Abstract
Tamoxifen therapy for estrogen receptor–positive breast cancer reduces the risk of recurrence by approximately one-half. Cytochrome P-450 2D6, encoded by the polymorphic cytochrome P-450 2D6 gene (CYP2D6), oxidizes tamoxifen to its most active metabolites. Steady-state concentrations of endoxifen (4-hydroxy-N-desmethyltamoxifen), the most potent antiestrogenic metabolite, are reduced in women whose CYP2D6 genotypes confer poor enzyme function. Thirty-one studies of the association of CYP2D6 genotype with breast cancer survival have yielded heterogeneous results. Some influential studies genotyped DNA from tumor-infiltrated tissues, and their results may have been susceptible to germline genotype misclassification from loss of heterozygosity at the CYP2D6 locus. We systematically reviewed 6 studies of concordance between genotypes obtained from paired nonneoplastic and breast tumor–infiltrated tissues, all of which showed excellent CYP2D6 genotype agreement. We applied these concordance data to a quantitative bias analysis of the subset of the 31 studies that were based on genotypes from tumor-infiltrated tissue to examine whether genotyping errors substantially biased estimates of association. The bias analysis showed negligible bias by discordant genotypes. Summary estimates of association, with or without bias adjustment, indicated no clinically important association between CYP2D6 genotype and breast cancer survival in tamoxifen-treated women.
Keywords: breast neoplasms, cytochrome P-450 2D6, tamoxifen
Tamoxifen has been used for almost 40 years to inhibit the progression of hormone-responsive breast tumors (1). Along with its metabolites, tamoxifen competes with estrogens for binding to the estrogen receptor (2). Five years of adjuvant tamoxifen therapy reduces the risk of breast cancer recurrence by approximately one-half in women with estrogen receptor–positive tumors (3, 4), and long-term survival benefit is evident after 10 years of tamoxifen treatment (5). Tamoxifen is currently the adjuvant endocrine therapy recommended for premenopausal women with estrogen receptor–positive breast tumors (6–8). It also remains an important alternate or sequential treatment to aromatase inhibitors for postmenopausal breast cancer patients (6).
Several cytochrome P-450 enzymes convert tamoxifen into more potent antiestrogenic metabolites, which include 4-hydroxytamoxifen and 4-hydroxy-N-desmethyltamoxifen (endoxifen) (9–11). Compared with tamoxifen, endoxifen has substantially lower steady-state concentrations in blood, but it has at least 100-fold higher affinity for the estrogen receptor (12, 13). Therefore, endoxifen concentration may play a key role in modulating tamoxifen's clinical effectiveness (14). Lower concentrations of endoxifen have been associated with an increased risk of recurrent or new primary breast cancer (14, 15). However, one prospective study found the opposite, suggesting that higher endoxifen concentrations promote recurrence (16). Endoxifen production depends primarily on the enzymatic activity of cytochrome P-450 2D6 (CYP2D6). Thus, it has been hypothesized that the clinical effectiveness of tamoxifen may depend on an individual's CYP2D6 activity (13, 17).
CYP2D6 is encoded by a polymorphic gene (CYP2D6) with at least 100 variants, some of which have relatively high allele frequencies in some populations and reduce or eliminate enzymatic activity (18). Based on CYP2D6 genotype combinations, patients with inferred absent, reduced, normal, or increased enzymatic activity have been classified as poor, intermediate, extensive, or ultrarapid metabolizers, respectively (19). CYP2D6 metabolic activity correlates with steady-state endoxifen concentrations (11, 20–22), and poor metabolizers are more likely to have plasma endoxifen levels below a hypothesized efficacious threshold (14). However, tamoxifen and its antiestrogenic metabolites may be present in sufficient excess to block estrogen receptor signaling regardless of CYP2D6 activity (2, 23).
More than 30 studies nested in clinical trials, in observational settings, or in clinical series have investigated the association between CYP2D6 genotype and tamoxifen effectiveness. The findings of these studies are highly heterogeneous, with relative risks ranging from 0.08 to 13.1 for the association between variant CYP2D6 genotypes and breast cancer recurrence or mortality (Table 1) (23–25). The heterogeneity of associations has been attributed to many different factors related to study design and analysis (23, 24). Although studies have had several limitations, which are comprehensively reviewed elsewhere (24, 25), no characteristic pattern fully explains the heterogeneity of reported results.
Table 1.
Summary of Variant/Varianta Data Used in Meta-Analyses of the Association Between Cytochrome P-450 2D6 (CYP2D6) Genotype and Risk of Breast Cancer Recurrence or Mortality
| First Author, Year (Reference No.) | Country/Region | Major Variant Allele | Relative Riskb | 95% CI | Weight,c % | DNAd | No. of Casese | No. of Personsf | ||
|---|---|---|---|---|---|---|---|---|---|---|
| V/V | W/W | V/V | W/W | |||||||
| Sirachainan, 2012 (55) | Thailand | *10 | 0.28 | 0.01, 2.6 | 1.7 | Nonneoplastic | 11 | 5 | 8 | 1 |
| Markkula, 2014 (56) | Sweden | *4 | 0.5 | 0.07, 3.82 | 2.0 | Nonneoplastic | 1.0* | 20 | 16.1* | 154 |
| Okishiro, 2009 (57) | Japan | *10 | 0.6 | 0.18, 1.98 | 4.1 | Nonneoplastic | 3 | 40 | ||
| Gor, 2010 (58) | Multicenter | *4 | 0.99 | 0.5, 1.99 | 6.6 | Nonneoplastic | 224 | 19 | ||
| Mwinyi, 2014 (59) | Switzerland | *4 | 1.0 | 0.14, 7.1 | 2.1 | Nonneoplastic | 1 | 6 | 5 | 30 |
| Abraham, 2010 (60) | United Kingdom | *4 | 1.13 | 0.84, 1.54 | 8.8 | Nonneoplastic | 23 | 302 | 130 | 1,950 |
| Schroth, 2007g (61) | Germany | *4 | 1.63 | 1.07, 2.46 | 8.3 | Nonneoplastic | 10 | 30 | 17 | 118 |
| Chamnanphon, 2013 (62) | Thailand | *10 | 1.68 | 0.60, 4.73 | 4.8 | Nonneoplastic | 7 | 18 | 7 | 13 |
| Goetz, 2013h (63) | Austria | *4 | 2.45 | 1.05, 5.73 | 5.7 | Nonneoplastic | ||||
| Bijl, 2009 (64) | The Netherlands | *4 | 4.10 | 1.10, 15.9 | 3.6 | Nonneoplastic | 3 | 17 | 4 | 52 |
| Xu, 2008 (65) | China | *10 | 4.70 | 1.10, 20 | 3.3 | Nonneoplastic | ||||
| Park, 2011 (66) | South Korea | *10 | 5.59 | 0.93, 33.5 | 2.4 | Nonneoplastic | 49 | 10 | 179 | 31 |
| Damodaran, 2012 (67) | India | *4, *10 | 7.29 | 2.92, 18.2 | 5.4 | Nonneoplastic | 8 | 121 | ||
| Kiyotani, 2010 (68) | Japan | *10 | 9.52 | 2.79, 32.5 | 4.0 | Nonneoplastic | 18 | 3 | 63 | 84 |
| Sukasem, 2012 (69) | Thailand | *10 | 10.5 | 1.56, 70.8 | 2.2 | Nonneoplastic | 10 | 8 | ||
| Teh, 2012 (70) | Malaysia | *10 | 13.1 | 1.54, 109 | 1.9 | Nonneoplastic | 12 | 33 | 1 | 24 |
| Regan, 2012 (27) | Multicenter | *4 | 0.57 | 0.26, 1.23 | 6.2 | Tumor | 7 | 60 | 76 | 609 |
| Rae, 2012 (26) | Multicenter | *4 | 0.99 | 0.48, 2.08 | 6.3 | Tumor | 24 | 38 | 58 | 317 |
| Dezentje, 2013 (72) | The Netherlands | *4 | 1.01 | 0.57, 1.78 | 7.4 | Tumor | 3.8* | 47 | 27.5* | 345 |
| Lash, 2011 (28) | Denmark | *4 | 1.4 | 0.84, 2.3 | 7.8 | Tumor | 41 | 299 | 30 | 308 |
| Goetz, 2005g (46) | United States | *4 | 1.85 | 0.76, 4.52 | 5.5 | Tumor | 6 | 13 | ||
Abbreviations: CI, confidence interval; V, variant; W, wild-type.
a As described in the text, wild-type was defined as a functional allele or inferred extensive metabolizer phenotype; variant was defined as a reduced or eliminated function allele or poor metabolizer phenotype. Therefore, wild-type/wild-type (the reference group) encompassed extensive metabolizers and ultrametabolizers, and variant/variant encompassed poor metabolizers.
b Adjusted hazard ratio, rate ratio, or odds ratio reported in the original publication.
c The study's relative weight, expressed as a percentage, in the random-effects meta-analysis.
d Nonneoplastic = DNA extracted from nonneoplastic tissue; tumor = DNA extracted from tumor-infiltrated tissue.
e Number of cases in genotype category. When reported as an integer, this number was abstracted from the manuscript. When reported as a fraction with an accompanying asterisk (*), this number was imputed from the total number of reported cases and the reported estimate of association. Where no number is shown, the number was not reported, and it was not imputed because the study used DNA extracted from nonneoplastic tissue and thus was not subjected to quantitative bias analysis.
f For cohort study designs, number of persons at risk within the genotype category. For case-control study designs, number of controls within the genotype category. When reported as an integer, this number was abstracted from the manuscript. When reported as a fraction with an accompanying asterisk (*), this number was imputed from the total number of reported subjects and reported genotype proportions. Where no number is shown, the number was not available from the original manuscript.
g Data from the studies by Schroth et al. (61) and Goetz et al. (46) were subsequently pooled (90). We used the results from the original studies.
h Goetz et al. extracted DNA from formalin-fixed, paraffin-embedded specimens but wrote that extraction and assay methods were specifically designed “to overcome the potential problems related to somatic deletion of the CYP2D6 chromosomal locus on 22q13” (i.e., loss of heterozygosity) (63, p. 501). Thus, we classified this study's results with those of the nonneoplastic group.
Among the most influential studies of the association between CYP2D6 genotype and tamoxifen effectiveness are 2 that were nested within prospective adjuvant treatment trials—the Breast International Group 1-98 (BIG 1-98) and Arimidex, Tamoxifen, Alone or in Combination (ATAC) clinical trials (26, 27). Neither study found a positive association between reduced CYP2D6 function and breast cancer recurrence. In a large case-control study, Lash et al. (28) also reported a near-null association between CYP2D6 genotype and recurrence risk. On the basis of current evidence, 3 independent guideline panels have explicitly recommend against implementing CYP2D6 genotype testing for patients who are candidates for tamoxifen therapy (6, 8, 29).
However, the validity of the evidence base—especially the BIG 1-98 and ATAC results—has been challenged because the summary CYP2D6 genotype frequencies departed from Hardy-Weinberg equilibrium (HWE) (30–33). More specifically, the BIG 1-98- and ATAC-nested studies were criticized for genotyping DNA extracted from formalin-fixed paraffin-embedded (FFPE) tumor tissue, potentially yielding genotype errors (signaled by HWE departure) due to somatic genetic alterations or low DNA quality (34). Departure from HWE has several potential causes other than genotyping error. These include sampling error, nonrandom mating pairs, effects of variants on breast cancer incidence, and—especially relevant to the BIG 1-98 and ATAC analyses—population admixture when multiethnic genotype frequencies are combined across trial sites (30–33, 35–41). Therefore, departure from HWE does not necessarily imply that a study is based on inaccurate genotyping. Rather than dwell on departure from HWE as a solitary measure of genotyping accuracy, we follow the advice of Berry (36) and focus our attention on whether genotype misclassification biased the estimates of association from studies that used DNA extracted from tumor-infiltrated tissue.
Breast tumors exhibit chromosomal instability—loss of heterozygosity (LOH)—near the CYP2D6 locus (34). LOH arises from a gross chromosomal event in somatic tissue (such as tumor tissue) resulting in loss of one of the alleles present in germline DNA. Genotyping DNA from tissues affected by LOH will generate a pseudohomozygote genotype for the remaining allele. While this would be an accurate genotype for germline homozygotes, it would not be an accurate genotype for germline heterozygotes. Because tamoxifen is metabolized primarily in the liver (42)—not in the tumor itself—it is the germline genotype that is pertinent to the CYP2D6/tamoxifen hypothesis.
Approximately 40% of estrogen receptor–positive breast tumors included in the Cancer Genome Atlas show indirect evidence of LOH at the CYP2D6 locus on chromosome 22 (33), but this proportion does not necessarily correspond to the rate of misclassification. Many tumors with chromosome rearrangements would have been germline homozygotic in any event. In addition, DNA extracted from tumor-infiltrated tissue should contain germline information from surrounding stromal and immune cells.
Nonetheless, misclassification of CYP2D6 genotype resulting from LOH could affect the validity of studies using DNA extracted from tumor-infiltrated tissues. As is reviewed in detail below, 6 validation studies (578 patients) have compared CYP2D6 genotype in DNA extracted from tumor-infiltrated tissues with CYP2D6 genotype in DNA extracted from nonneoplastic tissues (33, 39, 43–46). While in 5 of these 6 validation studies (388 patients) investigators reported near-perfect concordance (37), another study (190 patients) found only 88% agreement (33). The authors of the published report concluded that genotype measured from tumor-infiltrated tissues was not representative of germline genotype at the CYP2D6 locus (33). Quite remarkably, an accompanying editorial called for expunging from the evidence base all research findings in this area that were based on genotypes from tumor-infiltrated tissues (47).
We sought to quantify the impact of using DNA extracted from tumor-infiltrated tissues—and the potential concomitant genotype misclassification—on studies comprising the current evidence base for an association between CYP2D6 genotype and tamoxifen failure. We first comprehensively reviewed the studies assessing CYP2D6 genotype concordance between tumor-infiltrated tissues and paired nonneoplastic tissues. We then carried out a quantitative bias analysis to quantify the influence of such genotype misclassification on association estimates from vulnerable studies.
REVIEW OF THE CONCORDANCE STUDIES
Six studies have compared genotypes for the CYP2D6*4 allele (rs3892097) assayed in DNA extracted from both tumor-infiltrated tissues and paired nonneoplastic tissues; results are summarized in Table 2. Five of these studies found near-perfect genotype concordance between tissue sources (39, 43–46). Most recently, Goetz et al. (33) studied CYP2D6 genotype concordance in a series of 190 breast cancer patients enrolled in the North Central Cancer Treatment Group adjuvant breast cancer trial. CYP2D6*4 genotypes based on DNA extracted from tumor-enriched FFPE tissue (FFPE-T) were determined in these patients for a previous study and deviated from HWE (P ≤ 0.001) (46). For their reanalysis, Goetz et al. procured FFPE tissues from these patients that contained both tumor and nonneoplastic tissue (FFPE-TN) and reassayed CYP2D6*4 using the same TaqMan method (Applied Biosystems, Inc., Waltham, Massachusetts) as was used for the initial genotyping (33, 46). They found concordant genotypes in 163 out of 186 successfully assayed pairs (agreement = 88%, 95% confidence interval (CI): 82, 92), noting a higher frequency of heterozygotes in the genotypes from FFPE-TN compared with FFPE-T tissues. They then compared genotypes obtained from FFPE-TN with genotypes from buccal cells in 35 patients and found 100% concordance (95% CI: 92, 100) (33). The investigators did not test for LOH in the FFPE-T tissues, so the cause of discordant genotypes between FFPE-T and FFPE-TN tissues is unknown. However, these results do suggest that extraction of DNA from tumor-enriched tissue may lead to CYP2D6 germline genotype misclassification in some laboratories. Nonetheless, use of whole tumor sections and even cores from tumor-rich areas (39)—which harbor a substantial proportion of nonneoplastic cells—still yielded accurate genotype classification.
Table 2.
Technical Details and Results of CYP2D6*4 Genotype or CYP2D6 Activity Score Concordance Studies
| First Author, Year (Reference No.) | Call Rate, % | No. of Analyzed Pairs | No. of Pairs Assayed | Nonneoplastic Tissue Source | Genotyping Method | P for HWEa | Agreement, % | 95% CI |
|---|---|---|---|---|---|---|---|---|
| Rae, 2003 (44) | 100 | 10 | 10 | Peripheral blood lymphocytes | PCR/RFLP | 0.73 | 100 | 74, 100b |
| Goetz, 2005 (46) | 88 | 15 | 17 | Buccal cells | TaqMan allelic discrimination (Applied Biosystems, Inc., Waltham, Massachusetts) | Genotype distribution not reported | 100 | 82, 100b |
| Ahern, 2010 (43) | 99 | 105 | 106 | FFPE nonneoplastic lymphoid tissue | TaqMan allelic discrimination | 0.46 | 100 | 97, 100 |
| Thompson, 2011 (45) | 100 | 133 | 133 | Peripheral blood lymphocytes | AmpliChip CYP450 (Hoffmann-La Roche AG, Basel, Switzerland) | Genotype distribution not reported | 100 | 98, 100b |
| Rae, 2013 (39) | 99 | 121 | 122 | Peripheral blood lymphocytes | TaqMan allelic discrimination | 0.52c | 98 | 94, 100 |
| Goetz, 2015 (33) | 98 | 186 | 190 | FFPE sections containing nonmalignant tissue | TaqMan allelic discrimination | 0.0002c | 88 | 82, 92b |
Abbreviations: CI, confidence interval; CYP2D6, cytochrome P-450 2D6; FFPE, formalin-fixed, paraffin-embedded; HWE, Hardy-Weinberg equilibrium; PCR, polymerase chain reaction; RFLP, restriction fragment length polymorphism.
a P value from a χ2 test for departure of observed CYP2D6*4 genotype frequencies in DNA extracted from tumor-infiltrated tissues from genotype frequencies expected under HWE.
b Calculated from information presented in the published article; confidence limits are mid-P binomial.
c Calculated from information presented in the published article.
To summarize, 6 genotype concordance studies evaluated different tissue storage conditions (FFPE and fresh-frozen), different nonneoplastic tissue sources, different DNA extraction methods, and different genotyping methods (Table 2). Despite these differences, investigators in all 6 studies agreed that DNA extracted from tumor-infiltrated tissue is a suitable alternative to DNA extracted from nonneoplastic tissue as a source for pharmacogenetic studies of CYP2D6 activity. This could be explained by negligible LOH at the CYP2D6 locus or by contribution of germline genotype information by stromal and immune cells (44). These CYP2D6-specific concordance findings comport with similar work on different chromosomal loci with high LOH in breast cancer (48, 49).
QUANTITATIVE BIAS ANALYSIS
Objective and methods
We used well-established methods of quantitative bias analysis (50–52) to evaluate the impact of genotype misclassification arising from potential LOH in studies that extracted DNA from tumor-infiltrated tissue. Our complete protocol is described in Web Appendix 1 (available at http://aje.oxfordjournals.org/) and is only briefly summarized here. First, we identified estimates of association between CYP2D6 genotype and breast cancer outcomes in tamoxifen-treated women by adding findings from recently published studies to the results of an earlier meta-analysis (24). Studies were identified by searching PubMed (National Library of Medicine, Bethesda, Maryland) for the terms “tamoxifen” and “CYP2D6.” We did not impose language restrictions. We retrieved all manuscripts published up to June 1, 2015, concerning gene-induced inhibition of CYP2D6 activity and tamoxifen effectiveness as measured by breast cancer outcomes. Citations in retrieved manuscripts were cross-referenced to identify additional publications of relevance.
For each study, we abstracted information on the tissue source for the genotyped DNA and information about the estimate of association (Table 1 and Web Table 1). Next, for the studies that genotyped DNA from tumor-infiltrated tissue, we estimated and adjusted for the bias potentially introduced by genotype misclassification (50, 51, 53). We defined the sensitivity of genotype classification as the probability that a germline CYP2D6*4 variant homozygote was correctly classified as such by genotyping DNA from tumor-infiltrated tissue. We defined the specificity of genotype classification as the probability that a germline CYP2D6 wild-type homozygote was correctly classified as such by genotyping DNA from tumor-infiltrated tissue. We specified beta distributions for sensitivity and specificity using direct evidence from the 3 genotype concordance studies that published contingency tables (33, 43, 44) (see Web Figure 1). For each study based on tumor-infiltrated tissue, we randomly selected values of sensitivity and specificity from their respective beta distributions and used these to estimate frequencies of cases and persons-at-risk (or controls, for case-control designs) that would have been observed had nonneoplastic tissue been used instead. We calculated associations from these corrected data, and we simulated random error by resampling from a normal distribution with a mean value equal to the logarithm of the bias-adjusted point estimate and variance equal to that of the originally published association (50, 51, 54). We repeated this process 100,000 times for each study, which yielded a frequency distribution of corrected point estimates. Associations corrected for genotyping error were defined as the median of these distributions; 95% simulation intervals were characterized by the 2.5th and 97.5th percentiles of these distributions. We used random-effects meta-analysis methods to evaluate all of the studies simultaneously and also within groups defined by whether DNA had been derived from tumor-infiltrated tissue or nonneoplastic tissue. Studies genotyping from tumor-infiltrated tissues were included in the meta-analysis both with and without misclassification adjustment.
Results
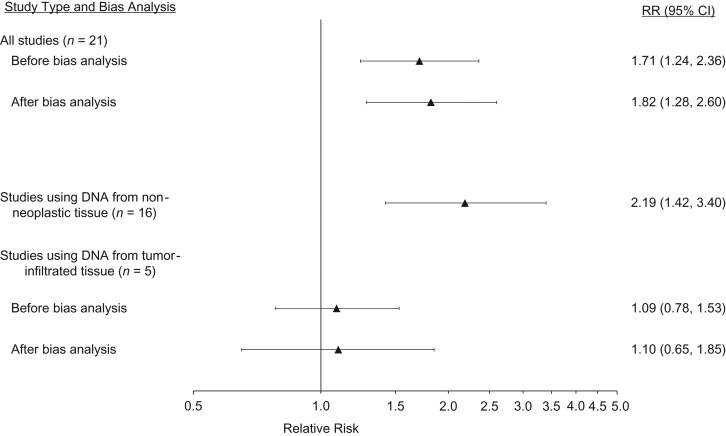
We identified 31 studies with results that were both pertinent to the meta-analysis and described in sufficient detail for inclusion (see Web references). DNA was extracted from nonneoplastic tissue in 21 of these 31 studies (68%) and from tumor-infiltrated tissue in the remaining 10 (32%). The meta-analysis of the association between the variant/variant genotype (or inferred poor metabolizer phenotype) and breast cancer recurrence or mortality, as compared with the wild-type/wild-type genotype (or inferred extensive or ultrametabolizer phenotype), included 21 of the 31 studies. In 16 (76%) of these 21 studies, researchers extracted DNA from nonneoplastic tissue (55–70), and in 5 of them (24%) they extracted DNA from tumor-infiltrated tissue (26–28, 46, 72). The summary estimates of association with and without bias analysis are reported in Table 3 and displayed in Figure 1. Before bias analysis, the summary association from the 5 studies that genotyped from tumor-infiltrated tissue showed an increase in breast cancer recurrence or mortality among CYP2D6*4 homozygotes compared with wild-type homozygotes (summary relative risk (RR) = 1.09, 95% CI: 0.78, 1.53). This null-centered summary association persisted after bias analysis, both when all concordance studies were used to inform bias parameters (summary RR = 1.11, 95% CI: 0.69, 1.80) and when only the Goetz et al. study (33), which showed the most extreme discordance among the 6 validation studies, was used to inform bias parameters (summary RR = 1.10, 95% CI: 0.65, 1.85). Results from meta-analysis of the association between heterozygote genotypes and breast cancer outcome, as compared with homozygote wild-type genotypes, are presented in Web Appendix 2 (see Web Table 2 and Web Figure 2).
Table 3.
Association Between Cytochrome P-450 2D6 (CYP2D6) Genotype and Tamoxifen Efficacy Before and After Quantitative Bias Analyses
| Before Quantitative Bias Analysis | Quantitative Bias Analysis Based on 3 Concordance Studies (33, 39, 43) | Quantitative Bias Analysis Based on the Goetz et al. (33) Study Only | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Point Estimate | 95% CI | P for Homogeneity | Point Estimate | 95% SI | Relative Bias, % | Point Estimate | 95% SI | Relative Bias, % | |
| All studies (n = 21) | 1.71 | 1.24, 2.36 | <0.001 | 1.80 | 1.28, 2.54 | −5.4 | 1.82 | 1.28, 2.60 | −6.8 |
| Studies using nonneoplastic tissue DNA (n = 16) | 2.19 | 1.42, 3.40 | <0.001 | NA | NA | NA | NA | NA | NA |
| Studies using tumor-infiltrated tissue DNA (n = 5) | 1.09 | 0.78, 1.53 | 0.27 | 1.11 | 0.69, 1.80 | −1.8 | 1.10 | 0.65, 1.85 | −0.3 |
Abbreviations: CI, confidence interval; NA, not applicable; SI, simulation interval.
Figure 1.
Relative risk (RR) of breast cancer recurrence or mortality according to cytochrome P-450 2D6 (CYP2D6) genotype (variant/variant vs. wild-type/wild-type) in meta-analyses. For particular studies included, see text and Table 1. CI, confidence interval.
The summary estimates of association were further from the null for the 16 studies that genotyped DNA extracted from nonneoplastic tissue than for the 5 studies that genotyped DNA extracted from tumor-infiltrated tissue. Comparison of the bias-adjusted summary estimate for these 5 studies with the original summary estimate showed a negligible difference (relative bias –5.4%). Under this bias model, genotyping error does not explain the difference in summary associations between studies with DNA extracted from nonneoplastic tissue and studies with DNA extracted from tumor-infiltrated tissue.
DISCUSSION
Summary of evidence
It has been suggested that studies using DNA extracted from tumor-infiltrated tissue may have been susceptible to genotyping error induced by LOH (33, 35, 47). The putative nondifferential misclassification of genotype may have biased these studies’ association estimates toward the null. Five of 6 genotype concordance studies showed near-perfect agreement between CYP2D6*4 genotype in nearly 400 samples of DNA extracted from tumor-infiltrated tissue paired with DNA extracted from nonneoplastic tissue. Bias analyses based on the point estimates from these 5 concordance studies would, of course, show no bias resulting from genotyping error. Combining the evidence from these studies with the evidence from the 1 study that showed genotype discordance (33) and applying standard good practices for bias analysis (52), we found little relative bias in the summary estimates of association. Results were virtually identical when we parameterized the bias analysis using only the evidence from the single study showing genotype discordance between tumor-infiltrated and nonneoplastic tissue.
These results are as expected, since even the 1 study with imperfect concordance showed high specificity (98% of wild-type/wild-type genotypes in DNA from nonneoplastic tissue were correctly classified as compared with paired tumor-infiltrated DNA (33) (see also Web Figure 1)). High specificity of classification generates little bias when the exposure—variant genotype in this case—is rare. The prevalence of CYP2D6*4 homozygotes is approximately 5% in populations of European descent, and the prevalence of CYP2D6*10 homozygotes is approximately 18% in populations of Asian descent (71). The bias analysis results are also consistent with evidence from a study in which Dezentje et al. (72) analyzed microsatellites flanking the CYP2D6 gene to detect LOH in DNA obtained from tumor-infiltrated tissue. Only 2.3% of the 731 tumor samples were excluded from statistical analysis because LOH could not be ruled out. Dezentje et al. reported a null association between variant/variant CYP2D6 genotype and the risk of breast cancer recurrence (hazard ratio = 1.01, 95% CI: 0.57, 1.78) (72).
In our main bias analysis, there were 2 groups of studies: 16 studies that genotyped DNA from nonneoplastic tissue (55–70) and 5 studies that genotyped DNA from tumor-infiltrated tissue (26–28, 46, 72). Summary estimates from these 2 groups of studies potentially lead to different conclusions about the association between genetic impairment of CYP2D6 activity and tamoxifen effectiveness. It is therefore imperative to evaluate whether characteristics of the 2 groups other than DNA source might explain the different summary associations.
First, studies using DNA extracted from nonneoplastic tissue tended to have smaller sample sizes and consequently higher variance. In the variant/variant meta-analysis, these studies accounted for 76% of the study number but only 67% of the relative weight in the random-effects meta-analysis.
Second, many of the studies using nonneoplastic tissue were susceptible to biases—notably immortal person-time bias and sparse-data bias—which have been extensively reviewed elsewhere (23, 24). None of the tumor-infiltrated tissue DNA studies were susceptible to these biases. Systematic errors like these could explain the observed statistical heterogeneity among the nonneoplastic tissue DNA studies (for homogeneity of the association estimates, P < 0.001) and the lack of statistical heterogeneity among the tumor-infiltrated tissue DNA studies (P for homogeneity = 0.27).
Third, we have previously presented evidence of publication bias in this topic area (24). This suggests that small, null studies may have been systematically excluded from publication and could not be included in our meta-analysis.
Furthermore, it appears biologically implausible that tamoxifen-treated women with poor CYP2D6 function would have a greater risk of recurrence than women who were not treated with tamoxifen. Given that 5 years of tamoxifen therapy reduces the risk of recurrence by about one-half, it is implausible that women with reduced-function metabolic phenotypes would have a relative risk of recurrence greater than approximately 2, in comparison with women who have full-function metabolic phenotypes. Lash et al. (23) previously published this plausibility limit on the basis of pooled clinical trial evidence of tamoxifen's effectiveness and the proportion of breast cancer patients expected to have fully functional or impaired alleles. In the variant/variant analysis, 8 of the 16 studies using nonneoplastic tissue DNA found ratio estimates of association greater than the plausibility limit of 2 (63–70), whereas none of the 5 studies based on tumor-infiltrated tissue DNA found a ratio estimate of association greater than the plausibility limit.
Finally, the CYP2D6*4 variant that is most common among Caucasians eliminates enzymatic activity, while the CYP2D6*10 variant that is most common among Asians only reduces enzymatic activity. Yet, counter to the biological rationale, the highest association estimates to date have been reported in Asian populations in studies evaluating the reduced-activity *10 variant (Table 1). Many of the studies that genotyped DNA from nonneoplastic tissues were studies of the reduced-function *10 variant with estimates of association implausibly larger than analogous studies of the loss-of-function *4 allele. These studies strongly influenced the meta-analytical estimate of association for the 16 studies using nonneoplastic tissue DNA. For these reasons, the summary estimates from the group of studies that genotyped DNA from tumor-infiltrated tissue are likely the most reliable for assessing the role of CYP2D6 genetic variation in the effectiveness of tamoxifen therapy.
In summary, the evidence base relied upon by guidelines (6, 8, 29) recommending against routine CYP2D6 genotyping to guide tamoxifen therapy in breast cancer remains robust. Efforts to strictly categorize one group of studies as invalid—in this case, based on the tissue source of the extracted DNA (33, 47)—are counterproductive in general (51) and in this topic area specifically (36). No study is ideal; efforts to dichotomize studies into those that are valid or invalid too often follow from prior beliefs about the expected association. Quantitative bias analysis, as implemented herein, avoids such unproductive dichotomization by quantitatively evaluating study imperfections using well-established good practices (51, 52). Our quantitative bias analysis showed little impact of genotype misclassification on the summary evidence, which follows directly from the excellent correspondence between CYP2D6 genotypes from paired tumor-infiltrated and nonneoplastic tissues. This remained true when modeled misclassification rates ignored the first 5 concordance studies and were based only on the concordance study that showed the greatest disagreement (33).
Implications for clinical practice
The enthusiasm for CYP2D6 testing has waned at all but a few treatment centers, probably because of the aforementioned robust evidence base supporting the guidelines that recommend against genotype-guided tamoxifen therapy. Other persuasive factors may include the inconsistent findings of studies interrogating the association between CYP2D6 genotype and tamoxifen efficacy and a concern about the potential to enhance treatment toxicity by dose-escalating patients with a low-activity CYP2D6 phenotype (73). Other strategies for optimizing the therapeutic benefit of tamoxifen therefore merit consideration.
First, both adherence (health-care behavior that abides by medical recommendations) and persistence (continuation of medication) have been long recognized as patient behaviors that affect tamoxifen's therapeutic benefit (74–77)—probably substantially more so than gene- or drug-induced CYP2D6 inhibition. It is estimated that 50% of patients do not persist with tamoxifen therapy through the entire 5-year course (75, 77, 78). Poor adherence to tamoxifen treatment, often defined as receiving fewer than 80% of scheduled doses, is strongly associated with less effective treatment (75, 79–81), increased medical costs (75), and increased mortality (75, 82).
The relative impact of adherence is particularly important given the survival benefits associated with extended use of tamoxifen therapy (5). Few of the pharmacogenomic studies reviewed herein incorporated information on tamoxifen adherence (45, 83, 84), and only 1 considered comprehensive genotype, adherence, and concomitant medication—with adherence being the dominant contributor (45). If genotype is a predictor of tamoxifen discontinuation (83), then adherence may be a key intermediate factor in the causal pathway between CYP2D6 genotype and breast cancer recurrence or mortality (23). Analyses of metabolic inhibition that treat adherence as if it were a confounder, rather than a mediator, could produce results that may be more biased than unadjusted estimates (85).
Menopausal side effects may be attributed to tamoxifen therapy (particularly hot flushes), which may in turn lead to prescriptions for drugs aimed at relief of these symptoms (86). Selective serotonin reuptake inhibitors are sometimes used to treat the side effects of tamoxifen and inhibit the activity of the CYP2D6 enzyme to varying degrees (20, 87–89). Substantial reductions in endoxifen plasma concentrations—comparable to those observed in homozygous carriers of the CYP2D6*4 variant allele—have been observed in patients using strong CYP2D6 inhibitors (i.e., paroxetine or sertraline) concurrently with tamoxifen, but not among those using the weaker CYP2D6 inhibitors (i.e., citalopram or venlafaxine) (20). Thus, inhibition of CYP2D6 by concomitantly administered drugs designed to ameliorate tamoxifen's side effects may also influence tamoxifen effectiveness, although the evidence for this is mixed (24). Citalopram or venlafaxine are perhaps more widely accepted as symptom-relieving agents and so, in reality, tamoxifen efficacy may not be greatly affected.
The interplay between genotype, side effects, patient behavior, and outcomes has been addressed retrospectively (45, 83). Prospective studies (particularly in premenopausal women) in both the preventive and therapeutic settings are required to dissect the relevant contributions. Further challenges include using the measurement of tamoxifen metabolites to direct therapy and/or effective service models to support adherence and improve survival. At present, there is an insufficient evidentiary basis to recommend routine CYP2D6 genotyping to guide tamoxifen therapy, a conclusion consistent with the current adjuvant therapy guidelines (6, 8, 29).
ACKNOWLEDGMENTS
Author affiliations: Departments of Surgery and Biochemistry, The Robert Larner, M.D. College of Medicine at The University of Vermont, Burlington, Vermont (Thomas P. Ahern); Department of Clinical Pharmacy, College of Pharmacy, University of Michigan, Ann Arbor, Michigan (Daniel L. Hertz); Department of Clinical Chemistry and Pharmacology, Odense University Hospital, Odense, Denmark (Per Damkier); Danish Breast Cancer Cooperative Group Secretariat, Rigshospitalet, Copenhagen University Hospital, Copenhagen, Denmark (Bent Ejlertsen); Institute of Pathology, Aarhus University Hospital, Aarhus, Denmark (Stephen J. Hamilton-Dutoit); Department of Internal Medicine, University of Michigan Medical Center, Ann Arbor, Michigan (James M. Rae); Department of Biostatistics and Computational Biology, Dana-Farber Cancer Institute and Harvard Medical School, Boston, Massachusetts (Meredith M. Regan); Department of Breast Surgical Oncology, University of Texas MD Anderson Cancer Center, Houston, Texas (Alastair M. Thompson); Department of Epidemiology, Rollins School of Public Health, Emory University, Atlanta, Georgia (Timothy L. Lash); and Department of Clinical Epidemiology, Aarhus University, Aarhus, Denmark (Deirdre P. Cronin-Fenton).
This work was supported by the National Institutes of Health (grants R01 CA118708 and R01 CA166825 to T.L.L. and grant R01GM099143 to J.M.R.); the Danish Cancer Society (grant DP06117 to S.H.D.); the Lundbeck Foundation (grant R167-2013-15861 to D.C.F.); Susan G. Komen for the Cure (grant CCR13264024 to T.P.A. and grant KG080081 to M.M.R.); the Breast Cancer Research Foundation (grant N003173 to J.M.R.); the Breast Cancer Campaign (A.M.T.); the Tayside Tissue Bank (A.M.T.); and the Mary Kay Foundation (grant 003-14 to T.P.A.).
We thank Helene Fevrier of Emory University for her assistance in extracting data from the published studies of CYP2D6 genotype and breast cancer recurrence.
No study sponsor had any role in study design; in the collection, analysis, and interpretation of data; in the writing of the report; or in the decision to submit the paper for publication.
Conflict of interest: none declared.
REFERENCES
- 1.Jordan VC. Tamoxifen: catalyst for the change to targeted therapy. Eur J Cancer. 2008;44(1):30–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jordan VC. Metabolites of tamoxifen in animals and man: identification, pharmacology, and significance. Breast Cancer Res Treat. 1982;2(2):123–138. [DOI] [PubMed] [Google Scholar]
- 3.Early Breast Cancer Trialists’ Collaborative Group Effects of chemotherapy and hormonal therapy for early breast cancer on recurrence and 15-year survival: an overview of the randomised trials. Lancet. 2005;365(9472):1687–1717. [DOI] [PubMed] [Google Scholar]
- 4.Early Breast Cancer Trialists’ Collaborative Group, Davies C, Godwin J, et al. . Relevance of breast cancer hormone receptors and other factors to the efficacy of adjuvant tamoxifen: patient-level meta-analysis of randomised trials. Lancet. 2011;378(9793):771–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davies C, Pan H, Godwin J, et al. . Long-term effects of continuing adjuvant tamoxifen to 10 years versus stopping at 5 years after diagnosis of oestrogen receptor-positive breast cancer: ATLAS, a randomised trial. Lancet. 2013;381(9869):805–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gradishar WJ, Anderson BO, Blair SL, et al. . Breast cancer version 3.2014. J Natl Compr Cancer Netw. 2014;12(4):542–590. [DOI] [PubMed] [Google Scholar]
- 7.Coates AS, Winer EP, Goldhirsch A, et al. . Tailoring therapies—improving the management of early breast cancer: St Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2015. Ann Oncol. 2015;26(8):1533–1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burstein HJ, Prestrud AA, Seidenfeld J, et al. . American Society of Clinical Oncology clinical practice guideline: update on adjuvant endocrine therapy for women with hormone receptor-positive breast cancer. J Clin Oncol. 2010;28(23):3784–3796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jordan VC, Collins MM, Rowsby L, et al. . A monohydroxylated metabolite of tamoxifen with potent antioestrogenic activity. J Endocrinol. 1977;75(2):305–316. [DOI] [PubMed] [Google Scholar]
- 10.Lien EA, Solheim E, Kvinnsland S, et al. . Identification of 4-hydroxy-N-desmethyltamoxifen as a metabolite of tamoxifen in human bile. Cancer Res. 1988;48(8):2304–2308. [PubMed] [Google Scholar]
- 11.Stearns V, Johnson MD, Rae JM, et al. . Active tamoxifen metabolite plasma concentrations after coadministration of tamoxifen and the selective serotonin reuptake inhibitor paroxetine. J Natl Cancer Inst. 2003;95(23):1758–1764. [DOI] [PubMed] [Google Scholar]
- 12.Lim YC, Desta Z, Flockhart DA, et al. . Endoxifen (4-hydroxy-N-desmethyl-tamoxifen) has anti-estrogenic effects in breast cancer cells with potency similar to 4-hydroxy-tamoxifen. Cancer Chemother Pharmacol. 2005;55(5):471–478. [DOI] [PubMed] [Google Scholar]
- 13.Johnson MD, Zuo H, Lee KH, et al. . Pharmacological characterization of 4-hydroxy-N-desmethyl tamoxifen, a novel active metabolite of tamoxifen. Breast Cancer Res Treat. 2004;85(2):151–159. [DOI] [PubMed] [Google Scholar]
- 14.Madlensky L, Natarajan L, Tchu S, et al. . Tamoxifen metabolite concentrations, CYP2D6 genotype, and breast cancer outcomes. Clinical Pharmacol Ther. 2011;89(5):718–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saladores P, Mürdter T, Eccles D, et al. . Tamoxifen metabolism predicts drug concentrations and outcome in premenopausal patients with early breast cancer. Pharmacogenomics J. 2015;15(1):84–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Love RR, Desta Z, Flockhart D, et al. . CYP2D6 genotypes, endoxifen levels, and disease recurrence in 224 Filipino and Vietnamese women receiving adjuvant tamoxifen for operable breast cancer. Springerplus. 2013;2(1):52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blankson EA, Ellis SW, Lennard MS, et al. . The metabolism of tamoxifen by human liver microsomes is not mediated by cytochrome P450IID6. Biochem Pharmacol. 1991;42(suppl):S209–S212. [DOI] [PubMed] [Google Scholar]
- 18.Hiratsuka M. In vitro assessment of the allelic variants of cytochrome P450. Drug Metab Pharmacokinet. 2012;27(1):68–84. [DOI] [PubMed] [Google Scholar]
- 19.Zanger UM, Raimundo S, Eichelbaum M. Cytochrome P450 2D6: overview and update on pharmacology, genetics, biochemistry. Naunyn Schmiedebergs Arch Pharmacol. 2004;369(1):23–37. [DOI] [PubMed] [Google Scholar]
- 20.Jin Y, Desta Z, Stearns V, et al. . CYP2D6 genotype, antidepressant use, and tamoxifen metabolism during adjuvant breast cancer treatment. J Natl Cancer Inst. 2005;97(1):30–39. [DOI] [PubMed] [Google Scholar]
- 21.Gjerde J, Hauglid M, Breilid H, et al. . Effects of CYP2D6 and SULT1A1 genotypes including SULT1A1 gene copy number on tamoxifen metabolism. Ann Oncol. 2008;19(1):56–61. [DOI] [PubMed] [Google Scholar]
- 22.Lim JS, Chen XA, Singh O, et al. . Impact of CYP2D6, CYP3A5, CYP2C9 and CYP2C19 polymorphisms on tamoxifen pharmacokinetics in Asian breast cancer patients. Br J Clin Pharmacol. 2011;71(5):737–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lash TL, Lien EA, Sørensen HT, et al. . Genotype-guided tamoxifen therapy: time to pause for reflection. Lancet Oncol. 2009;10(8):825–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cronin-Fenton DP, Damkier P, Lash TL. Metabolism and transport of tamoxifen in relation to its effectiveness: new perspectives on an ongoing controversy. Future Oncol. 2014;10(1):107–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hertz DL, McLeod HL, Irvin WJ Jr. Tamoxifen and CYP2D6: a contradiction of data. Oncologist. 2012;17(5):620–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rae JM, Drury S, Hayes DF, et al. . CYP2D6 and UGT2B7 genotype and risk of recurrence in tamoxifen-treated breast cancer patients. J Natl Cancer Inst. 2012;104(6):452–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Regan MM, Leyland-Jones B, Bouzyk M, et al. . CYP2D6 genotype and tamoxifen response in postmenopausal women with endocrine-responsive breast cancer: the Breast International Group 1-98 trial. J Natl Cancer Inst. 2012;104(6):441–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lash TL, Cronin-Fenton D, Ahern TP, et al. . CYP2D6 inhibition and breast cancer recurrence in a population-based study in Denmark. J Natl Cancer Inst. 2011;103(6):489–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goldhirsch A, Wood WC, Coates AS, et al. . Strategies for subtypes—dealing with the diversity of breast cancer: highlights of the St. Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2011. Ann Oncol. 2011;22(8):1736–1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stanton V., Jr Re: CYP2D6 genotype and tamoxifen response in postmenopausal women with endocrine-responsive breast cancer: the Breast International Group 1-98 trial [letter]. J Natl Cancer Inst. 2012;104(16):1265–1266. [DOI] [PubMed] [Google Scholar]
- 31.Pharoah PD, Abraham J, Caldas C. Re: CYP2D6 genotype and tamoxifen response in postmenopausal women with endocrine-responsive breast cancer: the Breast International Group 1-98 trial and Re: CYP2D6 and UGT2B7 genotype and risk of recurrence in tamoxifen-treated breast cancer patients [letter]. J Natl Cancer Inst. 2012;104(16):1263–1264. [DOI] [PubMed] [Google Scholar]
- 32.Nakamura Y, Ratain MJ, Cox NJ, et al. . Re: CYP2D6 genotype and tamoxifen response in postmenopausal women with endocrine-responsive breast cancer: the Breast International Group 1-98 trial [letter]. J Natl Cancer Inst. 2012;104(16):1264. [DOI] [PubMed] [Google Scholar]
- 33.Goetz MP, Sun JX, Suman VJ, et al. . Loss of heterozygosity at the CYP2D6 locus in breast cancer: implications for germline pharmacogenetic studies. J Natl Cancer Inst. 2015;107(2):dju401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ellsworth RE, Ellsworth DL, Lubert SM, et al. . High-throughput loss of heterozygosity mapping in 26 commonly deleted regions in breast cancer. Cancer Epidemiol Biomarkers Prev. 2003;12(9):915–919. [PubMed] [Google Scholar]
- 35.Goetz MP, Ingle JN. CYP2D6 genotype and tamoxifen: considerations for proper nonprospective studies. Clin Pharmacol Ther. 2014;96(2):141–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Berry DA. Response [letter]. J Natl Cancer Inst. 2014;106(5):dju065. [DOI] [PubMed] [Google Scholar]
- 37.Rae JM, Leyland-Jones B, Regan M, et al. . Re: Loss of heterozygosity at the CYP2D6 locus in breast cancer: implications for germline pharmacogenetic studies [letter]. J Natl Cancer Inst. 2015;107(5):djv065. [DOI] [PubMed] [Google Scholar]
- 38.Hertz DL, Kidwell KM, Thibert JN, et al. . Genotyping concordance in DNA extracted from formalin-fixed paraffin embedded (FFPE) breast tumor and whole blood for pharmacogenetic analyses. Mol Oncol. 2015;9(9):1868–1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rae JM, Regan MM, Thibert JN, et al. . Concordance between CYP2D6 genotypes obtained from tumor-derived and germline DNA. J Natl Cancer Inst. 2013;105(17):1332–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rae JM, Leyland-Jones B, Regan M. Response [letter]. J Natl Cancer Inst. 2014;106(5):dju064. [DOI] [PubMed] [Google Scholar]
- 41.Kidwell KM, Hertz DL, Leyland-Jones B, et al. . Analysis of the International Tamoxifen Pharmacogenetics Consortium (ITPC) dataset shows that genotyping DNA derived from tumor does not introduce CYP2D6 genotyping error or mask an association with tamoxifen efficacy [abstract PG-09-02]. Presented at the 2015 San Antonio Breast Cancer Symposium, San Antonio, Texas, December 8–12, 2015. http://www.abstracts2view.com/sabcs15/view.php?nu=SABCS15L_1373. Accessed March 1, 2016.
- 42.Borgna JL, Rochefort H. Hydroxylated metabolites of tamoxifen are formed in vivo and bound to estrogen receptor in target tissues. J Biol Chem. 1981;256(2):859–868. [PubMed] [Google Scholar]
- 43.Ahern TP, Christensen M, Cronin-Fenton D, et al. . Concordance of metabolic enzyme genotypes assayed from paraffin-embedded, formalin-fixed breast tumors and normal lymphatic tissue. Clin Epidemiol. 2010;2:241–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rae JM, Cordero KE, Scheys JO, et al. . Genotyping for polymorphic drug metabolizing enzymes from paraffin-embedded and immunohistochemically stained tumor samples. Pharmacogenetics. 2003;13(8):501–507. [DOI] [PubMed] [Google Scholar]
- 45.Thompson AM, Johnson A, Quinlan P, et al. . Comprehensive CYP2D6 genotype and adherence affect outcome in breast cancer patients treated with tamoxifen monotherapy. Breast Cancer Res Treat. 2011;125(1):279–287. [DOI] [PubMed] [Google Scholar]
- 46.Goetz MP, Rae JM, Suman VJ, et al. . Pharmacogenetics of tamoxifen biotransformation is associated with clinical outcomes of efficacy and hot flashes. J Clin Oncol. 2005;23(36):9312–9318. [DOI] [PubMed] [Google Scholar]
- 47.Johnson JA, Hamadeh IS, Langaee TY. Loss of heterozygosity at the CYP2D6 locus in breast cancer: implications for tamoxifen pharmacogenetic studies. J Natl Cancer Inst. 2015;107(2):dju437. [DOI] [PubMed] [Google Scholar]
- 48.Xie B, Freudenheim JL, Cummings SS, et al. . Accurate genotyping from paraffin-embedded normal tissue adjacent to breast cancer. Carcinogenesis. 2006;27(2):307–310. [DOI] [PubMed] [Google Scholar]
- 49.Schneider BP, Skaar TC, Sledge GW, et al. . Analysis of angiogenesis genes from paraffin-embedded breast tumor and lymph nodes. Breast Cancer Res Treat. 2006;96(3):209–215. [DOI] [PubMed] [Google Scholar]
- 50.Lash TL, Fox MP, Fink AK. Probabilistic bias analysis In: Applying Quantitative Bias Analysis to Epidemiologic Data. New York, NY: Springer Publishing Company; 2009:117–150. [Google Scholar]
- 51.Greenland S, Lash TL. Bias analysis In: Rothman KJ, Greenland S, Lash TL, eds. Modern Epidemiology. 3rd ed Philadelphia, PA: Lippincott Williams & Wilkins; 2008: 345–380. [Google Scholar]
- 52.Lash TL, Fox MP, MacLehose RF, et al. . Good practices for quantitative bias analysis. Int J Epidemiol. 2014;43(6):1969–1985. [DOI] [PubMed] [Google Scholar]
- 53.MacLehose RF, Gustafson P. Is probabilistic bias analysis approximately Bayesian. Epidemiology. 2012;23(1):151–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fox MP, Lash TL, Greenland S. A method to automate probabilistic sensitivity analyses of misclassified binary variables. Int J Epidemiol. 2005;34(6):1370–1376. [DOI] [PubMed] [Google Scholar]
- 55.Sirachainan E, Jaruhathai S, Trachu N, et al. . CYP2D6 polymorphisms influence the efficacy of adjuvant tamoxifen in Thai breast cancer patients. Pharmgenomics Pers Med. 2012;5:149–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Markkula A, Hjertberg M, Rose C, et al. . No association found between CYP2D6 genotype and early breast cancer events in tamoxifen-treated patients. Acta Oncol. 2014;53(2):195–200. [DOI] [PubMed] [Google Scholar]
- 57.Okishiro M, Taguchi T, Jin Kim S, et al. . Genetic polymorphisms of CYP2D6*10 and CYP2C19*2,*3 are not associated with prognosis, endometrial thickness, or bone mineral density in Japanese breast cancer patients treated with adjuvant tamoxifen. Cancer. 2009;115(5):952–961. [DOI] [PubMed] [Google Scholar]
- 58.Gor PP, Su HI, Gray RJ, et al. . Cyclophosphamide-metabolizing enzyme polymorphisms and survival outcomes after adjuvant chemotherapy for node-positive breast cancer: a retrospective cohort study. Breast Cancer Res Treat. 2010;12(3):R26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mwinyi J, Vokinger K, Jetter A, et al. . Impact of variable CYP genotypes on breast cancer relapse in patients undergoing adjuvant tamoxifen therapy. Cancer Chemother Pharmacol. 2014;73(6):1181–1188. [DOI] [PubMed] [Google Scholar]
- 60.Abraham JE, Maranian MJ, Driver KE, et al. . CYP2D6 gene variants: association with breast cancer specific survival in a cohort of breast cancer patients from the United Kingdom treated with adjuvant tamoxifen. Breast Cancer Res. 2010;12(4):R64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schroth W, Antoniadou L, Fritz P, et al. . Breast cancer treatment outcome with adjuvant tamoxifen relative to patient CYP2D6 and CYP2C19 genotypes. J Clin Oncol. 2007;25(33):5187–5193. [DOI] [PubMed] [Google Scholar]
- 62.Chamnanphon M, Pechatanan K, Sirachainan E, et al. . Association of CYP2D6 and CYP2C19 polymorphisms and disease-free survival of Thai post-menopausal breast cancer patients who received adjuvant tamoxifen. Pharmgenomics Pers Med. 2013;6:37–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Goetz MP, Suman VJ, Hoskin TL, et al. . CYP2D6 metabolism and patient outcome in the Austrian Breast and Colorectal Cancer Study Group trial (ABCSG) 8. Clin Cancer Res. 2013;19(2):500–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bijl MJ, van Schaik RH, Lammers LA, et al. . The CYP2D6*4 polymorphism affects breast cancer survival in tamoxifen users. Breast Cancer Res Treat. 2009;118(1):125–130. [DOI] [PubMed] [Google Scholar]
- 65.Xu Y, Sun Y, Yao L, et al. . Association between CYP2D6*10 genotype and survival of breast cancer patients receiving tamoxifen treatment. Ann Oncol. 2008;19(8):1423–1429. [DOI] [PubMed] [Google Scholar]
- 66.Park HS, Choi JY, Lee MJ, et al. . Association between genetic polymorphisms of CYP2D6 and outcomes in breast cancer patients with tamoxifen treatment. J Korean Med Sci. 2011;26(8):1007–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Damodaran SE, Pradhan SC, Umamaheswaran G, et al. . Genetic polymorphisms of CYP2D6 increase the risk for recurrence of breast cancer in patients receiving tamoxifen as an adjuvant therapy. Cancer Chemother Pharmacol. 2012;70(1):75–81. [DOI] [PubMed] [Google Scholar]
- 68.Kiyotani K, Mushiroda T, Imamura CK, et al. . Significant effect of polymorphisms in CYP2D6 and ABCC2 on clinical outcomes of adjuvant tamoxifen therapy for breast cancer patients. J Clin Oncol. 2010;28(8):1287–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sukasem C, Sirachainan E, Chamnanphon M, et al. . Impact of CYP2D6 polymorphisms on tamoxifen responses of women with breast cancer: a microarray-based study in Thailand. Asian Pac J Cancer Prev. 2012;13(9):4549–4553. [DOI] [PubMed] [Google Scholar]
- 70.Teh LK, Mohamed NI, Salleh MZ, et al. . The risk of recurrence in breast cancer patients treated with tamoxifen: polymorphisms of CYP2D6 and ABCB1. AAPS J. 2012;14(1):52–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Myrand SP, Sekiguchi K, Man MZ, et al. . Pharmacokinetics/genotype associations for major cytochrome P450 enzymes in native and first- and third-generation Japanese populations: comparison with Korean, Chinese, and Caucasian populations. Clin Pharmacol Ther. 2008;84(3):347–361. [DOI] [PubMed] [Google Scholar]
- 72.Dezentje VO, van Schaik RH, Vletter-Bogaartz JM, et al. . CYP2D6 genotype in relation to tamoxifen efficacy in a Dutch cohort of the Tamoxifen Exemestane Adjuvant Multinational (TEAM) trial. Breast Cancer Res Treat. 2013;140(2):363–373. [DOI] [PubMed] [Google Scholar]
- 73.Hertz DL, Snavely AC, Evans JP, et al. . Does increasing the daily tamoxifen dose in patients with diminished CYP2D6 activity increase toxicity? [abstract]. Presented at the 50th Annual Meeting of the American Society of Clinical Oncology, Chicago, Illinois, May 30–June 3, 2014. http://meetinglibrary.asco.org/content/129866-144. Accessed March 1, 2016.
- 74.Murphy CC, Bartholomew LK, Carpentier MY, et al. . Adherence to adjuvant hormonal therapy among breast cancer survivors in clinical practice: a systematic review. Breast Cancer Res Treat. 2012;134(2):459–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.McCowan C, Wang S, Thompson AM, et al. . The value of high adherence to tamoxifen in women with breast cancer: a community-based cohort study. Br J Cancer. 2013;109(5):1172–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Makubate B, Donnan PT, Dewar JA, et al. . Cohort study of adherence to adjuvant endocrine therapy, breast cancer recurrence and mortality. Br J Cancer. 2013;108(7):1515–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chlebowski RT, Kim J, Haque R. Adherence to endocrine therapy in breast cancer adjuvant and prevention settings. Cancer Prev Res (Phila). 2014;7(4):378–387. [DOI] [PubMed] [Google Scholar]
- 78.Partridge AH, Wang PS, Winer EP, et al. . Nonadherence to adjuvant tamoxifen therapy in women with primary breast cancer. J Clin Oncol. 2003;21(4):602–606. [DOI] [PubMed] [Google Scholar]
- 79.Dezentjé VO, van Blijderveen NJ, Gelderblom H, et al. . Effect of concomitant CYP2D6 inhibitor use and tamoxifen adherence on breast cancer recurrence in early-stage breast cancer. J Clin Oncol. 2010;28(14):2423–2429. [DOI] [PubMed] [Google Scholar]
- 80.Pagani O, Gelber S, Colleoni M, et al. . Impact of SERM adherence on treatment effect: International Breast Cancer Study Group Trials 13-93 and 14-93. Breast Cancer Res Treat. 2013;142(2):455–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ejlertsen B, Jensen MB, Mouridsen HT, et al. . Excess mortality in postmenopausal high-risk women who only receive adjuvant endocrine therapy for estrogen receptor positive breast cancer. Acta Oncol. 2014;53(2):174–185. [DOI] [PubMed] [Google Scholar]
- 82.Hershman DL, Shao T, Kushi LH, et al. . Early discontinuation and non-adherence to adjuvant hormonal therapy are associated with increased mortality in women with breast cancer. Breast Cancer Res Treat. 2011;126(2):529–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Rae JM, Sikora MJ, Henry NL, et al. . Cytochrome P450 2D6 activity predicts discontinuation of tamoxifen therapy in breast cancer patients. Pharmacogenomics J. 2009;9(4):258–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lorizio W, Wu AH, Beattie MS, et al. . Clinical and biomarker predictors of side effects from tamoxifen. Breast Cancer Res Treat. 2012;132(3):1107–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cole SR, Hernán MA. Fallibility in estimating direct effects. Int J Epidemiol. 2002;31(1):163–165. [DOI] [PubMed] [Google Scholar]
- 86.McCowan C, Thompson AM. The importance of nonpharmacogenetic factors in endocrine therapy. Pharmacogenomics. 2012;13(6):721–728. [DOI] [PubMed] [Google Scholar]
- 87.Borges S, Desta Z, Li L, et al. . Quantitative effect of CYP2D6 genotype and inhibitors on tamoxifen metabolism: implication for optimization of breast cancer treatment. Clin Pharmacol Ther. 2006;80(1):61–74. [DOI] [PubMed] [Google Scholar]
- 88.Desta Z, Ward BA, Soukhova NV, et al. . Comprehensive evaluation of tamoxifen sequential biotransformation by the human cytochrome P450 system in vitro: prominent roles for CYP3A and CYP2D6. J Pharmacol Exp Ther. 2004;310(3):1062–1075. [DOI] [PubMed] [Google Scholar]
- 89.Jeppesen U, Gram LF, Vistisen K, et al. . Dose-dependent inhibition of CYP1A2, CYP2C19 and CYP2D6 by citalopram, fluoxetine, fluvoxamine and paroxetine. Eur J Clin Pharmacol. 1996;51(1):73–78. [DOI] [PubMed] [Google Scholar]
- 90.Schroth W, Goetz MP, Hamann U, et al. . Association between CYP2D6 polymorphisms and outcomes among women with early stage breast cancer treated with tamoxifen. JAMA. 2009;302(13):1429–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]