Summary
The hydrolytic and cost efficiencies of five endopeptidases (Alcalase 2.4L, Corolase 7089, Neutrase 0.8L, Promod 671L and Protex 7L) to hydrolyze Atlantic salmon by-products were compared at standardized activity levels based on a casein assay. The substrate was characterized prior to the hydrolytic experiments (pH=6.5, t=50 °C) to obtain substrate--specific constants for nitrogen to protein mass (in g) ratio, i.e. conversion factor fN=5.23 and total amount of peptide bonds (htot)=9.3 mmol per g of protein. At low enzyme activity to substrate ratio, all enzymes were equally efficient in hydrolyzing the substrate. At highest enzyme activity to substrate ratio, Protex 7L, Alcalase 2.4L and Promod 671L gave higher degree of hydrolysis (DH=14.2–14.6%) than Corolase 7089 (13.2%) and Neutrase 0.8L (11.6%) after 120 min of hydrolysis. No differences were observed in protein recovery (yield of solubilized protein) relative to DH. Determination of DH was followed by the pH-STAT and o-phthaldialdehyde methods. Based on pH-STAT data, response surface regression models were established based on the combined effects of hydrolysis time and enzyme activity to substrate ratio on DH and protein recovery. The modelling approach was combined with enzyme cost to identify the most cost-efficient enzyme (Protex 7L).
Key words: endopeptidase, fish protein hydrolysate, degree of hydrolysis, pH-STAT, protein recovery, cost efficiency
Introduction
The world fisheries and fish farming industries generate large amounts of by-products with possible food and feed applications (1). In Norway alone, this accounted for more than 850 000 tonnes in 2013 (2). By-products from the fish filleting industry include blood, viscera, heads, bones, skins, trimmings and fins (3), which are mainly utilized in the manufacture of fish silage, fish meal and oil (2). All by-products from the fish filleting industry are food grade after the primary processing and represent raw material with a high protein level, including all essential amino acids and other valuable compounds (3–5). There is a great potential for better use of these by-products in food applications. A promising industrial food utilization of fish filleting by-products is through the manufacture of water-soluble protein hydrolysates, using exogenous enzymes. The development and availability of industrial food-grade proteases has opened up new possibilities for production of fish protein hydrolysates from these resources (6–8) with applications within the food and nutraceutical market (5). The main goal of the hydrolytic process is to obtain a high protein recovery, a target degree of hydrolysis (DH), good sensory and physicochemical properties and a high cost efficiency. Cost related to protease addition is one of the most important economic variables in the production of protein hydrolysates. To obtain a competitive production process, it is important that the chosen protease has a high substrate-specific hydrolytic activity and a high activity to cost ratio.
Protein recovery and DH are dependent on several factors including substrate composition, enzyme specificity, pH, temperature, reaction time and added water (8–11). The choice of protease and DH will also influence the chemical composition and functional properties of the resulting protein hydrolysates (12–14). The functional properties of the hydrolysate are linked to enzyme specificity through the generated size distribution and hydrophobic/hydrophilic balance of the peptides. Proteases can be divided into two groups, endopeptidases and exopeptidases, based on how they cleave the peptide bonds (15). While endopeptidases cleave the peptide bonds in the interior of the peptide chain, exopeptidases require the presence of an unsubstituted C- or N-terminus and only release free amino acids. Depending on the choice of enzyme and reaction conditions, different final products can be produced from the same substrate.
The pH of hydrolytic reactions is often adjusted and controlled at the optimum level of the specific protease (15). However, this causes additional processing cost and increases the level of salt in the final hydrolysate. In addition, the comparison of proteases is most frequently conducted based on equal mass to substrate ratio and reaction time (16–18). However, this will give different final DH and protein recovery levels of the hydrolysates. Comparing protease products based on equal hydrolytic activity to reach the same DH levels will give more insight in their actual efficiency in a given hydrolytic process (19).
Several techniques are available for monitoring the DH in a hydrolytic process (15, 20). The methods for determining DH are based on three main principles: the determination of trichloroacetic acid-soluble nitrogen (TCA-SN), the determination of free α-amino groups: 2,4,6-trinitrobenzenesulfonic acid (TNBS), o-phthaldialdehyde (OPA) and formol titration, and the titration of released protons (pH-STAT). The pH-STAT method is frequently used and enables continuous measurements of the hydrolytic process. However, due to titration of released protons by base this method adds high levels of cations to the final products. In industrial and upscale applications, the DH is preferably quantified by a determination of free α-amino groups.
Knowledge of basic principles within factors influencing a hydrolysis reaction, both substrate composition and protease performance, are crucial to obtain proper process control and an exact measure of the desirable final DH and protein recovery. The aim of this study is to establish a test protocol for the comparison of the cost efficiency of endopeptidases based on DH and protein recovery. Substrate (Atlantic salmon by-products) specific parameters for nitrogen conversion factor (fN) and total amount of peptide bonds (htot) were established based on the amino acid composition of the substrate. The performance of the enzymes was tested by the pH-STAT technique at standardized enzyme activity to substrate ratio and at the natural pH=6.5 of the substrate. In addition, the DH was quantified by the OPA method to assess the correlation between the two methods within the obtained DH range.
Materials and Methods
Materials
Food-grade farmed Atlantic salmon (Salmo salar) heads and backbones were provided by Sotra Fiskeindustri AS, Sotra, Norway. The raw material was coarsely precut using a Comitrol® Processor Model 1700 (Urschel Laboratories Inc., Valparaiso, IN, USA), vacuum packed in plastic bags and stored at –30 °C before use. The endopeptidases used were Alcalase 2.4L and Neutrase 0.8L (Novozymes, Bagsvćrd, Denmark), Corolase 7089 (AB Enzymes, Darmstadt, Germany), Promod 671L (Biocatalysts, Cardiff, UK) and Protex 7L (DuPont, Wilmington, DE, USA). All solvents and reagents for the chemical analysis were of analytical grade.
Chemical analysis
Total amino acid composition was measured by HPLC after hydrolysis of a freeze-dried sample in 6 M HCl for 22 h at 110 °C, using Waters AccQ·Tag method and fluorescence detection with excitation/emission at λ=250/395 nm (21). Cysteine and cystine were determined after performic acid oxidation. Asparagine and glutamine were estimated based on the release of ammonia in the HCl digest compared to a neutral control sample (22). Released ammonia was quantified by the method of Conway and Byrne (23). Tryptophan was chemically determined by the method of Miller (24). Nitrogen was analyzed by the Kjeldahl method (ISO 5983-2:2009) (25). Free fatty acid (FFA) content was determined by AOCS method Ca 5a-40 (26) and calculated in g per kg of oleic acid. Moisture and ash were determined gravimetrically according to ISO 6496: 1999 (27) and ISO 5984:2002 (28), respectively. Lipid content was determined by extraction with choloroform and methanol (29).
Calculation of nitrogen-to-protein conversion factor
A substrate-specific nitrogen-to-protein (N:P) conversion factor, fN, was determined based on the substrate amino acid composition according to Mossé (30) and Sriperm et al. (31):
where kA is the ratio of total mass of anhydrous amino acids to total nitrogen (N) mass of the amino acids and kP is the ratio of total mass of anhydrous amino acids to total N in the substrate:
where Sm(anhyd. AA)i was calculated according to the following equation:
where w(AA)i is the mass fraction of the individual amino acid residue (g/kg), Mr(AA)i is the molecular mass of the respective amino acid and Mr(AA)i−Mr(H2O) is the molecular mass of the respective amino acid minus the molecular mass of water.
Sm(NAA)i is the mass of total N in the amino acids. This was calculated by adding the sum of N in the amino acids to the sum of amide N released from asparagine and glutamine:
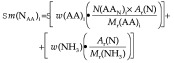 |
where N(AAN)i is the number of N atoms in the amino acid, Ar(N) is the relative atomic mass of nitrogen, w(NH3) is the amount of amide N in the HCl digest, and Mr(NH3) is the relative molecular mass of NH3.
Enzyme activity assay
Specific protease activity of the enzymes at pH=6.5 and t=50 °C was measured based on casein as substrate according to the procedure described by Cupp-Enyard (32). Casein solution (6.5 g/L), dissolved in 50 mM potassium phosphate buffer with pH=6.5, was subjected to digestion by varying concentrations of enzymes at 50 °C for 10 min, including a blank sample. The reaction was terminated by the addition of 1:1 of 110 mM trichloroacetic acid (TCA) and the sample was filtered through a 0.45- -µm polyethersulfone (PES) syringe filter (0.45µm; VWR Collection, Radnor, PA, USA) before the addition of Folin--Ciocalteu reagent and measurement of absorption at λ=660 nm in a Helios Beta spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). Absorbance values were compared to a tyrosine standard curve, in the concentration range of 0.00–0.55 µmol tyrosine. The activity of protease samples was determined in terms of units (U), defined as the µmol of tyrosine equivalents released from casein per min.
Enzymatic hydrolysis of salmon by-products
Precut salmon heads and backbones were thawed in water bath at 20 °C for 15 min and minced in a kitchen grinder (aperture 7 mm). The mince was diluted with water (1:1 by mass) and transferred to a 250-mL jacketed glass reactor connected to a heating circulator controlled by an external temperature sensor in direct contact with the fish slurry. The solution was agitated with a propeller stirrer (50 rpm) and equilibrated to 50 °C and pH=6.5, with 0.5 M NaOH, before the addition of enzyme. The amounts of enzyme of 5.5, 11, 22, 44 and 88 U per g of protein were added to achieve the desired activity for 120 min. Hydrolysate samples (approx. 20 mL) were taken at various time intervals (10, 20, 40, 80 and 120 min of hydrolysis) and enzyme activity was terminated by quickly heating to >90 °C in a microwave and resting at this temperature for 10 min. The samples were centrifuged in a Jouan C3i centrifuge (Jouan Inc., Winchester, VA, USA) for 15 min at 20 °C (5000×g) and the liquid phase was collected and filtered through a 0.45-µm PES syringe filter (VWR Collection). The liquid phases were stored at –20 °C until analysis.
Protein recovery determination
Protein recovery (PR) in the samples was determined based on the ratio of protein content in g (mL) in the liquid phase to that in the original substrate (mS):
Quantification of the degree of hydrolysis by the pH-STAT method
The degree of protein hydrolysis (DH) was determined by titration of liberated H+ ions using a Titrando 906 titrator (Metrohm, Herisau, Switzerland) and calculated as follows:
where VB is base consumption (mL) needed to maintain constant pH=6.5, cB is the concentration in mol/L of the base, α is the average degree of dissociation of the α-NH2 groups, mP is the mass of the protein being hydrolyzed (g), and htot the total amount of peptide bonds in the substrate (mmol of peptide bonds per g of protein). Base consumption was recorded every five seconds by the Titrando control software Tiamo (33). The htot is calculated based on the amount of amino acids in the protein:
The α constant is dependent on pH and temperature and can be computed by the following equation:
where pKa can be estimated according to Steinhardt and Beychok (34):
Correction of base consumption due to sampling of reaction mixture during a titration run was done according to Adler-Nissen (12):
 |
where VB,1, VB,2, etc. are the volumes of base consumption at the drawing of samples 1, 2, etc., m0 is the reaction mass at the start of the reaction and m is the mass of sample drawn from the reaction. Based on duplicate pH-STAT experiments (N=5), the average standard deviation of the calculated DH was estimated to be 0.08%.
Quantification of DH by the o-phthaldialdehyde method
The o-phthaldialdehyde (OPA) reagent was prepared according to Nielsen et al. (35). The leucine standard was prepared by dissolving l-leucine in water (0.13 mg/mL). Hydrolysate samples were diluted (10 mg/mL) with water and the OPA assay was carried out by the addition of 3 mL of OPA reagent to 400 µL of sample (or standard). The samples were equilibrated for 3 min before measurement of absorption at λ=340 nm, using distilled water as reference.
Statistical analysis
Statistical analysis was carried out using STATISTICA v. 12 software (36). The experimental pH-STAT data for 5.5, 11, 22, 44 and 88 U of enzyme per g of protein, i.e. base consumption every min between 0 and 10 min and every 5 min between 10 and 120 min of reaction time was used to calculate DH according to Eq. 7 and fitted to a second order polynomial equation using response surface methodology (37):
In the model, y is the estimated response, β0 the intercept, βi, βij and βii the regression coefficients of each factor, of each quadratic term and of the interaction term, respectively, ε the residual (error), and xi and xj the independent variables (amount of enzyme per g of protein and hydrolysis time). Protein recovery was based on sampling during the pH-STAT runs after 10, 20, 40, 80 and 120 min of hydrolysis, and calculated based on Eq. 6 before fitting to Eq. 12. The responses were fitted to the independent variables by multiple regressions, and the best subset model was identified based on backward removal of insignificant regressors (removal of the insignificant regressors at p>0.05). The quality of the fitted models was evaluated based on ANOVA, F-statistics and coefficient of multiple determination (R2). The linear correlations between variables were estimated by Pearson’s correlation coefficient (r), and p-value calculated by Student’s t-test.
Results and Discussion
Substrate characterization
The determination of DH during protein hydrolysis is based on calculations that include substrate-specific constants, i.e. htot and fN. The normally applied htot value for fish substrates (8.6 mmol of peptide bonds per g of protein) is based on cod myofibrillar protein (12, 38) and is not readily comparable to the salmon by-products used in this study. The universally used conversion factor (nitrogen to protein mass ratio in g) fN=6.25 is an old empirical value and only provides a rough estimate of the crude protein content in a sample (39). In this study, substrate- -specific values for htot and fN have been calculated to obtain a more accurate characterization of substrate properties and the hydrolysis process.
The proximate composition of the salmon substrate (N=3) revealed (267±3) g per kg of lipids, (429±3) g per kg of dry matter, (39±2) g per kg of ash and (24±0.4) g per kg of N. The amino acid composition revealed high levels of the essential amino acids. Enzymatic digests of proteins contain the original complement of amino acids present in the raw materials (4). Based on the amino acid composition (Table 1) a kA=5.49 and kP=4.96 were calculated as protein to nitrogen mass ratio. In general, kA is larger than kP because the denominator in Eq. 2 only corresponds to the N from the amino acids, while the denominator in Eq. 3 corresponds to total N in the substrate. In pure protein mixtures, without non-protein N, kA is equivalent to the true protein conversion factor. However, fish substrates will also contain other N-sources, e.g. non-protein amino acids, nucleotides, trimethylamine N-oxide (TMAO) and trimethylamine (TMA) (40). In real samples, fN is lower than kA, but higher than kP, and their average value is expected to give the best estimate of fN (30); for the used salmon substrate, the calculated fN was 5.23. The calculated fN deviates significantly from the universally used protein factor of 6.25. Use of this standard protein factor will give an overestimation of the crude protein content and consequently an underestimation of DH based on Eq. 7.
Table 1. Mass fractions of amino acids in protein and total nitrogen found in the salmon by-product substrate.
| Constituent | w/(g/kg) |
|---|---|
| Glycine | 42.9±0.4 |
| Alanine | 23.3±0.4 |
| Valine | 15.0±0.3 |
| Leucine | 20.4±0.4 |
| Isoleucine | 12.6±0.3 |
| Proline | 20.8±0.2 |
| Methionine | 10.5±0.1 |
| Serine | 15.0±0.1 |
| Threonine | 13.1±0.2 |
| Cysteine | 5.4±0.2 |
| Asparagine* | 14.6±0.3 |
| Glutamine* | 16.1±0.4 |
| Hydroxyproline | 11.6±0.5 |
| Glutamic acid | 25.0±0.4 |
| Aspartic acid | 14.0±0.5 |
| Lysine | 24.6±0.8 |
| Arginine | 22.8±0.2 |
| Histidine | 11.1±0.2 |
| Phenylalanine | 11.9±0.2 |
| Tyrosine | 9.2±0.2 |
| Tryptophan | 2.5±0.4 |
| Total amino acids | 342.4±1.6 |
| Total nitrogen | 58.7±0.3 |
The results are expressed as mean value±standard deviation (N=3) *estimated based on the release of NH3 after acid hydrolysis (0.37 g of N per g of sample)
The total amount of peptide bonds (htot) of the salmon substrate was estimated to 9.3 mmol per g of protein based on the total amino acid content (Table 1). The deviation from the normally applied literature value (8.6 mmol of peptide bonds per g of protein) (12) can mainly be attributed to the use of the substrate-specific fN. Use of the universal fN=6.25 will give systematic error in the estimation of htot, i.e. an underestimation of htot in substrates with real fN<6.25.
Enzyme activity
Comparison of protease efficiency is in most studies based on the addition of equal mass fraction of enzyme in g to the substrate (7, 16, 17, 41). This approach does not take into consideration differences in the specific hydrolytic activity of the respective protease formulations at the chosen pH and temperature conditions. The enzyme suppliers provide protease activities based on different assays and definitions of proteolytic units (Table 2; 42). This makes comparison of their proteolytic efficiency difficult to assess. In addition, the specified proteolytic activities are measured at the optimum pH and temperature conditions for the respective enzymes, conditions not necessarily relevant or in compliance with the application under study. To enable the comparison of how efficient different enzyme preparations are in hydrolyzing and solubilizing the substrate, there is a need for comparative studies based on a standardized activity measurement expressed in units per mass of substrate, as discussed by Kristinsson and Rasco (13, 19). In this study the specific activity of the included proteases (Table 2) was assayed based on a standardized substrate (casein) at the chosen experimental settings (pH=6.5 and t=50 °C).
Table 2. European Commission (EC) number, declared activity based on different methods, i.e. Anson units (AU), haemoglobin units (UHb), azocoll units (azo U) and casein units (casein U), measured activity at pH=6.5 and 50 °C in units (U) per mg of enzyme, enzyme cost, enzyme protein content, recommended enzyme dosage and optimum temperature and pH of commercial proteases.
| Enzyme | EC number |
Declared activitya |
Enzyme activity U/mg |
Enzyme costb EUR/106 U |
w(protein)c % |
w(recommended)a % |
t(optimum) °C |
pH (optimum) |
|---|---|---|---|---|---|---|---|---|
| Alcalase 2.4L | 3.4.21.62 | 2.4 AU/g | 1.4±0.20 | 22 | 7.1 | (0.5–2.0)d | 60f | 9.5f |
| Corolase 7089 | 3.4.24.28 | 840 UHb/g | 1.0±0.10 | 20 | 3.8 | (0.01–0.5)d | 55a | 7–8a |
| Protex 7L | 3.4.24.28 (3.4.21.62) | 1600 azo U/g | 0.8±0.07 | 17 | 3.8 | (0.1–1.0)e | 60a | 6.5a |
| Neutrase 0.8L | 3.4.24.28 | 0.8 AU/g | 0.6±0.06 | 29 | 2.6 | (0.5–2.0)d | 55f | 7.0f |
| Promod 671L | 3.4.21.X / 3.4.24.X | 80 casein U/g | 0.3±0.05 | 46 | 0.94 | Not provided | 60a | 6.5a |
aprovided by the respective enzyme suppliers given in the Materials and Methods section bprovided by the distributors based on DAP (delivered at place) terms cw(N)·6.25 dprotein basis esample wet mass basis faccording to Benjakul and Morrissey (42)
Several standardized methods are described in the literature to measure protease activity (43). The casein substrate used in this study is commonly used in enzyme assays and has shown comparable results with cod myofibrillar substrate when tested in different enzyme preparations (10). The observed specific enzyme activities (Table 2) demonstrate large differences between the included commercial endopeptidases in general, and also between enzymes with the same European Commission (EC) number (Corolase 7089, Neutrase 0.8L and Protex 7L). The enzyme activity is also highly correlated with the crude protein content (N×6.25) in the products (r=0.980, p=0.003), possibly reflecting the amount of purified enzyme incorporated in the respective product formulations. Generally, the observed protein content is low (Table 2) and any influence on the measurement of solubilized protein during hydrolysis can be considered negligible even at high enzyme activity to substrate ratio. In industrial applications, costs related to protease addition are of high importance and comparing enzymes on cost efficiency (EUR/U) rather than mass basis is of high relevance. Based on the measured activity levels and acquired price indication from the respective manufacturers, Promod 671L has the highest cost per 106 U followed by Neutrase 0.8L, Alcalase 2.4L, Corolase 7089 and Protex 7L (Table 2).
Assessment of endogenous enzyme activity
Endogenous enzyme activity might play a major role in the evaluation of protein hydrolysis using pH-STAT. A control sample (without added enzyme) was included to assess the potential hydrolysis contribution from endogenous enzymes that could influence the consumption of NaOH in the pH-STAT experiments. After two hours of hydrolysis only a small increase in DH (<0.5%) was observed, confirming an insignificant endogenous enzyme activity in the Atlantic salmon substrate. This is in agreement with Sovik and Rustad (44), who demonstrated low endogenous protease activity in cod cutoff at 50 °C and pH=7.
The concentration of FFA in the oil phase was evaluated to measure potential liberation of FFA by endogenous lipase activity during the hydrolysis experiments. It was proposed that this could potentially influence the consumption of NaOH by neutralization of the liberated FFA. However, the formation of FFA was very low and only increased from (11.0±1.5) g/kg before the hydrolysis to (12.0±1.0) g/kg after two hours of hydrolysis. These findings are in good agreement with reported level of FFA in fresh salmon (45) and confirm that the substrate does not contain significant amounts of endogenous lipase activity. The observation also confirms that there is no lipase activity in the Alcalase 2.4L product. The major fatty acids found in Atlantic salmon have a chain length ≥16 C, with pKa values above 8 (46). At pH=6.5 they are fully protonated and do not contribute to the consumption of NaOH if liberated by lipase activity in the substrate. However, at higher pH values normally used in pH-STAT measurements, the liberation of FFA by lipase activity may have influence on the titration curve. Based on these observations the endogenous enzyme activity can be considered negligible in our system and the measured proteolytic effect ascribed to the added exogenous proteases.
Enzymatic hydrolysis
The pH-STAT experiments were performed at pH=6.5 to obtain results as close as possible to an unbuffered system. The chosen pH is in the lower range applicable for pH-STAT experiments and gives a dissociation constant, α, of only 0.175. A low value of α implies that the determination of DH might be subject to slightly more uncertainty compared to higher pH values (12, 47).
Without the titration of liberated H+ by NaOH, the pH of the substrate hydrolysate was readily reduced from initial pH=6.5 to below pH=6.2 (Fig. 1). This confirms poor buffering capacity of the substrate in this pH range and verifies that the pH=6.5 is possible to use in pH-STAT experiments. The pH drop is rather steep in the first 15 min of hydrolysis at all activity levels followed by a slow asymptotic descent to a stationary level depending on the enzyme activity to substrate ratio (Fig. 1) and reflecting the degree of hydrolysis curves given in Fig. 2. The pKa (50 °C) of +H3N groups in polypeptides is estimated to be in the range of 6.9 to 7.2 (33). At a start of hydrolysis the pH=6.5 decreases due to release of protons from the liberated amino groups. As the hydrolysis progresses the rate of reaction is reduced because the reduced enzyme activity depends on pH, fewer peptide bonds available for cleavage, enzyme inactivation, substrate inhibition (48) and the possible protease inhibitors present in the substrate (49). The hydrolysis products might irreversibly inhibit a given amount of enzyme and this effect would be relatively large at low enzyme activity to substrate ratio and negligible at high enzyme activity to substrate ratio (12). The liberated small peptides have a higher pKa value than larger peptides (12, 50) and also slow down the reduction of pH. The lowest observed value of pH=6.1 is in good agreement with the upper range of pH=5–6 where the uptake and release of protons is expected to cancel out (12).
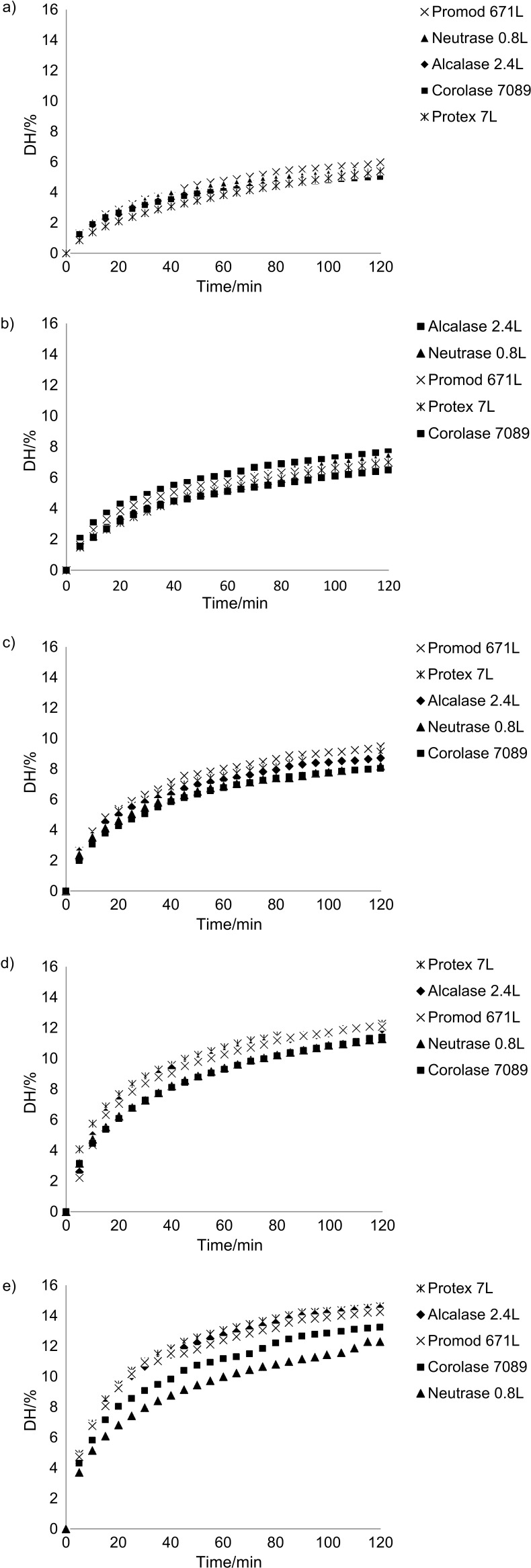
Fig. 1.
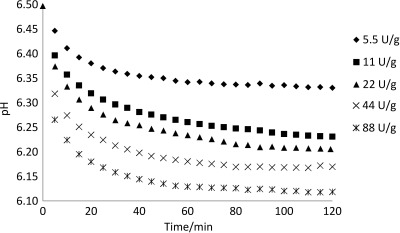
Effect of proteolytic enzyme activity on the reduction of pH at t=50 °C during hydrolysis by Alcalase 2.4L at enzyme activity in units (U) per g of substrate of 5.5–88 U/g
Fig. 2.
Degree of hydrolysis (DH) as a function of hydrolysis time measured by the pH-STAT at pH=6.5 and specific enzyme activity in units (U) per g of substrate of: a) 5.5 U/g, b) 11 U/g, c) 22 U/g, d) 44 U/g, and e) 88 U/g. The enzymes are listed in the respective legends based on hydrolytic efficiency
The enzymatic hydrolysis of the salmon substrate displayed typical curves for all activities, with an initial rapid phase, followed by a slower hydrolysis (Fig. 2) (51–53). Only minor differences between the enzymes in the progress of hydrolysis were observed at the lowest enzyme activity to substrate ratio (5.5 and 11 U/g), which confirms the measured activity levels determined by the casein assay and the comparability of activity based on the two substrates, casein and fish myofibrillar protein (10).
At higher enzyme activity to substrate ratios (22, 44 and 88 U/g), Alcalase 2.4L, Promod 671L and Protex 7L were found to be the most efficient enzymes, while Neutrase 0.8L and Corolase 7089 gave lower final DH after two hours of hydrolysis. At 88 U/g, a DH of 14.2–14.6% was achieved using Protex 7L, Alcalase 2.4L and Promod 671L, while Neutrase 0.8L and Corolase 7089 produced a final DH of 11.6 and 13.2%, respectively. The observed grouping can be attributed to a difference in enzyme specificity and peptide bonds available for hydrolysis. Corolase 7089, Neutrase 0.8L and Protex 7L have identical EC number and were expected to have similar efficiencies in hydrolyzing the substrate. However, Protex 7L also contains smaller amount of subtilisin-like protease kexin (EC 3.4.21.61), which may explain the better performance of this enzyme (Olsen, T.I., DuPont, personal communication). Promod 671L was included in this study to test a combination product of two endopeptidases with the potential to give a higher final DH than the other enzymes, due to more peptide bonds available for hydrolysis. It was one of the most efficient enzymes; however, it did not give higher DH compared to Alcalase 2.4L and Protex 7L.
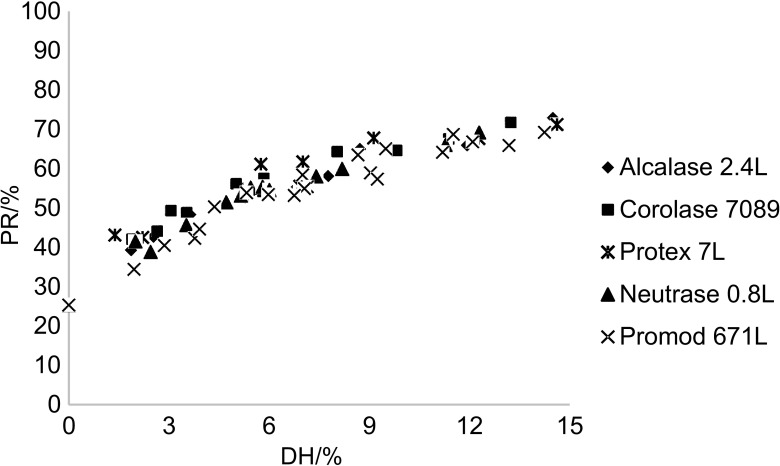
Protein recovery
Protein recovery, the ratio of protein content in the hydrolysate to that in the original substrate, was used as an index of protein solubilization and product yield. Even though DH was highly dependent on enzyme, protein recovery did not follow this relationship (Fig. 3). All enzymes demonstrated the same ability to solubilize the protein, to around 70% after extended hydrolysis. The correlation between the DH and protein recovery demonstrates a non-linear relationship, reaching a plateau at high DH, which is in agreement with Silva et al. (9). When enzyme activity was increased fourfold from 22 to 88 U/g, the DH increased from around 9 to 15% (Fig. 2). In comparison, the protein recovery response of all enzymes increased less than 10% (Fig. 3). The observation is in agreement with the results reported by Bhumiratana et al. (11), who demonstrated low increase in protein recovery when enzyme activity was increased fivefold. In the progress of the protein hydrolysis only part of the added enzyme is involved in the breakdown and solubilization of the insoluble high-molecular-mass proteins. The rest of the enzyme reacts with the already solubilized protein fragments. The net results of the latter activity will be an increase in DH without any increase in protein recovery with a progressively stronger effect at high degree of substrate solubilization (Fig. 3). On average (25±4) % of the protein was solubilized in a control sample incubated for 120 min at 50 °C without added enzyme, followed by heat treatment at >90 °C for 10 min. This can be ascribed to the content of small sarcoplasmic proteins, gelatin and free amino acids in the fish substrate (7, 54). On average, the protein recovery was increased to (54±1), (58±2), (64±3), (67±1) and (71±2) % after hydrolysis for two hours at enzyme activity to substrate ratio of 5.5, 11, 22, 44 and 88 U/g, respectively. Hence, if protein recovery is the main purpose of the hydrolysis process, the need to increase the enzyme concentration and DH should be carefully evaluated based on a cost-earnings calculation.
Fig. 3.
Relationship between protein recovery (PR) and degree of hydrolysis (DH) of the individual proteases
Response surface modelling
Response surface regression was used to model the combined effects of hydrolysis time and enzyme activity on DH and protein recovery. For all enzymes, DH models with high explained variance (R2>0.942, p<0.001) were established (Tables 3 and 4). Only insignificant differences were observed between the enzyme abilities to solubilize the substrate (Fig. 3), and a common protein recovery model (Table 3) could be established based on values from all the tested enzymes with R2=0.924 (p<0.001).
Table 3. Response surface models describing the effects of enzyme activity to substrate ratio (U/g) and hydrolysis time (min) on the degree of hydrolysis (DH) of the individual enzymes and protein recovery after backward removal of insignificant regressors.
| Variable | Alcalase 2.4L | Corolase 7089 | Protex 7L | Neutrase 0.8L | Promod 671L | Protein recovery |
|---|---|---|---|---|---|---|
| Intercept | –6.76·10–1** | –6.73·10–1*** | –1.32** | –6.31·10–1** | –7.70·10–1*** | 3.22·10–1*** |
| Time/min | 1.43·10–1*** | 1.22·10–1*** | 1.35·10–1*** | 1.30·10–1*** | 1.52·10–1*** | 3.67·10–1*** |
| U/g | 1.20·10–1*** | 1.15·10–1*** | 1.58·10–1*** | 1.24·10–1*** | 1.16·10–1*** | 5.67·10–1*** |
| Time/min·Time/min | –8.96·10–4*** | –7.31·10–4*** | –8.17·10–4*** | –7.44·10–4*** | –9.32·10–4*** | –1.70·10–3*** |
| U/g·U/g | 1.20·10–1*** | –8.24·10–4*** | –1.14·10–3*** | –9.30·10–4*** | –7.84·10–4*** | –4.01·10–3*** |
| Time/min·U/g | –8.34·10–4*** | 6.30·10–4*** | 7.13·10–4*** | 3.89·10–4*** | 6.32·10–4*** | NS |
| R2 | 0.942 | 0.960 | 0.948 | 0.944 | 0.944 | 0.924 |
NS=not significant *p<0.05, **p<0.01, ***p<0.001
Table 4. ANOVA for the fitted models given in Table 3.
| Source of variation |
Model | Sum of squares |
Degree of freedom |
Mean square |
F-ratio | p-value |
|---|---|---|---|---|---|---|
| Regression | Alcalase 2.4L | 2407.708 | 5 | 481.542 | 513.134 | <0.001 |
| Corolase 7089 | 1923.480 | 5 | 384.696 | 754.312 | <0.001 | |
| Neutrase 0.8L | 1653.687 | 5 | 330.737 | 536.937 | <0.001 | |
| Promod 671L | 2390.170 | 5 | 478.034 | 532.010 | <0.001 | |
| Protex 7L | 2650.714 | 5 | 530.143 | 576.339 | <0.001 | |
| Protein recovery | 6291.697 | 4 | 1572.924 | 172.987 | <0.001 | |
| Residual | Alcalase 2.4L | 149.211 | 159 | 0.938 | – | |
| Corolase 7089 | 80.069 | 159 | 0.510 | – | ||
| Neutrase 0.8L | 97.323 | 159 | 0.616 | – | ||
| Promod 671L | 141.970 | 159 | 0.899 | – | ||
| Protex 7L | 146.256 | 159 | 0.920 | – | ||
| Protein recovery | 518.285 | 57 | 9.093 | – | ||
| Total | Alcalase 2.4L | 2556.919 | 164 | 482.480 | ||
| Corolase 7089 | 2003.549 | 164 | 385.206 | |||
| Neutrase 0.8L | 1751.010 | 164 | 331.353 | |||
| Promod 671L | 2532.140 | 164 | 478.933 | |||
| Protex 7L | 2796.970 | 164 | 531.063 | |||
| Protein recovery | 6809.982 | 61 | 1582.017 |
The DH response could be fitted to a full model for all enzymes, while the interaction effect was not significant for the protein recovery response (Table 3). The main effects of hydrolysis time and enzyme activity to substrate ratio are both positive, reflecting that an increase of these factors gives higher DH and protein recovery levels. The square effects are both negative reflecting the descending effect on DH and protein recovery with increasing levels of the variables (Figs. 2 and 3). The positive interaction effect is higher on the response by increasing one of the two variables to a high level compared to a low level of the other.
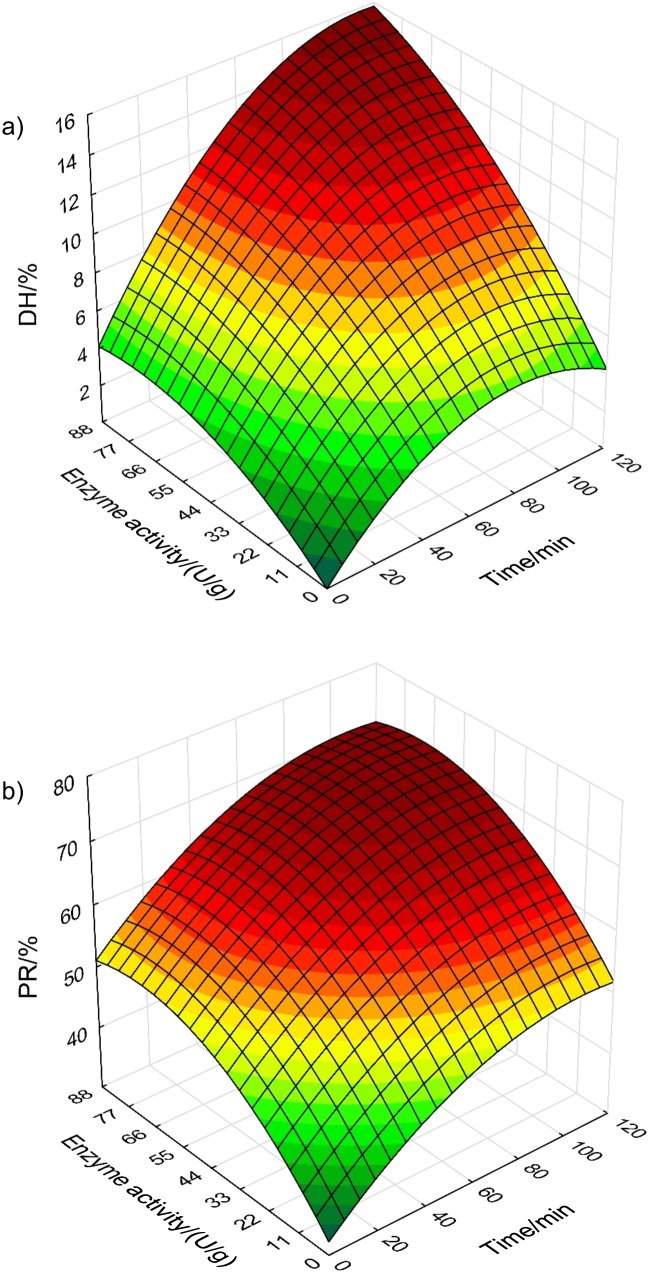
Effect of enzyme activity on the DH has commonly been reported in the literature based on a log-linear relationship at a fixed hydrolysis time (19, 42, 52). In the present modelling approach, the effect of enzyme concentration and hydrolysis time is combined in a common model. The obtained response surface models (Table 3) make it possible to define the needed enzyme activity to substrate ratio to reach a target DH at any reaction time, illustrated by the response surface plot of Alcalase 2.4L in Fig. 4a. The plot demonstrates a maximum DH when both hydrolysis time and enzyme activity are high. The curvature visualizes the higher effect of increasing enzyme activity to substrate ratio than to time within the studied range and the asymptotic descent to a maximum value (Fig. 2).
Fig. 4.
Response surface plots for: a) degree of hydrolysis (DH) of Alcalase 2.4L, and b) protein recovery (PR) based on regression models given in Table 3
The protein recovery response surface (Fig. 4b) demonstrates similar relationships, however, with a more equivalent effect of hydrolysis time compared to enzyme activity. Enzyme activity showed the highest main effect (Table 3), however, the higher squared negative effect gives a reduced response at increasing enzyme activity to substrate ratios. Combined, the established response surface models enable the estimation of the needed reaction time at a given enzyme activity to substrate ratio to reach a target DH and protein recovery of the studied substrate. The response surface regression approach was not able to model the initial equal level of DH and protein recovery at time zero at all enzyme activity to substrate ratios. In practice, the models should only be used for predictions at time >5 min.
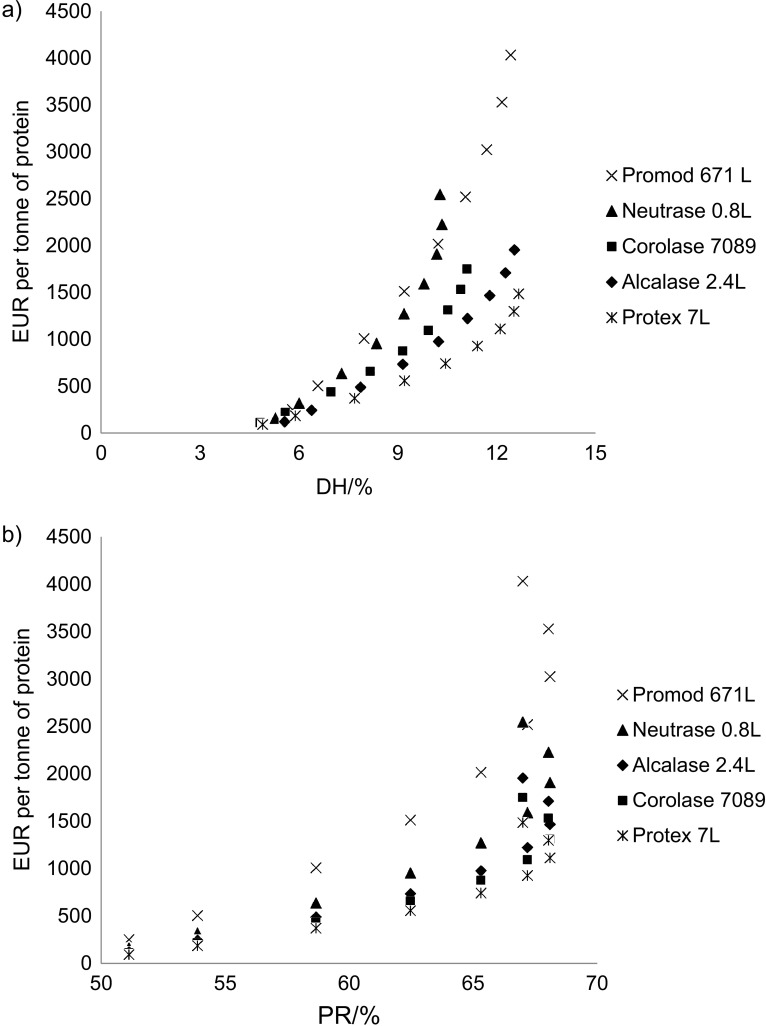
Cost efficiency assessment
The obtained DH and protein recovery regression models (Table 3) can easily be converted to the respective cost models by multiplying enzyme activity to substrate ratio by EUR/U (Table 2). This is illustrated in Fig. 5 where the cost of the respective enzymes to obtain different levels of DH (Fig. 5a) and protein recovery (Fig. 5b) after 60-minute reaction time is compared. The figures demonstrate large differences in cost efficiency between the enzymes to reach the same DH and protein recovery levels. Based on the acquired price indications, Protex 7L is the most cost-efficient enzyme closely followed by Alcalase 2.4L and Corolase 7089. Neutrase 0.8L and Promod 671L were found to be least cost efficient, mainly due to their higher price (Table 2). Similar relationships were also found for other reaction times (data not shown).
Fig. 5.
Enzyme cost as a function of model-based: a) degree of hydrolysis (DH), and b) protein recovery (PR) after a 60-minute reaction time using commercial endopeptidases
At the highest DH and protein recovery levels, the cost curves in Fig. 5 show a sharp increase; and in the case of protein recovery a backward bending. This is caused by the descending response and in the case of protein recovery also a small decline at the modelled highest enzyme activity to substrate ratios (Fig. 4b). The low and high enzyme activity to substrate ratios applied in this study were chosen to obtain a large response variance. This is important in response surface modelling. However, the highest enzyme activity to substrate levels (44 and 88 U/g) tested in this study demand the addition of significantly higher enzyme concentrations than recommended by the manufactures (Table 2) and are less applicable in industrial scale operations. The cost curves (Fig. 5) clearly show that the highest levels are not economically feasible at an industrial scale and should be reached by combining a lower enzyme activity to substrate ratio and longer reaction time.
Used in combination with other operation costs and variables, the modelling of DH, protein recovery and enzyme cost can be applied in optimizing the operation conditions for new and existing batch processing plants. The effect of temperature was not included in this study. An increased temperature will increase the reaction rate, but not influence the maximum obtainable DH or protein recovery at a fixed enzyme activity to substrate ratio (9, 55). Temperature will therefore not change the direct enzyme cost to obtain a target DH or protein recovery, but improve productivity in a batch processing plant if hydrolysis time is the limiting operation step.
Correlation between o-phthaldialdehyde and pH-STAT
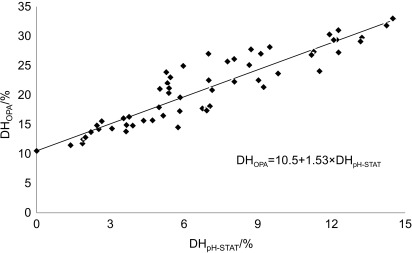
The pH-STAT method is an elegant and straightforward technique to monitor the DH in a hydrolysis process. The strict control of pH by base addition is, however, less attractive in industrial scale applications due to the increase of salt in the final hydrolysate. Several off-line analytical methods are described in the literature to follow DH in batch systems without pH control (20). The OPA method applied in this study showed a high correlation with the pH-STAT measurements (r=0.917, p<0.001; Fig. 6). The intercept of the y-axis at DHOPA=10.5 can be explained by the presence of small water-soluble proteins and free amino acids (54) that will react with the OPA reagent. The regression coefficient of 1.53 indicates a deviation between the two methods as also confirmed at other pH values (data not shown). Similar results have been reported by Himonides et al. (7), who compared the pH- -STAT and the 2,4,6-trinitrobenzenesulfonic acid (TNBS) techniques on salmon substrate. They observed, however, a nonlinearity at high base consumption equivalent to DH values >14%, above the range covered in this study (Fig. 6). The TNBS and OPA measurements are thought to be closer to the true value of cleaved peptide bonds because these techniques offer a direct assay of α-amino groups released during proteolytic breakdown. The underestimation of DH by the pH-STAT method may be explained by the higher pKa value of small peptides generated in the hydrolysis process. Tripeptides might have pKa values half a unit higher than polypeptides (12) and as a consequence liberate fewer protons after the hydrolytic step at a fixed pH, the net effect being reduced base consumption and an underestimation of DH. The observed linear correlation between the two methods within the studied DH range makes it possible to transfer the pH-STAT results to an unbuffered system and define an equivalent DH measured by the OPA method.
Fig. 6.
Relationship between the degree of hydrolysis (DH) measured by the pH-STAT technique (pH=6.5, t=50 °C) and the o-phthaldialdehyde (OPA) method
Conclusion
Atlantic salmon by-product-specific values for total amount of peptide bonds (htot) and nitrogen-to-protein conversion factor (fN) deviate from the commonly used and underline the importance of substrate characterization in order to achieve a precise estimation of protein content and degree of hydrolysis (DH). At low enzyme activity to substrate ratio all enzymes were equally efficient in hydrolyzing the substrate. At the higher enzyme to substrate ratios, Alcalase 2.4L, Protex 7L and Promod 671L were more efficient than Corolase 7089 and Neutrase 0.8L. The pH-STAT data can be combined with response surface regression to model the combined effects of enzyme activity to substrate ratio and hydrolysis time on DH and protein recovery. Combined with activity-specific enzyme cost the models can be transformed to cost models. Protex 7L was found to be the most cost-efficient enzyme, due to high hydrolytic efficiency and low price. All enzymes were equally able to solubilize the protein and protein recovery showed a nonlinear correlation with DH. A linear correlation between the pH-STAT and o-phthaldialdehyde (OPA) method was observed. The correlation can be used to define an equivalent DH level based on the OPA method in upscaling and industrial processes where the pH-STAT technique is not applicable. Combined with the response surface models, this can be an important tool in control of batch operations to obtain protein hydrolysates with desired DH and protein recovery.
Acknowledgements
The authors are grateful for the financial support from the Norwegian Ministry of Trade, Industry and Fisheries. We also want to thank the skillful and accurate laboratory personnel at Nofima BioLab.
References
- 1.FAO. The state of the world fisheries and aquaculture. Opportunities and challenges. Food and Agriculture Organization of the United Nations, Rome, Italy; 2014. [Google Scholar]
- 2.Report No SINTEF. A26097. Richardsen R, Nystřl R, Strandheim G, Kosmo JP. Analysis of marine by-products 2013. English summary. Trondheim, Norway: SINTEF Fisheries and Aquaculture; 2013. [Google Scholar]
- 3.Ramírez A. Salmon by-product proteins. Food and Agriculture Organization of the United Nations, Rome, Italy; 2007. [Google Scholar]
- 4.Liaset B, Espe M. Nutritional composition of soluble and insoluble fractions obtained by enzymatic hydrolysis of fish- -raw materials. Process Biochem. 2008;43:42–8. 10.1016/j.procbio.2007.10.007 [DOI] [Google Scholar]
- 5.Chalamaiah M, Kumar BD, Hemalatha R, Jyothirmayi T. Fish protein hydrolysates: proximate composition, amino acid composition, antioxidant activities and applications: a review. Food Chem. 2012;135:3020–38. 10.1016/j.foodchem.2012.06.100 [DOI] [PubMed] [Google Scholar]
- 6.Mackie IM. General review of fish-protein hydrolysates. Anim Feed Sci Technol. 1982;7:113–24. 10.1016/0377-8401(82)90045-1 [DOI] [Google Scholar]
- 7.Himonides AT, Taylor AKD, Morris AJ. A study of the enzymatic hydrolysis of fish frames using model systems. Food Nutr Sci. 2011;2:575–85. 10.4236/fns.2011.26081 [DOI] [Google Scholar]
- 8.Gbogouri GA, Linder M, Fanni J, Parmentier M. Influence of hydrolysis degree on the functional properties of salmon byproducts hydrolysates. J Food Sci. 2004;69:C615–22. 10.1111/j.1365-2621.2004.tb09909.x [DOI] [Google Scholar]
- 9.Silva VM, Park KJ, Hubinger MD. Optimization of the enzymatic hydrolysis of mussel meat. J Food Sci. 2010;75:C36–42. 10.1111/j.1750-3841.2009.01414.x [DOI] [PubMed] [Google Scholar]
- 10.Gildberg A, Arnesen JA, Carlehog M. Utilisation of cod backbone by biochemical fractionation. Process Biochem. 2002;38:475–80. 10.1016/S0032-9592(02)00103-6 [DOI] [Google Scholar]
- 11.Bhumiratana S, Hill CG, Jr, Amundson CH. Enzymatic solubilization of fish protein concentrate in membrane reactors. J Food Sci. 1977;42:1016–21. 10.1111/j.1365-2621.1977.tb12657.x [DOI] [Google Scholar]
- 12.Adler-Nissen J. Enzymatic hydrolysis of food proteins. New York, NY, USA: Elsevier Science Publishing Co., Inc.; 1986. [Google Scholar]
- 13.Kristinsson HG, Rasco BA. Biochemical and functional properties of Atlantic salmon (Salmo salar) muscle proteins hydrolyzed with various alkaline proteases. J Agric Food Chem. 2000;48:657–66. 10.1021/jf990447v [DOI] [PubMed] [Google Scholar]
- 14.Panyam D, Kilara A. Enhancing the functionality of food proteins by enzymatic modification. Trends Food Sci Technol. 1996;7:120–5. 10.1016/0924-2244(96)10012-1 [DOI] [Google Scholar]
- 15.Kunst T. Protein modification to optimize functionality. Protein hydrolsates. In: Whitaker JR, Voragen AGJ, Wong DWS, editors. Handbook of food enzymology. New York, NY, USA: Marcel Dekker, Inc.; 2003. pp. 221–36. http://dx.doi.org/ 10.1201/9780203910450.ch17 [DOI] [Google Scholar]
- 16.Aspmo SI, Horn SJ, Eijsink VGH. Enzymatic hydrolysis of Atlantic cod (Gadus morhua L.) viscera. Process Biochem. 2005;40:1957–66. 10.1016/j.procbio.2004.07.011 [DOI] [Google Scholar]
- 17.Hoyle NT, Merritt JH. Quality of fish-protein hydrolysates from herring (Clupea harengus). J Food Sci. 1994;59:76–9. 10.1111/j.1365-2621.1994.tb06901.x [DOI] [Google Scholar]
- 18.Seo WH, Lee HG, Baek HH. Evaluation of bitterness in enzymatic hydrolysates of soy protein isolate by taste dilution analysis. J Food Sci. 2008;73:S41–6. 10.1111/j.1750-3841.2007.00610.x [DOI] [PubMed] [Google Scholar]
- 19.Kristinsson HG, Rasco BA. Kinetics of the hydrolysis of Atlantic salmon (Salmo salar) muscle proteins by alkaline proteases and a visceral serine protease mixture. Process Biochem. 2000;36:131–9. 10.1016/S0032-9592(00)00195-3 [DOI] [Google Scholar]
- 20.Rutherfurd SM. Methodology for determining degree of hydrolysis of proteins in hydrolysates: a review. J AOAC Int. 2010;93:1515–22. [PubMed] [Google Scholar]
- 21.Cohen SA, Michaud DP. Synthesis of a fluorescent derivatization reagent, 6-aminoquinolyl-N-hydroxysuccinimidyl carbamate, and its application for the analysis of hydrolysate amino acids via high-performance liquid chromatography. Anal Biochem. 1993;211:279–87. 10.1006/abio.1993.1270 [DOI] [PubMed] [Google Scholar]
- 22.Mossé J, Huet JC, Baudet J. The amino acid composition of wheat grain as a function of nitrogen content. J Cereal Sci. 1985;3:115–30. 10.1016/S0733-5210(85)80022-9 [DOI] [Google Scholar]
- 23.Conway EJ, Byrne A. An absorption apparatus for the micro-determination of certain volatile substances. The micro-determination of ammonia. Biochem J. 1933;27:419–29. [PMC free article] [PubMed] [Google Scholar]
- 24.Miller EL. Determination of tryptophan content of feedingstuffs with particular reference to cereals. J Sci Food Agric. 1967;18:381–6. 10.1002/jsfa.2740180901 [DOI] [PubMed] [Google Scholar]
- 25.ISO 5983-2:2009. Animal feeding stuffs – Determination of nitrogen content and calculation of crude protein content. Part 2: Block digestion and steam distillation method. Geneva, Switzerland: International Organization for Standardization (ISO); 2009.
- 26.Official Method Ca AOCS. 5a-40. Determination of free fatty acids (FFA). Urbana, IL, USA: The American Oil Chemists’ Society (AOCS); 2012. [Google Scholar]
- 27.ISO 6496:1999. Animal feeding stuffs – Determination of moisture and other volatile matter content. Geneva, Switzerland: International Organization for Standardization (ISO); 1999.
- 28.ISO 5984:2002. Animal feeding stuffs – Determination of crude ash. Geneva, Switzerland: International Organization for Standardization (ISO); 2002.
- 29.Bligh EG, Dyer WJ. A rapid method for total lipid extraction and purification. Can J Biochem Physiol. 1959;37:911–7. 10.1139/o59-099 [DOI] [PubMed] [Google Scholar]
- 30.Mossé J. Nitrogen-to-protein conversion factor for ten cereals and six legumes or oilseeds. A reappraisal of its definition and determination. Variation according to species and to seed protein content. J Agric Food Chem. 1990;38:18–24. 10.1021/jf00091a004 [DOI] [Google Scholar]
- 31.Sriperm N, Pesti GM, Tillman PB. Evaluation of the fixed nitrogen-to-protein (N:P) conversion factor (6.25) versus ingredient specific N:P conversion factors in feedstuffs. J Sci Food Agric. 2011;91:1182–6. 10.1002/jsfa.4292 [DOI] [PubMed] [Google Scholar]
- 32.Cupp-Enyard C. Sigma’s non-specific protease activity assay – casein as a substrate. J Vis Exp. 2008;19:e899. 10.3791/899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.TiamoTM, v. 2.3, Metrohm, Herisau, Switzerland; 2011. Available from: http://www.metrohm.com/en/support-and-service/software-center/tiamo.
- 34.Steinhardt J, Beychok S. Interaction of protein with hydrogen ions and other small ions and molecules. In: Neurath H, editor. The Proteins. New York, NY, USA: Academic Press; 1964. pp. 139–304. http://dx.doi.org/ 10.1016/B978-0-12-395724-5.50012-0 [DOI] [Google Scholar]
- 35.Nielsen PM, Petersen D, Dambmann C. Improved method for determining food protein degree of hydrolysis. J Food Sci. 2001;66:642–6. 10.1111/j.1365-2621.2001.tb04614.x [DOI] [Google Scholar]
- 36.STATISTICA, v. 12, StatSoft, Inc, Tulsa, OK, USA; 2012. Available from: http://www.statsoft.com.
- 37.Meyers RJ, Montgomery DC, Anderson-Cook CM. Response surface methodology. Process and product optimization using designed experiments. New York, NY, USA: John Wiley & Sons, Inc. 2002. [Google Scholar]
- 38.Lalasidis G, Boström S, Sjöberg LB. Low molecular weight enzymatic fish protein hydrolysates: chemical composition and nutritive value. J Agric Food Chem. 1978;26:751–6. 10.1021/jf60217a045 [DOI] [PubMed] [Google Scholar]
- 39.Mariotti F, Tomé D, Patureau Mirand P. Converting nitrogen into protein – beyond 6.25 and Jones’ factors. Crit Rev Food Sci Nutr. 2008;48:177–84. 10.1080/10408390701279749 [DOI] [PubMed] [Google Scholar]
- 40.Van Waarde A. Biochemistry of non-protein nitrogenous compounds in fish including the use of amino-acids for anaerobic energy-production. Comp Biochem Physiol B. 1988;91:207–28. 10.1016/0305-0491(88)90136-8 [DOI] [Google Scholar]
- 41.Duan ZH, Wang JL, Yi MH, Yin AQ. Recovery of proteins from silver carp by-products with enzymatic hydrolysis and reduction of bitterness in hydrolysate. Food Process Eng. 2010;33:962–78. 10.1111/j.1745-4530.2008.00318.x [DOI] [Google Scholar]
- 42.Benjakul S, Morrissey MT. Protein hydrolysates from Pacific whiting solid wastes. J Agric Food Chem. 1997;45:3423–30. 10.1021/jf970294g [DOI] [Google Scholar]
- 43.Gupta R, Beg QK, Khan S, Chauhan B. An overview on fermentation, downstream processing and properties of microbial alkaline proteases. Appl Microbiol Biotechnol. 2002;60:381–95. 10.1007/s00253-002-1142-1 [DOI] [PubMed] [Google Scholar]
- 44.Sovik SL, Rustad T. Proteolytic activity in byproducts from cod species caught at three different fishing grounds. J Agric Food Chem. 2005;53:452–8. 10.1021/jf049350l [DOI] [PubMed] [Google Scholar]
- 45.Refsgaard HHF, Brockhoff PMB, Jensen B. Free polyunsaturated fatty acids cause taste deterioration of salmon during frozen storage. J Agric Food Chem. 2000;48:3280–5. 10.1021/jf000021c [DOI] [PubMed] [Google Scholar]
- 46.Kanicky JR, Poniatowski AF, Mehta NR, Shah DO. Cooperativity among molecules at interfaces in relation to various technological processes: effect of chain length on the pKa of fatty acid salt solutions. Langmuir. 2000;16:172–7. 10.1021/la990719o [DOI] [Google Scholar]
- 47.Kristinsson HG, Rasco BA. Fish protein hydrolysates: production, biochemical, and functional properties. Crit Rev Food Sci Nutr. 2000;40:43–81. 10.1080/10408690091189266 [DOI] [PubMed] [Google Scholar]
- 48.Marquez Moreno MC, Fernandez Cuadrado V. Enzymatic hydrolysis of vegetable proteins – mechanism and kinetics. Process Biochem. 1993;28:481–90. 10.1016/0032-9592(93)85032-B [DOI] [Google Scholar]
- 49.Hjelmeland K. Proteinase inhibitors in the muscle and serum of cod (Gadhus morhua). Isolation and characterization. Comp Biochem Physiol B. 1983;76:365–72. 10.1016/0305-0491(83)90084-6 [DOI] [Google Scholar]
- 50.Camacho F, González-Tello P, Páez-Dueńas MP, Guadix EM, Guadix A. Correlation of base consumption with the degree of hydrolysis in enzymic protein hydrolysis. J Dairy Res. 2001;68:251–65. 10.1017/S0022029901004824 [DOI] [PubMed] [Google Scholar]
- 51.Hevia P, Whitaker JR, Olcott HS. Solubilization of a fish protein concentrate with proteolytic enzymes. J Agric Food Chem. 1976;24:383–5. 10.1021/jf60204a048 [DOI] [PubMed] [Google Scholar]
- 52.Baek HH, Cadwallader KR. Enzymatic hydrolysis of crayfish processing by-products. J Food Sci. 1995;60:929–35. 10.1111/j.1365-2621.1995.tb06264.x [DOI] [Google Scholar]
- 53.Shahidi F, Han XQ, Synowiecki J. Production and characteristics of protein hydrolysates from capelin (Mallotus villosus). Food Chem. 1995;53:285–93. 10.1016/0308-8146(95)93934-J [DOI] [Google Scholar]
- 54.Oterhals Ĺ, Samuelsen TA. Plasticization effect of solubles in fishmeal. Food Res Int. 2015;69:313–21. 10.1016/j.foodres.2014.12.028 [DOI] [Google Scholar]
- 55.Hou H, Li B. Response surface optimization of enzymatic hydrolysis of silver sillago (Sillago sihama) with low bitter taste. Int J Food Eng. 2011;7(3): 10.2202/1556-3758.2007 [DOI] [Google Scholar]