Summary
The plant phytohormone abscisic acid (ABA) plays significant roles in integrating environmental signals with embryogenesis, germination, seedling establishment, the floral transition and the adaptation of plants to stressful environments by modulating stomatal movement and stress‐responsive gene expression. ABA signalling consists of ABA perception, signal transduction and ABA‐induced responses. ABA receptors such as members of the PYR/PYL family, group A type 2C protein phosphatases (as negative regulators), SnRK2 protein kinases (as positive regulators), bZIP transcription factors and ion channels are key components of ABA signalling. Post‐translational modifications, including dephosphorylation, phosphorylation and ubiquitination, play important roles in regulating ABA signalling. In this review, we focus on the roles of post‐translational modifications in ABA signalling. The studies presented provide a detailed picture of the ABA signalling network.
Keywords: abscisic acid, ABA signalling, post‐translational regulation, phosphorylation, dephosphorylation, ubiquitination, 26S proteasome system
Introduction
As sessile organisms, plants must adapt to their environment to survive, develop and propagate‐especially under stressful conditions. The phytohormone abscisic acid (ABA) is a key regulator of plant growth and development and of the responses of plants to biotic and abiotic stresses (Hirayama and Shinozaki, 2007; Melotto et al., 2006). ABA is involved in promoting embryo maturation, initiating and maintaining seed dormancy, repressing seed germination and postgermination growth, inhibiting the transition from vegetative to reproductive growth, modulating stomatal movement, inducing gene expression and improving plant adaptation to stressful environments (Cutler et al., 2010; Finkelstein, 2013). Much work has been carried out to discover the mechanisms underlying ABA signalling given the hormone's significance in plant development and its potential agricultural applications, and significant progress has been made. Five core components are involved in ABA signalling: ABA receptors, negative regulators, positive regulators, ABA‐responsive transcription factors and ABA‐responsive genes (Hauser et al., 2011).
The identification of intracellular ABA receptors, including those belonging to the pyrabactin resistance 1 (PYR1)/PYR1‐like (PYLs)/regulatory components of ABA receptors (RCAR) family (hereafter referred to as PYR/PYLs), has greatly enhanced our understanding of ABA signalling (Ma et al., 2009; Park et al., 2009; Santiago et al., 2009). PYR/PYLs belong to the steroidogenic acute regulatory protein‐related lipid transfer domain protein family, which is also called the Bet v 1 superfamily (Iyer et al., 2001; van Loon et al., 2006; Radauer et al., 2008). In Arabidopsis, fourteen PYR/PYLs have been identified (PYR1 and PYL1–PYL13), which are localized to the cytoplasm, plasma membrane or nucleus (McConnell et al., 2001; Park et al., 2009) (Table 1).
Table 1.
PYR/PYL ABA receptor and PP2C interactions identified in Arabidopsis
| ABA receptor | AGI annotation | PP2Cs interacted with | References |
|---|---|---|---|
| PYR1/RCAR11 | AT4G17870 | ABI1, ABI2, HAB1, HAB2, PP2CA | Fujii et al. (2009), Hao et al. (2011), Park et al. (2009) |
| PYL1/RCAR12 | AT5G46790 | ABI1, ABI2, HAB1, HAB2, PP2CA | Fujii et al. (2009), Hao et al. (2011), Ma et al. (2009), Park et al. (2009) |
| PYL2/RCAR14 | AT2G26040 | ABI1, HAB1, HAB2, PP2CA | (Fujii et al., 2009; Hao et al., 2011; Park et al., 2009) |
| PYL3/RCAR13 | AT1G73000 | ABI1, HAB1, HAB2, PP2CA | Fujii et al. (2009), Hao et al. (2011), Park et al. (2009) |
| PYL4/RCAR10 | AT2G38310 | ABI1, HAB1, HAB2, PP2CA | Fujii et al. (2009), Hao et al. (2011), Park et al. (2009), Pizzio et al. (2013) |
| PYL5/RCAR8 | AT5G05440 | ABI1, ABI2, HAB1, HAB2, PP2CA | Fujii et al. (2009), Hao et al. (2011), Ma et al. (2009), Santiago et al. (2009) |
| PYL6/RCAR9 | AT2G40330 | ABI1, HAB1, HAB2, PP2CA | Fujii et al. (2009), Hao et al. (2011), Santiago et al. (2009) |
| PYL7/RCAR2 | AT4G01026 | ABI1 | Fujii et al. (2009) |
| PYL8/RCAR3 | AT5G53160 | ABI1, ABI2, HAB1, HAB2, PP2CA | Fujii et al. (2009), Hao et al. (2011), Ma et al. (2009), Santiago et al. (2009) |
| PYL9/RCAR1 | AT1G01360 | ABI1, ABI2, HAB1, HAB2, PP2CA | Fujii et al. (2009), Hao et al. (2011), Ma et al. (2009), Park et al. (2009) |
| PYL10/RCAR4 | AT4G27920 | ABI1, HAB1, HAB2, PP2CA | Fujii et al. (2009), Hao et al. (2011) |
| PYL11/RCAR5 | AT5G45860 | ABI1 | Fujii et al. (2009) |
| PYL12/RCAR6 | AT5G45870 | PP2CA/AHG3 | Fujii et al. (2009), Park et al. (2009) |
| PYL13/RCAR7 | AT4G18620 | PP2CA/AHG3, ABI1, ABI2 | Fuchs et al. (2014), Zhao et al. (2013) |
The dephosphorylation activity of group A protein phosphatase type 2Cs (PP2Cs) and the phosphorylation activity of sucrose nonfermenting 1 (SNF1)‐related protein kinases 2 (SnRK2s) are required for ABA signalling (Ma et al., 2009; Park et al., 2009; Santiago et al., 2009). In the absence of ABA, a physical association exists between PP2Cs and C‐terminal subdomain II of SnRK2s; this inhibits the phosphorylation activity of SnRK2s and blocks ABA signal transduction. PP2Cs repress ABA signalling by dephosphorylating and inactivating SnRK2s (Fujii et al., 2007; Mustilli et al., 2002; Umezawa et al., 2009; Vlad et al., 2009; Yoshida et al., 2006a) (Figure 1). Following the induction of ABA synthesis by environmental or developmental factors, the hormone is perceived and bound by PYR/PYL family ABA receptors (Ma et al., 2009; Park et al., 2009; Santiago et al., 2009). The binding of ABA results in conformational changes in the receptor proteins, generating a platform for interaction with PP2Cs. The interaction between PYR/PYLs and PP2Cs suppresses the phosphatase activity of the enzymes and relieves the inhibition of SnRK2s (Nishimura et al., 2010; Park et al., 2009; Soon et al., 2012; Yin et al., 2009) (Figure 1). The released SnRK2s are then activated by autophosphorylation or phosphorylation via other kinases (Fujii et al., 2009; Ng et al., 2011; Soon et al., 2012). These activated SnRK2s are able to phosphorylate downstream proteins or transcription factors, including basic leucine zipper (bZIP) transcription factors called ABA‐responsive element binding factors (AREBs/ABFs) and S‐type anion channels (e.g. slow anion channel 1, SLAC1), to induce ABA responses (Brandt et al., 2012; Fujii et al., 2009; Geiger et al., 2009; Umezawa et al., 2009; Xue et al., 2011) (Figure 1).
Figure 1.
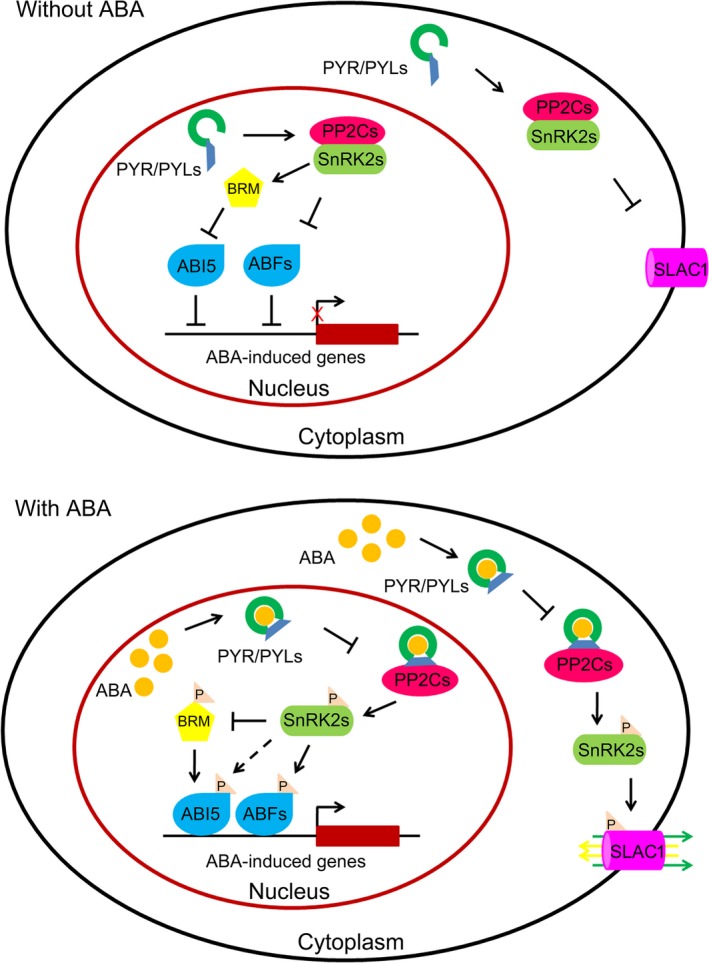
The ABA signalling cascade in Arabidopsis. The components involved in ABA signalling include the PYR/PYL family of ABA receptors, PP2Cs (negative regulator), SnRK2s (positive regulator), SWI/SNF chromatin‐remodelling ATPase BRM, transcription factors (AREB/ABFs, ABI5), ABA‐responsive genes and S‐type anion channels. ABA signalling occurs in the cytoplasm, nucleus and plasma membrane. In the presence of ABA, a complex is formed between ABA, receptor proteins and PP2Cs (ABA‐PYR/PYL/RCAR‐PP2C), which prevents PP2Cs from inhibiting the phosphorylation activity of SnRK2s. SnRK2s, phosphorylated via an unknown mechanism, then activate bZIP transcription factors and S‐type anion channels (e.g. SLAC1) and inactivate SWI/SNF chromatin‐remodelling ATPase BRM by phosphorylation. Once activated or inactivated, these proteins trigger the expression of ABA‐responsive genes or regulate the exchange of ions to confer plant tolerance to abiotic stresses.
In addition to dephosphorylation and phosphorylation, polyubiquitination mediated by E3 ubiquitin (Ub) ligases has been found to be important in ABA perception, ABA signal transduction and the induction of ABA responses. PYR/PRLs, PP2Cs, type 2A protein phosphatases (PP2As), transcription factors and proteins encoded by ABA‐responsive genes are targeted for E3 Ub ligase‐mediated polyubiquitination and degraded by the 26S proteasome. Several types of E3 Ub ligases are involved in ABA signalling. In this review, we focus on recent discoveries related to the post‐translational regulation of ABA signalling.
Dephosphorylation negatively regulates ABA signalling
PP2Cs and PP2As are negative regulators of ABA signalling. There are 76 PP2Cs in Arabidopsis, which are clustered into 10 groups (Fuchs et al., 2013). Nine PP2Cs involved in ABA signalling belong to group A: ABA‐insensitive (ABI) 1, ABI2, ABA‐hypersensitive germination (AHG) 1, AHG3/PP2CA, hypersensitive to ABA (HAB) 1, HAB2, highly ABA‐induced (HAI) 1, AKT1‐interacting PP2C 1 (AIP1)/HAI2 and HAI3 (Kuhn et al., 2006; Merlot et al., 2001; Robert et al., 2006; Rubio et al., 2009; Saez et al., 2004, 2006; Yoshida et al., 2006b). A common feature of clade A PP2Cs is that their expression is induced by high ABA levels or stressful conditions (Fujita et al., 2009, 2011). Loss‐of‐function mutations in genes encoding clade A PP2Cs cause ABA hypersensitivity, except for HAI1, AIP1/HAI2 and HAI3 (which encode the so‐called HAI PP2Cs) (Gosti et al., 1999; Merlot et al., 2001; Nishimura et al., 2007; Yoshida et al., 2006b). Single mutations in genes encoding HAI PP2Cs were not found to produce ABA‐responsive phenotypes (Yoshida et al., 2006b). The hai double or triple mutants show ABA‐hyposensitive phenotypes in seed germination, which contrast with the hypersensitive phenotypes of other group A PP2C mutants, and ABA‐hypersensitive responses in postgermination growth, which is similar to other group A PP2C mutants (Bhaskara et al., 2012). Furthermore, hai1‐1 mutation elevates the ABA response of pp2ca‐1/ahg3‐1 mutant. The pp2ca‐1hai1‐1 (or ahg3‐1hai1‐1) double mutant plants exhibit enhanced ABA‐mediated growth inhibition, increased ABA‐responsive gene induction and diminished water loss (Antoni et al., 2012). However, hai single mutant exhibits enhanced drought tolerance phenotypes, including the accumulation of proline and osmoregulatory solute and increased expression of abiotic stress‐responsive genes, which is not observed in other group A PP2C mutants. These results suggest that HAI PP2Cs have a greater role in ABA‐independent signalling than in ABA‐dependent signalling in response to drought stress (Bhaskara et al., 2012).
The selective inhibition of PP2Cs by a complex composed of PRY/PYLs and ABA has been reported (Antoni et al., 2012; Bhaskara et al., 2012) (Table 1). In the absence of ABA, clade A PP2Cs bind directly to and dephosphorylate SnRK2s, suppressing their kinase activity and blocking ABA signalling (Soon et al., 2012; Umezawa et al., 2009; Vlad et al., 2009). These results reveal the negative regulatory roles and functional differentiation of PP2Cs in the perception and transduction of ABA signals (Bhaskara et al., 2012; Cutler et al., 2010).
Recently, PP2As were shown to interact with SnRK2s and negatively regulate ABA signalling (Waadt et al., 2015) (Figure 2). PP2As are heterotrimeric holoenzyme complexes consisting of the following subunits: PP2AA, PP2AB and PP2AC (Shi, 2009; Xu et al., 2006). There are five catalytic subunits (PP2AC), three PP2AA‐regulatory subunits and 18 PP2AB‐regulatory subunits in Arabidopsis genome (DeLong, 2006; Farkas et al., 2007). The B subunits are classified into four subtypes, which comprise four subtypes: 2 B, 9 B′, 6 B″ and 1 TAP46 (Farkas et al., 2007). Three PP2A subunits, including PP2AA1/roots curl in NPA 1 (RCN1), PP2AA2 and PP2AA3, form ternary complexes to conduct their functions. Both PP2AAs and PP2ABs function as regulatory factors, whereas PP2ACs act as catalytic components of PP2A ternary complexes (DeLong, 2006; Farkas et al., 2007).
Figure 2.
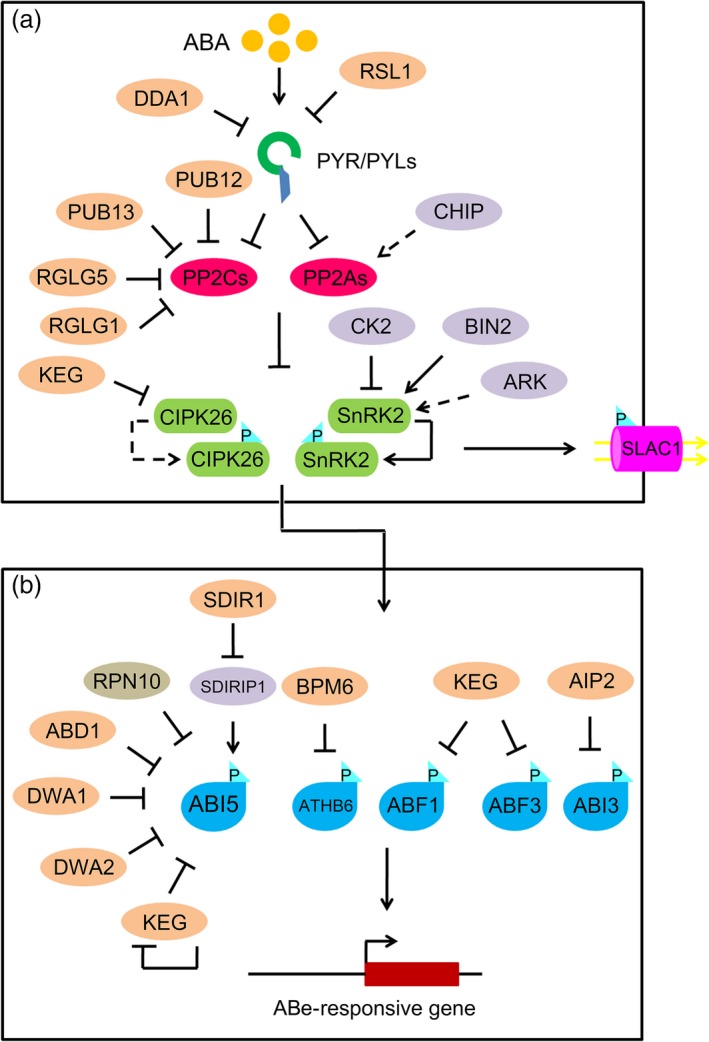
The regulatory roles of protein phosphorylation and ubiquitination in ABA signalling. (a) Post‐translational regulation of PYR/PYL ABA receptors, PP2Cs and PP2As phosphatases, SnRK2s and CIPK26 protein kinases in ABA signalling. DDA1 and RSL1, Cul4‐based and RING‐type E3 Ub ligases, are able to target PYR/PYL ABA receptors for degradation by the Ub‐mediated 26S proteasome pathway. Two U‐box E3 ligases (e.g. PUB12 and PUB13) and two RING E3 ligases (e.g. RGLG1 and RGLG5) target PP2Cs for degradation by the Ub‐mediated 26S proteasome pathway. Similar to the functions of PP2Cs, PP2AAs and PP2ABs regulatory subunits in PP2A complexes are negatively involved in ABA signalling. CHIP, a U‐box E3 ligase, uses it monoubiquitination activity to control the expression of RCN1 and PP2AA3 regulatory subunits in PP2A complexes. SnRK2s are able to be phosphorylated and activated by themselves, or by BIN2 or ARK to activate downstream ABA signalling. In contrast, OST1, a SnRK2 kinase involved in ABA signalling, is phosphorylated and inactivated by CK2 kinase to block downstream ABA signalling. CIPK26, a novel protein kinase, interacts with PP2Cs in the absence of ABA and phosphorylates downstream transcription factors such as ABI5 to trigger ABA responses. KEG, a RING‐type E3 ligase, can target CIPK26 for disruption. (b) Downstream transcription factors (e.g. ABI5, ABI3, ABFs and ATHB6) and ion channels are the substrates of SnRK2s or CIPK26, which are also the targets of E3 ligases. ABD1, DWA1 and DWA2, as Cul4‐based E3 ligases, interact with and ubiquitinate ABI5 for degradation by the 26 proteasome. RPN10, a core component of the 26S proteasome, targets ABI5 for degradation. In addition to targeting CIPK26 for degradation, KEG can target several substrates for disruption, including ABI5, ABF1 and ABF3. The self‐ubiquitination of KEG is triggered by a high level of ABA, causing the degradation of KEG and releasing ABI5. SDIR1, a RING finger E3 ligase, targets SDIRIP1 for degradation to regulate ABI5. In addition, AIP2 and BPM6, RING‐type and CUL3‐based E3 ligases, interact with and phosphorylate ABI3 and ATHB6, respectively, causing their degradation. Black arrows represent promotion. Black T‐shaped bars represent repression. Dashed lines represent unconfirmed relationships. P, phosphate.
Genetic studies have revealed the complicated roles of PP2As in ABA signalling (Charpentier et al., 2014; Kwak et al., 2002; Pernas et al., 2007; Saito et al., 2008). The regulatory PP2AA and PP2AB' subunits regulate ABA signalling through interacting with a SnRK2, open stomata 1 (OST1). The rcn1 mutant and pp2aa‐regulatory subunit double mutants exhibit ABA insensitivity during seed germination and stomatal closure (Kwak et al., 2002; Saito et al., 2008; Waadt et al., 2015). Further, the mutations pp2ac3, pp2ac4 and pp2ac5 confer ABA‐insensitive phenotypes in seed germination (Waadt et al., 2015). This suggests that PP2AA, PP2AB′, and PP2AC3, PP2AC4, PP2AC5 subunits might positively regulate ABA signalling in seed germination and stomatal closure. In contrast, the catalytic subunit mutant pp2ac2 shows ABA hypersensitivity during seed germination, root growth and seedling development (Pernas et al., 2007). These results suggest that the roles of PP2A regulatory and catalytic subunits in ABA response and ABA signalling might be different (Waadt et al., 2015). The exact mechanisms underlying the regulation of ABA signalling by PP2As and the relationship between clade A PP2Cs and PP2As are unclear.
Phosphorylation positively regulates ABA signalling
Several SnRK2s in Arabidopsis have been identified as ABA‐ or stress‐activated kinases (Mustilli et al., 2002; Yoshida et al., 2002). Specifically, there are 38 SnRKs, 10 of which are subgroup III SnRKs, or SnRK2s (Hrabak et al., 2003). These SnRK2s, including SnRK2.2/SRK2D, SnRK2.3/SRK2I and SnRK2.6/SRK2E/OST1, are activated by ABA in Arabidopsis protoplasts and perform as positive regulators of ABA signalling (Boudsocq et al., 2004, 2007; Fujii et al., 2007; Mustilli et al., 2002). Genomewide data indicate that the expression of a vast number of ABA‐ and osmotic stress‐responsive genes is impaired in the snrk2.2/snrk2.3/snrk2.6 triple mutant and the snrk2.2/2.3/2.6 triple mutant is practically bind to ABA, suggesting a positive role for SnRK2s in ABA signalling (Fujii and Zhu, 2009; Fujita et al., 2009). The phosphorylation activities of SnRK2s are suppressed via a physical association with PP2Cs in the absence of ABA (Soon et al., 2012; Umezawa et al., 2009; Vlad et al., 2009). However, in the presence of ABA, PYR/PYLs bind to the hormone and cause conformation changes to interact with PP2Cs; this interaction represses PP2C activity and releases SnRK2 suppression, triggering downstream ABA responses (Ma et al., 2009; Melcher et al., 2009; Miyazono et al., 2009; Nishimura et al., 2009; Park et al., 2009).
SnRK2s regulate a wide range of proteins through ABA‐mediated phosphorylation. AREB/ABFs, ABI3 and ABI5 are ABA response‐related transcription factors and targets of SnRK2s (Furihata et al., 2006; Kobayashi et al., 2005; Sirichandra et al., 2010). SnRK2s can also phosphorylate plasma membrane proteins. For example, the guard cell kinase SnRK2.6/OST1 has been shown to phosphorylate ion channels such as SLAC1 (Brandt et al., 2012; Geiger et al., 2009; Lee et al., 2009), quick anion channel 1/aluminium‐activated malate transporter 12 (Imes et al., 2013; Sasaki et al., 2010), the potassium channel KAT1 (Sato et al., 2009), NADPH oxidase respiratory burst oxidase homolog F (Sirichandra et al., 2009a) and the anion/proton exchanger CLCa (Wege et al., 2014), which play important roles in controlling stomatal aperture. Recently, SWI/SNF chromatin‐remodelling ATPase BRAHMA (BRM) in Arabidopsis was identified as the target protein of SnRK2s (Peirats‐Llobet et al., 2016). BRM is inactivated by SnRK2‐dependent phosphorylation and activated by PP2C‐dependent dephosphorylation. Phosphorylation of BRM by SnRK2s inhibits the activity of BRM, releasing the expression of ABI5. The brm‐3 mutant exhibits ABA‐hypersensitive phenotype through derepressing ABI5 (Han et al., 2012; Peirats‐Llobet et al., 2016). These findings suggest that BRM acts as a negative regulator of ABA signalling and connect ABA signalling to chromatin remodelling (Han et al., 2012; Peirats‐Llobet et al., 2016) (Figure 1). In addition, putative substrates have been identified through the mapping of peptide phosphorylation preferences and phosphoproteomic approaches (Shin et al., 2007; Sirichandra et al., 2010; Umezawa et al., 2013; Vlad et al., 2008; Wang et al., 2013). However, the functions of these putative substrates in ABA responses require validation.
In addition to SnRK2s, calcineurin B‐like interacting protein kinase 26 (CIPK26) has been found to interact with ABI1 and ABI2 during ABA signalling (Lyzenga et al., 2013) (Figure 2). CIPK26 also interacts with and phosphorylates ABI5 to activate downstream ABA responses. CIPK26 overexpression produces a hypersensitive phenotype in seed germination, suggesting a positive role for CIPK26 in ABA signalling (Lyzenga et al., 2013). Thus, CIPK26 regulates ABA signalling by phosphorylating and activating ABI5 (Lyzenga et al., 2013).
The regulation of SnRK2s has also been studied, and SnRK2 activity has been shown to be controlled by phosphorylation. The autophosphorylation activity levels of recombinant SnRK2.2 and SnRK2.3 were weaker than those of SnRK2.6, suggesting that some SnRK2s are activated by as yet unknown kinases in vivo (Ng et al., 2011). Recently, brassinosteroid (BR)‐insensitive 2 (BIN2), casein kinase 2 (CK2), and ABA and abiotic stress‐responsive Raf‐like kinase (ARK) were identified as kinases that regulate SnRK2 activity and stability (Cai et al., 2014; Saruhashi et al., 2015; Vilela et al., 2015).
BIN2 is a glycogen synthase kinase 3 (GSK3)‐like kinase that can phosphorylate and regulate the activity, stability and subcellular localization of a number of proteins in diverse systems (Grimes and Jope, 2001; Saidi et al., 2012). Previous studies showed that BIN2 functions as a negative regulator of BR signalling by phosphorylating transcription factors, including brassinazole‐resistant 1 and BRI1‐EMS‐suppressor 1 (He et al., 2002; Yin et al., 2002). In the ABA signalling pathway, BIN2 phosphorylates SnRK2.2 and SnRK2.3, enhancing their kinase activity levels. These data have provided significant insight into the modulation of ABA signalling by GSK3‐like kinases and crosstalk between BR signalling and ABA signalling in Arabidopsis (Cai et al., 2014; Yan et al., 2009) (Figure 2). Recently, ARK was identified as a novel regulatory component of SnRK2s in Physcomitrella patens (Saruhashi et al., 2015) (Figure 2). ARK is a group B3 Raf‐like MAP kinase kinase kinase (B3‐MAPKKK). A moss ark mutant showed reduced ABA sensitivity and reduced tolerance to hyperosmosis (Saruhashi et al., 2015). The mutant also showed impaired phosphorylation activity by SnRK2s, indicating that ARK is an essential signalling component regulating SnRK2 activity in basal land plants such as moss (Saruhashi et al., 2015). Six B3‐MAPKKKs are encoded in the Arabidopsis genome. B3‐MAPKKK genes from Arabidopsis can recover the ABA‐insensitive phenotype of the moss ark mutant, indicating that the functions of B3‐MAPKKKs in ABA signalling in Arabidopsis are similar to those in moss (Saruhashi et al., 2015). However, the functions of Arabidopsis B3‐MAPKKKs in ABA signalling are unclear. In addition, CK2 has been characterized as a regulator of SnRK2.6/SRK2E/OST1 protein stability (Vilela et al., 2015). CK2 is a highly conserved serine/threonine kinase with multiple subunits in eukaryotes (Meggio and Pinna, 2003). The CK2 holoenzyme has two catalytic ɑ‐subunits and two regulatory β‐subunits. There are four ɑ‐ and four β‐subunits in both the Arabidopsis and maize genomes (Riera et al., 2011; Salinas et al., 2006); CK2 phosphorylates ZmOST1 in planta to increase its binding to PP2C; this, in turn, triggers ZmOST1 protein degradation and affects its kinase activity (Vilela et al., 2015). These results indicate that CK2 is a negative regulator of ABA signalling and link CK2 directly to the core ABA signalling pathway (Vilela et al., 2015). However, ck2a mutants are hyposensitive to ABA during seed germination, cotyledon greening and stomatal opening due to the down‐regulated expression of OST1 and other ABA‐responsive genes (Mulekar et al., 2012; Wang et al., 2014). These contradictory results indicate that CK2 might have additional functions in regulating ABA signalling.
SnRK2s are key components of ABA signalling cascades. Understanding their activation, degradation mechanisms and substrates will increase our understanding of ABA signalling.
Ub‐mediated protein degradation regulates ABA signalling
Ubiquitination, a type of protein post‐translational modification, is involved in many aspects of plant development and environmental responses. Three enzymes are involved in ubiquitination: E1, as a Ub‐activating enzyme; E2, as a Ub‐conjugating enzyme; and E3, as a Ub‐protein ligase (Smalle and Vierstra, 2004). Upon polyubiquitination, substrate proteins undergo proteolysis by the 26S proteasome. Ub‐mediated protein degradation via the 26S proteasome thus provides a master mechanism for controlling the abundance of key regulators in Arabidopsis. E3 Ub ligases are responsible for the specificity of the system by interacting with the substrate; substrate recognition enables the transfer of the 76‐amino acid Ub molecule to a lysine residue within the target protein (Smalle and Vierstra, 2004). E3 Ub ligases are classified into four types: really interesting new gene (RING), cullin‐RING ligase (CRLs), U‐box and homologous to E6‐AP carboxyl terminus (Vierstra, 2009). The CRLs can be further divided into four subtypes: anaphase‐promoting complex, S‐phase kinase‐associated protein 1‐cullin 1‐F‐box, DNA damage‐binding protein (DDB) and bric‐a‐brac/tramtrack/broad complex (BTB) (Hua and Vierstra, 2011). Recently, CRLs, RING and U‐box E3 Ub ligases were found to be involved in ABA signalling (Yu et al., 2015) (Table 2 and Figure 2).
Table 2.
The roles of E3 ubiquitin ligases and the 26S proteasome in ABA signalling
| Type of E3 ligase | Name | Effect on ABA signalling | Target | References |
|---|---|---|---|---|
| CUL4‐DDB1 | DDA1 | Negativea | PYL4, PYL8, PYL9 | Irigoyen et al. (2014) |
| DWA1 | Negative | ABI5 | Lee et al. (2010) | |
| DWA2 | Negative | ABI5 | Lee et al. (2010) | |
| ABD1 | Negative | ABI5 | Seo et al. (2014) | |
| CUL3‐BTB | BPM6 | Negative | ATHB6 | Lechner et al. (2011) |
| RING | RSL1 | Negative | PYR1, PYL4 | Bueso et al. (2014) |
| KEG | Negative | CIPK26, ABI5, ABF1, ABF3 | Chen et al. (2013), Liu and Stone (2010, 2013), Lyzenga et al. (2013), Stone et al. (2006) | |
| SDIR1 | Positiveb | SDIRIP1 | Zhang et al. (2015, 2007) | |
| AIP2 | Negative | ABI3 | Kurup et al. (2000) | |
| AIRP3 | Positive | RD21 | Kim and Kim (2013), Pratelli et al. (2012) | |
| U‐box | PUB12 | Positive | ABI1 | Kong et al. (2015) |
| PUB13 | Positive | ABI1 | Kong et al. (2015) | |
| CHIP | Positive | PP2A | Luo et al. (2006) | |
| 26S proteasome | RPN10 | Negative | ABI5 | Smalle et al. (2003) |
Negative: inhibition of ABA signalling.
Positive: activation of ABA signalling.
Two E3 ligase complexes, CRL and RING, target ABA receptors for degradation
CRL and RING E3 ligase complexes were recently demonstrated to target PYR/PYLs for degradation via the Ub‐26S proteasome pathway (Bueso et al., 2014; Irigoyen et al., 2014). De‐etiolated 1 (DET1)‐damaged DNA binding protein 1 (DDB1)‐associated 1 (DDA1) is a component of COP10‐DET1‐DDB1‐related E3 ligase complexes via an interaction with DDB1. DDB1 is an essential member of the cullin 4 (CUL4)/DDB1 E3 ligase complex; it acts as the substrate receptor in CUL4 E3 ligases. DDA1 physically associates with PYL8, PYL4 and PYL9. It has been demonstrated that DDA1 targets PYL8 for degradation through 26S proteasome mediated by ubiquitination (Irigoyen et al., 2014). The presence of ABA blocks the ubiquitination of PYR/PYLs and stabilizes them (Irigoyen et al., 2014). Distinct subcellular localization patterns of E3 ligase complexes involved in ABA signalling have been observed. DDA1 controls the stability of PYL8 through nuclear colocalization (Irigoyen et al., 2014).
Ring finger of seed longevity 1 (RSL1), a single‐subunit RING‐type E3 Ub ligase, interacts directly with the ABA receptors PYR1 and PYL4 (Bueso et al., 2014). RSL1 catalyses the polyubiquitination of PYR1 and PYL4 to promote their degradation via the 26S proteasome (Bueso et al., 2014). Interestingly, RNA interference (RNAi) mutants in which at least three members of the RSL1‐like gene family were silenced showed ABA‐hypersensitive phenotypes in terms of the ABA‐mediated inhibition of seed germination and early seedling growth, whereas RSL1 overexpression reduced ABA sensitivity (Bueso et al., 2014). These findings suggest that RSL1 acts as a negative regulator of ABA signalling by mediating the degradation of ABA receptors (Bueso et al., 2014). The ubiquitination of PYR1 or PYL4 catalysed by RSL1 occurs at the plasma membrane (Bueso et al., 2014). Ubiquitinated plasma membrane proteins are usually delivered to the vacuolar degradation pathway by the endosomal sorting complex required for transport (ESCRT) machinery (MacGurn et al., 2012). Recently, the PYL4 was identified to interact with FYVE 1 (also termed as FYVE domain protein required for endosomal sorting 1, FREE1), a FYVE domain‐containing protein (Belda‐Palazon et al., 2016). FYVE1 is the only ESCRT component that binds to ubiquitin and mediates vacuolar sorting of proteins (Gao et al., 2015; Kolb et al., 2015). Mutations of fyve1 result in the accumulation of PYL4 and showed ABA‐hypersensitive phenotype in seed germination and root growth. The ubiquitylated PYL4 ABA receptor is the substrate for FYVE1 to mediate their delivery to the vacuolar degradation pathway (Belda‐Palazon et al., 2016). This is first discovery on the involvement of the ESCRT machinery in ABA signalling.
PP2C and PP2A are substrates of plant U‐box (PUB) and C‐terminus of Hsc70‐interacting protein (CHIP) E3 ligases
Group A PP2Cs have been found to be substrates of U‐box E3 ligases and RING E3 ubiquitin ligases in ABA signalling (Kong et al., 2015). The PUB E3 ligases PUB12 and PUB13 specifically target ABI1, but not ABI2, for degradation. It has been shown that PYR1 interacts with ABI1 in an ABA‐dependent manner (Park et al., 2009). In an in vitro assay, ABI1 was ubiquitinated by PUB12 and PUB13 only in the presence of ABA and PYR1, indicating that ABA and PYR1 facilitate the ubiquitination of ABI1 (Kong et al., 2015). The double mutant pub12 pub13 showed ABA insensitivity; however, introducing the abi1‐3 loss‐of‐function mutation into that mutant recovered its ABA‐insensitive phenotype. The accumulation of ABI1 in pub12 pub13 mutant plants has also been observed (Kong et al., 2015). These results suggest that ABI1 stability, controlled by PUB12 and PUB13, may contribute to the efficient release of ABI1 repression in the presence of ABA or under conditions of stress (Kong et al., 2015).
The ring domain ligase (RGLG) 1 and RGLG5 have also been demonstrated to be involved in ABA signalling, which belong to RING E3 ligases (Wu et al., 2016; Yin et al., 2007). They physically interact with PP2CA, ABI2 and HAB2, which is enhanced by ABA. RGLG1 and RGLG5 catalyse ubiquitination of PP2CA, possibly ABI2 and HAB2, and target them for degradation through 26S proteasome (Wu et al., 2016). Loss‐of‐function mutants of RGLG1 and RGLG5 showed ABA‐insensitive phenotypes in germination and postgermination growth. These results suggest that both RGLG1 and RGLG5 are positive regulators of ABA signalling through disrupting repressors of ABA signalling to activate ABA pathway (Wu et al., 2016).
CHIP is an E3 Ub ligase possessing monoubiquitination activity in Arabidopsis. Both RCN1 and PP2AA3 regulatory subunits of PP2A are substrates of AtCHIP (Luo et al., 2006). The overexpression of AtCHIP causes ABA hypersensitivity during seed germination and ABA‐induced stomatal closure (Luo et al., 2006). AtCHIP can monoubiquitinate PP2AA3 or RCN1 regulatory subunits of PP2A and enhance PP2A activity under stressful conditions, but it does not affect the enzyme's stability (Luo et al., 2006). However, the position and function of AtCHIP‐mediated PP2AA3 or RCN1 monoubiquitination must be resolved.
Although both PP2Cs and PP2As are regulated by ubiquitination, the regulatory mechanisms are different. PUB12 and PUB13 E3 ligases target ABI1 for degradation via the 26S proteasome (Kong et al., 2015). However, the CHIP E3 ligase monoubiquitinates PP2As, enhancing their phosphatase activity (Luo et al., 2006). These results suggest different roles for ubiquitination in the regulation of ABA signalling.
CRL and RING E3 ligase complexes regulate ABA signalling by targeting transcription factors for turnover
Several transcription factors involved in ABA signalling (ABI5, ABF1, ABF3, ABI3, ATHB6, and salt‐ and drought‐induced RING finger 1 [SDIR1]‐interacting protein 1 [SDIRIP1]) can be ubiquitinated and degraded via Ub‐mediated proteolysis. Further, several specific E3 ligases have been discovered (Chen et al., 2013; Lee et al., 2010; Liu and Stone, 2013; Lyzenga et al., 2013; Seo et al., 2014; Stone et al., 2006).
DWD hypersensitive to ABA1 and ABA2 (DWA1 and DWA2, respectively) and ABA‐hypersensitive DCAF 1 (ABD1), the substrate receptors in CUL4‐RING E3 ligase complexes, are involved in ABA signalling (Lee et al., 2010). DWA1, DWA2 and ABD1 bind to ABI5 and promote its degradation by the 26S proteasome (Lee et al., 2010; Seo et al., 2014). Both dwa1‐1/dwa2‐1 double mutant and abd1 single mutant plants were found to be hypersensitive to ABA during seed germination and seedling growth and to exhibit retarded degradation of ABI5, indicating that DWA1, DWA2 and ABD1 are negative regulators of ABA signalling (Lee et al., 2010; Seo et al., 2014). In addition, ABI5 binding protein (AFP) has been identified as a negative regulator of ABI5 (Lopez‐Molina et al., 2003). AFP represses ABA signalling by promoting ABI5 degradation via the Ub‐mediated 26S proteasome pathway (Lopez‐Molina et al., 2003). However, whether AFP represses signalling by promoting ABI5 degradation is subject to debate. DWA1, DWA2, ABD1 and AFP are localized to the nucleus, where they target ABI5 for Ub‐mediated degradation (Lee et al., 2010; Lopez‐Molina et al., 2003; Seo et al., 2014).
Keep on going (KEG), a RING‐type E3 ligase, negatively regulates ABA signalling by targeting bZIP transcription factors, including ABI5, ABF1 and ABF3, and the kinase CIPK26 for Ub‐mediated degradation (Chen et al., 2013; Liu and Stone, 2013; Lyzenga et al., 2013; Stone et al., 2006). The ubiquitination activity of KEG has been demonstrated in vitro (Stone et al., 2006). A T‐DNA insertion mutant of KEG was shown to be extremely sensitive to ABA in the inhibition of root growth, suggesting that KEG is a negative regulator of ABA signalling (Lyzenga et al., 2013). KEG interacts directly with ABI5, ABF1 and ABF3, which are essential for ABA‐mediated responses. In the absence of ABA, KEG targets ABI5, ABF1 and ABF3 for degradation to maintain low levels of these proteins (Chen et al., 2013; Liu and Stone, 2013; Lyzenga et al., 2013; Stone et al., 2006). The presence of ABA increases the abundance of ABI5, ABF1 and ABF3 and positively regulates their stability (Chen et al., 2013; Lopez‐Molina et al., 2001). ABA promotes KEG self‐ubiquitination and degradation via Ub‐mediated proteolysis, resulting in the turnover of KEG and the accumulation of ABI5, ABF1 and ABF3 leading to downstream ABA responses (Chen et al., 2013; Liu and Stone, 2010). KEG is localized to the trans‐Golgi network/early endosome and interacts with ABI5. It is thought that KEG targets ABI5 for degradation in the cytoplasm or trans‐Golgi network before it reaches the nucleus (Liu and Stone, 2013). Moreover, KEG interacts physically with and ubiquitinates CIPK26, causing its degradation by the 26S proteasome. KEG overexpression enhances the turnover of CIPK26 (Lyzenga et al., 2013).
Similar to ABI5, the level of ABI3 is regulated by proteasomal degradation mediated by ABI3‐interacting protein 2 (AIP2), a C3H2C3‐type RING‐motif protein that functions as an E3 ligase. ABI3 is an unstable protein containing four functional domains: A1, B1, B2 and B3 (Kurup et al., 2000). AIP2 interacts physically with ABI3 and acts as a negative regulator of ABA signalling by targeting ABI3 for destruction through the 26S proteasome. The null mutant aip2‐1 was shown to possess increased levels of ABI3 compared with wild type and to be hypersensitive to ABA. Further work revealed that exogenous or endogenous ABA facilitates the accumulation of ABI3 mRNA and protein, and it triggers downstream ABA responses by directly or indirectly interrupting the interaction of AIP2 with ABI3 (Kurup et al., 2000).
As substrate‐binding adaptors of CUL3‐based Ub E3 ligases, meprin and TRAF homology‐BTB proteins (termed BPMs) target a class I homeobox‐leucine zipper transcription factor, ATHB6, for degradation (Lechner et al., 2011). ATHB6 functions as a negative regulator of ABA responses (Himmelbach et al., 2002). In bpm RNAi lines, the level of ATHB6 was increased while the ubiquitinated form of ATHB6 was decreased compared to wild type. However, overexpression of BPM6 accelerated the turnover of ATHB6, suggesting that BPMs influence ATHB6 stability (Lechner et al., 2011). Further, bpm RNAi transgenic lines exhibited an ABA‐insensitive phenotype in stomatal closure, indicating that BPMs play a positive role in ABA signalling (Johannesson et al., 2003; Lechner et al., 2011).
SDIR1, a RING finger E3 ligase, is also involved in ABA signal transduction (Zhang et al., 2007, 2015). The expression of SDIR1 is universal, especially in stomatal guard cells and leaf mesophyll cells under drought stress. The RING finger domain of SDIR1 is required for its E3 Ub ligase activity (Zhang et al., 2007). Overexpression of SDIR1 leads to ABA‐hypersensitive phenotypes in seed germination and stomatal closure. In contrast, the sdir1‐1 mutant exhibited an ABA‐insensitive phenotype (Zhang et al., 2007). The results of genetic studies show that SDIR1 acts upstream of bZIP family genes, including ABI5, ABF3 and ABF4, and that it positively regulates ABA signalling (Zhang et al., 2007). SDIRIP1, a direct target of SDIR1, was recently identified using cell biological, molecular biological and biochemical approaches (Zhang et al., 2015). SDIR1 interacts with and ubiquitinates SDIRIP1, marking it for degradation by the 26S proteasome. SDIRIP1‐RNAi lines showed ABA‐hypersensitive phenotypes, indicating that SDIRIP1 acts as a negative regulator of ABA signalling. SDIRIP1 selectively regulates the expression of ABI5, rather than ABF3 or ABF4, to regulate ABA‐mediated seed germination and responses to salt (Zhang et al., 2015). Overall, the SDIR1/SDIRIP1 complex plays a vital role in ABA signalling via the ubiquitination pathway (Zhang et al., 2007, 2015).
RPN10, a component of the 26S proteasome, regulates ABI5 stability
The selectivity of the Ub‐mediated 26S proteasome pathway is achieved not only at the level of ubiquitination, but also by the proteasome itself (Smalle and Vierstra, 2004). The 26S proteasome is an ATP‐dependent, self‐compartmentalized protease that can be divided into two particles: the 20S core protease (CP) and 19S regulatory particle (RP). The RP confers both ATP dependence and substrate specificity to the holoenzyme, especially with respect to those substrates bearing a polyubiquitin tag, by binding to each end of the CP (Smalle and Vierstra, 2004). RPN10 is a subunit of the RP in Arabidopsis. The mutant rpn10‐1 showed increased ABA sensitivity and ABI5 accumulation, suggesting that RPN10 is essential for the degradation of ABI5 by the proteasome. The kinetics of ABI5 accumulation with and without ABA suggests that ABI5 is normally short‐lived. Although multiple mechanisms could explain this, the simplest explanation is that ABI5 turnover is achieved through the interaction of ABI5 with RPN10. This interaction may be direct or indirect, dependent or independent of previous ubiquitination events, and may involve adaptor proteins such as radiation sensitive (RAD) 23, the RAD23/ABI3 complex or DSK2 to promote the association of ABI5 with the protease. ABA may stabilize ABI5 by preventing this association. Clearly, identifying the proteins that interact with RPN10 will be critical in defining how it helps to deliver appropriate cargo to the 26S proteasome (Smalle et al., 2003).
AtAIRP3/LOG2, a RING E3 ligase, targets RESPONSIVE TO DEHYDRATION 21 (RD21), an ABA‐responsive gene, for disruption
Besides the aforementioned post‐translational regulation of upstream factors involved in ABA perception and signal transduction, downstream ABA‐responsive genes are subject to ubiquitination (Kim and Kim, 2013). AtAIRP3/LOG2, a RING‐type E3 ligase, is involved in ABA signalling. Both the knockout mutant atairp3/log2‐2 and AtAIRP3‐RNAi knockdown transgenic plants exhibited ABA‐insensitive phenotypes in the inhibition of seed germination and stomatal closure, indicating a positive role for AIRP3 in ABA responses (Kim and Kim, 2013; Pratelli et al., 2012). In Arabidopsis, AtAIRP3/LOG2 interacts with RD21, a drought‐inducible cysteine proteinase belonging to the papain family (Kim and Kim, 2013). AIRP3 ubiquitinates RD21, targeting it for degradation by the 26S proteasome. These data indicate that AIPR3 functions as an E3 ligase towards RD21 and that it acts positively in ABA signalling (Kim and Kim, 2013).
Perspectives
ABA plays important roles in regulating plant development and tolerance to biotic and abiotic stresses. As the characterization of PYR/PYLs as ABA receptors, great progress has been made in understanding the regulation (especially the post‐translational regulation) of ABA signalling. Dephosphorylation, phosphorylation and ubiquitination have all been reported to be involved in ABA signalling. After binding to ABA, PYR/PYLs interact physically with group A PP2Cs, disrupting the interaction between PP2Cs and SnRK2s, blocking the dephosphorylation activity of PP2Cs and activating SnRK2s to trigger downstream ABA responses. Multiple substrates of SnRK2s have been identified, including transcription factors, ion channel proteins, a potassium channel protein and a chromatin‐remodelling factor. The ubiquitination of ABA receptors and signalling molecules was recently reported, implying the importance of this modification in ABA signalling. However, certain problems must be addressed in order for us to fully understand the molecular networks involved in ABA signalling.
First, studies are needed to dissect the transcriptional, post‐transcriptional and post‐translational regulatory mechanisms of PYR/PYLs. Although it has been shown that PYL4, PYL8 and PYL9 are targeted by DDA1 for degradation by the 26S proteasome, PYR1 and PYL4 are substrates of RSL1, an E3 ubiquitin ligase; thus, the regulation of ABA receptors is far from clear (Bueso et al., 2014; Irigoyen et al., 2014). Second, even though autophosphorylation is a possible means of regulation for SnRK2s, the mechanisms underlying the internal phosphorylation of SnRK2 are obscure (Sirichandra et al., 2009b). In addition, other types of kinases, similar to SnRK2s and CIPK26, are likely involved in ABA signalling. Third, the targets of a great number of E3 Ub ligases involved in ABA signalling should be clarified in the future. Numerous E3 ligases, including DOR (Zhang et al., 2008), EID1‐like protein 3 (Koops et al., 2011), MAX2 (Bu et al., 2014), DWA3 (Lee et al., 2011), PUB9 (Samuel et al., 2008), PUB18 (Bergler and Hoth, 2011), PUB19 (Liu et al., 2011), AIRP1 (Ryu et al., 2010), AIRP2 (Cho et al., 2011), AIRP4 (Yang et al., 2016), ATL43 (Serrano et al., 2006), ECERIFERUM 9 (CER9) (Zhao et al., 2014), RDUF1/2 (Kim et al., 2012), RHA2a/2b (Bu et al., 2009; Li et al., 2011) and TLP3/9 (Bao et al., 2014), have been shown to play positive or negative roles in ABA signalling. However, their specificities are unknown.
The identification of their substrates will deepen our understanding of the mechanisms underlying ABA signalling networks.
Conflict of Interest
There is no any conflict of interest being declared.
Funding
This work was supported by National Natural Science Foundation of China (NSFC) [grant number 31371222] and Doctoral Fund of Ministry of Education of China (RFDP) [grant number 20132103110004].
Acknowledgements
We thank Dr. Jessica Habashi for her help in English editing of the manuscript. We also apologize to colleagues whose work was not cited due to space limitations.
References
- Antoni, R. , Gonzalez‐Guzman, M. , Rodriguez, L. , Rodrigues, A. , Pizzio, G.A. and Rodriguez, P.L. (2012) Selective inhibition of clade A phosphatases type 2C by PYR/PYL/RCAR abscisic acid receptors. Plant Physiol. 158, 970–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao, Y. , Song, W.M. , Jin, Y.L. , Jiang, C.M. , Yang, Y. , Li, B. , Huang, W.J. et al (2014) Characterization of Arabidopsis Tubby‐like proteins and redundant function of AtTLP3 and AtTLP9 in plant response to ABA and osmotic stress. Plant Mol. Biol. 86, 471–483. [DOI] [PubMed] [Google Scholar]
- Belda‐Palazon, B. , Rodriguez, L. , Fernandez, M.A. , Castillo, M.C. , Anderson, E.A. , Gao, C. , Gonzalez‐Guzman, M. et al (2016) FYVE1/FREE1 interacts with the PYL4 ABA receptor and mediates its delivery to the vacuolar degradation pathway. Plant Cell, 28, 2291–2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergler, J. and Hoth, S. (2011) Plant U‐box armadillo repeat proteins AtPUB18 and AtPUB19 are involved in salt inhibition of germination in Arabidopsis. Plant Biol. (Stuttg) 13, 725–730. [DOI] [PubMed] [Google Scholar]
- Bhaskara, G.B. , Nguyen, T.T. and Verslues, P.E. (2012) Unique drought resistance functions of the highly ABA‐induced clade A protein phosphatase 2Cs. Plant Physiol. 160, 379–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudsocq, M. , Barbier‐Brygoo, H. and Lauriere, C. (2004) Identification of nine sucrose nonfermenting 1‐related protein kinases 2 activated by hyperosmotic and saline stresses in Arabidopsis thaliana. J. Biol. Chem. 279, 41758–41766. [DOI] [PubMed] [Google Scholar]
- Boudsocq, M. , Droillard, M.J. , Barbier‐Brygoo, H. and Lauriere, C. (2007) Different phosphorylation mechanisms are involved in the activation of sucrose non‐fermenting 1 related protein kinases 2 by osmotic stresses and abscisic acid. Plant Mol. Biol. 63, 491–503. [DOI] [PubMed] [Google Scholar]
- Brandt, B. , Brodsky, D.E. , Xue, S. , Negi, J. , Iba, K. , Kangasjarvi, J. , Ghassemian, M. et al (2012) Reconstitution of abscisic acid activation of SLAC1 anion channel by CPK6 and OST1 kinases and branched ABI1 PP2C phosphatase action. Proc. Natl. Acad. Sci. USA, 109, 10593–10598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bu, Q. , Li, H. , Zhao, Q. , Jiang, H. , Zhai, Q. , Zhang, J. , Wu, X. et al (2009) The Arabidopsis RING finger E3 ligase RHA2a is a novel positive regulator of abscisic acid signaling during seed germination and early seedling development. Plant Physiol. 150, 463–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bu, Q. , Lv, T. , Shen, H. , Luong, P. , Wang, J. , Wang, Z. , Huang, Z. et al (2014) Regulation of drought tolerance by the F‐box protein MAX2 in Arabidopsis. Plant Physiol. 164, 424–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bueso, E. , Rodriguez, L. , Lorenzo‐Orts, L. , Gonzalez‐Guzman, M. , Sayas, E. , Munoz‐Bertomeu, J. , Ibanez, C. et al (2014) The single‐subunit RING‐type E3 ubiquitin ligase RSL1 targets PYL4 and PYR1 ABA receptors in plasma membrane to modulate abscisic acid signaling. Plant J. 80, 1057–1071. [DOI] [PubMed] [Google Scholar]
- Cai, Z. , Liu, J. , Wang, H. , Yang, C. , Chen, Y. , Li, Y. , Pan, S. et al (2014) GSK3‐like kinases positively modulate abscisic acid signaling through phosphorylating subgroup III SnRK2s in Arabidopsis. Proc. Natl. Acad. Sci. USA, 111, 9651–9656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charpentier, M. , Sun, J. , Wen, J. , Mysore, K.S. and Oldroyd, G.E. (2014) Abscisic acid promotion of arbuscular mycorrhizal colonization requires a component of the PROTEIN PHOSPHATASE 2A complex. Plant Physiol. 166, 2077–2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y.T. , Liu, H. , Stone, S. and Callis, J. (2013) ABA and the ubiquitin E3 ligase KEEP ON GOING affect proteolysis of the Arabidopsis thaliana transcription factors ABF1 and ABF3. Plant J. 75, 965–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho, S.K. , Ryu, M.Y. , Seo, D.H. , Kang, B.G. and Kim, W.T. (2011) The Arabidopsis RING E3 ubiquitin ligase AtAIRP2 plays combinatory roles with AtAIRP1 in abscisic acid‐mediated drought stress responses. Plant Physiol. 157, 2240–2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler, S.R. , Rodriguez, P.L. , Finkelstein, R.R. and Abrams, S.R. (2010) Abscisic acid: emergence of a core signaling network. Annu. Rev. Plant Biol. 61, 651–679. [DOI] [PubMed] [Google Scholar]
- DeLong, A. (2006) Switching the flip: protein phosphatase roles in signaling pathways. Curr. Opin. Plant Biol. 9, 470–477. [DOI] [PubMed] [Google Scholar]
- Farkas, I. , Dombradi, V. , Miskei, M. , Szabados, L. and Koncz, C. (2007) Arabidopsis PPP family of serine/threonine phosphatases. Trends Plant Sci. 12, 169–176. [DOI] [PubMed] [Google Scholar]
- Finkelstein, R. (2013) Abscisic Acid synthesis and response. Arabidopsis Book, 11, e0166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs, S. , Grill, E. , Meskiene, I. and Schweighofer, A. (2013) Type 2C protein phosphatases in plants. FEBS J. 280, 681–693. [DOI] [PubMed] [Google Scholar]
- Fuchs, S. , Tischer, S.V. , Wunschel, C. , Christmann, A. and Grill, E. (2014) Abscisic acid sensor RCAR7/PYL13, specific regulator of protein phosphatase coreceptors. Proc. Natl. Acad. Sci. USA, 111, 5741–5746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii, H. and Zhu, J.K. (2009) Arabidopsis mutant deficient in 3 abscisic acid‐activated protein kinases reveals critical roles in growth, reproduction, and stress. Proc. Natl. Acad. Sci. USA, 106, 8380–8385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii, H. , Verslues, P.E. and Zhu, J.K. (2007) Identification of two protein kinases required for abscisic acid regulation of seed germination, root growth, and gene expression in Arabidopsis. Plant Cell, 19, 485–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii, H. , Chinnusamy, V. , Rodrigues, A. , Rubio, S. , Antoni, R. , Park, S.Y. , Cutler, S.R. et al (2009) In vitro reconstitution of an abscisic acid signalling pathway. Nature, 462, 660–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita, Y. , Nakashima, K. , Yoshida, T. , Katagiri, T. , Kidokoro, S. , Kanamori, N. , Umezawa, T. et al (2009) Three SnRK2 protein kinases are the main positive regulators of abscisic acid signaling in response to water stress in Arabidopsis. Plant Cell Physiol. 50, 2123–2132. [DOI] [PubMed] [Google Scholar]
- Fujita, Y. , Fujita, M. , Shinozaki, K. and Yamaguchi‐Shinozaki, K. (2011) ABA‐mediated transcriptional regulation in response to osmotic stress in plants. J. Plant. Res. 124, 509–525. [DOI] [PubMed] [Google Scholar]
- Furihata, T. , Maruyama, K. , Fujita, Y. , Umezawa, T. , Yoshida, R. , Shinozaki, K. and Yamaguchi‐Shinozaki, K. (2006) Abscisic acid‐dependent multisite phosphorylation regulates the activity of a transcription activator AREB1. Proc. Natl. Acad. Sci. USA, 103, 1988–1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, C. , Zhuang, X. , Cui, Y. , Fu, X. , He, Y. , Zhao, Q. , Zeng, Y. et al (2015) Dual roles of an Arabidopsis ESCRT component FREE1 in regulating vacuolar protein transport and autophagic degradation. Proc. Natl. Acad. Sci. USA, 112, 1886–1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger, D. , Scherzer, S. , Mumm, P. , Stange, A. , Marten, I. , Bauer, H. , Ache, P. et al (2009) Activity of guard cell anion channel SLAC1 is controlled by drought‐stress signaling kinase‐phosphatase pair. Proc. Natl. Acad. Sci. USA, 106, 21425–21430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosti, F. , Beaudoin, N. , Serizet, C. , Webb, A.A. , Vartanian, N. and Giraudat, J. (1999) ABI1 protein phosphatase 2C is a negative regulator of abscisic acid signaling. Plant Cell, 11, 1897–1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimes, C.A. and Jope, R.S. (2001) The multifaceted roles of glycogen synthase kinase 3beta in cellular signaling. Prog. Neurobiol. 65, 391–426. [DOI] [PubMed] [Google Scholar]
- Han, S.K. , Sang, Y. , Rodrigues, A. , Wu, M.F. , Rodriguez, P.L. and Wagner, D. (2012) The SWI2/SNF2 chromatin remodeling ATPase BRAHMA represses abscisic acid responses in the absence of the stress stimulus in Arabidopsis. Plant Cell, 24, 4892–4906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao, Q. , Yin, P. , Li, W. , Wang, L. , Yan, C. , Lin, Z. , Wu, J.Z. et al (2011) The molecular basis of ABA‐independent inhibition of PP2Cs by a subclass of PYL proteins. Mol. Cell, 42, 662–672. [DOI] [PubMed] [Google Scholar]
- Hauser, F. , Waadt, R. and Schroeder, J.I. (2011) Evolution of abscisic acid synthesis and signaling mechanisms. Curr. Biol. 21, R346–R355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, J.X. , Gendron, J.M. , Yang, Y. , Li, J. and Wang, Z.Y. (2002) The GSK3‐like kinase BIN2 phosphorylates and destabilizes BZR1, a positive regulator of the brassinosteroid signaling pathway in Arabidopsis. Proc. Natl. Acad. Sci. USA, 99, 10185–10190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himmelbach, A. , Hoffmann, T. , Leube, M. , Hohener, B. and Grill, E. (2002) Homeodomain protein ATHB6 is a target of the protein phosphatase ABI1 and regulates hormone responses in Arabidopsis. EMBO J. 21, 3029–3038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirayama, T. and Shinozaki, K. (2007) Perception and transduction of abscisic acid signals: keys to the function of the versatile plant hormone ABA. Trends Plant Sci. 12, 343–351. [DOI] [PubMed] [Google Scholar]
- Hrabak, E.M. , Chan, C.W. , Gribskov, M. , Harper, J.F. , Choi, J.H. , Halford, N. , Kudla, J. et al (2003) The Arabidopsis CDPK‐SnRK superfamily of protein kinases. Plant Physiol. 132, 666–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua, Z. and Vierstra, R.D. (2011) The cullin‐RING ubiquitin‐protein ligases. Annu. Rev. Plant Biol. 62, 299–334. [DOI] [PubMed] [Google Scholar]
- Imes, D. , Mumm, P. , Bohm, J. , Al‐Rasheid, K.A. , Marten, I. , Geiger, D. and Hedrich, R. (2013) Open stomata 1 (OST1) kinase controls R‐type anion channel QUAC1 in Arabidopsis guard cells. Plant J. 74, 372–382. [DOI] [PubMed] [Google Scholar]
- Irigoyen, M.L. , Iniesto, E. , Rodriguez, L. , Puga, M.I. , Yanagawa, Y. , Pick, E. , Strickland, E. et al (2014) Targeted degradation of abscisic acid receptors is mediated by the ubiquitin ligase substrate adaptor DDA1 in Arabidopsis. Plant Cell, 26, 712–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer, L.M. , Koonin, E.V. and Aravind, L. (2001) Adaptations of the helix‐grip fold for ligand binding and catalysis in the START domain superfamily. Proteins, 43, 134–144. [DOI] [PubMed] [Google Scholar]
- Johannesson, H. , Wang, Y. , Hanson, J. and Engstrom, P. (2003) The Arabidopsis thaliana homeobox gene ATHB5 is a potential regulator of abscisic acid responsiveness in developing seedlings. Plant Mol. Biol. 51, 719–729. [DOI] [PubMed] [Google Scholar]
- Kim, J.H. and Kim, W.T. (2013) The Arabidopsis RING E3 ubiquitin ligase AtAIRP3/LOG2 participates in positive regulation of high‐salt and drought stress responses. Plant Physiol. 162, 1733–1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, S.J. , Ryu, M.Y. and Kim, W.T. (2012) Suppression of Arabidopsis RING‐DUF1117 E3 ubiquitin ligases, AtRDUF1 and AtRDUF2, reduces tolerance to ABA‐mediated drought stress. Biochem. Biophys. Res. Commun. 420, 141–147. [DOI] [PubMed] [Google Scholar]
- Kobayashi, Y. , Murata, M. , Minami, H. , Yamamoto, S. , Kagaya, Y. , Hobo, T. , Yamamoto, A. et al (2005) Abscisic acid‐activated SNRK2 protein kinases function in the gene‐regulation pathway of ABA signal transduction by phosphorylating ABA response element‐binding factors. Plant J. 44, 939–949. [DOI] [PubMed] [Google Scholar]
- Kolb, C. , Nagel, M.K. , Kalinowska, K. , Hagmann, J. , Ichikawa, M. , Anzenberger, F. , Alkofer, A. et al (2015) FYVE1 is essential for vacuole biogenesis and intracellular trafficking in Arabidopsis. Plant Physiol. 167, 1361–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong, L. , Cheng, J. , Zhu, Y. , Ding, Y. , Meng, J. , Chen, Z. , Xie, Q. et al (2015) Degradation of the ABA co‐receptor ABI1 by PUB12/13 U‐box E3 ligases. Nat. Commun. 6, 8630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koops, P. , Pelser, S. , Ignatz, M. , Klose, C. , Marrocco‐Selden, K. and Kretsch, T. (2011) EDL3 is an F‐box protein involved in the regulation of abscisic acid signalling in Arabidopsis thaliana. J. Exp. Bot. 62, 5547–5560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn, J.M. , Boisson‐Dernier, A. , Dizon, M.B. , Maktabi, M.H. and Schroeder, J.I. (2006) The protein phosphatase AtPP2CA negatively regulates abscisic acid signal transduction in Arabidopsis, and effects of abh1 on AtPP2CA mRNA. Plant Physiol. 140, 127–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurup, S. , Jones, H.D. and Holdsworth, M.J. (2000) Interactions of the developmental regulator ABI3 with proteins identified from developing Arabidopsis seeds. Plant J. 21, 143–155. [DOI] [PubMed] [Google Scholar]
- Kwak, J.M. , Moon, J.H. , Murata, Y. , Kuchitsu, K. , Leonhardt, N. , DeLong, A. and Schroeder, J.I. (2002) Disruption of a guard cell‐expressed protein phosphatase 2A regulatory subunit, RCN1, confers abscisic acid insensitivity in Arabidopsis. Plant Cell, 14, 2849–2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechner, E. , Leonhardt, N. , Eisler, H. , Parmentier, Y. , Alioua, M. , Jacquet, H. , Leung, J. et al (2011) MATH/BTB CRL3 receptors target the homeodomain‐leucine zipper ATHB6 to modulate abscisic acid signaling. Dev. Cell, 21, 1116–1128. [DOI] [PubMed] [Google Scholar]
- Lee, S.C. , Lan, W. , Buchanan, B.B. and Luan, S. (2009) A protein kinase‐phosphatase pair interacts with an ion channel to regulate ABA signaling in plant guard cells. Proc. Natl. Acad. Sci. USA, 106, 21419–21424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J.H. , Yoon, H.J. , Terzaghi, W. , Martinez, C. , Dai, M. , Li, J. , Byun, M.O. et al (2010) DWA1 and DWA2, two Arabidopsis DWD protein components of CUL4‐based E3 ligases, act together as negative regulators in ABA signal transduction. Plant Cell, 22, 1716–1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J.H. , Terzaghi, W. and Deng, X.W. (2011) DWA3, an Arabidopsis DWD protein, acts as a negative regulator in ABA signal transduction. Plant Sci. 180, 352–357. [DOI] [PubMed] [Google Scholar]
- Li, H. , Jiang, H. , Bu, Q. , Zhao, Q. , Sun, J. , Xie, Q. and Li, C. (2011) The Arabidopsis RING finger E3 ligase RHA2b acts additively with RHA2a in regulating abscisic acid signaling and drought response. Plant Physiol. 156, 550–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, H. and Stone, S.L. (2010) Abscisic acid increases Arabidopsis ABI5 transcription factor levels by promoting KEG E3 ligase self‐ubiquitination and proteasomal degradation. Plant Cell, 22, 2630–2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, H. and Stone, S.L. (2013) Cytoplasmic degradation of the Arabidopsis transcription factor abscisic acid insensitive 5 is mediated by the RING‐type E3 ligase KEEP ON GOING. J. Biol. Chem. 288, 20267–20279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y.C. , Wu, Y.R. , Huang, X.H. , Sun, J. and Xie, Q. (2011) AtPUB19, a U‐box E3 ubiquitin ligase, negatively regulates abscisic acid and drought responses in Arabidopsis thaliana. Mol Plant, 4, 938–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Loon, L.C. , Rep, M. and Pieterse, C.M. (2006) Significance of inducible defense‐related proteins in infected plants. Annu. Rev. Phytopathol. 44, 135–162. [DOI] [PubMed] [Google Scholar]
- Lopez‐Molina, L. , Mongrand, S. and Chua, N.H. (2001) A postgermination developmental arrest checkpoint is mediated by abscisic acid and requires the ABI5 transcription factor in Arabidopsis. Proc. Natl. Acad. Sci. USA, 98, 4782–4787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez‐Molina, L. , Mongrand, S. , Kinoshita, N. and Chua, N.H. (2003) AFP is a novel negative regulator of ABA signaling that promotes ABI5 protein degradation. Genes Dev. 17, 410–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, J. , Shen, G. , Yan, J. , He, C. and Zhang, H. (2006) AtCHIP functions as an E3 ubiquitin ligase of protein phosphatase 2A subunits and alters plant response to abscisic acid treatment. Plant J. 46, 649–657. [DOI] [PubMed] [Google Scholar]
- Lyzenga, W.J. , Liu, H. , Schofield, A. , Muise‐Hennessey, A. and Stone, S.L. (2013) Arabidopsis CIPK26 interacts with KEG, components of the ABA signalling network and is degraded by the ubiquitin‐proteasome system. J. Exp. Bot. 64, 2779–2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, Y. , Szostkiewicz, I. , Korte, A. , Moes, D. , Yang, Y. , Christmann, A. and Grill, E. (2009) Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science, 324, 1064–1068. [DOI] [PubMed] [Google Scholar]
- MacGurn, J.A. , Hsu, P.C. and Emr, S.D. (2012) Ubiquitin and membrane protein turnover: from cradle to grave. Annu. Rev. Biochem. 81, 231–259. [DOI] [PubMed] [Google Scholar]
- McConnell, J.R. , Emery, J. , Eshed, Y. , Bao, N. , Bowman, J. and Barton, M.K. (2001) Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature, 411, 709–713. [DOI] [PubMed] [Google Scholar]
- Meggio, F. and Pinna, L.A. (2003) One‐thousand‐and‐one substrates of protein kinase CK2? FASEB J. 17, 349–368. [DOI] [PubMed] [Google Scholar]
- Melcher, K. , Ng, L.M. , Zhou, X.E. , Soon, F.F. , Xu, Y. , Suino‐Powell, K.M. , Park, S.Y. et al (2009) A gate‐latch‐lock mechanism for hormone signalling by abscisic acid receptors. Nature, 462, 602–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melotto, M. , Underwood, W. , Koczan, J. , Nomura, K. and He, S.Y. (2006) Plant stomata function in innate immunity against bacterial invasion. Cell, 126, 969–980. [DOI] [PubMed] [Google Scholar]
- Merlot, S. , Gosti, F. , Guerrier, D. , Vavasseur, A. and Giraudat, J. (2001) The ABI1 and ABI2 protein phosphatases 2C act in a negative feedback regulatory loop of the abscisic acid signalling pathway. Plant J. 25, 295–303. [DOI] [PubMed] [Google Scholar]
- Miyazono, K. , Miyakawa, T. , Sawano, Y. , Kubota, K. , Kang, H.J. , Asano, A. , Miyauchi, Y. et al (2009) Structural basis of abscisic acid signalling. Nature, 462, 609–614. [DOI] [PubMed] [Google Scholar]
- Mulekar, J.J. , Bu, Q. , Chen, F. and Huq, E. (2012) Casein kinase II alpha subunits affect multiple developmental and stress‐responsive pathways in Arabidopsis. Plant J. 69, 343–354. [DOI] [PubMed] [Google Scholar]
- Mustilli, A.C. , Merlot, S. , Vavasseur, A. , Fenzi, F. and Giraudat, J. (2002) Arabidopsis OST1 protein kinase mediates the regulation of stomatal aperture by abscisic acid and acts upstream of reactive oxygen species production. Plant Cell, 14, 3089–3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng, L.M. , Soon, F.F. , Zhou, X.E. , West, G.M. , Kovach, A. , Suino‐Powell, K.M. , Chalmers, M.J. et al (2011) Structural basis for basal activity and autoactivation of abscisic acid (ABA) signaling SnRK2 kinases. Proc. Natl. Acad. Sci. USA, 108, 21259–21264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura, N. , Yoshida, T. , Kitahata, N. , Asami, T. , Shinozaki, K. and Hirayama, T. (2007) ABA‐Hypersensitive Germination1 encodes a protein phosphatase 2C, an essential component of abscisic acid signaling in Arabidopsis seed. Plant J. 50, 935–949. [DOI] [PubMed] [Google Scholar]
- Nishimura, N. , Hitomi, K. , Arvai, A.S. , Rambo, R.P. , Hitomi, C. , Cutler, S.R. , Schroeder, J.I. et al (2009) Structural mechanism of abscisic acid binding and signaling by dimeric PYR1. Science, 326, 1373–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura, N. , Sarkeshik, A. , Nito, K. , Park, S.Y. , Wang, A. , Carvalho, P.C. , Lee, S. et al (2010) PYR/PYL/RCAR family members are major in‐vivo ABI1 protein phosphatase 2C‐interacting proteins in Arabidopsis. Plant J. 61, 290–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, S.Y. , Fung, P. , Nishimura, N. , Jensen, D.R. , Fujii, H. , Zhao, Y. , Lumba, S. et al (2009) Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science, 324, 1068–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peirats‐Llobet, M. , Han, S.K. , Gonzalez‐Guzman, M. , Jeong, C.W. , Rodriguez, L. , Belda‐Palazon, B. , Wagner, D. et al (2016) A direct link between abscisic acid sensing and the chromatin‐remodeling ATPase BRAHMA via core ABA signaling pathway components. Mol. Plant, 9, 136–147. [DOI] [PubMed] [Google Scholar]
- Pernas, M. , Garcia‐Casado, G. , Rojo, E. , Solano, R. and Sanchez‐Serrano, J.J. (2007) A protein phosphatase 2A catalytic subunit is a negative regulator of abscisic acid signalling. Plant J. 51, 763–778. [DOI] [PubMed] [Google Scholar]
- Pizzio, G.A. , Rodriguez, L. , Antoni, R. , Gonzalez‐Guzman, M. , Yunta, C. , Merilo, E. , Kollist, H. et al (2013) The PYL4 A194T mutant uncovers a key role of PYR1‐LIKE4/PROTEIN PHOSPHATASE 2CA interaction for abscisic acid signaling and plant drought resistance. Plant Physiol. 163, 441–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratelli, R. , Guerra, D.D. , Yu, S. , Wogulis, M. , Kraft, E. , Frommer, W.B. , Callis, J. et al (2012) The ubiquitin E3 ligase LOSS OF GDU2 is required for GLUTAMINE DUMPER1‐induced amino acid secretion in Arabidopsis. Plant Physiol. 158, 1628–1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radauer, C. , Lackner, P. and Breiteneder, H. (2008) The Bet v 1 fold: an ancient, versatile scaffold for binding of large, hydrophobic ligands. BMC Evol. Biol. 8, 286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riera, M. , Irar, S. , Velez‐Bermudez, I.C. , Carretero‐Paulet, L. , Lumbreras, V. and Pages, M. (2011) Role of plant‐specific N‐terminal domain of maize CK2beta1 subunit in CK2beta functions and holoenzyme regulation. PLoS ONE, 6, e21909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert, N. , Merlot, S. , N'Guyen, V. , Boisson‐Dernier, A. and Schroeder, J.I. (2006) A hypermorphic mutation in the protein phosphatase 2C HAB1 strongly affects ABA signaling in Arabidopsis. FEBS Lett. 580, 4691–4696. [DOI] [PubMed] [Google Scholar]
- Rubio, S. , Rodrigues, A. , Saez, A. , Dizon, M.B. , Galle, A. , Kim, T.H. , Santiago, J. et al (2009) Triple loss of function of protein phosphatases type 2C leads to partial constitutive response to endogenous abscisic acid. Plant Physiol. 150, 1345–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu, M.Y. , Cho, S.K. and Kim, W.T. (2010) The Arabidopsis C3H2C3‐type RING E3 ubiquitin ligase AtAIRP1 is a positive regulator of an abscisic acid‐dependent response to drought stress. Plant Physiol. 154, 1983–1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saez, A. , Apostolova, N. , Gonzalez‐Guzman, M. , Gonzalez‐Garcia, M.P. , Nicolas, C. , Lorenzo, O. and Rodriguez, P.L. (2004) Gain‐of‐function and loss‐of‐function phenotypes of the protein phosphatase 2C HAB1 reveal its role as a negative regulator of abscisic acid signalling. Plant J. 37, 354–369. [DOI] [PubMed] [Google Scholar]
- Saez, A. , Robert, N. , Maktabi, M.H. , Schroeder, J.I. , Serrano, R. and Rodriguez, P.L. (2006) Enhancement of abscisic acid sensitivity and reduction of water consumption in Arabidopsis by combined inactivation of the protein phosphatases type 2C ABI1 and HAB1. Plant Physiol. 141, 1389–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saidi, Y. , Hearn, T.J. and Coates, J.C. (2012) Function and evolution of ‘green’ GSK3/Shaggy‐like kinases. Trends Plant Sci. 17, 39–46. [DOI] [PubMed] [Google Scholar]
- Saito, N. , Munemasa, S. , Nakamura, Y. , Shimoishi, Y. , Mori, I.C. and Murata, Y. (2008) Roles of RCN1, regulatory A subunit of protein phosphatase 2A, in methyl jasmonate signaling and signal crosstalk between methyl jasmonate and abscisic acid. Plant Cell Physiol. 49, 1396–1401. [DOI] [PubMed] [Google Scholar]
- Salinas, P. , Fuentes, D. , Vidal, E. , Jordana, X. , Echeverria, M. and Holuigue, L. (2006) An extensive survey of CK2 alpha and beta subunits in Arabidopsis: multiple isoforms exhibit differential subcellular localization. Plant Cell Physiol. 47, 1295–1308. [DOI] [PubMed] [Google Scholar]
- Samuel, M.A. , Mudgil, Y. , Salt, J.N. , Delmas, F. , Ramachandran, S. , Chilelli, A. and Goring, D.R. (2008) Interactions between the S‐domain receptor kinases and AtPUB‐ARM E3 ubiquitin ligases suggest a conserved signaling pathway in Arabidopsis. Plant Physiol. 147, 2084–2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago, J. , Rodrigues, A. , Saez, A. , Rubio, S. , Antoni, R. , Dupeux, F. , Park, S.Y. et al (2009) Modulation of drought resistance by the abscisic acid receptor PYL5 through inhibition of clade A PP2Cs. Plant J. 60, 575–588. [DOI] [PubMed] [Google Scholar]
- Saruhashi, M. , Kumar Ghosh, T. , Arai, K. , Ishizaki, Y. , Hagiwara, K. , Komatsu, K. , Shiwa, Y. et al (2015) Plant Raf‐like kinase integrates abscisic acid and hyperosmotic stress signaling upstream of SNF1‐related protein kinase2. Proc. Natl. Acad. Sci. USA, 112, E6388–E6396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki, T. , Mori, I.C. , Furuichi, T. , Munemasa, S. , Toyooka, K. , Matsuoka, K. , Murata, Y. et al (2010) Closing plant stomata requires a homolog of an aluminum‐activated malate transporter. Plant Cell Physiol. 51, 354–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato, A. , Sato, Y. , Fukao, Y. , Fujiwara, M. , Umezawa, T. , Shinozaki, K. , Hibi, T. et al (2009) Threonine at position 306 of the KAT1 potassium channel is essential for channel activity and is a target site for ABA‐activated SnRK2/OST1/SnRK2.6 protein kinase. Biochem. J. 424, 439–448. [DOI] [PubMed] [Google Scholar]
- Seo, K.I. , Lee, J.H. , Nezames, C.D. , Zhong, S. , Song, E. , Byun, M.O. and Deng, X.W. (2014) ABD1 is an Arabidopsis DCAF substrate receptor for CUL4‐DDB1‐based E3 ligases that acts as a negative regulator of abscisic acid signaling. Plant Cell, 26, 695–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano, M. , Parra, S. , Alcaraz, L.D. and Guzman, P. (2006) The ATL gene family from Arabidopsis thaliana and Oryza sativa comprises a large number of putative ubiquitin ligases of the RING‐H2 type. J. Mol. Evol. 62, 434–445. [DOI] [PubMed] [Google Scholar]
- Shi, Y. (2009) Serine/threonine phosphatases: mechanism through structure. Cell, 139, 468–484. [DOI] [PubMed] [Google Scholar]
- Shin, R. , Alvarez, S. , Burch, A.Y. , Jez, J.M. and Schachtman, D.P. (2007) Phosphoproteomic identification of targets of the Arabidopsis sucrose nonfermenting‐like kinase SnRK2.8 reveals a connection to metabolic processes. Proc. Natl. Acad. Sci. USA, 104, 6460–6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirichandra, C. , Gu, D. , Hu, H.C. , Davanture, M. , Lee, S. , Djaoui, M. , Valot, B. et al (2009a) Phosphorylation of the Arabidopsis AtrbohF NADPH oxidase by OST1 protein kinase. FEBS Lett. 583, 2982–2986. [DOI] [PubMed] [Google Scholar]
- Sirichandra, C. , Wasilewska, A. , Vlad, F. , Valon, C. and Leung, J. (2009b) The guard cell as a single‐cell model towards understanding drought tolerance and abscisic acid action. J. Exp. Bot. 60, 1439–1463. [DOI] [PubMed] [Google Scholar]
- Sirichandra, C. , Davanture, M. , Turk, B.E. , Zivy, M. , Valot, B. , Leung, J. and Merlot, S. (2010) The Arabidopsis ABA‐activated kinase OST1 phosphorylates the bZIP transcription factor ABF3 and creates a 14‐3‐3 binding site involved in its turnover. PLoS ONE, 5, e13935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalle, J. and Vierstra, R.D. (2004) The ubiquitin 26S proteasome proteolytic pathway. Annu. Rev. Plant Biol. 55, 555–590. [DOI] [PubMed] [Google Scholar]
- Smalle, J. , Kurepa, J. , Yang, P. , Emborg, T.J. , Babiychuk, E. , Kushnir, S. and Vierstra, R.D. (2003) The pleiotropic role of the 26S proteasome subunit RPN10 in Arabidopsis growth and development supports a substrate‐specific function in abscisic acid signaling. Plant Cell, 15, 965–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soon, F.F. , Ng, L.M. , Zhou, X.E. , West, G.M. , Kovach, A. , Tan, M.H. , Suino‐Powell, K.M. et al (2012) Molecular mimicry regulates ABA signaling by SnRK2 kinases and PP2C phosphatases. Science, 335, 85–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone, S.L. , Williams, L.A. , Farmer, L.M. , Vierstra, R.D. and Callis, J. (2006) KEEP ON GOING, a RING E3 ligase essential for Arabidopsis growth and development, is involved in abscisic acid signaling. Plant Cell, 18, 3415–3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umezawa, T. , Sugiyama, N. , Mizoguchi, M. , Hayashi, S. , Myouga, F. , Yamaguchi‐Shinozaki, K. , Ishihama, Y. et al (2009) Type 2C protein phosphatases directly regulate abscisic acid‐activated protein kinases in Arabidopsis. Proc. Natl. Acad. Sci. USA, 106, 17588–17593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umezawa, T. , Sugiyama, N. , Takahashi, F. , Anderson, J.C. , Ishihama, Y. , Peck, S.C. and Shinozaki, K. (2013) Genetics and phosphoproteomics reveal a protein phosphorylation network in the abscisic acid signaling pathway in Arabidopsis thaliana. Sci. Signal. 6, rs8. [DOI] [PubMed] [Google Scholar]
- Vierstra, R.D. (2009) The ubiquitin‐26S proteasome system at the nexus of plant biology. Nat. Rev. Mol. Cell Biol. 10, 385–397. [DOI] [PubMed] [Google Scholar]
- Vilela, B. , Najar, E. , Lumbreras, V. , Leung, J. and Pages, M. (2015) Casein kinase 2 negatively regulates abscisic acid‐activated SnRK2s in the core abscisic acid‐signaling module. Mol. Plant, 8, 709–721. [DOI] [PubMed] [Google Scholar]
- Vlad, F. , Turk, B.E. , Peynot, P. , Leung, J. and Merlot, S. (2008) A versatile strategy to define the phosphorylation preferences of plant protein kinases and screen for putative substrates. Plant J. 55, 104–117. [DOI] [PubMed] [Google Scholar]
- Vlad, F. , Rubio, S. , Rodrigues, A. , Sirichandra, C. , Belin, C. , Robert, N. , Leung, J. et al (2009) Protein phosphatases 2C regulate the activation of the Snf1‐related kinase OST1 by abscisic acid in Arabidopsis. Plant Cell, 21, 3170–3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waadt, R. , Manalansan, B. , Rauniyar, N. , Munemasa, S. , Booker, M.A. , Brandt, B. , Waadt, C. et al (2015) Identification of open stomata1‐interacting proteins reveals interactions with sucrose non‐fermenting1‐related protein kinases2 and with Type 2A protein phosphatases that function in abscisic acid responses. Plant Physiol. 169, 760–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, P. , Xue, L. , Batelli, G. , Lee, S. , Hou, Y.J. , Van Oosten, M.J. , Zhang, H. et al (2013) Quantitative phosphoproteomics identifies SnRK2 protein kinase substrates and reveals the effectors of abscisic acid action. Proc. Natl. Acad. Sci. USA, 110, 11205–11210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y. , Chang, H. , Hu, S. , Lu, X. , Yuan, C. , Zhang, C. , Wang, P. et al (2014) Plastid casein kinase 2 knockout reduces abscisic acid (ABA) sensitivity, thermotolerance, and expression of ABA‐ and heat‐stress‐responsive nuclear genes. J. Exp. Bot. 65, 4159–4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wege, S. , De Angeli, A. , Droillard, M.J. , Kroniewicz, L. , Merlot, S. , Cornu, D. , Gambale, F. et al (2014) Phosphorylation of the vacuolar anion exchanger AtCLCa is required for the stomatal response to abscisic acid. Sci. Signal. 7, ra65. [DOI] [PubMed] [Google Scholar]
- Wu, Q. , Zhang, X. , Peirats‐Llobet, M. , Belda‐Palazon, B. , Wang, X. , Cui, S. , Yu, X. et al (2016) Ubiquitin Ligases RGLG1 and RGLG5 Regulate Abscisic Acid Signaling by Controlling the Turnover of Phosphatase PP2CA. Plant Cell, 28, 2178–2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, Y. , Xing, Y. , Chen, Y. , Chao, Y. , Lin, Z. , Fan, E. , Yu, J.W. et al (2006) Structure of the protein phosphatase 2A holoenzyme. Cell, 127, 1239–1251. [DOI] [PubMed] [Google Scholar]
- Xue, S. , Hu, H. , Ries, A. , Merilo, E. , Kollist, H. and Schroeder, J.I. (2011) Central functions of bicarbonate in S‐type anion channel activation and OST1 protein kinase in CO2 signal transduction in guard cell. EMBO J. 30, 1645–1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, Z. , Zhao, J. , Peng, P. , Chihara, R.K. and Li, J. (2009) BIN2 functions redundantly with other Arabidopsis GSK3‐like kinases to regulate brassinosteroid signaling. Plant Physiol. 150, 710–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, L. , Liu, Q. , Liu, Z. , Yang, H. , Wang, J. , Li, X. and Yang, Y. (2016) Arabidopsis C3HC4‐RING finger E3 ubiquitin ligase AtAIRP4 positively regulates stress‐responsive abscisic acid signaling. J. Integr. Plant Biol. 58, 67–80. [DOI] [PubMed] [Google Scholar]
- Yin, Y. , Wang, Z.Y. , Mora‐Garcia, S. , Li, J. , Yoshida, S. , Asami, T. and Chory, J. (2002) BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell, 109, 181–191. [DOI] [PubMed] [Google Scholar]
- Yin, X.J. , Volk, S. , Ljung, K. , Mehlmer, N. , Dolezal, K. , Ditengou, F. , Hanano, S. et al (2007) Ubiquitin lysine 63 chain forming ligases regulate apical dominance in Arabidopsis. Plant Cell, 19, 1898–1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin, P. , Fan, H. , Hao, Q. , Yuan, X. , Wu, D. , Pang, Y. , Yan, C. et al (2009) Structural insights into the mechanism of abscisic acid signaling by PYL proteins. Nat. Struct. Mol. Biol. 16, 1230–1236. [DOI] [PubMed] [Google Scholar]
- Yoshida, R. , Hobo, T. , Ichimura, K. , Mizoguchi, T. , Takahashi, F. , Aronso, J. , Ecker, J.R. et al (2002) ABA‐activated SnRK2 protein kinase is required for dehydration stress signaling in Arabidopsis. Plant Cell Physiol. 43, 1473–1483. [DOI] [PubMed] [Google Scholar]
- Yoshida, R. , Umezawa, T. , Mizoguchi, T. , Takahashi, S. , Takahashi, F. and Shinozaki, K. (2006a) The regulatory domain of SRK2E/OST1/SnRK2.6 interacts with ABI1 and integrates abscisic acid (ABA) and osmotic stress signals controlling stomatal closure in Arabidopsis. J. Biol. Chem. 281, 5310–5318. [DOI] [PubMed] [Google Scholar]
- Yoshida, T. , Nishimura, N. , Kitahata, N. , Kuromori, T. , Ito, T. , Asami, T. , Shinozaki, K. et al (2006b) ABA‐hypersensitive germination3 encodes a protein phosphatase 2C (AtPP2CA) that strongly regulates abscisic acid signaling during germination among Arabidopsis protein phosphatase 2Cs. Plant Physiol. 140, 115–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, F. , Wu, Y. and Xie, Q. (2015) Precise protein post‐translational modifications modulate ABI5 activity. Trends Plant Sci. 20, 569–575. [DOI] [PubMed] [Google Scholar]
- Zhang, Y. , Yang, C. , Li, Y. , Zheng, N. , Chen, H. , Zhao, Q. , Gao, T. et al (2007) SDIR1 is a RING finger E3 ligase that positively regulates stress‐responsive abscisic acid signaling in Arabidopsis. Plant Cell, 19, 1912–1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y. , Xu, W. , Li, Z. , Deng, X.W. , Wu, W. and Xue, Y. (2008) F‐box protein DOR functions as a novel inhibitory factor for abscisic acid‐induced stomatal closure under drought stress in Arabidopsis. Plant Physiol. 148, 2121–2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, H. , Cui, F. , Wu, Y. , Lou, L. , Liu, L. , Tian, M. , Ning, Y. et al (2015) The RING finger ubiquitin E3 ligase SDIR1 targets SDIR1‐INTERACTING PROTEIN1 for degradation to modulate the salt stress response and ABA signaling in Arabidopsis. Plant Cell, 27, 214–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, Y. , Chan, Z. , Xing, L. , Liu, X. , Hou, Y.J. , Chinnusamy, V. , Wang, P. et al (2013) The unique mode of action of a divergent member of the ABA‐receptor protein family in ABA and stress signaling. Cell Res. 23, 1380–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, H. , Zhang, H. , Cui, P. , Ding, F. , Wang, G. , Li, R. , Jenks, M.A. et al (2014) The putative E3 ubiquitin ligase ECERIFERUM9 regulates abscisic acid biosynthesis and response during seed germination and postgermination growth in arabidopsis. Plant Physiol. 165, 1255–1268. [DOI] [PMC free article] [PubMed] [Google Scholar]


