G.D. , Whitaker, J. , Chapman, K.D. , Horn, H. and Grey, T.L. (2014) Seed size and oil content are key determinants of seedling vigor in Gossypium hirsutum . J. Cotton Sci. 18, 1–9. [Google Scholar]
- de Souza, R.J. , Mente, A. , Maroleanu, A. , Cozma, A.I. , Ha, V. , Kishibe, T. , Uleryk, E. et al. (2015) Intake of saturated and trans unsaturated fatty acids and risk of all cause mortality, cardiovascular disease, and type 2 diabetes: systematic review and meta‐analysis of observational studies. BMJ, 351, h3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoltzfus, D.L. , Fehr, W.R. , Welke, G.A. , Hammond, E.G. and Cianzio, S.R. (2000) A fap5 allele for elevated palmitate in soybean. Crop Sci. 40, 647–650. [Google Scholar]
- Sun, J.‐Y. , Hammerlindl, J. , Forseille, L. , Zhang, H. and Smith, M.A. (2014) Simultaneous over‐expressing of an acyl‐ACP thioesterase (FatB) and silencing of acyl‐acyl carrier protein desaturase by artificial microRNAs increases saturated fatty acid levels in Brassica napus seeds. Plant Biotech. J. 12, 624–637. [Google Scholar]
- Turley, R.B. and Chapman, K.D. (2010) Ontogeny of cotton seeds: gametogenesis, embryogenesis, germination, and seedling growth. In Physiology of Cotton ( Stewart, J.M. , Oosterhuis, D.M. , Heitholt, J.J. and Mauney, J.R. , eds), pp. 332–341. New York: Springer. [Google Scholar]
- Williams, J.P. , Khan, M.U. and Wong, D. (1995) A simple technique for the analysis of positional distribution of fatty acids on di‐ and triacylglycerols using lipase and phospholipase A2. J. Lipid Res. 36, 1407–1412. [PubMed] [Google Scholar]
- Wu, J. , James, D.W. Jr , Dooner, H.K. and Browse, J. (1994) A mutant of Arabidopsis deficient in the elongation of palmitic acid. Plant Physiol. 106, 143–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaliha, O. , Chong, C.L. , Cheow, C.S. , Norizzah, A.R. and Kellens, M.J. (2004) Crystallization properties of palm oil by dry fractionation. Food Chem. 86, 245–250. [Google Scholar]
- Zhang, Y. , Wang, X.F. , Ding, Z.G. , Ma, Q. , Zhang, G.R. , Zhang, S.L. , Li, Z.K. et al. (2013) Transcriptome profiling of Gossypium barbadense inoculated with Verticillium dahliae provides a resource for cotton improvement. BMC Genom. 14, 637. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Two way ANOVA of oil content in WT, KIR‐1, KIR‐10 across two generations (T4 and T5).
Table S2. Two way ANOVA of germination rate in WT, KIR‐1, KIR‐10 at cool (18 °C) and warm (28 °C) temperatures.


