Summary
In order to detect thermotolerant Campylobacter spp., 241 samples of fresh chicken meat, at retail in Croatia, were analysed according to a standard method, followed by biochemical test and molecular polymerase chain reaction/restriction enzyme analysis for exact species determination. Campylobacter spp. prevalence was 73.86%. Campylobacter jejuni and Campylobacter coli were isolated from 53.53 and 15.35% of the samples, respectively. In 4.98% of isolates thermotolerant Campylobacter spp. were not determined. The multilocus sequence typing method was used to evaluate genetic diversity of eight Campylobacter jejuni and four Campylobacter coli isolates. To our knowledge, these results of genotyping provided the first data on the presence of sequence types (STs) and clonal complexes (CCs) of Campylobacter jejuni and C. coli isolates in Croatia. By applying the multilocus sequence typing, a new allele of tkt gene locus was discovered and marked tkt508. The C. jejuni ST 6182 and C. coli ST 6183 genotypes were described for the first time, and all other identified genotypes were clustered in the previously described sequence types and clonal complexes. These findings provide useful information on the prevalence and epidemiology of Campylobacter jejuni and C. coli in Croatia.
Key words: thermotolerant Campylobacter, chicken meat in Croatia, prevalence, multilocus sequence typing (MLST) method
Introduction
Thermotolerant species of the genus Campylobacter, namely Campylobacter jejuni, Campylobacter coli, Campylobacter lari and Campylobacter upsaliensis cause campylobacteriosis, a zoonosis transmitted by food in most cases and the most common zoonosis in the majority of industrialized countries (1, 2). In humans, the disease is caused by C. jejuni in approx. 90% of cases, making this species most relevant from the public health viewpoint, while 5–10% of the cases are caused by C. coli, a small percentage by C. lari, and even smaller by C. upsaliensis. In the Republic of Croatia, campylobacteriosis has been a notifiable disease since 2007, with 1642 cases reported in 2014, ranking it the leading zoonosis in the country (3). Campylobacter is a genus of ubiquitous bacteria that have been isolated from a great variety of sources including water, soil, insects, domestic and wild mammals, domestic poultry, and wild birds (4). Meat from intensive poultry production, primarily broilers, is the main source of campylobacteriosis in humans. During specific slaughterhouse processing of chicken carcasses, the surface parts of the carcasses are frequently contaminated with Campylobacter from the intestine, which survive the production line and pose a risk for human health. According to the data from the official laboratories for food control in Croatia, the presence of Campylobacter spp. was found in 5% of total samples (5).
The main problem encountered in the classical method of phenotypic characterization of the C. jejuni and C. coli bacteria using the hippurate hydrolysis test are the hippurate-negative C. jejuni strains (6–8), as well as the false-positive hippurate hydrolysis results recorded for strains that do not belong to C. jejuni (9, 10). In such cases, species can only be differentiated by use of molecular methods.
A considerable number of polymerase chain reaction (PCR) protocols have been developed to date for identification of the Campylobacter genus, species and subspecies in samples of food, faeces or environment; however, a gold standard in the PCR diagnosis of Campylobacter is still lacking. One of the methods to identify thermotolerant Campylobacter spp., named polymerase chain reaction/restriction enzyme analysis (PCR/REA) and based on enzymatic digestion of PCR product with AluI and Tsp509I restriction enzymes, has been described by Fermér and Engvall (11).
Development of the molecular genotyping methods revealed a great genomic diversity of the bacteria of the genus Campylobacter. Due to their genomic variability, Campylobacter display characteristics of a weakly clonal population where lineages of related bacterial isolates can be distinguished. Genotyping by the multilocus sequence typing (MLST) method is currently preferred, because it offers a possibility of simple and unambiguous comparison of the results among laboratories all over the world. The method is based on single nucleotide differences in nucleotide sequences of the seven housekeeping genes encoding the enzymes responsible for different types of the bacterial intermediary metabolism. In Croatia, there are no data on genetic diversity of the most frequently isolated causative agents of campylobacteriosis, i.e. C. jejuni and C. coli characterized by the MLST method.
The aims of the study are as follows: (i) to test samples of fresh chicken meat originating from Croatian poultry productions using a standardized method for isolation of bacterial cultures of thermotolerant Campylobacter spp., (ii) to identify the C. jejuni, C. coli, C. lari and C. upsaliensis species by a commercial biochemical assay and by the PCR/REA molecular method, and (iii) to perform genotyping of the selected C. jejuni and C. coli isolates by the MLST method for the first time in Croatia, and to compare the obtained results with the pubMLST C. jejuni/C. coli database of world isolates, as well as with the respective literature reports.
Materials and Methods
Sampling and isolation of thermotolerant Campylobacter spp.
The study included 241 samples of fresh chicken meat for laboratory routine analysis, i.e. clean carcasses free from offal, head and lower legs from various Croatian retailers. Each test sample contained parts of the skin, neck and trunk, total mass of 25 g. Isolation of Campylobacter species colonies was performed according to the EN ISO 10272-1:2006 method (12). Test sample was diluted in 225 mL of buffered peptone water (Merck, Darmstadt, Germany), representing initial dilution from which 10 mL were taken and added to 90 mL of selective Bolton broth (Merck) for bacterial growth. Upon completion of 4- to 6-hour incubation at 37 °C and (44±4) h at 41.5 °C, samples were inoculated on one modified charcoal cefoperazone deoxycolate agar plate (Merck) and one CampyFood ID® agar plate (bioMérieux, Marcy l’Etoile, France). Both agars were incubated for (44±4) h at 41.5 °C. The growth of pure colony cultures on Columbia agar (Merck) with 5% sheep blood for confirmation of the bacteria of the genus Campylobacter was followed by microscopic verification of the respective morphology (spirilla, curved rod-shaped bacteria) and motility (spiral corkscrew-like motility), microaerophilic incubation without growth at 25 °C for (44±4) h, aerobic incubation without growth at 41.5 °C, and positive oxidase test (Bactident® Oxidase, Merck) to demonstrate the cytochrome oxidase enzyme. Pure cultures of detected bacteria were taken by inoculation loop and stored in plastic microtubes in 200 µL of UltraPure™ DNase/RNase-free distilled water (Invitrogen, Hilden, Germany) at –20 °C until PCR/REA and MLST testing. Bacterial isolates were permanently stored in plastic microtubes with 1 mL of Tryptic Soy Broth (Merck) with the addition of 15% glycerol at –80 °C. The CampyGen (Oxoid, Basingstoke, UK), Anaerocult® C set and Anaerocult® C mini set (Merck), and Anoxomat™ Mark II device (Mart Microbiology B.V., Drachten, The Netherlands) were used to ensure microaerophilic conditions necessary for bacterial culture growth. The next procedure of biochemical verification of isolated bacteria was performed on a VITEK2 device (bioMérieux) with the use of specific Neisseria-Haemophilus (NH) colorimetric cards, according to the manufacturer’s instructions. The following reference strains were used as positive controls on bacterial isolation, biochemical and molecular identification: C. jejuni ATCC 33560, C. coli CCUG 11283, C. lari CCUG 23947 and C. upsaliensis CCUG 14913.
PCR/REA
Thermotolerant Campylobacter spp. isolates were tested by the PCR method based on the amplification of 23S rRNA gene variable region and digestion with two restriction enzymes, AluI and Tsp509I (11, 13) on QIAxcel (Qiagen, Hilden, Germany) device. The highly polymorphic part of 23S rRNA gene, which is specific for the thermotolerant species C. jejuni, C. coli, C. lari and C. upsaliensis, was amplified first. Then, for further species differentiation, the PCR product was submitted to cleavage with the AluI and Tsp5091 restriction enzymes (Fermentas, St. Leon-Rot, Germany) in separate reactions.
MLST genotyping
Multilocus sequence typing according to Dingle et al. (14, 15) was carried out on 12 randomly selected isolates including eight C. jejuni and four C. coli species. The PCR products thus prepared were sent for sequencing to Macrogen (Amsterdam, The Netherlands). Using the pubMLST C. jejuni/C. coli database (16), each identified allele was assigned allelic number and one type of nucleotide sequence marked sequence type (ST) was recognized on the basis of a combination of seven allelic numbers. Each ST was classified in the respective clonal complex (CC), which represents a group of two or more independent isolates sharing identical alleles at five or more gene loci.
Statistical analysis
Statistical analysis was performed using an online program EpiTools epidemiological calculator (17). All obtained results were calculated by Wilson score interval and expressed as prevalence estimate plus upper and lower limits of specified 95% confidence interval (95% CI).
Results
Thermotolerant bacteria of the genus Campylobacter were isolated from 178 of 241 samples, whereas the presence of these bacteria was not found in 63 samples. The prevalence of thermotolerant Campylobacter spp. was 73.86% (95% CI=67.97–79.00%).
Three of 178 positive isolates were biochemically identified as Neisseria spp., which did not correspond to phenotypic indicators determined by light microscopy (morphology and motility characteristic of Campylobacter). Another 25 isolates were biochemically characterized as C. coli/C. jejuni ssp. jejuni and two isolates as C. coli/C. fetus ssp. fetus. The rest of 148 isolates were biochemically confirmed as C. jejuni ssp. jejuni and C. coli. Reference cultures were used as positive controls in the VITEK2 system, showing that it differentiated correctly C. jejuni and C. coli. The results of biochemical identification of C. lari and C. upsaliensis strains from culture collections were positive, although expressed as C. coli/C. jejuni ssp. jejuni, which confirmed the test sensitivity for the four thermotolerant Campylobacter species investigated in the study.
The isolates characterized as Neisseria spp. were identified by the PCR/REA procedure as follows: one as C. jejuni and another one as C. coli, whereas the third one was confirmed by PCR method to belong to thermotolerant Campylobacter; however, the species could not be identified by REA (positive PCR, negative REA). Twenty-five isolates that could not be species-characterized biochemically due to dubious C. coli/C. jejuni ssp. jejuni reading were also submitted to the PCR/REA procedure. Of these 25 samples, 14 (seven C. jejuni and seven C. coli) isolates were identified successfully, whereas species could not be determined for the rest of 11 isolates (positive PCR, negative REA). Two strains biochemically recognized as C. coli/C. fetus ssp. fetus were identified as C. coli. Two of the 148 isolates that were biochemically confirmed beyond doubt as C. jejuni and C. coli could not be identified even after multiple PCR/REA repeat procedures because the reading indicated unreadable pattern of the REA procedure (positive PCR, negative REA). Bacterial species C. lari and C. upsaliensis were not identified by the PCR/REA molecular testing.
In total, results of testing confirmed 129 C. jejuni isolates (53.53%; 95% CI=47.22–59.72%), 37 C. coli isolates (15.35%; 95% CI=11.35–20.44%) and 12 isolates of unverified species (4.98%; 95% CI=2.87–8.50%). The C. lari and C. upsaliensis species were not confirmed (Figs. 1 and 2).
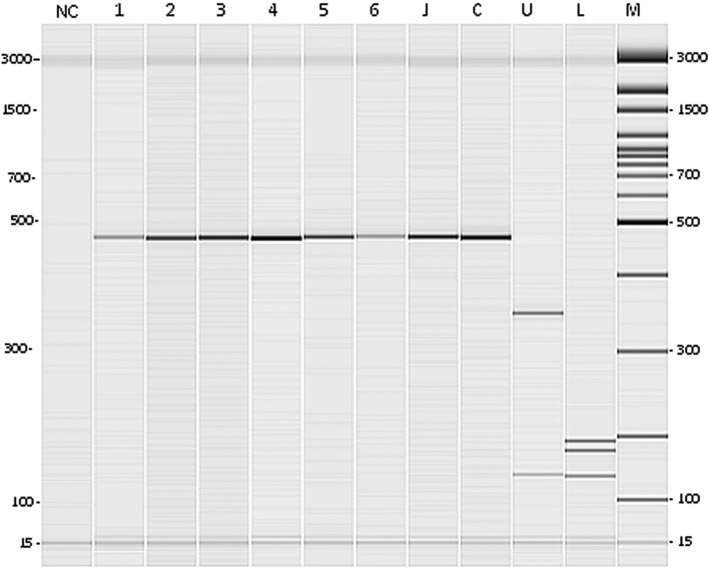
Fig. 1.
Results of thermotolerant Campylobacter spp. identification by the restriction enzyme analysis procedure: digestion with Tsp509I enzyme (gel obtained by capillary electrophoresis). NC=negative control, lanes 1 to 6=C. jejuni or C. coli isolates, J=C. jejuni ATCC 33560, C=C. coli CCUG 11283, U=C. upsaliensis CCUG 14913, L=C. lari CCUG 23947, and M=100 bp molecular size marker
Fig. 2.
Results of thermotolerant Campylobacter spp. identification by the restriction enzyme analysis procedure: digestion with AluI enzyme (gel obtained by capillary electrophoresis). NC=negative control, lanes 1, 2, 4 and 6=C. jejuni isolates, lanes 3 and 5=C. coli isolates, J=C. jejuni ATCC 33560, C=C. coli CCUG 11283, U=C. upsaliensis CCUG 14913, L=C. lari CCUG 23947, and M=100 bp molecular size marker
Sequencing was performed on seven gene loci of 12 different isolates (Table 1). Ten different sequence types (STs) were identified, with two STs confirmed twice in different isolates. Forty-five different alleles, i.e. allelic numbers, were identified in the study. One novel allele (tkt508) previously unknown in the database was described, while a novel C. jejuni ST (ST 6182) and novel C. coli ST (ST 6183) were determined on the basis of a new combination of the known alleles. Isolate testing by the MLST method revealed the following STs: C. jejuni isolates: ST 25, ST 51, ST 918, ST 2036, ST 4878 and novel ST 6182; and C. coli isolates: ST 1750, ST 1769, ST 854 and novel ST 6183. Identified ST of the isolates were located within seven clonal complexes (CCs) as follows: C. jejuni isolates: CC 460, CC 443, CC 45, CC 48, CC 353 and CC 464; and C. coli isolates where all ST belonged to CC 828.
Table 1. Results of seven gene locus sequencing by multilocus sequence typing method, the obtained sequence type (ST) and clonal complex (CC) of all Campylobacter isolates.
| Tested isolate | aspA | glnA | gltA | glyA | pgm | tkt | uncA | ST | CC |
|---|---|---|---|---|---|---|---|---|---|
| C. jejuni | 14 | 30 | 2 | 2 | 42 | 59 | 6 | 6182* | 460 |
| C. jejuni | 7 | 17 | 2 | 15 | 23 | 3 | 12 | 51 | 443 |
| C. jejuni | 4 | 7 | 10 | 1 | 1 | 7 | 1 | 25 | 45 |
| C. jejuni | 2 | 4 | 1 | 4 | 19 | 1 | 5 | 918 | 48 |
| C. jejuni | 7 | 17 | 52 | 10 | 11 | 3 | 6 | 2036 | 353 |
| C. jejuni | 4 | 7 | 10 | 1 | 1 | 7 | 1 | 25 | 45 |
| C. jejuni | 7 | 2 | 2 | 2 | 10 | 3 | 1 | 4878 | 464 |
| C. jejuni | 7 | 17 | 52 | 10 | 11 | 3 | 6 | 2036 | 353 |
| C. coli | 33 | 39 | 30 | 115 | 113 | 43 | 17 | 1750 | 828 |
| C. coli | 33 | 38 | 30 | 82 | 104 | 43 | 17 | 854 | 828 |
| C. coli | 33 | 429 | 30 | 82 | 113 | 508** | 17 | 6183* | 828 |
| C. coli | 33 | 176 | 30 | 115 | 113 | 43 | 17 | 1769 | 828 |
aspA=aspartase A, glnA=glutamine synthetase, gltA=citrate synthase, glyA=serine hydroxymethyltransferase, pgm=phosphoglucomutase, tkt=transketolase, uncA=ATP synthase α subunit *novel ST, **novel allelic number
Discussion
Results of the study showed a high prevalence of thermotolerant Campylobacter (73.86%) in fresh chicken meat in Croatia, with C. jejuni as the predominant species (53.53%), followed by C. coli (15.35%). These findings are consistent with the results of studies conducted in European Union (EU) member countries and worldwide. Bardoň et al. (18) report on the Campylobacter prevalence of 75% in fresh broiler meat in Czech Republic, with C. jejuni isolated in 70%, C. coli in 18% and mixed contamination with both bacterial species in 12% of cases. Prencipe et al. (19) found the prevalence of thermotolerant Campylobacter at 40.8% in their study of retail chicken meat in the Abruzzi e Molise region in Italy, confirming the presence of C. jejuni in 68.3%, C. coli in 21.5%, C. upsaliensis and C. mucosalis in 10.2%, and of more than one Campylobacter species in 23.1% of samples. Using data from the available literature on the prevalence of Campylobacter spp. in retail poultry meat, Suzuki and Yamamoto (20) provided a review of Campylobacter spp. prevalence in fresh poultry meat across the world (in %): North America 63.8, Central and South America 82.3, Europe 53.3, Africa 73.1, Asia 60.3, Oceania 90.4 and overall mean prevalence 58.0. Results of our study are also consistent with the study carried out in 2008 at the EU level, including 26 member countries, along with Switzerland and Norway, where the mean all-countries Campylobacter prevalence of 75.8% was determined in fresh broiler carcasses taken from slaughterhouses. C. jejuni was isolated in two-thirds and C. coli in the majority of the rest of isolates, C. lari was identified in 0.4% of cases, C. upsaliensis was not isolated at all, whereas 4.1% of isolates were not identified (21).
In contrast to the prevalence of Campylobacter spp. recorded in the present study, Finland (5.5%), Sweden (14.6%) and Norway (5.1%) have low prevalence rates of these bacteria in the samples of fresh broiler carcasses (21), possibly attributable to climatic conditions (22), along with implementation of the Scandinavian biosafety measure approach (23) and strict slaughterhouse hygienic measures (24).
Uzunović-Kamberović (25) determined a higher prevalence of C. coli than C. jejuni in fresh poultry meat in Bosnia and Herzegovina, which is in contrast to our findings. Also, the results of this study differ from the results of other studies conducted in Croatia, in which the prevalence of Campylobacter in fresh chicken meat was 13.3 (26) and 14.6% (27), i.e. considerably lower than in our study. This variation can be explained by different sampling and testing methods used in particular studies.
In our study, 98.31% of Campylobacter spp. were confirmed by the VITEK2 identification system (bioMérieux). Valenza et al. (28) reported the 100% Campylobacter spp. identification by the same system, with 100% accuracy for C. coli and 88.8% accuracy for C. jejuni, whereas other C. jejuni strains could not be differentiated from C. coli. We can speculate that in our study, inaccurate culture identification by VITEK2 procedure may have been due to absorbance of the testing culture suspension that was at the lower limit of acceptability, or the culture vital properties had been lost beyond microaerophilic conditions prior to testing. Namely, these cultures are recommended to be readily submitted to biochemical testing upon exposure to aerobic atmosphere. During clinical strain testing by the VITEK2 system, Rennie et al. (29) reported the 96.5% rate of correct identification, including 10.2% of low discrimination of strains of undeterminable species difference, 2.7% of incorrect identification and 0.8% of the results classified as unidentified, which is consistent with our study.
Thermotolerant bacteria of the genus Campylobacter were identified in 100% of cases by the PCR/REA method, whereby identification at the species level failed in 14 (7.87%) isolates (positive PCR, negative REA). These were presumably mixed bacterial cultures, which is in agreement with other literature reports (30, 31). Out of the remaining 148 isolates confirmed by the VITEK2 system, two (1.4%) samples confirmed as C. jejuni and C. coli could not be identified by the PCR/REA method because they yielded unreadable REA pattern (negative REA).
To our knowledge, this study results are the first report of the presence of C. jejuni and C. coli isolate STs and CCs in fresh retail chicken meat from domestic production in Croatia. The newly characterized ST 6182 and ST 6183 were identified for the first time and included in the pubMLST C. jejuni/C. coli database. Searching the entire pubMLST C. jejuni/C. coli database revealed data on the total number of entered isolates, country of origin, year, disease, source and epidemiology of STs obtained in the study. It should be noted that all identified STs, with the exception of ST 4878 and ST 6183, are proven causes of gastroenteritis caused by Campylobacter that were isolated from patients’ faeces in different parts of the world. The ST 4878 genotype was isolated only from environmental waters in The Netherlands and from chicken in Slovenia, and the ST 6183 from environmental waters in Luxemburg; yet the CC 464 and CC 828 isolates, which include these strains, have been demonstrated to cause campylobacteriosis. Isolates belonging to CC 464 were isolated from chicken meat samples (32), and CC 464 and CC 353 from patients’ faeces and chicken in China (33). ST 51 was confirmed as the cause of campylobacteriosis in Scotland, which involved 165 individuals attending a party and was caused by inadequate thermal processing of chicken liver (34). Kärenlampi et al. (35) investigated isolates of human, cattle and poultry origin in Finland and found the CC 45 isolates to predominate in the overall number of them and to be mostly related to poultry and contact with cats and dogs. They also confirmed the association of CC 828 isolates and patients aged ≥60, whereas CC 48 isolates were associated with consuming raw minced meat. CC 353 was isolated from humans, CC 45 from humans, cattle and poultry, CC 48 from humans and cattle, and CC 828 from humans and poultry. In the present study, C. coli ST 854, member of CC 828, was isolated from retail chicken meat, supporting the findings reported by Kärenlampi et al. (35) and Dingle et al. (15), who also isolated ST 854 from live chickens and fresh chicken meat. However, they did not find association with other potential sources such as humans or, for instance, pigs kept at the same farm; thus, they presumed this genotype to probably have tendency for a particular source type, in that case chicken, as the host. In the studies of isolates originating from different sources (humans and animals) in Switzerland, CC 21 and CC 45 were most frequently identified, with the exception of samples originating from pigs (36). Also, CC 21, CC 45 and CC 48 were the most common CCs identified in isolates originating from broilers in Sweden (37). Results of this study are consistent with those reported by other authors (38, 39), where CC 828 was most prevalent with different C. coli STs.
Conclusions
The results obtained in the present study pointed to a high prevalence of thermotolerant bacteria of the genus Campylobacter in fresh chicken meat from domestic production on the Croatian market. C. jejuni was most prevalent, followed by C. coli, whereas C. lari and C. upsaliensis were not confirmed. Using the MLST method, a novel allele for the tkt gene locus, named tkt508, was detected and a novel C. coli ST characterized. Based on the new allele combination, a novel C. jejuni ST (ST 6182) and novel C. coli ST (ST 6183) were determined. At the moment, ST 6182 is a unique ST that has been identified for the first time and exclusively in the Republic of Croatia. The STs of analyzed isolates, except for ST 4878, ST 6182 and ST 6183, as well as each of the seven CCs have been demonstrated to cause gastroenteritis. However, these genotypes have been isolated from patients’ faeces in different parts of the world, suggesting that they are potentially infective and can cause campylobacteriosis in humans.
The present study provides foundation for further investigation of a greater number of isolates originating from different sources in Croatia in order to assess their epidemiological association originating from the environment, animals and food with isolates obtained from humans suffering from campylobacteriosis.
References
- 1.Foodborne diseases active surveillance network (FoodNet) annual report. Atlanta, GA, USA: Centers for Disease Control and Prevention (CDC); 2013. Available from: http://www. cdc.gov/foodnet/reports/annual-reports-2013.html.
- 2.European Food Safety Authority (EFSA) and European Centre for Disease Prevention and Control (ECDC). The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2013. EFSA J. 2015;1:3991 10.2903/j.efsa.2015.3991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.World Animal Health Information Database (WAHID) Interface. Paris, France: World Organisation for Animal Health (OIE). Available from: http://www.oie.int/wahis_2/public/wahid. php/Countryinformation/Zoonoses.
- 4.Levin RE. Campylobacter jejuni: a review of its characteristics, pathogenicity, ecology, distribution, subspecies characterization and molecular methods of detection. Food Biotechnol. 2007;21:271–347. 10.1080/08905430701536565 [DOI] [Google Scholar]
- 5.The Ministry of Agriculture in cooperation with the Croatian Food Agency. Annual report on the results of analyses of official food and feed controls in 2012; 2013 (in Croatian). Available from: http://www.hah.hr/pdf/izvjesce_laboratorija_za_2012_godinu.pdf.
- 6.Morris GK, El Sherbeeny MR, Patton CM, Kodaka H, Lombard GL, Edmonds P, et al. Comparison of four hippurate hydrolysis methods for identification of thermophilic Campylobacter spp. J Clin Microbiol. 1985;22:714–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.On SLW. Taxonomy of Campylobacter, Arcobacter, Helicobacter and related bacteria: current status, future prospects and immediate concerns. Symp Ser Soc Appl Microbiol. 2001;90:1S–15S. 10.1046/j.1365-2672.2001.01349.x [DOI] [PubMed] [Google Scholar]
- 8.Zorman T, Smole Možina S. Classical and molecular identification of thermotolerant campylobacters from poultry meat. Food Technol Biotechnol. 2002;40:177–84. [Google Scholar]
- 9.Denis M, Soumet C, Rivoal K, Ermel G, Blivet D, Salvat G, et al. Development of a m-PCR assay for simultaneous identification of Campylobacter jejuni and C. coli. Lett Appl Microbiol. 1999;29:406–10. 10.1046/j.1472-765X.1999.00658.x [DOI] [PubMed] [Google Scholar]
- 10.Nakari UM, Puhakka A, Siitonen A. Correct identification and discrimination between Campylobacter jejuni and C. coli by a standardized hippurate test and species-specific polymerase chain reaction. Eur J Clin Microbiol Infect Dis. 2008;27:513–8. 10.1007/s10096-008-0467-9 [DOI] [PubMed] [Google Scholar]
- 11.Fermér C, Engvall EO. Specific PCR identification and differentiation of the thermophilic campylobacters, Campylobacter jejuni, C. coli, C. lari, and C. upsaliensis. J Clin Microbiol. 1999;37:3370–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.EN ISO 10272-1:2006. Microbiology of food and animal feeding stuffs – Horizontal method for detection and enumeration of Campylobacter spp. – Part 1: Detection method. Geneva, Switzerland: International Organization for Standardization (ISO); 2006.
- 13.Engvall EO, Brändström B, Gunnarsson A, Mörner T, Wahlström H, Fermér C. Validation of a polymerase chain reaction/restriction enzyme analysis method for species identification of thermophilic campylobacters isolated from domestic and wild animals. J Appl Microbiol. 2002;92:47–54. 10.1046/j.1365-2672.2002.01491.x [DOI] [PubMed] [Google Scholar]
- 14.Dingle KE, Colles FM, Wareing DRA, Ure R, Fox AJ, Bolton FE, et al. Multilocus sequence typing system for Campylobacter jejuni. J Clin Microbiol. 2001;39:14–23. 10.1128/JCM.39.1.14-23.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dingle KE, Colles FM, Falush D, Maiden MCJ. Sequence typing and comparison of population biology of Campylobacter coli and Campylobacter jejuni. J Clin Microbiol. 2005;43:340–7. 10.1128/JCM.43.1.340-347.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jolley KA, Maiden MCJ. BIGSdb: Scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics. 2010;11:595. 10.1186/1471-2105-11-595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sergeant ESG. EpiTools epidemiological calculators. Bruce, Australia: Ausvet Pty Ltd; 2016. Available from: http://epitools.ausvet.com.au. [Google Scholar]
- 18.Bardoň J, Kolář M, Karpíšková R, Hricová K. Prevalence of thermotolerant Campylobacter spp. in broilers at retail in the Czech Republic and their antibiotic resistance. Food Contr. 2011;22:328–32. 10.1016/j.foodcont.2010.08.001 [DOI] [Google Scholar]
- 19.Prencipe V, Parisciani G, Calistri P, Caporale CM, Iannitto G, Morelli D, et al. Thermotolerant Campylobacter in poultry meat marketed in the Abruzzo and Molise regions of Italy: prevalence and contamination levels. Vet Ital. 2007;43:167–74. [PubMed] [Google Scholar]
- 20.Suzuki H, Yamamoto S. Campylobacter contamination in retail poultry meats and by-products in the world: a literature survey. J Vet Med Sci. 2009;71:255–61. 10.1292/jvms.71.255 [DOI] [PubMed] [Google Scholar]
- 21.European Food Safety Authority (EFSA) Analysis of the baseline survey on the prevalence of Campylobacter in broiler batches and of Campylobacter and Salmonella on broiler carcasses in the EU, 2008, Part A: Campylobacter and Salmonella prevalence estimates. EFSA J. 2010;3:1503 10.2903/j.efsa.2010.1503 [DOI] [Google Scholar]
- 22.Patrick ME, Christiansen LE, Wainř MW, Ethelberg S, Madsen H, Wegener HC. Effects of climate on incidence of Campylobacter spp. in humans and prevalence in broiler flocks in Denmark. Appl Environ Microbiol. 2004;70:7474–80. 10.1128/AEM.70.12.7474-7480.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Second Report on Campylobacter. London, UK: Advisory Committee on the Microbiological Safety of Food (ACMSF), Food Standards Agency (FSA); 2005. Available from: https://www.food.gov.uk/sites/default/files/mnt/drupal_data/sources/files/multimedia/pdfs/acmsfcampylobacter.pdf.
- 24.Rosenquist H, Sommer HM, Nielsen NL, Christensen BB. The effect of slaughter operations on the contamination of chicken carcasses with thermotolerant Campylobacter. Int J Food Microbiol. 2006;108:226–32. 10.1016/j.ijfoodmicro.2005.12.007 [DOI] [PubMed] [Google Scholar]
- 25.Uzunović-Kamberović S. Epidemiology of Campylobacter jejuni and Campylobacter coli infections in the Zenica-Doboj Canton, Bosnia and Herzegovina – a laboratory based surveillance in the 1999–2001 period. Coll Antropol. 2005;29:655–9. [PubMed] [Google Scholar]
- 26.Granić K, Krčar D, Uhitil S, Jakšić S. Determination of Campylobacter spp. in poultry slaughterhouses and poultry meat. Vet Arh. 2009;79:491–7. [Google Scholar]
- 27.Kovačić A, Listeš I, Vučica C, Kozačinski L, Tripković I, Šiško-Kraljević K. Distribution and genotypic characterization of Campylobacter jejuni isolated from poultry in Split and Dalmatia county, Croatia. Zoonoses Public Health. 2013;60:269–76. 10.1111/j.1863-2378.2012.01519.x [DOI] [PubMed] [Google Scholar]
- 28.Valenza G, Ruoff C, Vogel U, Frosch M, Abele-Horn M. Microbiological evaluation of the new VITEK 2 Neisseria-Haemophilus identification card. J Clin Microbiol. 2007;45:3493–7. 10.1128/JCM.00953-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rennie RP, Brosnikoff C, Shokoples S, Reller LB, Mirrett S, Janda W, et al. Multicenter evaluation of the new Vitek 2 Neisseria-Haemophilus identification card. J Clin Microbiol. 2008;46:2681–5. 10.1128/JCM.00449-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Englen MD, Fedorka-Cray PJ. Evaluation of a commercial diagnostic PCR for the identification of Campylobacter jejuni and Campylobacter coli. Lett Appl Microbiol. 2002;35:353–6. 10.1046/j.1472-765X.2002.01193.x [DOI] [PubMed] [Google Scholar]
- 31.Best EL, Powell EJ, Swift C, Grant KA, Frost JA. Applicability of a rapid duplex real-time PCR assay for speciation of Campylobacter jejuni and Campylobacter coli directly from culture plates. FEMS Microbiol Lett. 2003;229:237–41. 10.1016/S0378-1097(03)00845-0 [DOI] [PubMed] [Google Scholar]
- 32.Wang X, Zhao S, Harbottle H, Tran T, Blickenstaff K, Abbott J, et al. Antimicrobial resistance and molecular subtyping of Campylobacter jejuni and Campylobacter coli from retail meats. J Food Prot. 2011;74:616–21. 10.4315/0362-028X.JFP-10-432 [DOI] [PubMed] [Google Scholar]
- 33.Zhang M, Gu Y, He L, Ran L, Xia S, Han X, et al. Molecular typing and antimicrobial susceptibility profiles of Campylobacter jejuni isolates from north China. J Med Microbiol. 2010;59:1171–7. 10.1099/jmm.0.022418-0 [DOI] [PubMed] [Google Scholar]
- 34.The molecular epidemiology of Scottish Campylobacter isolates from human cases of infection using multilocus sequence typing (MLST). CaMPS – Campylobacter MLST project in Scotland. Aberdeen, UK: University of Aberdeen; 2009. Available from: http://www.foodstandards.gov.scot/sites/default/files/339-1-595_CaMPS_S14006_Final_Report.pdf.
- 35.Kärenlampi R, Rautelin H, Schönberg-Norio D, Paulin L, Hänninen ML. Longitudinal study of Finnish Campylobacter jejuni and C. coli isolates from humans, using multilocus sequence typing, including comparison with epidemiological data and isolates from poultry and cattle. Appl Environ Microbiol. 2007;73:148–55. 10.1128/AEM.01488-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Korczak BM, Zurfluh M, Emler S, Kuhn-Oertli J, Kuhnert P. Multiplex strategy for multilocus sequence typing, fla typing, and genetic determination of antimicrobial resistance of Campylobacter jejuni and Campylobacter coli isolates collected in Switzerland. J Clin Microbiol. 2009;47:1996–2007. 10.1128/JCM.00237-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Griekspoor P, Engvall EO, Olsen B, Waldenström J. Multilocus sequence typing of Campylobacter jejuni from broilers. Vet Microbiol. 2010;140:180–5. 10.1016/j.vetmic.2009.07.022 [DOI] [PubMed] [Google Scholar]
- 38.Thakur S, Morgan Morrow WE, Funk JA, Bahnson PB, Gebreyes WA. Molecular epidemiologic investigation of Campylobacter coli in swine production systems, using multilocus sequence typing. Appl Environ Microbiol. 2006;72:5666–9. 10.1128/AEM.00658-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cody AJ, McCarthy NM, Wimalarathna HL, Colles FM, Clark L, Bowler ICJW, et al. A longitudinal 6-year study of the molecular epidemiology of clinical Campylobacter isolates in Oxfordshire, United Kingdom. J Clin Microbiol. 2012;50:3193–201. 10.1128/JCM.01086-12 [DOI] [PMC free article] [PubMed] [Google Scholar]