Abstract
Prospective cohort studies are needed to assess the relationship between the fecal microbiome and human health and disease. To evaluate fecal collection methods, we determined technical reproducibility, stability at ambient temperature, and accuracy of 5 fecal collection methods (no additive, 95% ethanol, RNAlater Stabilization Solution, fecal occult blood test cards, and fecal immunochemical test tubes). Fifty-two healthy volunteers provided fecal samples at the Mayo Clinic in Rochester, Minnesota, in 2014. One set from each sample collection method was frozen immediately, and a second set was incubated at room temperature for 96 hours and then frozen. Intraclass correlation coefficients (ICCs) were calculated for the relative abundance of 3 phyla, 2 alpha diversity metrics, and 4 beta diversity metrics. Technical reproducibility was high, with ICCs for duplicate fecal samples between 0.64 and 1.00. Stability for most methods was generally high, although the ICCs were below 0.60 for 95% ethanol in metrics that were more sensitive to relative abundance. When compared with fecal samples that were frozen immediately, the ICCs were below 0.60 for the metrics that were sensitive to relative abundance; however, the remaining 2 alpha diversity and 3 beta diversity metrics were all relatively accurate, with ICCs above 0.60. In conclusion, all fecal sample collection methods appear relatively reproducible, stable, and accurate. Future studies could use these collection methods for microbiome analyses.
Keywords: feces, microbiota, specimen collection
The human microbiome (i.e., the collection of microbial genes found in and on the human body) has been observed to be associated with a number of health conditions, such as obesity (1), inflammatory bowel disease (2), and cancer (3). However, the observed microbial associations with specific outcomes tend to vary between studies. For example, in studies of the association of the fecal microbiome with colorectal cancer, alpha diversity (i.e., within-sample diversity) has been found to be increased (4), the same (5–7), and decreased (8, 9) for colorectal cancer cases compared with controls. Variability between studies could result from real biological differences between studies but might also be due to artefacts induced by variations in sample collection and storage, DNA extraction, sequencing technology, or other unknown factors.
Most of the previous human microbiome research studies have been cross-sectional, particularly fecal microbiome studies, because few (if any) cohorts have collected fecal samples. In order to facilitate human microbiome research, there is a need for investigators in prospective cohort studies to collect samples, including fecal samples, that can be used to evaluate the role of the microbiome in disease etiology. For new biospecimen collections, identification of methods that are stable at room temperature for a period of time (e.g., the time a sample might take to travel from a study participant's home to a central biorepository) and that adequately represent the microbial content of the sample is essential.
We recently completed a study in which we evaluated 7 fecal sampling methods: no additive, RNAlater Stabilization Solution (Ambion, Austin, Texas), 70% ethanol, ethylenediaminetetraacetic acid, dry swab, and pre- and postdevelopment fecal occult blood test (FOBT) card. These methods were used to determine technical reproducibility (i.e., consistency of replicates stored in the same manner), stability at ambient temperature over 1 and 4 days, and accuracy (i.e., comparison of each method to a putative “gold standard”). We found that the FOBT cards and samples stored in RNAlater had the highest stability when stored at room temperature over the course of 4 days and that the swab, FOBT card, and 70% ethanol samples were the most correlated with the no-additive samples (10). However, that study had a limited sample size of 20 participants, had generally poor results for 70% ethanol, and did not include the fecal immunochemical test (FIT), which is now used in some colorectal cancer screening programs. Therefore, we designed a larger study to evaluate 5 fecal collection methods: no additive, 95% ethanol, RNAlater, postdevelopment FOBT cards, and FIT tubes.
METHODS
Study participants
Fifty-two healthy volunteers were recruited from among Mayo Clinic employees in Rochester, Minnesota. To be eligible, participants had to be 18 years of age or older, not have used antibiotics or probiotics within the past 2 weeks, have no history of pelvic radiation, and not currently be undergoing chemotherapy. All participants provided informed consent, and the study was approved by the Mayo Clinic Studies Institutional Review Board and the National Cancer Institute Office of Human Subjects Research.
Fecal specimen collection
At recruitment, participants provided oral samples using an OMNIgene DISCOVER kit (DNA Genotek Inc., Ottawa, Ontario, Canada), which collected saliva, and a Scope mouthwash (Procter & Gamble Co., Cincinnati, Ohio) and then filled out a questionnaire regarding tobacco use, alcohol consumption, oral health habits, recent antibiotic exposure, and demographic characteristics. The participants were then invited to return at a later date to provide the fecal specimen. Upon arrival for collection, an Exakt Pak canister (Inmark Packaging, Austell, Georgia) was provided to each participant. The participant collected the feces, and the study coordinator delivered it to the laboratory for immediate processing. The participant then completed an abbreviated questionnaire that was used to obtain information about diet, colon health, menstrual history, and recent antibiotic exposure.
The fecal specimens were mixed manually using a spatula, and aliquots for the different collection methods were generated in a random order for each participant. For each participant, 16 aliquots of feces, 4 triple-slide FOBT cards, and 4 FIT tubes were created. Approximately 1–2 grams of feces, representing a full scoop, was placed in a Sarstedt feces tube (Numbrecht, Germany) containing no additive (4 aliquots), 2.5 mL of RNAlater Stabilization Solution (4 aliquots), or 2.5 mL of 95% ethanol (Sigma-Aldrich, St. Louis, Missouri; 8 aliquots). Four triple-slide Hemoccult II Elite Dispensapak Plus for FOBT (Beckman Coulter, Brea, California) were smeared thinly with feces and the flap was closed. Four FIT tubes (Polymedco, Inc., Cortlandt Manor, New York) were created by dipping into the fecal specimen with the FIT probe, and the tube was shaken. Two aliquots from each FIT tube were created and stored in cryovials.
Four replicates of the no-additive, 95% ethanol, and FIT cryovials samples and 2 replicates of the RNAlater samples were frozen immediately at −80°C (day 0). Two FOBT cards were developed using 2 drops of Hemoccult Sensa Developer (Beckman Coulter) applied to the guaiac paper on the back of the card (i.e., the testing strategy for occult blood in colorectal cancer screening) and frozen immediately at −80°C (day 0). The remaining samples were left at ambient temperature for 96 hours. The remaining FOBT cards were then developed, and all remaining samples were frozen at −80°C (day 4).
DNA extraction and sequencing
An outline of the samples used for this study is presented in Table 1. For each participant, 2 aliquots of the day-0 no-additive samples were used and considered the gold standard. No day-4 no-additive samples were collected because the no-additive samples had poor stability at ambient temperature (10). Two aliquots each of the 95% ethanol, RNAlater, and FIT cryovials of the day-0 and day-4 samples were included. One day-0 and 1 day-4 triple-slide FOBT card were included. The remaining aliquots were stored for future analyses.
Table 1.
Collection Methods for Fecal Samples and Number of Aliquots Used for Microbiome Analyses, Mayo Clinic, Rochester, Minnesota, 2014
| Collection Method | No. Frozen Immediately (Day 0) | No. Frozen After 4 Days (Day 4) |
|---|---|---|
| No additive | 2 | 0 |
| RNAlater | 2 | 2 |
| 95% ethanol | 2 | 2 |
| FOBT carda | 1 | 1 |
| FIT tube | 2 | 2 |
Abbreviations: FIT, fecal immunochemical test; FOBT, fecal occult blood test.
a The FOBT card was a triple-slide (3-window) card that was developed with peroxide at day 0 or day 4. Three windows were used per card, and each window was considered a separate aliquot.
The samples were shipped on dry ice to the University of California, San Diego (La Jolla, California), thawed at 4°C, and kept on ice during plating. The fecal samples were swabbed using wooden swabs (Puritan Cotton Tipped Applicators, Puritan Medical Products, Guilford, Maine), which were then used for the DNA extraction. The no-additive, 95% ethanol, and RNAlater aliquots were sampled by pulling out the fecal material and swabbing. The FOBT cards were swabbed vigorously with a dry swab. A swab was dipped into each aliquot from the FIT tubes.
DNA extraction, polymerase chain reaction amplification, and amplicon preparation for sequencing were performed as described by Caporaso et al. (11) using the universal bacterial primer set 515F/806R (11, 12). Negative controls included no-template controls for DNA extraction and polymerase chain reaction amplification. All barcoded amplicons were pooled in equal concentrations for sequencing on the Illumina HiSeq (San Diego, California). After removing singletons and reads with read errors, the average coverage was approximately 37,000 reads per sample.
Bioinformatic data processing
Reads were demultiplexed and quality filtered using QIIME 1.9 (13). Each sample was independently cleaned by removing all candidate read-errors using deblur (https://github.com/biocore/deblur; A.A. and R.K., unpublished data, 2016). The cleaned read files were joined to make a single biological observation matrix table, with each operational taxonomic unit (OTU) representing a unique 150–base pair sequence. Taxonomy was assigned to the OTUs using Greengenes database, version 13.8 (14) and RDP classifier 2.2 (15). The data were rarefied to 9,500 reads per sample. Alpha diversity measures (observed OTUs and the Shannon diversity index) were calculated using the R phyloseq package (16), the Bray-Curtis distance was calculated using the R vegan package, and unweighted, generalized, and weighted UniFrac were calculated using the R GUniFrac package (17).
Statistical analysis
Descriptive characteristics of the population based on the questionnaire data provided by the participants are presented. To identify potential outliers, we calculated the Partitioning Around Medoids algorithm based on the unweighted UniFrac distance. First, we used the Partitioning Around Medoids algorithm set with clusters to 2, which included data from the oral and fecal samples. Fecal samples that clustered with the oral samples were excluded. We then set the clusters to the number of subjects. Samples that did not cluster around the correct subject were identified as suspicious. These suspicious samples were viewed using principal coordinate analysis plots and genus-level abundance barplots. Samples that did not appear to be similar to the rest of the samples from that individual were then excluded.
A distance-based coefficient of determination (R2) was calculated from the beta diversity estimates from unweighted UniFrac, generalized UniFrac, weighted UniFrac, and Bray-Curtis distance to evaluate the percentage of microbial variability explained by subject, sample collection type, and day of freezing. We compared measures of alpha diversity (observed OTUs and the Shannon diversity index) between each fecal collection method at day 0 and day 4 and created a linear mixed-effects model to test for differences between methods compared with the gold-standard immediately frozen no-additive fecal sample with adjustment for the freezing time point.
We calculated intraclass correlation coefficients (ICCs) from a mixed-effects model to evaluate the technical reproducibility, stability at ambient temperature, and accuracy of the different fecal collection methods. The ICCs were calculated based on the square root of the relative abundances of the 3 most dominant phyla (Actinobacteria, Bacteroidetes, and Firmicutes), 2 alpha diversity metrics (observed OTUs and the Shannon diversity index), and the top principal coordinate analysis component of 4 beta diversity metrics (unweighted UniFrac, generalized UniFrac, weighted UniFrac, and the Bray-Curtis distance). The top principal coordinate analysis component explained 15.3%, 13.5%, 28.4%, and 10.2% of the variability for unweighted UniFrac, generalized UniFrac, weighted UniFrac, and the Bray-Curtis distance, respectively. We also calculated the ICCs for the square root of the relative abundance of the major genera (i.e., prevalence in the population >50% and a mean relative abundance >0.2%). For technical reproducibility, we calculated the ICCs for the duplicate fecal samples for each method at each freezing time point. For stability at ambient temperature, we randomly selected 1 replicate for each collection type from samples frozen immediately and 1 replicate from samples frozen after 4 days at ambient temperature and then calculated the ICC. For the accuracy calculation, we considered the fecal sample with no additive that was frozen immediately to be the gold standard. We then randomly selected 1 replicate from each of the collection methods frozen immediately and calculated an ICC compared with the gold standard. We calculated 95% confidence intervals for samples with replicates based on resampling (i.e., stability and accuracy of fecal samples) with 50 random samplings. For samples without replicates (i.e., reproducibility of fecal samples), the 95% confidence interval was estimated using the R ICC package (confidence interval = “Smith”). For the accuracy calculation, we also calculated the Spearman correlation coefficient to determine whether the rank order compared with the gold standard was preserved. We calculated P values for between-methods comparisons using a t test. Given the multiple testing, a P value < 0.018 would be statistically significant for a false discovery rate of 5%. All statistical analyses were conducted using R, version 3.1.2.
RESULTS
Descriptive characteristics
The participants in this sample were predominantly female (65.4%) and ranged in age from 22 to 56 years, with a mean age of 35.8 (standard deviation, 10.2) years. The majority had at least a bachelor degree (69.2%) and were non-Hispanic white (90.4%). Because participants did not typically collect fecal samples on the same day as recruitment, 1 participant had used antibiotics in the month before the fecal sample collection (Table 2).
Table 2.
Descriptive Statistics of Study Participants (n = 52), Mayo Clinic, Rochester, Minnesota, 2014
| Characteristic | No. | % |
|---|---|---|
| Age, years | 35.8 (10.2)a | |
| Sex | ||
| Male | 18 | 34.6 |
| Female | 34 | 65.4 |
| Educational level | ||
| Associate degree or less | 16 | 30.8 |
| Bachelor degree | 21 | 40.4 |
| Master or Doctoral degree | 15 | 28.8 |
| Race/ethnicity | ||
| Non-Hispanic white | 47 | 90.4 |
| Other | 5 | 9.6 |
| Used antibiotics past month | 1 | 1.9 |
a Data are expressed as mean (standard deviation).
In the outlier analysis, we identified 2 no-additive, three 95% ethanol, 3 RNAlater, 5 FOBT, and 12 FIT samples to either incorrectly cluster with oral samples or to not cluster with the other samples from that individual. The outlier samples represented only 1 of the duplicates for all samples except for 1 FIT tube for which both duplicates were considered outliers and removed. After removal of the outliers, the percentage of microbial variability related to between subject variability was high for all beta diversity estimates (Web Figure 1, available at http://aje.oxfordjournals.org/). The differences by sample collection type and time at ambient temperature were substantially lower than the between-subject variability.
When comparing alpha diversity between collection methods, in general, overall alpha diversity metrics at day 0 and day 4 were similar for both observed OTUs (Web Figure 2A) and the Shannon diversity index (Web Figure 2B). In the linear mixed-effects model, compared with what was seen in the gold-standard no-additive samples that were frozen immediately, on average, 7 more OTUs (95% confidence interval: 2.98, 11.01) were detected on FOBT cards, and those samples had an increase in the Shannon diversity index of 0.16 (95% confidence interval: 0.12, 0.20). In contrast, 9 fewer OTUs (95% confidence interval: −4.55, −13.07) were detected in samples stored in 95% ethanol, and those samples had a decrease in the Shannon diversity index of 0.10 (95% confidence interval: −0.07, −0.14) compared with the gold standard. No statistically significant differences were detected in alpha diversity for the FIT tube and RNAlater samples compared with the gold standard. When comparing the relative abundances at the phylum level, the patterns by sample collection type seemed generally similar for day-0 (Web Figure 3A) and day-4 (Web Figure 3B) fecal samples, although samples preserved using an FOBT card, RNAlater, and 95% ethanol appeared to have a higher relative abundance of Actinobacteria and a lower relative abundance of Verrucomicrobia compared with the samples with no additives and those in the FIT tubes.
Technical reproducibility
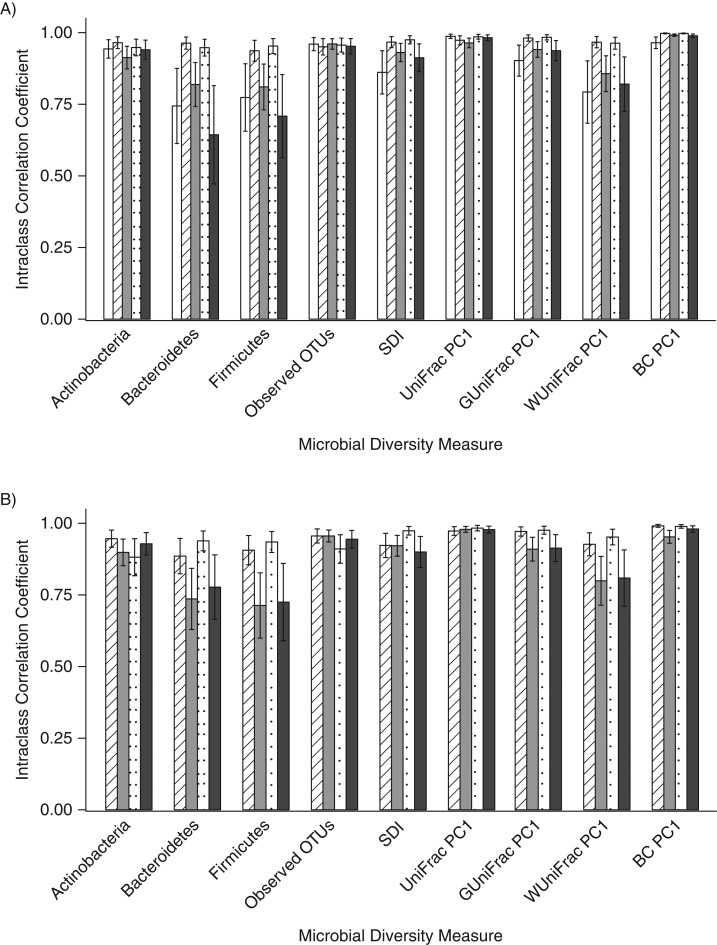
The ICCs for the duplicate day-0 fecal samples collected with the different methods were generally high (range, 0.64–1.00) for the measurement of 3 phyla, 2 alpha diversity metrics, and the first principal coordinate for 4 beta diversity metrics. However, for the relative abundance of Bacteroidetes and Firmicutes, the ICCs were slightly lower for 95% ethanol (for Bacteroidetes, ICC = 0.64; for Firmicutes, ICC = 0.71) and no-additive (for Bacteroidetes, ICC = 0.74; for Firmicutes, ICC = 0.77) samples (Figure 1A; Web Table 1). For the day-4 samples, the ICCs for the duplicate fecal samples were again relatively high (range, 0.71–0.99), but the ICCs for the 95% ethanol samples (for Bacteroidetes, ICC = 0.78; for Firmicutes, ICC = 0.73) and the FOBT card samples (for Bacteroidetes, ICC = 0.74; for Firmicutes, ICC = 0.71) were lower for the relative abundances of Bacteroidetes and Firmicutes than they were for the other methods (Figure 1B; Web Table 2). For both the day-0 and day-4 samples, there were statistically significant differences (P < 0.018) between methods for some of the metrics (Web Tables 3 and 4), and the genus-level ICCs were generally high for both day-0 and day-4 samples (Web Tables 5 and 6).
Figure 1.
Technical reproducibility of replicates frozen immediately (A) and frozen after incubation at room temperature for 4 days (B) for the evaluation of the relative abundance of 3 phyla, 2 alpha diversity metrics, and the first principal coordinate of 4 beta diversity metrics using intraclass correlation coefficients, Mayo Clinic, Rochester, Minnesota, 2014. White columns indicate replicates stored in no solution, striped columns indicate those stored in fecal immunochemical test tubes, gray columns indicate those stored on fecal occult blood test cards, dotted columns indicate those stored in RNAlater Stabilization Solution (Ambion, Austin, Texas), and black columns indicate those stored in 95% ethanol. BC, Bray-Curtis distance; OTU, operational taxonomic unit; PC1, principal coordinate analysis component 1; SDI, Shannon diversity index.
Stability at ambient temperature
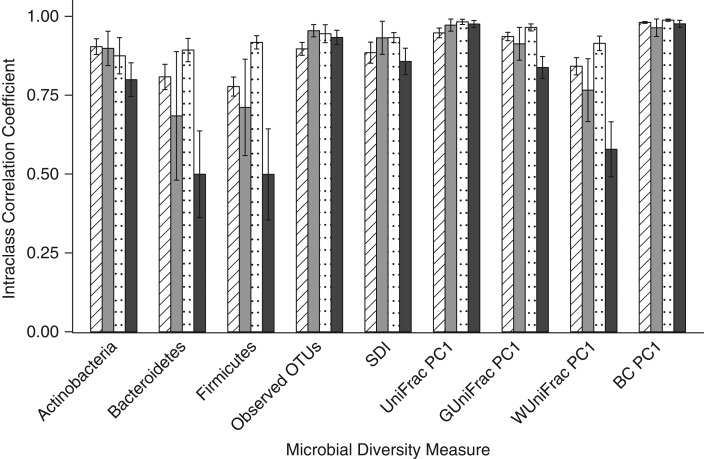
We compared the day-0 fecal samples to the day-4 samples held at room temperature in order to simulate mailed-in samples. The samples preserved in RNAlater tended to have the highest ICCs for the relative abundance of the 3 phyla (range, 0.88–0.92), and the 95% ethanol samples had the lowest ICCs for the 3 phyla (range, 0.50–0.80). All methods had relatively high ICCs for the observed OTUs (range, 0.90–0.93), the Shannon diversity index (range, 0.86–0.93), and 3 of the 4 beta diversity metrics (for unweighted UniFrac, the ICC range was 0.95–0.98; for generalized UniFrac, the ICC range was 0.84–0.97; and for Bray-Curtis distance, the ICC range was 0.96–0.99). Fecal samples stored in 95% ethanol had an ICC of 0.58 for weighted UniFrac (Figure 2; Web Table 7). A number of statistically significant differences (P < 0.018) were detected for stability between methods (Web Table 8); however, the genus-level ICCs were generally high for all methods (Web Table 9).
Figure 2.
Stability of fecal samples incubated at room temperature for 4 days and then frozen compared with that of samples frozen immediately for the evaluation of relative abundance of 3 phyla, 2 alpha diversity metrics, and the first principal coordinate of 4 beta diversity metrics using intraclass correlation coefficients, Mayo Clinic, Rochester, Minnesota, 2014. Striped columns indicate those stored in fecal immunochemical test tubes, gray columns indicate those stored on fecal occult blood test cards, dotted columns indicate those stored in RNAlater Stabilization Solution (Ambion, Austin, Texas), and black columns indicate those stored in 95% ethanol. BC, Bray-Curtis distance; OTU, operational taxonomic unit; PC1, principal coordinate analysis component 1; SDI, Shannon diversity index.
Accuracy compared with gold standard
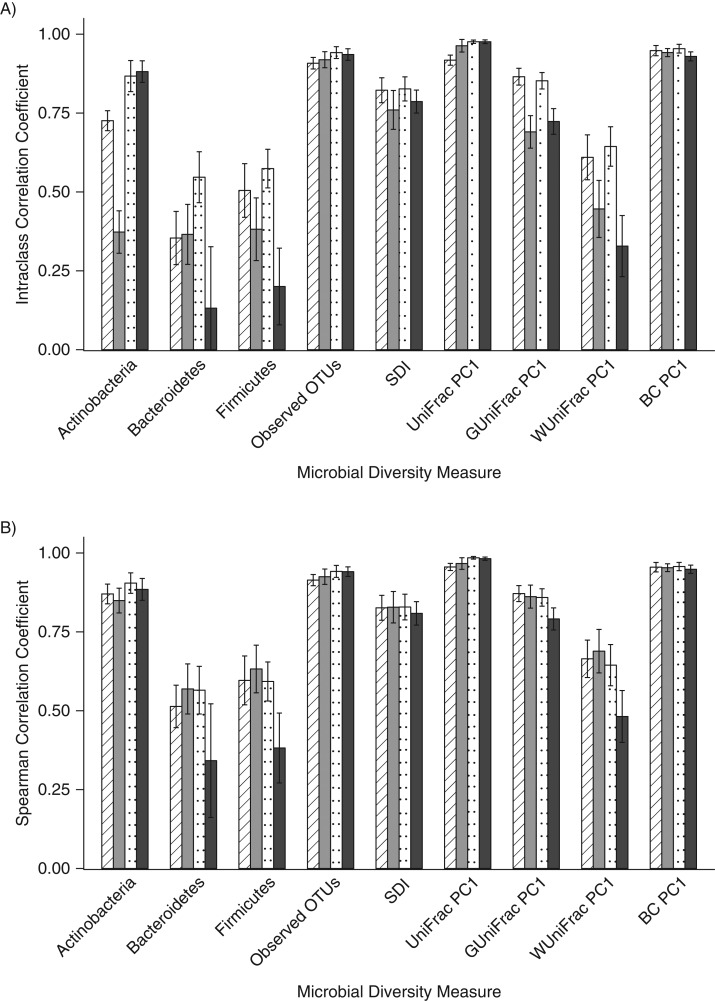
When compared with the gold-standard no-additive fecal samples that had been frozen immediately, in general, the ICCs were low for the relative abundance of Bacteroidetes (range, 0.13–0.55) and Firmicutes (range, 0.20–0.57) and for weighted UniFrac (range, 0.33–0.64), but the ICCs were higher for observed OTUs (range, 0.76–0.83), the Shannon diversity index (range, 0.76–0.83), unweighted UniFrac (range, 0.92–0.98), and the Bray-Curtis distance (range, 0.93–0.95) (Figure 3A, Web Table 10). When accuracy was measured using the Spearman correlation to assess rank order, the correlation coefficients were stronger than the ICCs detected for most metrics (Figure 3B, Web Table 11). When comparing the ICCs, there were many statistically significant differences (P < 0.018) between collection methods (Web Table 12), but there were fewer statistically significant differences in Spearman correlation coefficients for the collection methods (Web Table 13). The genus-level ICCs and Spearman correlation coefficients had similar patterns, with lower values for ICCs and higher values for the Spearman correlations (Web Tables 14 and 15).
Figure 3.
Accuracy of fecal samples frozen immediately compared with the gold standard, which was considered to be the no-additive sample, Mayo Clinic, Rochester, Minnesota, 2014. Intraclass correlation coefficients (A) and Spearman correlation coefficients (B) were calculated for the relative abundance of 3 phyla, 2 alpha diversity metrics, and the first principal coordinate of 4 beta diversity metrics. Striped columns indicate those stored in fecal immunochemical test tubes, gray columns indicate those stored on fecal occult blood test cards, dotted columns indicate those stored in RNAlater Stabilization Solution (Ambion, Austin, Texas), and black columns indicate those stored in 95% ethanol. BC, Bray-Curtis distance; OTU, operational taxonomic unit; PC1, principal coordinate analysis component 1; SDI, Shannon diversity index.
DISCUSSION
In the present study, we found that the between-subject differences greatly outweighed any differences by sample collection type and time spent at ambient temperature. All methods appeared to have similar alpha diversity estimates, although FOBT cards had statistically significantly greater counts of OTUs and Shannon diversity indices than did the no-additive gold-standard sample. Using ICCs to estimate reproducibility, stability, and accuracy, we observed that all of the collection methods tended to perform relatively well compared with the no-additive gold-standard sample, although there was some variability by diversity metric. Specifically, the metrics that were sensitive to relative abundance measures, including the relative abundances for the top 3 phyla and weighted UniFrac, tended to have lower ICCs than did other alpha diversity measures or the beta diversity measures. This was likely due to the fact that the relative abundances are highly interrelated because of the normalization (i.e., all relative abundance values will sum to 1), and therefore if 1 of the preservation methods slightly changes the relative abundance of 1 phylum, all other phyla will be altered. However, overall, it appears that all of the included methods were acceptable and could be utilized for future fecal sample collections in microbiome studies.
Investigators in a number of previous studies have assessed different collection methods for fecal samples and the impact of leaving a sample at room temperature on the fecal microbiome (10, 18–33). The FOBT card, or a similar Whatman FTA card (GE Healthcare Life Sciences, Pittsburgh, Pennsylvania), has been tested in a few studies (10, 21, 26, 32). The samples on these cards tend to perform well in these studies compared with immediately frozen fecal samples, although in 1 study, researchers detected lower DNA yields from the Whatman FTA card (26). Similar to our findings, increased alpha diversity in Whatman FTA cards was observed a recent study; this was hypothesized to be related to increased cell lysis through the card, because most of the blank cards did not have detectable microbes (32). RNAlater has also been tested in a number of studies (10, 20–24, 26, 30, 32), but the results varied. The majority of studies concluded that RNAlater was an acceptable preservative for microbial analyses of fecal samples (10, 22, 23, 26, 30), although in some studies, researchers detected decreased alpha diversity (20, 21, 30), decreased DNA purity or yields (21, 24), or lower stability at room temperature for longer periods (33) compared with immediately frozen fecal samples. Storage of fecal samples in 70% ethanol does not appear to be suitable for microbiome analyses (10), but 95% ethanol has been previously observed to adequately preserve fecal samples (23, 33). In 1 study, researchers evaluated FIT tubes for microbiome analysis and found the FIT samples to be similar to fecal samples with no additive (33). In both this study and previous work, it appeared that all of the included methods were appropriate for microbiome studies, which was expected because we eliminated methods from previous studies that did not appear to have good reproducibility, stability, or accuracy. Although the interindividual differences were seen to outweigh the sample collection method and time at ambient temperature, any new study should utilize only 1 collection method for comparisons, because we detected statistically significant differences between methods with regard to technical reproducibility, stability, and accuracy. However, short differences in the time taken to freeze unique individuals’ samples will be unlikely to greatly affect results as long as these differences were not related to the outcome of interest.
The present study is not without limitations. Our study included predominantly non-Hispanic white participants. It is possible that the technical reproducibility, stability, and accuracy of these methods could vary within other racial/ethnic groups because of differing exposures, such as diet or differing microbial composition; therefore, these collection methods should be tested in other populations. In addition, participants were generally healthy, and the effect of collection methods on samples may vary for people with highly dysbiotic microbiota. However, previous studies have included patients with irritable bowel syndrome and inflammatory bowel disease (19, 29), and in general, researchers did not observe distinct differences by collection method between disease groups. In addition, we only conducted 16S ribosomal RNA gene amplicon analyses; future studies should look at how collection methods affect downstream multi-omic analyses, such as whole-genome shotgun metagenomic sequencing and metabolomics. We only preserved the samples in 2.5 mL of RNAlater, which may not be a sufficient volume to adequately preserve the sample. However, we were focused on methods that may be feasible for large epidemiologic studies, and as we described previously, larger volumes of RNAlater would likely be cost prohibitive (34). Finally, the gold-standard immediately frozen, no-additive sample may not entirely represent the microbial composition compared with an immediately extracted sample, because sample freezing has been observed to increase the relative abundance of Firmicutes and decrease the relative abundance of Bacteroidetes compared with immediate extraction (35). However, immediate DNA extraction in large epidemiologic studies is unlikely to be feasible, and given this difficulty, it has been suggested that studies instead ensure that the same protocol for freezing is followed for all samples (35).
The present study also has a number of strengths. To our knowledge, this study has the largest sample of participants of all studies comparing fecal collection methods. Previously, the number of participants included ranged from 1 (20) to 28 participants (29). Because there is high interindividual variability in the gut microbiome (36) and variable microbial composition may be differentially affected by collection method, it is important to evaluate fecal collection methods in larger groups of people. In addition, this is the second study in which the feasibility of collecting fecal samples using FIT tubes has been demonstrated. A number of health plans in the United States and around the world are screening for colorectal cancer using FITs, so a new cohort could be created using those samples after screening.
In conclusion, all of the fecal sample collection methods appear to be relatively reproducible, stable, and accurate. For future studies, the selection of a fecal sample collection method will depend on the feasibility of each method. For example, the FOBT card can be shipped in the mail, which may make the use of FOBT cards appropriate for large, geographically diverse cohorts. It is important that investigators in large epidemiologic studies collect fecal samples for microbiome analysis in order to determine the prospective nature of currently observed cross-sectional microbiome associations with health and disease.
ACKNOWLEDGMENTS
Author affiliations: Metabolic Epidemiology Branch, Division of Cancer Epidemiology and Genetics, National Cancer Institute, National Institutes of Health, Bethesda, Maryland (Emily Vogtmann, Christian C. Abnet, Rashmi Sinha); Cancer Prevention Fellowship Program, Division of Cancer Prevention, National Cancer Institute, Bethesda, Maryland (Emily Vogtmann); Microbiome Program, Center for Individualized Medicine, Mayo Clinic, Rochester, Minnesota (Jun Chen, Heidi Nelson, Nicholas Chia); Health Sciences Research, Mayo Clinic, Rochester, Minnesota (Jun Chen, Nicholas Chia); Department of Pediatrics, University of California, San Diego, La Jolla, California (Amnon Amir, Rob Knight); Biostatistics Branch, Division of Cancer Epidemiology and Genetics, National Cancer Institute, Bethesda, Maryland (Jianxin Shi); Department of Surgery, Mayo Clinic, Rochester, Minnesota (Heidi Nelson, Nicholas Chia); Department of Computer Science & Engineering, University of California, San Diego, La Jolla, California (Rob Knight); and Biomedical Engineering and Physiology, Mayo College, Rochester, Minnesota (Nicholas Chia).
This work was supported by the Intramural Research Program of the National Cancer Institute at the National Institutes of Health; the Gerstner Family Career Development Awards and Mayo Clinic Center for Individualized Medicine (to J.C.); a grant from the National Institutes of Health (1R01CA179243 to N.C.); and the Howard Hughes Medical Institute and the Sloan Foundation awards (to R.K.).
Data have been submitted to the European Nucleotide Archive under accession number PRJEB13895 (ERP015478).
We thank Dr. Xianfeng Chen (Department of Health Sciences Research, Mayo Clinic), Dr. Adam Robbins-Pianka (Department of Pediatrics, University of California San Diego), Yoshiki Vazquez Baeza (Department of Pediatrics, University of California San Diego), Grant Gogul (Department of Pediatrics, University of California San Diego), James Gaffney (Department of Pediatrics, University of California San Diego), and Greg Humphrey (Department of Pediatrics, University of California San Diego) for their technical assistance and Dr. Patricio Jeraldo (Department of Surgery, Mayo Clinic) for his insightful discussions.
This work was presented in part as a poster at ASM Microbe 2016, June 16–20, 2016, Boston, Massachusetts.
Conflict of interest: R.K. is chief science officer at Biota Technology, Inc.; has received honoraria from the Speakers Bureau of American Academy of Anti-Aging Medicine and Zurich Insurance Company; has ownership interest (including patents) in Biota Technology, Inc.; and is a consultant/advisory board member for Commense, Inc., Johnson & Johnson/Janssen, and Prometheus Therapeutics & Diagnostics. The other authors report no conflicts.
REFERENCES
- 1.Ley RE. Obesity and the human microbiome. Curr Opin Gastroenterol. 2010;26(1):5–11. [DOI] [PubMed] [Google Scholar]
- 2.Kostic AD, Xavier RJ, Gevers D. The microbiome in inflammatory bowel disease: current status and the future ahead. Gastroenterology. 2014;146(6):1489–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vogtmann E, Goedert JJ. Epidemiologic studies of the human microbiome and cancer. Br J Cancer. 2016;114(3):237–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Feng Q, Liang S, Jia H, et al. Gut microbiome development along the colorectal adenoma-carcinoma sequence. Nat Commun. 2015;6:6528. [DOI] [PubMed] [Google Scholar]
- 5.Weir TL, Manter DK, Sheflin AM, et al. Stool microbiome and metabolome differences between colorectal cancer patients and healthy adults. PLoS One. 2013;8(8):e70803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu N, Yang X, Zhang R, et al. Dysbiosis signature of fecal microbiota in colorectal cancer patients. Microb Ecol. 2013;66(2):462–470. [DOI] [PubMed] [Google Scholar]
- 7.Zeller G, Tap J, Voigt AY, et al. Potential of fecal microbiota for early-stage detection of colorectal cancer. Mol Syst Biol. 2014;10:766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ahn J, Sinha R, Pei Z, et al. Human gut microbiome and risk for colorectal cancer. J Natl Cancer Inst. 2013;105(24):1907–1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu J, Feng Q, Wong SH, et al. Metagenomic analysis of faecal microbiome as a tool towards targeted non-invasive biomarkers for colorectal cancer [published online ahead of print September 25, 2015]. Gut. (doi:10.1136/gutjnl-2015-309800). [DOI] [PubMed] [Google Scholar]
- 10.Sinha R, Chen J, Amir A, et al. Collecting fecal samples for microbiome analyses in epidemiology studies. Cancer Epidemiol Biomarkers Prev. 2016;25(2):407–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Caporaso JG, Lauber CL, Walters WA, et al. Ultra-high- throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012;6(8):1621–1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Walters WA, Caporaso JG, Lauber CL, et al. PrimerProspector: de novo design and taxonomic analysis of barcoded polymerase chain reaction primers. Bioinformatics. 2011;27(8):1159–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Caporaso JG, Kuczynski J, Stombaugh J, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7(5):335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DeSantis TZ, Hugenholtz P, Larsen N, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72(7):5069–5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cole JR, Wang Q, Cardenas E, et al. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 2009;37(Database issue):D141–D145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McMurdie PJ, Holmes S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One. 2013;8(4):e61217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen J, Bittinger K, Charlson ES, et al. Associating microbiome composition with environmental covariates using generalized UniFrac distances. Bioinformatics. 2012;28(16):2106–2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cardona S, Eck A, Cassellas M, et al. Storage conditions of intestinal microbiota matter in metagenomic analysis. BMC Microbiol. 2012;12:158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carroll IM, Ringel-Kulka T, Siddle JP, et al. Characterization of the fecal microbiota using high-throughput sequencing reveals a stable microbial community during storage. PLoS One. 2012;7(10):e46953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Choo JM, Leong LE, Rogers GB. Sample storage conditions significantly influence faecal microbiome profiles. Sci Rep. 2015;5:16350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dominianni C, Wu J, Hayes RB, et al. Comparison of methods for fecal microbiome biospecimen collection. BMC Microbiol. 2014;14:103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Flores R, Shi J, Yu G, et al. Collection media and delayed freezing effects on microbial composition of human stool. Microbiome. 2015;3:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Franzosa EA, Morgan XC, Segata N, et al. Relating the metatranscriptome and metagenome of the human gut. Proc Natl Acad Sci USA. 2014;111(22):E2329–E2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gorzelak MA, Gill SK, Tasnim N, et al. Methods for improving human gut microbiome data by reducing variability through sample processing and storage of stool. PLoS One. 2015;10(8):e0134802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lauber CL, Zhou N, Gordon JI, et al. Effect of storage conditions on the assessment of bacterial community structure in soil and human-associated samples. FEMS Microbiol Lett. 2010;307(1):80–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nechvatal JM, Ram JL, Basson MD, et al. Fecal collection, ambient preservation, and DNA extraction for PCR amplification of bacterial and human markers from human feces. J Microbiol Methods. 2008;72(2):124–132. [DOI] [PubMed] [Google Scholar]
- 27.Ott SJ, Musfeldt M, Timmis KN, et al. In vitro alterations of intestinal bacterial microbiota in fecal samples during storage. Diagn Microbiol Infect Dis. 2004;50(4):237–245. [DOI] [PubMed] [Google Scholar]
- 28.Roesch LF, Casella G, Simell O, et al. Influence of fecal sample storage on bacterial community diversity. Open Microbiol J. 2009;3:40–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tedjo DI, Jonkers DM, Savelkoul PH, et al. The effect of sampling and storage on the fecal microbiota composition in healthy and diseased subjects. PLoS One. 2015;10(5):e0126685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Voigt AY, Costea PI, Kultima JR, et al. Temporal and technical variability of human gut metagenomes. Genome Biol. 2015;16:73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu GD, Lewis JD, Hoffmann C, et al. Sampling and pyrosequencing methods for characterizing bacterial communities in the human gut using 16S sequence tags. BMC Microbiol. 2010;10:206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Song SJ, Amir A, Metcalf JL, et al. Preservation methods differ in fecal microbiome stability, affecting suitability for field studies. mSystems. 2016;1(3):e00021–00016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baxter NT, Koumpouras CC, Rogers MA, et al. DNA from fecal immunochemical test can replace stool for detection of colonic lesions using a microbiota-based model. Microbiome. 2016;4(1):59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sinha R, Vogtmann E, Chen J, et al. Fecal microbiome in epidemiologic studies-response. Cancer Epidemiol Biomarkers Prev. 2016;25(5):870–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bahl MI, Bergström A, Licht TR. Freezing fecal samples prior to DNA extraction affects the Firmicutes to Bacteroidetes ratio determined by downstream quantitative PCR analysis. FEMS Microbiol Lett. 2012;329(2):193–197. [DOI] [PubMed] [Google Scholar]
- 36.Human Microbiome Project Consortium Structure, function and diversity of the healthy human microbiome. Nature. 2012;486(7402):207–214. [DOI] [PMC free article] [PubMed] [Google Scholar]