Abstract
We investigated the evolution of resistance to the antifungal drug itraconazole in replicate populations of Aspergillus fumigatus that were founded from a strain with a genotype of sensitivity to a single drug and then propagated under uniform conditions. For each population, conidia were serially transferred 10 times to agar medium either with or without itraconazole. After 10 transfers in medium supplemented with itraconazole, 10 itraconazole-resistant mutant strains were isolated from two populations. These mutant strains had different growth rates and different levels of itraconazole resistance. Analysis of the ergosterol contents of these mutants showed that they accumulate ergosterol when they are grown in the presence of itraconazole. The replacement of the CYP51A gene of the wild-type strain changed the susceptibility pattern of this strain to one of itraconazole resistance only when CYP51A genes with N22D and M220I mutations were used as selectable marker genes. Real-time quantitative reverse transcription-PCR was used to assess the levels of expression of the Afumdr1, Afumdr2, Afumdr3, Afumdr4, AtrF transporter, CYP51A, and CYP51B genes in these mutant strains. Most mutants showed either constitutive high-level expression or induction upon exposure of Afumdr3, Afumdr4, and AtrF to itraconazole. Our results suggest that overexpression of drug efflux pumps and/or selection of drug target site mutations are at least partially responsible for itraconazole resistance and could be considered mechanisms for the emergence of clinical resistance to this drug.
Aspergillus fumigatus is the most common species of Aspergillus and causes life-threatening pulmonary disease in severely immunocompromised patients (6). The treatment of these patients has largely been limited to therapy with the polyene drug amphotericin B; the broad-spectrum triazoles, such as itraconazole or voriconazole; and/or the echinocandin caspofungin (12, 24, 33). Amphotericin B therapy can be highly toxic and can result in nephrotoxicity, whereas triazoles are fungistatic and their activities can be limited by drug resistance (7). Despite its safety profile and good therapeutic performance, the continuous use of triazoles can result in the development of drug resistance; and a number of itraconazole-resistant clinical isolates (7) as well as spontaneous and induced mutants of A. fumigatus (4, 18, 19) have been documented. The azoles block the ergosterol biosynthesis pathway by inhibiting the enzyme 14α-demethylase, the product of CYP51 (8). Fungal azole resistance involves both amino acid changes in the target site that alter drug-target interactions and those that decrease the net level of azole accumulation (20, 28, 36). Compensatory pathways have been documented for the mechanisms of resistance to the azole and polyene classes and involve alterations of specific steps in ergosterol biosynthesis (17, 29). For example, analysis of the sterol compositions of two separate azole-resistant Candida albicans clinical isolates revealed the accumulation of ergosta-7,22-dienol, which is a feature consistent with the absence of sterol Δ5,6-desaturase activity, which is encoded by ERG3 (14, 15, 25, 29). The reduced intracellular accumulation has also been correlated with overexpression of multidrug resistance (MDR) efflux transporter genes of the ATP-binding cassette (ABC) and the major facilitator superfamily (MFS) classes (2, 16, 17).
The emergence of drug resistance in all pathogenic microorganisms is an evolutionary process initiated by exposure to antimicrobial agents (3). Since the antifungal agents rarely eradicate the pathogen population completely, survivors are subject to strong directional selection for resistance. The emergence of resistance is a function of the rate of mutation to resistance and the size of the surviving population. Fungistatic drugs, such as the triazoles, have the potential to leave more survivors than fungicidal drugs, and this larger effective population size can contribute to a higher probability of resistance in the pathogen. In this study, our main objective was to investigate the emergence of itraconazole resistance in replicate populations of A. fumigatus that were founded from a strain with a genotype of sensitivity to a single drug and that were propagated under uniform conditions. We found that itraconazole resistance in these populations of isolates emerges rapidly and involves multiple resistance mechanisms.
MATERIALS AND METHODS
Strains, culture conditions, and in vitro evolution of drug resistance.
A. fumigatus strain CEA17 (pyrG mutant) (5) was used in this work. The media used were YAG medium (2% glucose, 0.5% yeast extract, 2% agar, trace elements), YUU medium (YAG medium supplemented with 1.2 g each of uracil and uridine per liter), YG medium (which has the same composition as YAG medium, but without agar), and minimal medium (1% glucose, nitrate salts, trace elements, 2% agar [pH 6.5]). Trace elements, vitamins, and nitrate salts are described in the appendix to the work of Kafer (13).
Eight independent populations of A. fumigatus were founded from a single colony of strain CEA17 (5). Conidia (1.0 × 106) from each population were serially propagated for 10 transfers (approximately 48 to 72 h incubation for each transfer) in YUU medium. In each transfer, 100 μl of a suspension with 1.0 × 106 conidia was inoculated onto YUU medium plates, and the plates were incubated at 37°C. Four populations (populations A to D) were grown in YUU medium without drug, while four populations (populations E to H) were grown in YUU medium with 1.0 μg of itraconazole/ml. After the 5th and the 10th transfers, approximately 1.0 × 106 conidia were inoculated onto YUU medium with 10 or 50 μg of itraconazole/ml, respectively. The strains that grew on these plates were isolated for further molecular characterization.
In vitro antifungal susceptibility testing.
Testing of the susceptibilities of the mutant strains to antifungal agents was performed by assessing the MICs of the drugs by the M38-A microdilution technique proposed by the National Committee for Clinical Laboratory Standards (24). Conidia from YAG agar cultures grown for 72 h at 37°C were pooled in approximately 1 ml of sterile 0.85% saline-0.1% Tween 20, and the resulting conidial suspensions were transferred to a sterile tube. After spore dispersion with a vortex mixer, the optical densities of the conidial suspensions were read at 590 nm. Since strain CEA17 is auxotrophic for uridine and uracil (pyrG mutant), the suspensions were diluted in RPMI 1640 broth supplemented with 1.2 g each of uracil and uridine (Life Technologies) per liter to approximately 104 conidia ml−1. Concentrated (two times) antifungal solutions were prepared in RPMI 1640 broth supplemented with 1.2 g each of uracil and uridine per liter, and a serial dilution series was prepared in a 96-well microtiter plate. The plates were inoculated by plating 0.1 ml of the diluted conidial suspension in each well, which contained 0.1 ml of drug at a concentration in the following range: amphotericin B (Sigma), 0.25 to 16 μg/ml, and itraconazole (Janssen Pharmaceutica), 0.25 to 16 μg/ml. Dimethyl sulfoxide (Sigma) was used to dissolve the drugs. The plates were prepared in duplicate and were incubated at 37°C for 48 h. The MIC was the lowest drug concentration that induced the complete inhibition of growth. Quality control strains Candida parapsilosis ATCC 22019 and Candida krusei ATCC 6258 were used to validate the susceptibility testing results (9).
RNA isolation, real-time RT-PCR, and sequencing of CYP51A and CYP51B genes.
Approximately 1.0 × 105 conidia of each A. fumigatus isolate/ml were inoculated in 250-ml Erlenmeyer flasks containing 50 ml of YG medium supplemented with 1.2 g each of uracil and uridine per liter either in the absence or in the presence of 10 ng of itraconazole/ml (parental strain) or 5 μg of itraconazole/ml (mutant strains). These cultures were grown for 24 h at 37°C, and total RNA was extracted with the Trizol reagent (Invitrogen, Carlsbad, Calif.), as described previously (30). RNase-free DNase treatment was done as previously described by Semighini et al. (30). The absence of DNA contamination after the RNase-free DNase treatment was verified by PCR amplification of the GPDH gene. cDNA was synthesized by using SuperScript reverse transcriptase (Invitrogen). All reverse transcription-PCRs (RT-PCRs) were performed with an ABI Prism 7700 sequence detection system (Perkin-Elmer Applied Biosystems, Foster City, Calif.), and a Taq-Man PCR reagent kit was used for the PCRs, as described by Semighini et al. (31). The primer and probe sequences used in the PCRs are described in Table 1.
TABLE 1.
Primers and fluorescent probes used in this work
| Primer or probe | Sequencea |
|---|---|
| GPDH-1F | 5′-GCCAAGGTCATCAACGACAA-3′ |
| GPDH-1R | 5′-GAGTGGACGGTGGTCATG-3′ |
| GPDH-1M2 | 5′-FAM-ACCCTCAACAATGCCG-TAMRA-3′ |
| ATRF-3F | 5′-GGTCATTTGGAACGGAGAACGA-3′ |
| ATRF-3R | 5′-TGTTTGAACAAGACACCGACCTT-3′ |
| ATRF-3M2 | 5′-FAM-TTCGCCGGTTCTCCTG-TAMRA-3′ |
| MDR1-9F | 5′-GAACGCACCACGAGTTGATT-3′ |
| MDR1-9R | 5′-CTGCAGATTGACCAGCTCGTAATA-3′ |
| MDR1-9M2 | 5′-FAM-CCGGCCCTTGTTTCG-TAMRA-3′ |
| MDR2-2F | 5′-CTTGTTATCCGCCAATGTCTGTAGT-3′ |
| MDR2-2R | 5′-GGAGGTAGAAAACAGCCTGACT-3′ |
| MDR2-2M2 | 5′-FAM-TTGAACGGCCGAAACC-TAMRA-3′ |
| MDR3-2F | 5′-CATCCTCATTCCCTTGCATATCGT-3′ |
| MDR3-2R | 5′-TCAAGCTATAACCGCCCACATG-3′ |
| MDR3-2M2 | 5′-FAM-CATCCGGGTGGCCTC-TAMRA-3′ |
| MDR4-4F | 5′-TTGCTGGTGTTTGGTGAGTGA-3′ |
| MDR4-4R | 5′-GCCTCCTGTTTGATAATGCTCTCA-3′ |
| MDR4-4M2 | 5′-FAM-CACGCCGCTACCGTC-TAMRA-3′ |
| CYPA-6F | 5′-GGAACAGCTGGCCAATCTTG-3′ |
| CYPA-6R | 5′-ACTGGAGCGGAGGAAGACT-3′ |
| CYPA-6M2 | 5′-FAM-CCCCGCCGGGCCAG-TAMRA-3′ |
| CYPB-10F | 5′-CGCTGGACGGCATAGGT-3′ |
| CYPB-10R | 5′-AGTGATTGTGCCAAGCTGAAGAT-3′ |
| CYPB-10M2 | 5′-FAM-TTGGCGAGCAATTTGCAT-TAMRA-3′ |
| P450-A1 | 5′-ATGGTGCCGATGCTATGG-3′ |
| P450-A2 | 5′-CTGTCTCACTTGGATGTG-3′ |
| P450-B1 | 5′-ATGGGTCTCATCGCGTTC-3′ |
| P450-B2 | 5′-TCAGGCTTTGGTAGCGG-3′ |
| CYP51AF | 5′-ACATGATATGGAACCTGATGA-3′ |
| CYP51AR | 5′-TCATCAGGTTCCATCTCATGT-3′ |
| CYP51BF | 5′-TCGCTCCTATCAACTTCATGT-3′ |
| CYP51BR | 5′-ACATGAAGTTGATAGCAGCGA-3′ |
Abbreviations: FAM, 6-carboxyfluorescein; TAMRA, 6-carboxy-N,N,N′,N′-tetramethylrhodamine.
The entire open reading frames of the CYP51A and CYP51B genes, which encode sterol 14α-demethylase, of all A. fumigatus isolates were sequenced. Total genomic DNA was extracted as described previously (30). The open reading frames were amplified by PCR by using high-fidelity Platinum Taq DNA polymerase (Invitrogen) and the primers described in Table 1. The PCR conditions for the CYP51 genes were 94°C for 2 min and 35 cycles of 94°C for 1 min, 49°C (for the CYP51A gene) and 52°C (for the CYP51B gene) for 1 min, and 68°C for 2 min, followed by an extension step at 68°C for 10 min. After the reaction, the 1,618- and 1,730-bp PCR products for the CYP51A and CYP51B genes, respectively, were purified with a Qiagen PCR cleanup kit and inserted into TOPO TA cloning kit (Invitrogen), according to the instructions of the manufacturer. The sequencing reaction mixtures were prepared as described by Semighini et al. (30). Sequence data were compared with the published sequences of the CYP51A and CY51B genes (21).
SQM.
The in vitro itraconazole susceptibilities of the A. fumigatus isolates were determined by quantification of total intracellular ergosterol by the sterol quantification method (SQM), as described previously (1), with slight modifications. To prepare the inocula, conidia from isolates grown on YUU agar at 35°C for 4 days were harvested with 3 ml of sterile 0.85% saline. The conidial suspensions were transferred to a sterile tube and mixed with a vortex mixer. The optical densities were read at 590 nm and were adjusted to 80% transmittance. For each isolate, 100 μl of the conidial suspension was inoculated into 50 ml of YUU broth containing 0, 4, 16, or 64 μg of itraconazole/ml. The cultures were grown at 35°C for 24 to 48 h with constant shaking. Fungal cells were collected by vacuum filtration, weighed, and transferred to sterile glass screw-cap tubes. Five milliliters of 25% alcoholic potassium hydroxide solution (25 g of KOH and 35 ml of sterile distilled water brought to 100 ml with 100% ethanol) was added to each tube, and the samples were mixed on a vortex mixer for 2 min. The cell suspensions were incubated in an 85°C water bath for 3 h and allowed to cool to room temperature. Sterols were extracted by addition of 2 ml of sterile distilled water and 5 ml of spectrophotometry-grade n-heptane (Sigma), followed by vigorous mixing in a vortex mixer for 5 min. The samples were kept at room temperature for 1 to 2 h to allow the phases to separate or were stored at 4°C until the next day. One milliliter of the heptane layer (containing ergosterol) was transferred to a 1.5-ml quartz cuvette and analyzed spectrophotometrically by scanning at wavelengths between 200 and 300 nm. If necessary, the samples were diluted fivefold with 100% ethanol and reanalyzed. The ergosterol content as a percentage of the wet cell weight was calculated by the following equations: value 1 = [(A281.5/290) × F]/wet cell weight, value 2 = [(A230/518) × F]/wet cell weight, and percent ergosterol = value 1 − value 2. F is the factor for dilution in ethanol, and 290 and 518 are fixed values determined for crystalline ergosterol and 24(28)dihydroergosterol, respectively. The presence of a detectable level of ergosterol in cells grown in itraconazole was indicative of decreased itraconazole susceptibility.
DNA-mediated transformation.
The itraconazole sensitivity of wild-type strain CEA17 was complemented by DNA-mediated transformation with a PCR-amplified CYP51A gene. Transformations were performed as described by Osmani et al. (27). After transformation and incubation of the plates at 37°C for 24 h, 10 ml of melted YAG medium containing 1 μg of itraconazole/ml was poured over the medium in the plates, and the plates were incubated at 37°C for 1 week. The uridine and uracil requirements of strain CEA17 were complemented by DNA-mediated transformation, and transformants were transferred to minimal medium with different concentrations of itraconazole.
RESULTS AND DISCUSSION
In vitro evolution of itraconazole resistance in A. fumigatus.
Ten itraconazole-resistant mutants appeared in two of the four populations (populations G and H) that were propagated in the presence of itraconazole at a concentration that inhibits radial growth by 50% (1 μg of itraconazole/ml; populations E to H). No resistant mutations were detected in the four populations (populations A to D) that were propagated in the absence of the drug. The mutant colonies were able to grow on YUU medium with 10 or 50 μg of itraconazole/ml, while the progenitor colony was not. Two mutants were isolated from population H at the 5th transfer (H05 mutants) in the presence of 10 μg of itraconazole/ml (frequency, 2 × 10−6). These mutant strains displayed reduced growth rates (see Table 3), and mutant strain H5-1-10 did not conidiate (data not shown). Three mutants were isolated from population H at the 10th transfer (H10 mutants) in the presence of 50 μg of itraconazole/ml (frequency, 3 × 10−6); two of these mutants displayed the same growth rate (50A mutants), while one of them displayed an altered morphology with a reduced growth rate (50B mutant) (see Table 3). Four mutants were isolated from population G at the 10th transfer in the presence of 50 μg of itraconazole/ml (frequency, 4 × 10−6); again, three of them displayed the same growth rate (50A mutants), while one of them showed a reduced growth rate (50B mutant) (see Table 3). All these mutant strains have the pyrG mutant marker phenotype, strongly indicating that they are descendants of the original parental strain and were not contaminants (data not shown). The genetic stability of itraconazole resistance was tested by repeated subculturing of the resistant isolates in the absence of the antifungal agent, followed by MIC determinations. The mutants displayed different levels of resistance and growth rates (Tables 2 and 3). For instance, in the absence of itraconazole, mutants G10-1-50B, H05-1-10, H05-3-10, and H10-1-50B had much slower growth rates than the parental strain (Table 3). Interestingly, mutant strains H10-1-50A, H10-2-50A, and H10-3-50A, G10-1-50A, G10-2-50A, and G10-3-50A had slightly faster growth rates than the parental strain (Table 3).
TABLE 3.
Radial growth of A. fumigatus itraconazole-resistant mutants
| Strain | Radial growth (mm) ata:
|
|
|---|---|---|
| 24 h | 48 h | |
| CEA17 | 7.50 ± 0.57 | 36.75 ± 1.70 |
| G10-1-50A | 8.25 ± 0.50 | 39.75 ± 1.00 |
| G10-2-50A | 8.50 ± 0.57 | 41.50 ± 1.00 |
| G10-3-50A | 8.00 ± 0.00 | 39.50 ± 1.00 |
| G10-1-50B | 5.50 ± 0.57 | 32.25 ± 0.50 |
| H05-1-10 | 2.00 ± 0.00 | 15.25 ± 0.50 |
| H05-3-10 | 1.00 ± 0.00 | 22.75 ± 2.06 |
| H10-1-50A | 7.75 ± 0.95 | 39.25 ± 0.95 |
| H10-2-50A | 8.25 ± 0.50 | 39.25 ± 1.50 |
| H10-3-50A | 9.50 ± 0.57 | 41.00 ± 2.00 |
| H10-1-50B | 1.00 ± 0.00 | 20.75 ± 1.70 |
The results are the averages of four repetitions.
TABLE 2.
MICs for A. fumigatus itraconazole-resistant mutants
| Strain | MIC (μg/ml)a
|
|
|---|---|---|
| Itraconazole | Amphotericin B | |
| A. fumigatus | ||
| CEA17 (wild type) | 0.25 | 1.0 |
| G10-1-50A | >16 | 1.0 |
| G10-2-50A | >16 | 0.5 |
| G10-3-50A | >16 | 1.0 |
| G10-1-50B | 1.0 | 1.0 |
| H05-3-10 | >16 | 0.5 |
| H10-1-50A | >16 | 1.0 |
| H10-2-50A | >16 | 1.0 |
| H10-3-50A | >16 | 1.0 |
| H10-1-50B | >16 | 1.0 |
| C. parapsilosis ATCC 22019 | 0.12 | 0.5 |
| C. krusei ATCC 6258 | 0.25 | 1.0 |
The results are the averages of three repetitions.
All mutant strains except G10-1-50B achieved a high level of resistance to itraconazole (MIC > 16 μg/ml) (Table 2). The MIC for mutant strain G10-1-50B was 1 μg/ml. We also tested these mutant strains for cross-resistance to amphotericin B. All mutants were fully susceptible to amphotericin B, but curiously, mutants G10-250A and H05-3-10 showed increased susceptibility (Table 2). Strain H05-1-10 does not conidiate, and consequently, it was not possible to determine the MIC for this strain.
Analysis of ergosterol contents and sequencing of CYP51A and CYP51B genes.
The primary mechanism of action by which azole antifungal drugs inhibit fungal cell growth is through the disruption of the normal sterol biosynthetic pathway, leading to a reduction in ergosterol biosynthesis (36). We therefore decided to quantify the membrane sterols in the mutants by using a modification of SQM (1). Table 4 shows the percent inhibition of ergosterol biosynthesis in the A. fumigatus itraconazole-resistant mutants. Mutant strains G10-1-50A, G10-2-50A, G10-3-50A, H05-3-10, H10-1-50A, and H10-2-50A all behaved similarly, in that they grew in the presence of all itraconazole concentrations tested (i.e., 4, 16, and 64 μg/ml) after 24 h. By SQM mutant strains G10-1-50B, H10-3-50A, and H10-1-50B required 48 h to achieve sufficient growth in the presence of any of the drug concentrations. After 48 h, ergosterol biosynthesis in strains H10-3-50A and H10-1-50B was less sensitive to itraconazole than ergosterol biosynthesis in strain G10-1-50B. Wild-type strain CEA17 did not grow in the presence of any drug concentration even after 48 h. These results indicate all the mutant strains accumulated ergosterol when they were grown in the presence of itraconazole and that mutations that prevent the accumulation of ergosterol, such as mutations in the ERG3 gene, possibly do not occur in these mutants.
TABLE 4.
Inhibition of ergosterol biosynthesis in A. fumigatus itraconazole-resistant mutants
| Strain | % Inhibitiona |
|---|---|
| CEA17 (wild type)b | 100 |
| G10-1-50A | 0 |
| G10-2-50A | 62 |
| G10-3-50A | 0 |
| G10-1-50Bc | 91 |
| H05-3-10 | 42 |
| H10-1-50A | 76 |
| H10-2-50A | 35 |
| H10-3-50Ad | 0 |
| H10-1-50Be | 37 |
Percent inhibition of ergosterol biosynthesis at 64 μg/ml relative to that for the drug-free growth control. Based on the studies with C. albicans, the MIC by SQM was defined as the drug concentration which inhibited ergosterol biosynthesis by ≥80% relative to that for the drug-free control. However, studies have not been done with A. fumigatus to determine what MIC end point is most appropriate. However, by use of the guidelines for C. albicans, the MICs for all mutants except strain G10-1-50B would be >16 μg/ml; that for strain G10-1-50B would be 1 μg/ml.
No growth was detected at any drug concentration, even after 48 h of growth.
Dramatically reduced growth was detected at all drug concentrations after 24 h of incubation. Adequate growth was detected at 48 h, and 61% inhibition of ergosterol biosynthesis was detected at 16 μg/ml.
Dramatically reduced growth was detected at all drug concentrations after 24 h of incubation. Adequate growth was detected at 48 h.
Dramatically reduced growth was detected at 16 and 64 μg/ml after 24 h of incubation. Adequate growth was detected at 48 h.
Since most of the mutant strains showed ergosterol in their cells when they were grown in the presence of itraconazole, we decided to verify if point mutations were present in the CYP51 genes from these strains. Thus, sequence analysis of the entire CYP51A and CYP51B genes from our isolates was performed, and the sequences were compared to the published CYP51 gene sequences (21). Several nonsilent mutations were observed in the CYP51A genes of all isolates (Table 5) except isolate H05-1-10. Isolate G10-1-50A contained a P394L mutation; and isolates G10-1-50A, G10-2-50A, and G10-3-50A contained G54R mutations. Isolate G10-1-50B contained a Y491H mutation, and isolate H05-3-10 contained a T440A mutation. Isolate H10-1-50A contained an N22D mutation, while isolates H10-2-50A and H10-3-50A contained M220I mutations. Only isolates G10-1-50B, G10-3-50A, and H10-2-50A contained mutations in the CYP51B gene: S505P, S177F, and F59L, respectively.
TABLE 5.
Amino acid substitutions in CYP51A and CYP51B genes from A. fumigatus itraconazole-resistant mutants
| Mutant strain | Amino acid substitution ina:
|
|
|---|---|---|
| CYP51A | CYP51B | |
| G10-1-50A | G54R, P394L | None |
| G10-2-50A | G54R | None |
| G10-3-50A | G54R, S393S | S177F, P178P |
| G10-1-50B | Y491H | S505P |
| H05-1-10 | L20L | None |
| H05-3-10 | T440A | None |
| H10-1-50A | N22D | None |
| H10-2-50A | M220I | F59L |
| H10-3-50A | M220I | NDb |
Amino acids are numbered from the translation start amino acid methionine of CYP51A and CYP51B. The numbers indicate the position at which an amino acid change occurs.
ND, not determined.
To verify if the CYP51A mutations could really confer itraconazole resistance, we performed gene replacement experiments. The wild-type and mutated CYP51A genes were amplified by PCR and introduced into A. fumigatus strain CEA17 by DNA-mediated transformation, as described previously (26). The replacement of the CYP51A gene of the A. fumigatus wild-type strain changed the susceptibility pattern of CEA17 to one of the itraconazole resistance only when the mutated cyp51A genes of H10-1-50A (N22D) and H10-2-50A (M220I) were used as selectable markers (data not shown). Mann et al. (19) demonstrated that mutations in CYP51A, specifically, mutations in codon 54, are associated with posaconazole and itraconazole resistance in A. fumigatus. Diaz-Guerra et al. (8) analyzed the sequences of the CYP51 genes of 12 itraconazole-resistant clinical isolates and 3 itraconazole-susceptible clinical isolates of A. fumigatus. Those investigators showed that six itraconazole-resistant strains exhibited a substitution of another amino acid for glycine at position 54. When the mutated CYP51A gene replaced the CYP51A gene in the A. fumigatus wild-type strain or was constitutively expressed in a posaconazole-susceptible A. fumigatus strain, the resultant transformants exhibited decreases in itraconazole and posaconazole susceptibilities, respectively (8, 19). In addition to this mutation in codon 54, codon 236 in CYP51A and codons 35, 42, 187, 387, and 394 were also reported to be mutated in A. fumigatus pocanazole-resistant clinical isolates (8). Recently, Mellado et al. (22) reported that substitutions at methionine 220 in the A. fumigatus CYP51A gene are responsible for resistance to azole antifungal drugs in vitro. Surprisingly, and in contrast to what was reported by Diaz-Guerra et al. (8), in our hands transformation of the wild type by using the CYP51A gene carrying a point mutation that leads to an amino acid change at G54R did not yield any itraconazole-resistant transformants.
Expression of drug efflux transporter and CYP51 genes.
Decreased intracellular accumulation of itraconazole has been reported for A. fumigatus drug-resistant clinical isolates and isolates of A. fumigatus generated in vitro (7, 18), suggesting that the drug efflux pumps can contribute significantly to azole resistance. However, no particular pump has been identified to be responsible for efflux in A. fumigatus. The ABC and the MFS comprise the two major classes of efflux pumps known to contribute to drug resistance (35). The A. fumigatus genome contains 37 ABC family transporters and 251 MFS family transporters, as currently annotated (Ian Paulsen, personal communication). To explore the involvement of these genes in the itraconazole resistance phenotypes displayed by our mutant strains, real-time RT-PCR was used to assess gene expression levels. Taq-Man probes for five A. fumigatus transporters already described in the literature as possibly being involved in drug resistance (23, 32, 34) were designed. Four of these transporters, Afumdr1, Afumdr2, Afumdr4, and AtrF, are ABC transporters, while Afumdr3 is an MFS transporter. We also tested the expression of these genes in a previously isolated itraconazole-resistant mutant, RIT13 (23).
Table 6 shows that the highest levels of constitutively expressed Afumdr1 mRNA were observed in the G10-1-50B and H10-3-50A mutant strains, resulting in fivefold more copies of mRNA than the number in parental strain CEA17; RIT13 did not display increased levels of Afumdr1 mRNA constitutive expression compared to that of the wild-type strain. The highest levels of Afumdr2 were obtained for the H10-3-50A mutant strain, which displayed 28-fold more mRNA than the parental strain; again, the RIT13 mutant did not show any constitutive expression of this gene (Table 6). All mutants except H05-3-10 and H10-1-50B displayed increased levels of constitutive mRNA expression of the Afumdr3 and Afumdr4 genes, ranging from 2- to 324-fold and 2.8- to 2,700-fold, respectively, for both genes; the RIT13 mutant displayed high levels of expression of the mRNAs of both genes (Table 6). For the AtrF gene, mutants G10-1-50B and H10-3-50A showed the highest levels of constitutive mRNA expression, resulting in 7.6- and 17-fold more mRNA copies than the number detected in the parental strain; the RIT13 mutant showed increased levels of AtrF expression (Table 6).
TABLE 6.
CYP51 and efflux transporter gene mRNA expression levels in the mutant strains assessed by real-time RT-PCR
| Strain | cDNA levela
|
cDNA level
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Afumdr1
|
Afummdr2
|
Afumdr3
|
Afumdr4
|
AtrF
|
CYP51A
|
CYP51B
|
||||||||
| Const. expres.b | Itra treatedb | Const. expres. | Itra treated | Const. expres. | Itra treated | Const. expres. | Itra treated | Const. expres. | Itra treated | Const. expres. | Itra treated | Const. expres. | Itra treated | |
| CEA17 | 1.00 (1.00-1.00) | 3.67 (317-4.17) | 1.00 (1.00-1.00) | 0.33 (0.30-0.37) | 1.00 (0.75-1.25) | 1.50 (1.25-1.75) | 1.00 (0.97-1.03) | 23.85 (23.38-24.31)b | 1.00 (0.80-1.20) | 0.20 (0.20-0.20)b | 1.00 (1.00-1.00) | 7.20 (6.72-7.69)b | 1.00 (0.97-1.03) | 4.27 (3.94-4.61)b |
| G10-1-50A | 0.50 (0.50-0.50) | 12.33 (11.67-13.00) | 1.00 (0.98-1.02) | 2.89 (2.94-2.84) | 13.25 (13.00-13.50) | 20.17 (18.56-21.72) | 5.03 (4.85-5.21) | 11.55 (11.53-11.57) | 1.00 (1.00-1.00) | 17.60 (17.40-17.80) | 1.69 (1.67-1.70) | 2.82 (2.69-2.96) | 1.18 (1.08-1.28) | 2.31 (2.34-2.28) |
| G10-1-50B | 5.00 (5.00-5.00) | 2.53 (2.50-2.57) | 1.78 (1.64-1.91) | 3.38 (3.33-3.41) | 174.50 (160.75-188.25) | 3.56 (3.55-3.56) | 28.46 (27.18-29.74) | 6.25 (6.09-6.40) | 7.60 (6.80-8.40) | 5.61 (6.24-5.10) | 0.72 (0.70-0.74) | 7.13 (7.21-7.05) | 0.55 (0.52-0.57) | 4.17 (3.89-4.43) |
| G10-2-50A | 0.83 (0.83-0.83) | 6.40 (3.80-9.00) | 0.89 (0.88-0.90) | 5.38 (3.03-7.67) | 6.50 (6.25-6.75) | 60.58 (33.40-85.74) | 2.82 (2.79-2.85) | 35.69 (21.06-50.05) | 0.60 (0.60-0.60) | 35.00 (22.00-48.00) | 1.19 (1.07-1.30) | 3.70 (2.52-4.69) | 0.55 (0.54-0.55) | 2.83 (1.68-3.95) |
| G10-3-50A | 0.67 (0.67-0.67) | 5.25 (5.25-5.25) | 0.89 (0.87-0.91) | 5.50 (4.33-6.61) | 40.75 (37.25-44.25) | 0.15 (0.13-0.16) | 7.87 (7.69-8.05) | 2.49 (2.53-2.46) | 2.40 (2.40-2.40) | 0.25 (0.25-0.25) | 1.65 (1.65-1.65) | 4.06 (3.85-4.26) | 1.18 (1.17-1.19) | 2.54 (2.15-2.92) |
| H05-3-10 | 0.18 (0.17-0.18) | 0.29 (0.30-0.28) | 0.16 (0.16-0.16) | 0.41 (0.13-0.68) | 0.02 (0.02-0.02) | 0.60 (0.04-1.13) | 0.52 (0.10-0.94) | 0.05 (0.19-0.04) | 2.22 (2.19-2.26) | 0.18 (0.18-0.19) | 4.48 (4.13-4.83) | 2.43 (2.52-2.35) | 1.08 (0.94-1.22) | 0.60 (0.58-0.62) |
| H10-1-50A | 0.83 (0.83-0.83) | 6.40 (6.00-6.80) | 1.78 (1.07-2.49) | 1.88 (3.01-1.39) | 324.25 (315.25-333.25) | 1.09 (1.03-1.14) | 7.97 (7.90-8.05) | 8.77 (7.77-9.75) | 2.00 (1.80-2.20) | 12.60 (13.44-11.91) | 1.17 (1.09-1.24) | 3.63 (3.88-3.42) | 0.82 (0.81-0.83) | 2.22 (1.38-3.04) |
| H10-2-50A | 0.83 (0.83-0.83) | 4.00 (3.80-4.20) | 0.67 (0.62-0.71) | 3.33 (3.41-3.27) | 2.00 (2.00-2.25) | 214.38 (201.13-202.33) | 2,769 (2,717-2,821) | 15.52 (14.71-16.30) | 0.20 (0.20-0.20) | 108.0 (104.0-112.0) | 1.65 (1.63-1.67) | 2.75 (2.77-2.73) | 0.82 (0.82-0.82) | 3.00 (2.89-3.11) |
| H10-3-50A | 5.00 (4.83-5.17) | 0.53 (0.52-0.55) | 28.00 (27.83-28.17) | 0.03 (0.03-0.03) | 178.25 (118.25-238.25) | 0.29 (0.41-0.23) | 170.87 (168.97-172.77) | 0.06 (0.06-0.06) | 17.20 (15.40-19.00) | 0.21 (0.22-0.20) | 0.26 (0.24-0.28) | 24.71 (25.62-23.93) | 1.45 (1.41-1.50) | 1.50 (1.54-1.47) |
| H10-1-50B | 0.18 (0.18-0.18) | 0.71 (0.68-0.74) | 0.10 (0.09-0.11) | 1.49 (1.60-1.40) | 0.0017 (0.0011-0.0023) | 74.75 (107.10-58.98) | 0.18 (0.16-0.19) | 4.13 (3.89-4.32) | 3.38 (3.19-3.58) | 2.57 (2.53-2.60) | 6.05 (5.84-6.25) | 1.10 (1.09-1.12) | 0.54 (0.53-0.54) | 0.67 (0.53-0.80) |
| RIT13 | 0.33 (0.33-0.33) | 16.00 (14.50-17.50) | 0.89 (0.89-0.89) | 38.25 (37.70-38.80) | 23.75 (20.75-26.75) | 258.33 (257.73-258.79) | 7.41 (7.31-7.51) | 249.3 (237.0-261.30) | 1.80 (1.60-2.00) | 162.22 (180.00-148.00) | 1.20 (1.09-1.33) | 2.81 (2.50-3.13) | 0.64 (0.64-0.64) | 2.86 (2.70-3.01) |
Const. expres., constitutive expression, cDNA levels were calculated relative to that of the untreated parental strain, CEA17. The data represent the averages of three replicates, with the ranges shown in parentheses.
Values were obtained by using the cDNA copy number of each untreated strain as the baseline for strains in the presence of drug following normalization based on the GPDH cDNA copy number. Cells were grown in the presence of 10 ng of itraconazole (Itra)/ml (CEA17) or 5 μg of itraconazole/ml for 24 h. Ranges are shown in parentheses.
To explore whether gene expression responded to itraconazole, cells were grown in the presence of itraconazole at 10 ng/ml (parental strain) or 5 μg/ml (mutant strains) for 24 h at 37°C. The Afumdr1, Afumdr2, Afumdr3, Afumdr4, and AtrF genes in the parental strain grown in the presence of itraconazole were found to be induced 3.6-, 0.33-, 1.5-, 23-, and 0.20-fold more, respectively, than in the parental strain grown in the absence of itraconazole (Table 6). In the RIT13 mutant, the same genes were induced 16-, 38-, 258-, 249-, and 162-fold more, respectively, in the presence of itraconazole than in the absence of the drug (Table 6). Following exposure to itraconazole, the levels of Afumdr1 transcripts increased 12-fold in mutant G10-1-50A; Afumdr2 gene mRNA levels were increased from 1.5- to 5.5-fold in all mutants except H5-3-10 and H10-3-50A; the levels of Afumdr3 mRNA expression were increased from 3.5- to 214-fold in G10-1-50A, G10-1-50B, G10-2-50A, H10-2-50A, and H10-1-50B; the levels of Afumdr4 mRNA expression were increased only in mutant G10-2-50A (35-fold); and the levels of AtrF mRNA expression were also increased from 2.5- to 108-fold in all mutants except G10-3-50A, H05-3-10, and H10-3-50A. In the parental strain the CYP51A and CYP51B genes were induced 7.20- and 4.27-fold more, respectively, in the presence of itraconazole than in its absence. The CYP51A and CYP51B genes showed variable but increased levels of expression in the mutant strains; only mutant strain H10-3-50A showed higher levels of CYP51A mRNA expression (Table 6).
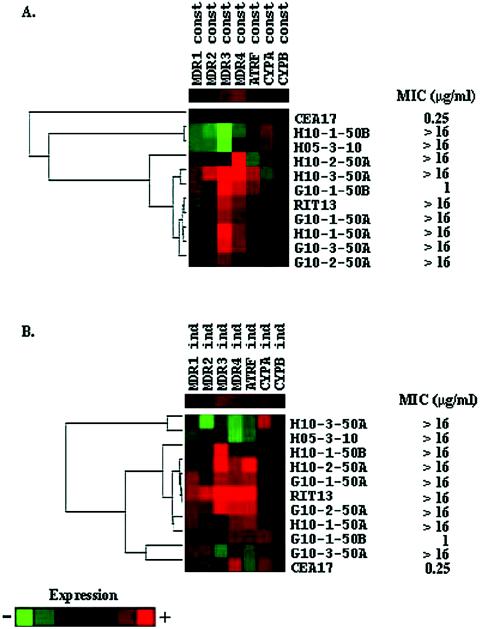
Hierarchical clustering analysis of the mRNA expression patterns obtained from cells grown in the absence of itraconazole produced four different groups of mutant strains according to their mRNA expression pattern (Fig. 1A): (i) CEA17; (ii) H10-1-50B and H5-3-10; (iii) H10-2-50A; and (iv) H10-3-50A, G10-1-50B, RIT13, G10-1-50A, H10-1-50A, G10-3-50A, and G10-2-50A. The first group showed a low constitutive mRNA expression, while the second group showed a high correlation (Pearson correlation coefficient [PCC] = 0.91) in low constitutive mRNA expression of all transporter genes. The third group showed a high constitutive expression of the Afumdr4 gene, while the fourth group displayed a high correlation (PCC = 0.91) of increased mRNA expression in the Afumdr3 and -mdr4 genes. In this last group, increased constitutive mRNA expression of Afumdr1 and -mdr2 also correlates with increased constitutive mRNA expression of Afumdr3 and -mdr4 genes in mutant strain H10-3-50A.
FIG. 1.
Hierarchical clustering showing the gene expression profiles of seven different A. fumigatus genes during growth in the absence (A) and in the presence (B) of itraconazole. In both experiments, the relative levels of each transcript were monitored by real-time RT-PCR of mRNA extracted after 24 h of growth. These values were log2 transformed and submitted to a hierarchical clustering algorithm by using the default parameters of CLUSTER software, kindly provided by M. Eisen and available at http://www.microarrays.org/software.html. In both cases, as shown by the scales on the right, a color scheme was used to designate genes that were down-regulated (green) or up-regulated (red). The clustering, along with the resulting dendrogram, was displayed by using TREEVIEW software, also available at the URL mentioned above (10).
When the mRNA expression pattern of the strains grown in the presence of itraconazole was clustered, three different groups were observed (Fig. 1B): (i) H10-3-50A and H5-3-10; (ii) H10-1-50B, H10-2-50A, G10-1-50A, rit13, G10-2-50A, H10-1-50A, and G10-1-50B; and (iii) G10-3-50A and CEA17. In the first group, low induction mRNA levels showed a high correlation, except for CYP51A in the mutant strain H10-3-50A, while the second group showed a high correlation (PCC = 0.78) among all genes, but the transporter genes are expressed at higher levels. The third group displayed a low correlation (0.49) with increased mRNA levels for all the genes, except in mutant strain G10-3-50A (low mRNA levels in Afumdr3, -mdr4, and AtrF) and CEA17 (low mRNA levels in Afumdr2, -mdr3, and AtrF).
The constitutive and itraconazole-induced gene expression profiles indicate that the drug efflux transporter genes are overexpressed in many mutants, including those with target site mutations. In addition, overexpression of the CYP51A and CYP51B genes seems to play a minor role in these mutants. These results suggest that the mutations that have evolved have resulted in a gain of function of either alleles for the transporter genes or their common activator (or a loss of function of a repressor of a common activator). In Saccharomyces cerevisiae, pleiotropic drug resistance can be brought about by the alteration or amplification of several different genes whose products can act either as membrane transporters or as transcriptional regulators (1). This performance is well known for the yeast transcriptional activators PDR1 and PDR3, which positively influence the expression of drug transporter genes such as PDR5 (12). The results presented here suggest that Afumdr3, Afumdr4, and AtrF, as well as possibly other unidentified drug efflux pumps, likely contribute to itraconazole resistance in our mutant strains. These transporter genes have already been shown to be induced by itraconazole (17, 22), and Afumdr3 and Afumdr4 have been shown to be overexpressed in itraconazole-resistant mutants induced in vitro (17). Interestingly, there was no correlation between the MICs and the level of mRNA expression, since mutant strains (for instance, strain G10-1-50B) for which MICs were low (1 μg/ml) showed high correlations among their levels of mRNA expression, while mutant strains (for instance, strain H05-3-10) for which MICs were high (>16 μg/ml) showed the opposite behavior (Table 2 and Fig. 1).
Final remarks.
Our interests in this study were (i) to recover itraconazole-resistant strains when A. fumigatus was propagated in the presence of subinhibitory concentrations of the drug and (ii) to describe the mechanisms of drug resistance that could be operating in these mutants. Since the replicate populations were founded from a single genotype, any variation among or within populations must have been due to a mutation that occurred during the experiment. Each population was propagated in a controlled, homogeneous environment in which the drug concentration was adjusted to offer the fungus the best opportunity to develop resistance. The 10 mutant strains isolated were all from two of the four populations maintained in the presence of itraconazole. These 10 mutants had variable levels of resistance to itraconazole, variable growth rates in the absence of drug, and variable patterns of altered gene expression, implying multiple underlying mechanisms of resistance. Although certain mutations appeared in multiple isolates (CYP51A G54R in population G and M220I in population H after 10 transfers), the resistant strains in populations G and H were not merely exact clonal copies of the same genotype. Each strain had a unique genotype and was distinguished from all others by one or more mutational events.
If the presence of itraconazole imposes strong directional selection on populations of A. fumigatus, then why did none of the 10 mutant types rise at high frequencies in populations G and H? The two most plausible explanations are as follows. First, the mutations in each strain may have been only partially exposed to selection. On germination of the 106 conidia used to initiate each transfer, heterokaryons would form and any drug-resistant mutant nuclei would likely share the same cytoplasm as drug-sensitive, nonmutant nuclei. If the resistance determinants range more toward recessive than dominant, then the frequency of mutant types would not be expected to rise quickly in response to selection. Second, the physically structured filamentous mycelium of A. fumigatus may retard the adaptive sweeps that occur rapidly within planktonic, well-mixed populations of microorganisms in which mutations conferring increased reproductive output rise to a frequency of 100%, to the exclusion of other genotypes. Clonal competition (11) may even be more of a factor in A. fumigatus cultures than in planktonic cultures.
Although all mutant types remained present at low frequencies in their respective populations, it is striking that (i) 3 of the 10 resistant strains showed more than one nonsynonymous mutation in CYP51A and CYP51B and (ii) all resistant strains showed altered patterns of gene expression, implying that mutations occurred in other genes as well. The multiple mutational events leading to the resistant genotypes are unlikely to have all occurred at one time. Instead, an initial resistance mutation must have been passed on to increasing numbers of descendants until a second mutation occurred, and so on. Although there was no successive fixation of mutations as in well-mixed cultures, selection must have played at least some role in increasing the numbers of mutant types over time. Selection must be a necessary factor because in its absence no itraconazole resistance mutations were recovered in the populations propagated without the drug. The aconidial strain H05-1-10 represents a special case; this genotype would not be expected to be carried into the next transfer among the conidia harvested.
In summary, in vitro evolution of itraconazole resistance in A. fumigatus is linked to mutations in both the CYP51A and the CYP51B genes and to the overexpression of several drug efflux transporters. These genes were constitutively overexpressed in some mutants, while in others they were induced by the presence of drug. Itraconazole resistance emerged rapidly, at the fifth transfer, in the presence of subinhibitory concentrations of itraconazole. However, the MICs for these mutants were low and the mutants had reduced growth rates. Interestingly, most of the mutants isolated in the 10th transfer showed not only high levels of itraconazole resistance but also higher growth rates than the parental strain. The most important observation was the fact that these mutants concomitantly presented with multiple resistance mechanisms, and this surely will be an important consideration for the emergence of clinical resistance to azoles.
Acknowledgments
This research was supported by the Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP), Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) of Brazil.
We thank Gregory S. May and the two anonymous reviewers for critical reading of the manuscript.
REFERENCES
- 1.Arthington-Skaggs, B. A., H., Jradi, T. Desai, and C. J. Morrison. 1999. Quantitation of ergosterol content: novel method for determination of fluconazole susceptibility of Candida albicans. J. Clin. Microbiol. 37:3332-3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balzi, E., and A. Goffeau. 1995. Yeast multidrug resistance: the PDR network. J. Bioerneg. Biomembr. 27:71-76. [DOI] [PubMed] [Google Scholar]
- 3.Cowen, L. E., J. B. Anderson, and L. M. Kohn. 2002. Evolution of drug resistance in Candida albicans. Annu. Rev. Microbiol. 56:139-165. [DOI] [PubMed] [Google Scholar]
- 4.Dannaoui, E., E. Borel, M. F. Monier, M. A. Piens, S. Picot, and F. Persat. 2001. Acquired itraconazole resistance in Aspergillus fumigatus. J. Antimicrob. Chemother. 47:333-340. [DOI] [PubMed] [Google Scholar]
- 5.D'Enfert, C. 1996. Selection of multiple disruption events in Aspergillus fumigatus using the orotidine-5′-decarboxylase gene, pyrG, as a unique transformation marker. Curr. Genet. 30:76-82. [DOI] [PubMed] [Google Scholar]
- 6.Denning, D. W. 1996. Diagnosis and management of invasive aspergillosis. Curr. Clin. Top. Infect. Dis. 16:277-299. [PubMed] [Google Scholar]
- 7.Denning, D. W., K. Venkateswarlu, K. L. Oakley, M. J. Anderson, N. J. Manning, D. A. Stevens, D. A. Warnock, and S. L. Kelly. 1997. Itraconazole resistance in Aspergillus fumigatus. Antimicrob. Agents Chemother. 41:1364-1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Diaz-Guerra, T. M., E. Mellado, M. Cuenca-Estrella, and J. L. Rodríguez-Tudela. 2003. A point mutation in the 14α-sterol demethylase gene cyp51A contributes to itraconazole resistance in Aspergillus fumigatus. Antimicrob. Agents Chemother. 47:1120-1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Edwards, J. E., Jr. 2001. Management of severe candidal infections: integration and review of current guidelines for treatment and prevention. Curr. Clin. Top. Infect. Dis. 21:135-147. [PubMed] [Google Scholar]
- 10.Eisen, M. B., P. T. Spellman, P. O. Brown, and D. Botstein. 1998. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 95:14863-14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elena, S. F., and R. E. Lenski. 2003. Evolution experiments with microorganisms: the dynamics and genetic bases of adaptation. Nat. Rev. Genet. 4:457-489. [DOI] [PubMed] [Google Scholar]
- 12.Herbrecht, R., D. Denning, T. F. Patterson, J. E. Bennett, R. E. Greene, J. W. Oestmann, W. V. Kern, K. A. Marr, P. Ribaud, O. Lortholary, R. Silvestre, R. H. Rubin, J. R. Wingard, P. Stark, C. Durand, D. Caillot, E. Thiel, P. H. Chandrasekar, M. R. Hodges, H. T. Schlamm, P. F. Troke, and B. de Pauw. 2002. Voriconazole versus amphotericin B for primary therapy of invasive aspergillosis. N. Engl. J. Med. 347:408-415. [DOI] [PubMed] [Google Scholar]
- 13.Kafer, E. 1977. Meiotic and mitotic recombination in Aspergillus and its chromosomal aberrations. Adv. Genet. 19:33-131. [DOI] [PubMed] [Google Scholar]
- 14.Kelly, S. L., D. C. Lamb, D. E. Kelly, J. Loeffler, and H. Einsele. 1996. Resistance to fluconazole and amphotericin in Candida albicans from AIDS patients. Lancet 348:1523-1524. [DOI] [PubMed] [Google Scholar]
- 15.Kelly, S. L., D. C. Lamb, D. E. Kelly, N. J. Manning, N. J. Manning, J. Loeffler, H. Hebart, U. Schumacher, and H. Einsele. 1997. Resistance to fluconazole and cross-resistance to amphotericin B in Candida albicans from AIDS patients caused by defective sterol delta 5,6-desaturation. FEBS Lett. 400:80-82. [DOI] [PubMed] [Google Scholar]
- 16.Kolaczkowski, M., J. Kolaczowska, J. Lucynski, S. Witek, and A. Goffeau. 1998. In vivo characterization of the drug resistance profile of the major ABC transporters and other components of the yeast pleiotropic drug resistance network. Microb. Drug Resist. 4:143-158. [DOI] [PubMed] [Google Scholar]
- 17.Lupetti, A., R. Danesi, M. Campa, M. del Tacca, and S. Kelly. 2002. Molecular basis of resistance to azole antifungals. Trends Mol. Med. 8:76-81. [DOI] [PubMed] [Google Scholar]
- 18.Manavathu, E. K., J. A. Vazquez, and P. H. Chandrasekar. 1999. Reduced susceptibility in laboratory-selected mutants of Aspergillus fumigatus to itraconazole due to decreased intracellular accumulation of the antifungal agent. Int. J. Antimicrob. Agents 12:213-219. [DOI] [PubMed] [Google Scholar]
- 19.Mann, P. A., R. M. Parmegiani, S.-Q. Wei, C. A. Mendrick, X. Li, D. Loenberg, B. DiDomenico, R. S. Hare, S. S. Walker, and P. M. McNicholas. 2003. Mutations in Aspergillus fumigatus resulting in reduced susceptibility to posaconazole appear to be restricted to a single amino acid in the cytochrome P-450 14α-demethylase. Antimicrob. Agents Chemother. 47:577-581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marichal, P., L. Koymas, S. Willlemsens, D. Bellens, P. Verhasselt, W. Luyten, M. Borgers, F. C. S. Ramaekers, F. C. Odds, and H. VandenBossche. 1999. Contribution of mutations in the cytochrome P-450 14-α-demethylase (Erg11p, Cyp51p) to azole resistance in Candida albicans. Microbiology 145:2701-2713. [DOI] [PubMed] [Google Scholar]
- 21.Mellado, E., T. M. Diaz-Guerra, M. Cuenca-Estrella, and J. L. Rodríguez-Tudela. 2001. Identification of two different 14-alpha sterol demethylase-related genes (Cyp51A and Cyp51B) in Aspergillus fumigatus and other Aspergillus species. J. Clin. Microbiol. 39:2431-2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mellado, E., G. Garcia-Effron, L. Alcazar-Fuoli, M. Cuenca-Estrella, and J. L. Rodriguez-Tudela. 2004. Substitutions at methionine 220 in the 14α-sterol demethylase (Cyp51A) of Aspergillus fumigatus are responsible for resistance in vitro to azole antifungal drugs. Antimicrob. Agents Chemother. 48:2747-2750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nascimento, A. M., G. H. Goldman, S. Park, S. A. Marras, G. Delmas, U. Oza, K. Lolans, M. N. Dudley, P. A. Mann, and D. S. Perlin. 2003. Multiple resistance mechanisms among Aspergillus fumigatus mutants with high-level resistance to itraconazole. Antimicrob. Agents Chemother. 47:1519-1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.National Committee for Clinical Laboratory Standards. 2002. Reference method for broth dilution antifungal susceptibility testing of conidium-forming filamentous fungi. Proposed standard M38-A. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 25.Nolte, F. S., T. Parkinson, D. J. Falconer, S. Dix, J. Williams, C. Gilmore, R. Geller, and J. R. Wingard. 1997. Isolation and characterization of fluconazole- and amphotericin B-resistant Candida albicans from blood of two patients with leukemia. Antimicrob. Agents Chemother. 41:196-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Osherov, N., D. P. Kontoyannis, A. Romans, and G. S. May. 2001. Resistance to itraconazole in Aspergillus nidulans and Aspergillus fumigatus is conferred by extra copies of the A. nidulans P-450 14α-demethylase gene, pdmA. J. Antimicrob. Chemother. 48:75-81. [DOI] [PubMed] [Google Scholar]
- 27.Osmani, S. A., G. S. May, and N. R. Morris. 1987. Regulation of the mRNA levels of nimA, a gene required for the G2-M transition in Aspergillus nidulans. J. Cell Biol. 104:1495-1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sanglard, D., and F. C. Odds. 2002. Resistance of Candida species to antifungal agents: molecular mechanisms and clinical consequences. Lancet Infect. Dis. 2:73-85. [DOI] [PubMed] [Google Scholar]
- 29.Sanglard, D., F. Ischer, T. Parkinson, D. Falconer, and J. Bille. 2003. Candida albicans: mutations in the ergosterol biosynthetic pathway and resistance to several antifungal agents. Antimicrob. Agents Chemother. 47:2404-2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Semighini, C. P., M. Marins, M. H. S. Goldman, and G. H. Goldman. 2002. Quantitative analysis of the relative transcript levels of ABC transporter Atr genes in Aspergillus nidulans by real-time reverse transcription-PCR assay. Appl. Environ. Microbiol. 68:1351-1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Semighini, C. P., G. Delmas, S. Park, D. Amstrong, D. Perlin, and G. H. Goldman. 2001. New restriction fragment length polymorphism (RFLP) markers for Aspergillus fumigatus. FEMS Immunol. Med. Microbiol. 31:15-19. [DOI] [PubMed] [Google Scholar]
- 32.Slaven, J. W., M. J. Anderson, D. Sanglard, G. K. Dixon, J. Bille, I. S. Roberts, and D. W. Denning. 2002. Increased expression of a novel Aspergillus fumigatus ABC transporter gene, AtrF, in the presence of itraconazole in an itraconazole resistant clinical isolate. Fungal Genet. Biol. 36:199-206. [DOI] [PubMed] [Google Scholar]
- 33.Stone, E. A., H. B. Fung, and H. L. Kirschenbaum. 2002. Caspofungin: an echinocandin antifungal agent. Clin. Ther. 24:351-377. [DOI] [PubMed] [Google Scholar]
- 34.Tobin, M. B., R. B. Peery, and P. L. Skatrud. 1997. Genes encoding multiple drug resistance-like proteins in Aspergillus fumigatus and Aspergillus flavus. Gene 200:11-23. [DOI] [PubMed] [Google Scholar]
- 35.Van Bambeke, F., E. Balzi, and P. M. Tulkens. 2000. Antibiotic efflux pumps. Biochem. Pharmacol. 60:457-470. [DOI] [PubMed] [Google Scholar]
- 36.White, T. C., K. A. Marr, and R. A. Bowden. 1998. Clinical, cellular, and molecular factors that contribute to antifungal drug resistance. Clin. Microbiol. Rev. 11:382-402. [DOI] [PMC free article] [PubMed] [Google Scholar]