Abstract
The gram-positive bacterium Streptomyces noursei ATCC 11455 produces a complex mixture of polyene macrolides generally termed nystatins. Although the structures for nystatins A1 and A3 have been reported, the identities of other components of the nystatin complex remain obscure. Analyses of the culture extract from the S. noursei wild type revealed the presence of several nystatin-related compounds for which chemical structures could be suggested on the basis of their molecular weights, their UV spectra, and knowledge of the nystatin biosynthetic pathway. Nuclear magnetic resonance (NMR) studies with one of these polyene macrolides identified it as a nystatin analogue containing a mycarose moiety at C-35. A similar investigation was performed with the culture extract of the ERD44 mutant, which has a genetically altered polyketide synthase (PKS) NysC and which was previously shown to produce a heptaene nystatin analogue. The latter compound, tentatively named S44HP, and its derivative, which contains two deoxysugar moieties, were purified; and their structures were confirmed by NMR analysis. Nystatin analogues with an expanded macrolactone ring were also observed in the extract of the ERD44 mutant, suggesting that the altered PKS can “stutter” during the polyketide chain assembly. These data provide new insights into the biosynthesis of polyene macrolide antibiotics and the functionalities of PKSs and post-PKS modification enzymes.
Glycosylated polyene macrolides are very efficient antifungal agents widely used for the treatment of both topical and invasive fungal infections in humans (50). The main advantages of polyene macrolides over other antifungal drugs, such as azoles and flucytosines, are their fungicidal actions and the extremely low incidence of resistant pathogens. The fungicidal actions of polyene macrolide antibiotics are strictly dependent on the presence of sterols in the membranes of the sensitive cells (8). The selective action of these types of antibiotics is based on their higher affinities to the ergosterol present in fungal membranes compared to that of cholesterol, the structural component of mammalian cell membranes. The mode of action of glycosylated polyene macrolides is based on their ability to interact with sterols and to form channels that perforate the membrane, which allows the leakage of ions and other small molecules out of the cell, ultimately resulting in cell death (9). It is presumed that conjugated double bonds present on the macrolactone rings of these molecules (hence the term polyene) are responsible for antibiotic-sterol interactions. Unfortunately, the relatively high toxicities of polyene antibiotics for the mammalian cells and the poor distributions of these compounds in tissues due to low water solubility limit their usefulness for antifungal therapy. Novel polyene antibiotic analogues with reduced toxicities and increased water solubilities might help circumvent these problems.
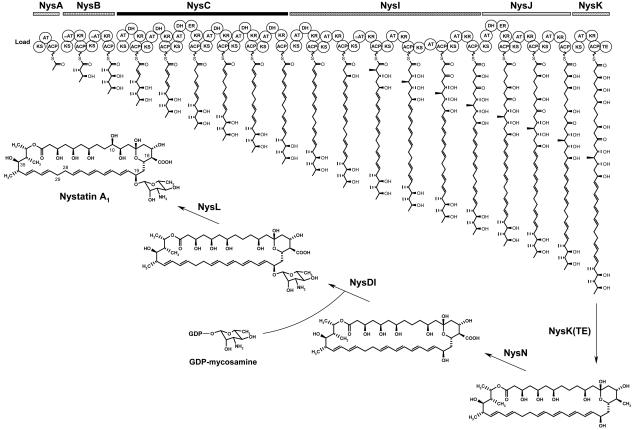
Modification of the antibiotic structure through engineering of its biosynthetic genes is a promising strategy for the production of novel pharmaceuticals (28). The gene cluster from the gram-positive bacterium Streptomyces noursei ATCC 11455 that governs the biosynthesis of the polyene macrolide antibiotic nystatin has recently been cloned and analyzed (13). Six polyketide synthase (PKS) proteins were implicated in the assembly of the 38-membered nystatin macrolactone ring, while other proteins were apparently required for post-PKS modification of the nystatin aglycone (Fig. 1). The modular PKS proteins NysA, NysB, NysC, NysI, NysJ, and NysK perform a decarboxylative condensation of the carboxylic acids to form a polyketide chain, which is then released from the PKS and cyclized via the action of a thioesterase domain. In addition, modules in the PKS contain enzymatic domains with reductive activities which modify the polyketide chain during synthesis. The macrolactone ring of nystatin assembled by the PKS is then further modified via the actions of monooxygenases NysN and NysL and a glycosyltransferase, NysDI. The formidable complexity of the biosynthetic machinery involved in nystatin synthesis suggests that this system might be error prone and, thus, might eventually produce many nystatin-related molecules. It is well known that the polyene macrolide producers simultaneously synthesize several structurally related compounds (33). Indeed, preliminary analysis of the culture extracts from S. noursei revealed the presence of related polyene macrolides, with nystatin A1 (see Fig. 4A) being the major component. Although the nystatin molecule has two sets of conjugated double bonds (2 + 4) on its macrolactone ring, it is classified as a tetraene (33). Recently, the nystatin PKS NysC was genetically manipulated, which resulted in the synthesis of novel nystatin analogues (14). One of the mutants, ERD44, was shown to produce a heptaene nystatin analogue in small quantities.
FIG. 1.
Model for biosynthesis of nystatin A1 in S. noursei ATCC 11455. PKS domains: KS, ketosynthase; AT, acyltransferase (acetate specific); mAT, acyltransferase (propionate specific); KR, ketoreductase; DH, dehydratase; ER, enoyl reductase; TE, thioesterase; ACP, acyl carrier protein. Post-PKS modifying enzymes: NysN, P450 monooxygenase (oxidation of methyl group at C-16); NysDI, mycosaminyl transferase; NysL, P450 monooxygenase (hydroxylation at C-10).
FIG. 4.
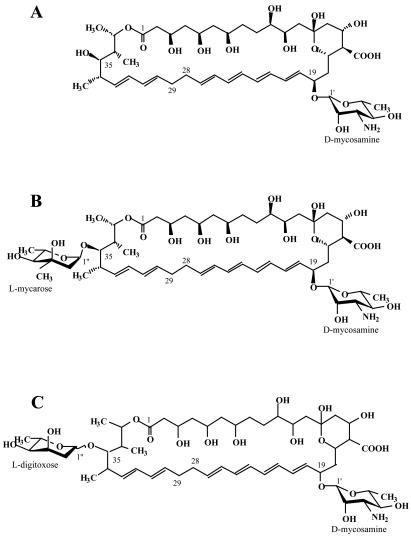
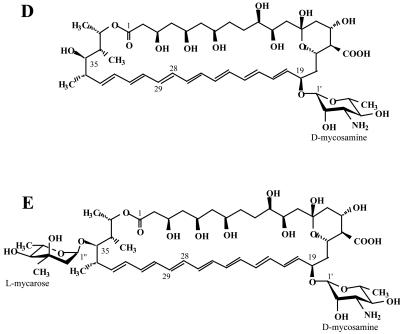
Structures of the various forms of nystatin A and its analogues. (A) Nystatin A1 (10, 29); (B) NYST1070 (this work); (C) nystatin A3 (49); (D) S44HP (this work); (E) NYST1068 (this work).
In the present study, we have identified several nystatin-related polyene macrolides that are produced by wild-type S. noursei using liquid chromatography with diode array peak detection coupled with mass spectrometry (LC-MS). In addition, major fractions of polyene macrolides produced by the ERD44 recombinant strain with a mutated NysC PKS have been identified and characterized. Several newly identified compounds produced by the wild-type strain and strain ERD44 have been purified and subjected to nuclear magnetic resonance (NMR) structural studies and bioactivity testing.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
S. noursei strains ATCC 11455 and ERD44 (14) were maintained on ISP2 agar medium (Difco, Detroit, Mich.). Liquid cultures were grown in 500-ml shake flasks with 100 ml of semidefined SAO-23 medium (41) at 28°C for 5 days. For the large-scale production of polyene macrolides, cultures were grown in the 3-liter fermentors essentially as described previously (41). The polyene macrolides produced have low water solubilities, and the major fraction of the polyenes is therefore precipitated in the culture medium at the end of the fermentation. After 5 days of cultivation the culture was centrifuged in aliquots. The pellet containing bacterial cells, undissolved medium components, and polyenes was frozen (−20°C) until extraction with methanol (MeOH), and analyses were then performed.
LC-MS analysis and purification of polyene macrolides.
Analytical LC-MS analysis of the S. noursei wild-type and recombinant strains was performed on an Agilent 1100 high-pressure liquid chromatograph connected to an Agilent mass selective detector (MSD) time of flight (TOF) mass spectrometer system with electrospray ionization in the positive mode. Automatic calibration of the mass axis was performed by continuous infusion of Agilent electrospray (ES)-TOF tuning through a second nebulizer needle in the ion source. A Waters NovaPak C18 column (dimensions, 150 by 2.1 mm) was used for high-pressure liquid chromatography (HPLC), and the flow rate was 300 μl/min. The mobile phase consisted of 10 mM ammonium acetate (pH 4.0) and acetonitrile (ACN). For analysis of the extract from the wild type, the ACN concentration was increased linearly from 37 to 60% for the first 10 min and was then kept at a concentration of 60% for the rest of the run. The strain ERD44 extract was run with a linear gradient from 40 to 70% ACN in 10 mM ammonium acetate (pH 4.0) for the first 10 min, and the concentration was then kept at 70% for the rest of the run. A nystatin A1 standard (Sigma, St. Louis, Mo.) was used as a reference compound in all HPLC and LC-MS experiments.
For preparative isolation of the polyene macrolides, the macrolides were extracted from the cell pellets with 10 ml of MeOH per 2 g of cell pellet, and the samples were run on a preparative Agilent 1100 system with an online diode array detector (DAD)-Agilent MSD Trap mass spectrometer monitoring with flow split to the MSD and a fraction collector. A preparative Waters NovaPak C18 column (300 by 9 mm) run isocratically at 30°C with a flow rate of 8 ml/min and a 1-ml injection volume was used. The mobile phase consisted of 0.2 g of EDTA per liter, 16 ml of tetrahydrofuran per liter, and ACN (275 g/liter for tetraenes, 300 g/liter for heptaenes, and 355 g/liter for octaenes). The volume was adjusted to 1 liter with purified water (MilliQ). The pH was adjusted to 4.0 with acetic acid.
The collected fractions were pooled, and an aliquot was always reanalyzed by analytical LC-MS for determination of the purity of the isolated compound. Purity was calculated by comparing areas of UV peaks (309 nm for tetraenes, 392 for heptaenes, and 418 for octaenes). Concentrations were determined by using the experimentally determined extinction coefficients and comparing them with that for the nystatin A1 standard. The purities were >97% for all purified compounds. The rest of the sample was dried with a Speed-Vac centrifuge after the mobile phase was exchanged for a water-MeOH mixture (10 to 90%) by solid-phase extraction (Oasis HLB SPE kit; Waters). The dried samples were kept in the dark at −20°C until NMR and bioactivity testing.
NMR experiments.
Samples of 1 to 2 mM for NMR were obtained by dissolving S44HP and NYST1068 in dimethyl sulfoxide (DMSO)-d6 and NYST1070 in methanol-d4 under dry argon. All NMR spectra were recorded at 25°C on a Bruker Avance DRX 500 (1H = 500.125 MHz; 13C = 125.76 MHz) spectrometer with a 5-mm triple-resonance (1H, 13C, 15N) probe head equipped with a supplementary self-shielded z-gradient coil.
Homonuclear two-dimensional experiments by double-quantum filtered correlation spectroscopy (DQF-COSY) (38), total correlation spectroscopy (TOCSY), Hartmann-Hann spectroscopy (12, 22), and rotating Overhauser effect (ROE) spectroscopy (5, 11) were recorded with a 1.5-s recovery delay in the phase-sensitive mode by the States-TPPI (time proportional phase incrementation) method (30) with 512 (t1) × 1,024 (t2) complex datum points (where t1 and t2 are the indirect and direct dimension, respectively). They were recorded at 32 scans per increment and spectral widths of 4,500 Hz in both dimensions. A mixing time of 80 ms was used for the TOCSY experiments, and a mixing time of 250 ms was used for ROE spectroscopy. Spectra were processed with GIFA (version 4) software (36). After zero filling in the t1 dimension, the data were apodized with shifted sine-bell and Gaussian window functions in both dimensions to obtain a final matrix of 1,024 (F1) × 1,024 (F2) real datum points (where F1 and F2 are the phased indirect dimension and the phased direct dimension, respectively. Chemical shifts were referenced relative to the solvent chemical shifts (1H = 2.49 ppm for DMSO and 3.31 ppm for MeOH).
1H-13C heteronuclear two-dimensional spectra were obtained by 13C-heteronuclear multiple bound connectivity spectroscopy (HMBC) (6) and phase-sensitive 13C-heteronuclear single-quantum coherence spectroscopy (HSQC) (7), recorded by the echo-antiecho method (3). The coherence pathway selection was achieved by applying pulsed-field gradients as coherence filters (18, 35). Spectra were typically collected with 128 (t1, 13C) and 1,024 (t2, 1H) complex points and 360 scans per t1 increment. Spectral widths were 17,605 Hz in F1 and 4,496 Hz in F2, with carrier frequencies at 70 and 3.5 ppm, respectively. The data were processed with PIPP software (24). They were apodized with shifted square sine-bell window and Gaussian functions in the F1 and F2 dimensions, respectively, after linear prediction and zero filling in the t1 dimension to obtain a final matrix of 512 (F1) × 1,024 (F2) real datum points.
Bioassay.
The test organism used for the polyene macrolide bioactivity assays was Candida albicans ATCC 10231, grown in 120 μl of standard M19 medium without NaCl (with an inoculum of 1,000 CFU per well) and 30 μl of diluted polyene macrolide samples. The antibiotics were diluted in MeOH in series to ensure that concentrations yielded from complete growth inhibition to no growth inhibition of the test organism. The test organism cultures with antibiotic dilutions were then incubated in 96-well microtiter plates at 30°C without shaking; and after 12, 14, and 16 h, growth was measured as the optical density at 490 nm on a SpectraMax Plus microtiter plate reader. The optical density was plotted against the antibiotic concentration, and the MIC at which 50% of isolates are inhibited (MIC50) was estimated from the regression curve at 50% growth inhibition.
RESULTS
Nystatin-related polyene macrolides produced by wild-type S. noursei.
According to the literature, S. noursei ATCC 11455 produces a complex mixture of polyene macrolides containing nystatins A1, A2, and A3, which were assumed to be structurally related (21, 32, 33). The chemical structures of nystatins A1 and A3 have been solved (21, 37, 49), while no structural or mass spectrometry data on nystatin A2 could be unraveled. In order to gain a better insight into the process of nystatin biosynthesis, we have attempted to identify the major polyene components produced by S. noursei during fermentation.
Polyene macrolides exhibit characteristic three-peak UV spectra, with λmax being a function of the number of conjugated double bonds (33). Diode-array (DAD) HPLC analysis of the culture extract after fermentation under controlled conditions (see Materials and Methods) revealed the presence of several tetraene (λ = 280 to 320) and heptaene (λ = 370 to 410) compounds with different retention times (Fig. 2A). In order to test whether all these compounds were synthesized by the actions of the nystatin biosynthetic enzymes, similar metabolite profiling was performed for strain NDA59, which contains a deletion that affects the PKS loading module NysA, which is required for the initiation of nystatin biosynthesis (15). No polyene macrolides could be detected in the extract of mutant NDA59 (data not shown), implying that all the compounds detected in the extract from the wild-type strain with UV spectra characteristic of polyenes (33) are related to nystatin.
FIG. 2.
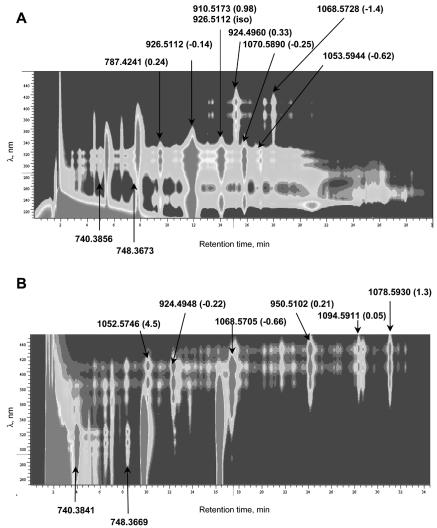
HPLC-DAD-MS analyses of polyene macrolides produced by S. noursei strains ATCC 11455 (A) and ERD44 (B). MWs (M+H)+ for monoisotopic peaks are indicated, with the errors (in parts per million) in the MWs of the proposed compounds given in parentheses.
Once that we were convinced of the common origin of the detectable polyene compounds produced by the wild-type S. noursei strain, we set up analyses with combined diode array and MS detection to determine their molecular weights (MWs) in order to assign chemical structures to these polyene macrolides. Apparently, nystatin A1 (MW = 926.5112) represented the major component in the polyene mixture produced by the wild-type strain. The errors in the proposed MWs of the compounds (in parts per million) are given in parentheses in Fig. 2. An error of 1 ppm represents uncertainty in the third decimal for the MWs of compounds at about 1,000. The accurate atomic mass of the monoisotopic nystatin A1 peak is 925.5035, and the MW of the (M+H)+ ion is 926.5113. The error was only −0.14 ppm, underlining the very high degree of accuracy of the MW determination in our analysis. In this case, of all C, H, N, and O combinations, the nystatin (M+H)+ formula (C47H76NO17) had the closest hit to the experimentally determined value when it was tested with Analyst mass calculator software. Interestingly, under the fermentation conditions without pH control (batch fermentation), we have observed the accumulation of two nystatin-related macrolides that had MWs of 926.5112 but that showed different HPLC retention times (Fig. 2A). It seems possible that the second compound represents the isomeric form of nystatin previously observed by Ostrosky-Zeichner et al. (34), the appearance of which depends on the pH. This putative isomeric form of nystatin, which represented the second-largest peak, was eluted from the HPLC column almost simultaneously with the nystatin analogue with an MW of 910.5173. We suggest that the latter analogue represents 10-deoxy nystatin, which accumulates due to the eventual failure of P450 monooxygenase NysL to perform hydroxylation. C-10 is the only position where a hydroxyl group is assumed to be added by a separate modifying protein rather than originates during macrolactone synthesis.
The third major tetraene component detected in the extract had an MW of 1,070.5890. The structural identity of this nystatin analogue, named NYST1070, could not be proposed on the basis of the nystatin biosynthetic pathway, prompting us to purify this compound for NMR analysis (see below).
A considerable amount of a polyene compound with an MW of 787.4241 was detected in the extract. The molecular mass of this nystatin analogue corresponded to the sodium adduct ion of the nystatin molecule lacking both the C-10 hydroxyl group and the C-19-linked mycosamine moiety. The mobility of the latter compound during HPLC was strongly affected by the pH of the eluent (data not shown), presumably due to the presence of only one ionizable group (COO− at C-16) instead of the two ionizable groups normally present on the glycosylated molecule (NH3+ on the mycosamine moiety and COO− at C-16). Apparently, this compound accumulated due to the failure of the glycosyltransferase NysDI and monooxygenase NysL to modify this precursor.
A tetraene with an MW of 1,053.5944 was detected as a relatively minor fraction in the culture extract (Fig. 2A). The MW of this compound suggests that this polyene macrolide represents deoxy NYST1070, an analogue of NYST1070 without a hydroxyl group at C-10. We have observed that fermentation conditions can significantly affect the production of this polyene macrolide, sometimes making it one of the major components (data not shown). No major polyene macrolide fraction with an MW of 1,056.5743, which would correspond to nystatin A3, was identified in the extract of wild-type S. noursei, suggesting that synthesis and accumulation of this component might depend on the growth conditions. A peak for a tetraene with an MW of 1,056.5733 was identified in the shoulder of the large nystatin A1 peak at a 12-min retention time. The intensity of this ion was 104 lower than that for the nystatin A1 peak. This peak probably represents nystatin A3 since the error from the MW of the latter was only −0.96 ppm. However, this assignment and postulation of the presence of nystatin A3 remain uncertain since we were not able to completely separate this peak.
Several heptaene compounds were detected in the culture extract of the wild-type strain, and the MWs for two of them were identified (Fig. 2A). One compound, with an MW of 924.9460, corresponded to the fully post-PKS-modified nystatin analogue with a double bond between C-28 and C-29. Most likely, this analogue is produced as a result of a failure of the enoyl reductase in module 5 of the NysC PKS to perform the reduction. The second heptaene identified had an MW of 1,068.5728, which would correspond to the heptaene nystatin analogue NYST1070 with a C-28-C-29 double bond. We have purified this compound, named NYST1068, for structural analysis, which confirmed the identity of this metabolite (see below).
Mutant ERD44 produces tetraene, heptaene, and octaene nystatin analogues.
S. noursei ERD44 was constructed previously in an attempt to create a producer of a heptaene nystatin analogue by inactivating the enoyl reductase domain in module 5 of NysC PKS (14). However, presumably due to the rather large deletion introduced into the NysC PKS, the overall functionality of the resulting PKS, NysC-44, was affected, leading to severely reduced levels of polyene production. Several peaks corresponding to the polyene macrolides were identified upon analysis of the ERD44 culture extract by DAD-HPLC and LC-MS (Fig. 2B). It should be noted that a different solvent system was used in this case in order to achieve better separation (see Materials and Methods), and therefore, the retention times of the compounds from the extracts of the wild-type strain and ERD44 cannot be compared. However, chromatographic peaks with equal MWs and UV spectra presumably represent the same compounds.
Two tetraene macrolides in the extract from the ERD44 culture were identified as having MWs of 740.3841 and 748.3669, respectively (Fig. 2B). Minor amounts of these compounds were also found in the extract of the wild-type strain (MWs, 748.3673 and 740.3856) (Fig. 2A). We were not able to correlate these masses to components of the nystatin biosynthesis pathway or to isolate these compounds by preparative HLPC for NMR analysis. The ERD44 extract did not contain a peak at 926.5112 ± 200 ppm, strongly suggesting that the ERD44 mutant does not produce nystatin A1.
The MWs of three major heptaene macrolides produced by ERD44 were determined. Two of them, with MWs of 924.4948 and 1,068.5705, respectively, represented the heptaene macrolides produced by the wild-type strain as minor components. The heptaene macrolide with an MW of 924.4948, named S44HP, has been purified, and its identity as a nystatin analogue with a C-28-C-29 double bond was confirmed by NMR (see below). The third heptaene, which had an MW of 1,052.5746, presumably represents the NYST1068 analogue lacking the C-10 hydroxyl group.
Surprisingly, a relatively large portion of the polyene macrolides produced by ERD44 was represented by octaenes. Compounds with UV spectra characteristic of octaenes (λ = 390 to 430) and MWs of 950.5102, 1,078.5930, and 1,094.5911 were identified (Fig. 2B). The latter suggested the incorporation of an additional acetate extender into the macrolactone ring of nystatin, followed by a round of reduction by ketoreductase and dehydratase domains, which result in the addition of a component with an MW of ca. 26.02 (C2H2), which adds to the MWs of the compounds. It seems likely that the octaene analogue with an MW of 950.5102 is a completely modified nystatin derivative with an expanded macrolactone ring which is produced by ERD44 due to the “stuttering” of the PKS NysC-44, similar to those described for the erythromycin PKS (47) and the aureothin PKS (27). The polyene macrolide with an MW of 1,094.5911 corresponds to the NYST1070 analogue with an expanded macrolactone ring, while the compound with an MW of 1,078.5930 is presumably a precursor of the latter that lacks the C-10 hydroxyl.
Structures and biological activities of the newly identified polyene macrolides. (i) S44HP.
Assignment of 1H and 13C resonances of S44HP in DMSO-d6 was performed by using methods previously applied to nystatin A1 (29, 45). The NMR spectra were very similar to those of nystatin A1 in DMSO-d6 (data not shown) (29) except for the C-26-C-30 fragment, whose signals overlapped with those of the ethylenes C-23-C-25 (Table 1). This is therefore consistent with a nystatin A1 structure (see Fig. 4A), including the stereostructure of the chiral carbons, for which the C-28-C-29 saturated carbons that separate the diene and tetraene would be dehydrogenated, resulting in a heptaenic structure with an MW of 924, as confirmed by mass spectrometry. The loss of those two hydrogens was clearly demonstrated by the disappearance of the 27-H/28-H and 29-H/30-H correlation peaks from the ethylene/methylene (6 to 6.5 ppm/2 to 2.5 ppm) region of the DQF-COSY (Fig. 3) or TOCSY spectra. Overlapping of the 28-H and 29-H signals hampered the observation of any 3J coupling constant or ROE spectroscopy correlation that could have assessed the Z or E configuration of the C-28-C-29 double bond. However, analysis of the UV-visible spectrum indicates an E configuration, leading therefore to an all-trans set of seven conjugated double bonds (Fig. 4D).
TABLE 1.
1H and 13C chemical shifts of S44HP and NYST1068 in DMSO-d6 and of NYST1070 in methanol-d4 at 25°Ca
| Position | S44HP
|
NYST1068
|
NYST1070
|
|||
|---|---|---|---|---|---|---|
| δ1H (ppm) | δ 13C (ppm) | δ 1H (ppm) | δ 13C (ppm) | δ 1H (ppm) | δ 13C (ppm) | |
| 2 | 2.14 | 42.9 | 2.20 | 43.0 | 2.46 | 42.3 |
| 3 (3-OH) | 4.04 (4.69) | 66.2 | 4.04 (4.75) | 66.6 | 4.21 | 67.2 |
| 4 | 1.41 | 45.3 | 1.42 | 45.4 | 1.64 | 43.9 |
| 5 (5-OH) | 3.77 (5.08) | 70.2 | 3.81 (5.06) | 70.5 | 4.03 | 70.0 |
| 6 | 1.34 | 45.4 | 1.35 | 45.4 | 1.58 | 44.0 |
| 7 (7-OH) | 3.57 (4.83) | 71.1 | 3.60 (4.81) | 71.1 | 3.81 | 71.1 |
| 8-9 | 1.42 | 28.1 | 1.42 | 28.4 | 1.64 | 29.2 |
| 10 (10-OH) | 3.03 (4.36) | 74.1 | 3.07 (4.40) | 74.4 | 3.34 | 74.4 |
| 11 (11-OH) | 3.78 (4.91) | 71.6 | 3.79 (4.89) | 71.7 | 4.06 | 71.1 |
| 12ax; 12eq | 1.38; 1.81 | 43.1 | 1.42; 1.77 | 43.8 | 1.64; 1.87 | |
| 13 (13-OH) | (6.07) | (6.02) | ||||
| 14ax; 14eq | 1.12; 1.85 | 45.1 | 1.14; 1.87 | 45.3 | 1.32; 2.04 | |
| 15 (15-OH) | 3.98 | 66.1 | 3.99 (4.83) | 66.2 | 4.34 | 77.0 |
| 16 | 1.92 | 57.9 | 2.00 | 57.9 | 2.04 | 60.6 |
| 17 | 4.19 | 65.9 | 4.17 | 66.1 | 4.29 | |
| 18ax; 18eq | 1.52; 2.03 | 37.3 | 1.58; 1.98 | 1.73; 2.15 | ||
| 19 | 4.38 | 75.5 | 4.41 | 75.0 | 4.44 | 78.2 |
| 20 | 5.90 | 5.91 | 5.91 | |||
| 21 | 6.07 | 6.11 | 6.21 | |||
| 22 | 6.34 | 6.34 | 6.31 | |||
| 23-25 | ||||||
| 26 | 6.14 | |||||
| 27 | 5.72 | |||||
| 28 | 2.21 | 32.6 | ||||
| 29 | 2.24 | 32.5 | ||||
| 30 | 5.59 | |||||
| 31 | 6.14 | 6.15 | 5.98 | |||
| 32 | 6.04 | 6.01 | 5.99 | |||
| 33 | 5.41 | 5.51 | 5.46 | |||
| 34 | 2.26 | 43.3 | 2.42 | 2.48 | 39.1 | |
| 34-CH3 | 1.01 | 19.4 | 0.94 | 18.7 | 1.03 | 17.5 |
| 35 (35-OH) | 3.06 (4.78) | 77.9 | 3.36 | 3.60 | 85.3 | |
| 36 | 1.70 | 40.5 | 1.88 | 2.14 | 42.9 | |
| 36-CH3 | 0.89 | 12.9 | 0.92 | 12.0 | 0.99 | 10.9 |
| 37 | 5.19 | 69.7 | 4.98 | 71.9 | 4.98 | 72.5 |
| 37-CH3 | 1.08 | 17.8 | 1.15 | 18.7 | 1.22 | 17.8 |
| 1′ | 4.38 | 96.6 | 4.38 | 96.3 | 4.59 | 98.4 |
| 2′ (2′-OH) | 3.75 | 68.2 | 3.77 (5.44) | 68.0 | 4.01 | 68.1 |
| 3′ (3′-NH2) | 2.89 | 56.0 | 2.95 (7.96) | 55.9 | 3.17 | 56.0 |
| 4′ (4′-OH) | 3.19 (5.60) | 69.8 | 3.25 (5.65) | 69.5 | 3.37 | 69.5 |
| 5′ | 3.17 | 73.6 | 3.17 | 73.8 | 3.31 | 73.6 |
| 6′ | 1.16 | 18.5 | 1.17 | 18.6 | 1.29 | 16.7 |
| 1′′ | 4.75 | 100.8 | 4.90 | 100.1 | ||
| 2′′ | 1.93; 1.72 | 43.1 | 2.03; 1.84 | 41.5 | ||
| 3′′ (3′′-OH) | (3.65) | 70.0 | 70.3 | |||
| 3′′-CH3 | 1.11 | 28.3 | 1.21 | 25.2 | ||
| 4′′ (4′′-OH) | 2.83 (4.43) | 78.0 | 2.96 | 76.8 | ||
| 5′′ | 3.81 | 67.0 | 3.92 | 65.9 | ||
| 6′′ | 1.13 | 19.6 | 1.26 | 17.3 | ||
The accuracies of the chemical shifts measured are ±0.02 ppm for 1H and ±0.2 ppm for 13C. Ax and eq, the pseudo-axial and pseudo-equatorial orientations relative to the average plane of the macrocycle, respectively.
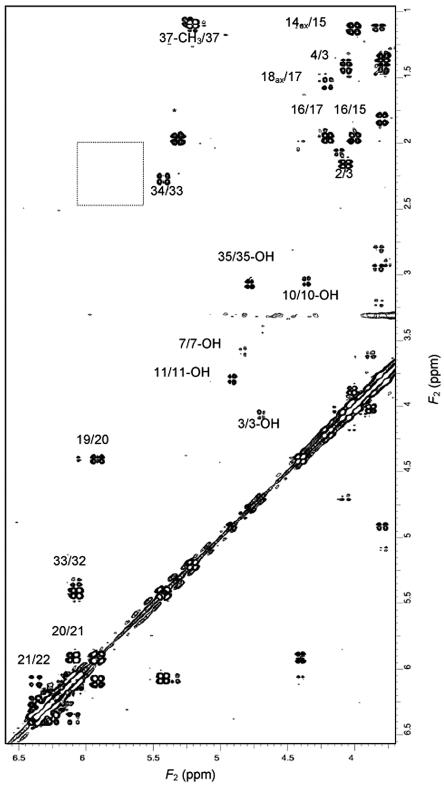
FIG. 3.
Region of the spectrum of S44HP determined by DQF-COSY spectroscopy in DMSO-d6 at 25°C. The empty square shows the absence of 27-H/28-H and 29-H/30-H correlations in the ethylene/methylene region. The asterisk indicates a peak from an impurity.
Some exchangeable protons were also observed. They could be easily identified, as they tended to disappear upon presaturation of the residual water signal or addition of a small amount of D2O. They were unambiguously assigned to 3-OH, 5-OH, 7-OH, 10-OH, 11-OH, 13-OH, 35-OH, and 4′-OH protons (Table 1) thanks to the correlation peaks with the proton attached to the same carbon observed in the spectra obtained by DQF-COSY (Fig. 3), TOCSY, and ROE spectroscopy.
(ii) NYST1068.
NYST1068 was studied as described above in DMSO-d6 at 25°C. The NMR spectra of NYST1068 exhibited features very similar to those of S44HP, with chemical shifts variations within ±0.1 ppm for 1H and ±1 ppm for 13C for most resonances of the aglycone and the mycosamine (Table 1; Fig. 5). 1H chemical shift variations larger than 0.1 ppm were observed only for 34-H, 35-H, 36-H, and 37-H, while the C-34, C-35, C-36, and C-37 13C chemical shifts could not be determined due to the absence of a correlation peak in the spectrum obtained by 13C-HSQC. Exchangeable proton resonances of 3-OH, 5-OH, 7-OH, 10-OH, 11-OH, 13-OH, 15-OH, 2′-OH, 4′-OH, and 3′-NH2 (Table 1) could be identified in the two-dimensional homonuclear spectra. The only missing hydroxyl proton was 35-OH, which was clearly observed in the S44HP spectra. These findings indicate that NYST1068 conserves the S44HP aglycone stereostructure with the mycosamine at C-19 and is likely to be glycosylated at 35-OH by a second sugar moiety that gives an extra MW of 144, according to mass spectroscopy data.
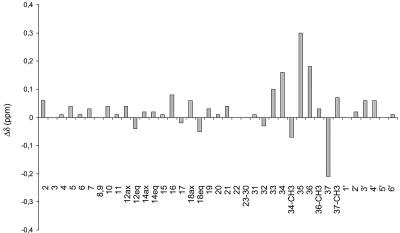
FIG. 5.
1H chemical shift variations between NYST1068 and S44HP for the aglycone and mycosamine nonexchangeable protons. ax and eq, pseudo-axial and pseudo-equatorial orientations relative to the average plane of the macrocycle, respectively.
Two additional spin systems were indeed observed in the spectra obtained by TOCSY and COSY for the NYST1068 compound. The first spin system consisted of three protons, a CH proton, a1 (1H = 4.75 ppm; 13C = 100.8 ppm), coupled to two CH2 geminal protons, a2 and a3 (1H = 1.93, 1.72 ppm; 13C = 43.1 ppm), as inferred from the spectra obtained by 13C-HSQC and their coupling constant of 14 Hz. The second spin system contained one CH proton, b1 (1H = 3.81 ppm; 13C = 67.0 ppm), coupled both to a methyl group, b2 (1H = 1.13 ppm; 13C = 19.6 ppm), and to a CH proton, b3 (1H = 2.83 ppm; 13C = 78.0 ppm), which was itself coupled to an exchangeable proton, b4 (1H = 4.43 ppm). Two additional signals, one methyl, d (1H = 1.11 ppm; 13C = 28.3 ppm), and one exchangeable proton, e (1H = 3.65 ppm), which did not belong to any spin systems were observed.
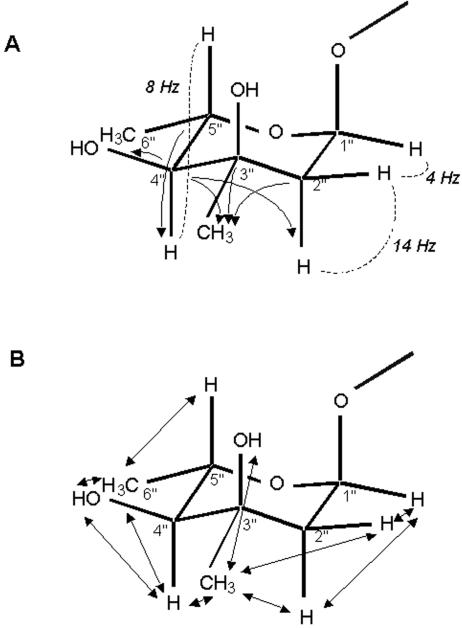
The 13C-HMBC experiment, which correlated 1H and 13C separated by two or more bonds, showed that the two spin systems, a and b, could be linked by a quaternary carbon, c (13C = 72.0 ppm), and that d was correlated to all of them. Together with the spectrum obtained by ROE spectroscopy that particularly enabled the correlation of e to d, the following sugar moiety was established (Fig. 6): the anomeric proton 1" (a1) is covalently linked to a 2"-CH2 (a2 and a3), followed by a quaternary carbon 3" (c) replaced by a methyl 3"-CH3 (d) and a substituent bearing the exchangeable proton e. Then, 4"-CH (b3), which bears the exchangeable proton b4, is linked to 5"-CH (b1), which itself is replaced by 6"-CH3 (b2). The 1H and 13C chemical shifts, as well as the MW determined by mass spectrometry, enabled identification of the exchangeable protons as hydroxyl protons for the 3"-OH (e) and 4d′-OH (b4) substituents.
FIG. 6.
Elucidation of the structure of the l-mycarose moiety. (A) Single-headed arrows show correlations going from 13C to 1H, as determined by 13C-HMBC spectroscopy. Dotted lines indicate some 1H-1H coupling constants. (B) Double-headed arrows refer to interproton ROEs.
Relative configurations were provided by analysis of the peaks obtained by ROE spectroscopy and the coupling constants. The coupling constant of 8.4 Hz observed between 4"-CH and 5"-CH indicated an axial-axial configuration. The 4"-CH/3"-CH3 peak obtained by ROE spectroscopy therefore suggested that 3"-CH3 is in an equatorial position. Thus, the second sugar was finally identified as mycarose (2,6-dideoxy-3-C-methyl-ribohexopyranose) (Fig. 6), which was confirmed by the good agreement of our NMR data with the data in the literature (26, 39, 42). An α configuration for the glycosidic bond was suggested by the weak coupling of 4 Hz observed between the 1"-CH and 2"-CHax (where ax indicates a pseudo-axial orientation) protons and by the absence of correlations with 3"-OH or 5"-CH by ROE spectroscopy. Nevertheless, the question of whether it was an l- or a d-mycarose could not be answered directly. A nondecoupled experiment by 13C-HSQC provided an approximate value of 160 Hz for the 1" 1JC,H coupling constant, which is more consistent with an l configuration than with a d configuration, given the α-type glycosylic linkage (23). However, this remains to be confirmed, possibly by modeling of the NYST1068 structure by ROE spectroscopy between mycarose and the aglycone, which is in progress.
Several correlations between the sugar and the aglycone were indeed observed by ROE spectroscopy: 1"-CH/35-H, 1"-CH/34-CH3, 3"-OH/36-H, 3"-OH/36-CH3, 5"-H/36-CH3, and 5"-H/37-CH3. This perfectly correlates with the absence of the 35-OH proton from the NMR spectra and with the chemical shift variations relative to the spectrum of S44HP observed around C-35 (Fig. 5), confirming that the mycarose is attached at the 35-OH (Fig. 4E). The absence of C-34-C-37 in the 13C-HSQC could be due to signal broadening arising from conformational exchange at this glycosylation site.
(iii) NYST1070.
NYST1070 in MeOH-d4 was studied similarly by NMR. All 1H and 13C chemical shifts were assigned (Table 1) and compared to the spectrum of nystatin A1 in the same solvent (45). As was the case for NYST1068 compared to S44HP, the only significant spectral changes were 1H and 13C chemical shift variations in the C-34-C-37 region (data not shown). C-34-C-37 CH signals were broad but were observable in the 13C-HSQC peaks, which allowed the assignment of the 13C chemical shifts that were missing in NYST1068. The largest variations were for C-35 CH 1H (0.37 ppm) and 13C (6.7 ppm).
Spectral analysis also revealed the presence of a second sugar moiety that displayed the same NMR features as NYST1068, apart from the exchangeable hydroxyl protons that were not observable. Thus, NYST1070 was proved to have the nystatin A1 structure with an additional glycosylation by mycarose at 35-OH (Fig. 4B), which precisely corresponds to the MW of 1,070.5890 determined by MS.
Antifungal activities of newly identified polyene macrolides.
The test for activity against C. albicans was performed with the following polyene macrolides: S44HP (28,29-didehydro nystatin), NYST1068 (28,29-didehydro 35-mycarosyl nystatin), NYST1070 (35-mycarosyl nystatin), and MW 950.5102 (octaene analogue with an expanded macrolactone ring). The data obtained in this experiment (Table 2) showed that the antifungal activities of S44HP, NYST1068, and the octaene nystatin analogue are superior to that of nystatin.
TABLE 2.
MICs50 of some of the polyene macrolides characterized for C. albicans ATCC 10231 as the test organism
| Polyene macrolide | MIC50 (μg/ml) |
|---|---|
| Nystatin A1 | 0.45 |
| NYST1070 | 0.86 |
| NYST1068 | 0.23 |
| S44HP | 0.07 |
| Octaene (MW, 950) | 0.12 |
DISCUSSION
The PKSs involved in the biosynthesis of polyene macrolides are one of the most complex molecular machines responsible for the assembly of secondary metabolites in bacteria. Indeed, PKSs that provide close to 90 enzymatic activities for the synthesis of such polyene macrolides as nystatin and amphotericin B have been described (1, 13, 17). The complexity of the system for the biosynthesis of glycosylated polyene macrolides is further exemplified by post-PKS modifications, such as oxidation, glycosylation, epoxidation, and hydroxylation, which further increase the potential for chemical diversity (2). It is thus not surprising that bacteria producing polyene macrolides were reported to synthesize a complex mixture of related compounds (33). Recent analysis of the recombinant strains of S. nodosus, the producer of the amphotericin B complex, revealed that the chemical diversity of polyenes can be further increased by genetic manipulation targeted to post-PKS modification steps in producer strains (16). In the present study we have attempted to assess the diversity of the nystatin-related polyene macrolides produced by both wild-type S. noursei and its mutant with an altered PKS.
We have been able to determine the accurate MWs of at least eight nystatin-related polyene macrolides produced by the wild-type strain. The latter data, together with the knowledge of the nystatin biosynthetic pathway, allowed us to predict the structural identities of these metabolites. As reported earlier (14), a heptaene macrolide with an MW of 924.4960, which corresponds to the nystatin analogue with a C-28-C-29 double bond, was identified. It seems plausible that such an analogue is produced due to the malfunction of the enoyl reductase domain in module 5 of the nystatin PKS, the situation reminiscent of that in S. nodosus, which produces a mixture of amphotericin A (tetraene) and amphotericin B (heptaene) (17). One of the major components in the nystatin complex, NYST1070, for which no structural data have been available so far, was purified and analyzed by NMR.
The molecular structure of NYST1070 has been elucidated (Fig. 4B). From our NMR data, the stereostructure of nystatin A1 is conserved in NYST1070 and, presumably, in nystatin A3. However, both NYST1070 and A3 (49) are additionally glycosylated by a 2,6-dideoxy sugar at C-35 on the side opposite that of the d-mycosamine attached at C-19 (Fig. 4B and C). The extra sugar is either l-mycarose or l-digitoxose (49) for NYST1070 and nystatin A3, respectively. Interestingly, these two 2,6-dideoxy sugars differ only by the absence of a methyl substituent at 3" in digitoxose. This, as well as our failure to identify considerable amounts of nystatin A3 in the extract from the wild-type strain, suggests that the latter metabolite might accumulate due to the failure of a specific 3-C-methyltransferase to modify the digitoxose moiety. Apparently, this was not the case for the cultures grown under the conditions used in this study.
The presence of a mycarose moiety on the polyene macrolides has never been described before, at least to our knowledge. However, l- or d-mycarose has been found in other polyketide antibiotic molecules with antibacterial activities, such as mithramycin A (25, 48), tylosin (4), and platenolide glycosides isolated from S. hygroscopicus (26). Studies of erythromycin A biosynthesis also showed the presence of l-mycarose in some intermediates (erythromycins C and D), which is later converted to l-cladinose (44). Biosynthesis of mycarose has been investigated in detail for erythromycin (40, 44), mithramycin (25), and tylosin (4, 19). Biosynthetic pathways to mycarose were proposed, and the genes involved in the various steps were identified in gene clusters, particularly (S)-adenosylmethionine-dependent 3-C-methyltransferases for tylosin (tylCIII) (20) and erythromycin (eryBIII) (40). Genes for the biosynthesis of l-digitoxose, a sugar moiety of jadomycin B produced by S. venezuelae (46), showed similarities with those involved in mycarose biosynthesis in the erythromycin, mithramycin, and tylosin pathways. It thus seems likely that in S. noursei mycarose biosynthesis and digitoxose biosynthesis share common steps and digitoxose may be a shunt product of mycarose synthesis, as has been suggested for platenolide glycosides that are glycosylated with either α-l-mycarose or β-l-digitoxose (26). It remains unclear which glycosyltransferase is responsible for the appearance of the mycarose moiety on the NYST1070 molecule. This question will be answered, at least with regard to NysDI, by inactivation of the corresponding mycosaminyl transferase gene in S. noursei, which is under way.
The ERD44 mutant of S. noursei, which contained a deletion affecting the enoyl reductase domain in module 5 of the PKS NysC, was also shown to produce a complex mixture of nystatin-related polyene macrolides. We were able to identify six nystatin analogues produced by this mutant. As expected, the heptaene analogue S44HP was one of the major polyene products produced by this mutant. Remarkably, ERD44 also produced other polyene macrolides that apparently contained four and eight conjugated double bonds.
The structures of two nystatin analogues produced by mutant ERD44 have been determined. Apparently, in S44HP and NYST1068 the C-28-C-29 double bond appears to be due to the absence of enoyl reduction after the fifth condensation step in the nystatin polyketide chain assembly, which leads to heptaenic versions of nystatin A1 and NYST1070, respectively. This is parallel to the case for amphotericin B (31), which is the heptaenic version of amphotericin A (43). Amphotericin A is coproduced with amphotericin B and differs from nystatin A1 only by the positions of two hydroxyl groups in the polyol region.
Our analysis of an array of the polyene macrolides synthesized by S. noursei by using the nystatin biosynthetic apparatus highlights the potential of polyene producers in terms of making structurally related but diverse metabolites with significantly different antimicrobial activities. In addition, this information provides new insights into the functionalities of PKSs that can be used for rational metabolic engineering aimed at the efficient synthesis of certain nystatin analogues. The new polyene macrolides discovered during this study, especially those with high levels of antifungal activity, might be useful for the generation of novel anti-infective agents for human therapy.
Acknowledgments
This work was supported by the Research Council of Norway. The NMR studies were supported by the French Ministère Délégué à la Recherche et aux Nouvelles Technologies-CNRS (UMR 5180) and Région Rhône-Alpes of France (Thématique Prioritaire de Recherche 2003-2006 Sciences Analytiques Appliquées, operations 03 01 386701/702).
REFERENCES
- 1.Aparicio, J. F., R. Fouces, M. V. Mendes, N. Olivera, and J. F. Martín. 2000. A complex multienzyme system encoded by five polyketide synthase genes is involved in the biosynthesis of the 26-membered polyene macrolide pimaricin in Streptomyces natalensis. Chem. Biol. 7:895-905. [DOI] [PubMed] [Google Scholar]
- 2.Aparicio, J. F., P. Caffrey, J. A. Gil, and S. B. Zotchev. 2003. Polyene antibiotic biosynthesis gene clusters. Appl. Microbiol. Biotechnol. 61:179-188. [DOI] [PubMed] [Google Scholar]
- 3.Bachmann, P., W. P. Aue, L. Müller, and R. R. Ernst. 1977. Phase separation in two-dimensional spectroscopy. J. Magn. Reson. 28:29-39. [Google Scholar]
- 4.Bate, N., A. R. Butler, I. P. Smith, and E. Cundliffe. 2000. The mycarose-biosynthetic genes of Streptomyces fradiae, producer of tylosin. Microbiology 146:139-146. [DOI] [PubMed] [Google Scholar]
- 5.Bax, A., and D. G. Davis. 1985. Practical aspects of two-dimensional transverse NOE spectroscopy. J. Magn. Reson. 63:207-213. [Google Scholar]
- 6.Bax, A., and M. F. Summers. 1986. 1H and 13C assignments from sensitivity-enhanced detection of heteronuclear multi-bound connectivity by 2D multiple quantum NMR. J. Am. Chem. Soc. 108:2093-2094. [Google Scholar]
- 7.Bodenhausen, G., and D. J. Ruben. 1980. Natural abundance nitrogen-15 NMR by enhanced heteronuclear spectroscopy. J. Phys. Lett. 69:185-189. [Google Scholar]
- 8.Bolard, J. 1986. Interaction of polyene antibiotics with membrane lipids: physicochemical studies of the molecular basis of selectivity. Drugs Exp. Clin. Res. 12:613-618. [PubMed] [Google Scholar]
- 9.Bolard, J. 1986. How do the polyene macrolide antibiotics affect the cellular membrane properties? Biochim. Biophys. Acta 864:257-304. [DOI] [PubMed] [Google Scholar]
- 10.Borowski, E., J. Zielinski, L. Falkowski, T. Ziminski, J. Golik, P. Kolodziejczyk, E. Jereczek, M. Gdulewicz, Y. Shenin, and T. Kotientko. 1971. The complete structure of the polyene macrolide nystatin A1. Tetrahedron Lett. 60:685-692. [Google Scholar]
- 11.Bothner-By, A., R. L. Stephens, J. M. Lee, C. D. Warren, and R. W. Jeanloz. 1984. Structure determination of a tetrasaccharide: transient nuclear Overhauser effects in the rotating frame. J. Am. Chem. Soc. 106:811-813. [Google Scholar]
- 12.Braunschweiler, L., and R. R. Ernst. 1983. Coherence transfer by isotropic mixing: application to proton correlation spectroscopy. J. Magn. Reson. 53:521-528. [Google Scholar]
- 13.Brautaset, T., O. N. Sekurova, H. Sletta, T. E. Ellingsen, A. R. Strøm, S. Valla, and S. B. Zotchev. 2000. Biosynthesis of the polyene antifungal antibiotic nystatin in Streptomyces noursei ATCC 11455: analysis of the gene cluster and deduction of the biosynthetic pathway. Chem. Biol. 7:395-403. [DOI] [PubMed] [Google Scholar]
- 14.Brautaset, T., P. Bruheim, H. Sletta, L. Hagen, T. E. Ellingsen, A. R. Strøm, S. Valla, and S. B. Zotchev. 2002. Hexaene derivatives of nystatin produced as a result of an induced rearrangement within the nysC polyketide synthase gene in S. noursei ATCC 11455. Chem. Biol. 9:367-373. [DOI] [PubMed] [Google Scholar]
- 15.Brautaset, T., S. E. Borgos, H. Sletta, T. E. Ellingsen, and S. B. Zotchev. 2003. Site-specific mutagenesis and domain substitutions in the loading module of the nystatin polyketide synthase, and their effects on nystatin biosynthesis in Streptomyces noursei. J. Biol. Chem. 278:14913-14919. [DOI] [PubMed] [Google Scholar]
- 16.Byrne, B., M. Carmody, E. Gibson, B. Rawlings, and P. Caffrey. 2003. Biosynthesis of deoxyamphotericins and deoxyamphoteronolides by engineered strains of Streptomyces nodosus. Chem. Biol. 10:1215-1224. [DOI] [PubMed] [Google Scholar]
- 17.Caffrey, P., S. Lynch, E. Flood, S. Finnan, and M. Oliynyk. 2001. Amphotericin biosynthesis in Streptomyces nodosus: deductions from analysis of polyketide synthase and late genes. Chem. Biol. 8:713-723. [DOI] [PubMed] [Google Scholar]
- 18.Cavanagh, J., and M. Rance. 1990. Sensitivity enhancement in isotropic mixing (TOCSY) experiments. J. Magn. Reson. 88:72-85. [Google Scholar]
- 19.Chen, H., G. Agnihotri, Z. Guo, N. L. S. Que, X. H. Chen, and H.-W. Liu. 1999. Biosynthesis of mycarose: isolation and characterization of the enzymes involved in the C-2 deoxygenation. J. Am. Chem. Soc. 121:8124-8125. [Google Scholar]
- 20.Chen, H., Z. Zhao, T. M. Hallis, Z. Guo, and H.-W. Liu. 2001. Insights into the branched-chain formation of mycarose: methylation catalyzed by an (S)-adenosylmethionine-dependent methyltransferase. Angew. Chem. Int. Ed. 40:607-610. [DOI] [PubMed] [Google Scholar]
- 21.Chong, C. N., and R. W. Rickards. 1970. Macrolide antibiotics studies. XVI. The structure of nystatin. Tetrahedron Lett. 59:5145-5148. [DOI] [PubMed] [Google Scholar]
- 22.Davies, D. G., and A. Bax. 1985. Assignment of complex 1H NMR spectra via two-dimensional homonuclear Hartmann-Hahn spectroscopy. J. Am. Chem. Soc. 107:2820-2821. [Google Scholar]
- 23.Duus, J., C. H. Gotfredsen, and K. Bock. 2000. Carbohydrate structural determination by NMR spectroscopy: modern methods and limitations. Chem. Rev. 100:4589-4614. [DOI] [PubMed] [Google Scholar]
- 24.Garrett, D. S., R. Powers, A. M. Gronenborn, and G. M. Clore. 1991. A common-sense approach to peak picking in 2-dimensional, 3-dimensional, and 4-dimensional spectra using automatic computer analysis of contour diagrams. J. Magn. Res. 95:214-220. [DOI] [PubMed] [Google Scholar]
- 25.González, A., L. L. Remsing, F. Lombó, M. J. Fernández, L. Prado, A. F. Braña, E. Künzel, J. Rohr, C. Méndez, and J. A. Salas. 2001. The mtmVUV genes of the mithramycin gene cluster in Streptomyces argillaceus are involved in the biosynthesis of the sugar moieties. Mol. Gen. Genet. 264:827-835. [DOI] [PubMed] [Google Scholar]
- 26.Gräfe, U., W. Schade, W. Ihn, G. Reinhardt, K. Dornberger, H. Thrum, and L. Radics. 1980. Isolation and structures of nitrogen-free platenolide glycosides. II. The 5-O-(α-mycarosyl)-5-O-(3′-demethyl-β-mycarosyl)-platenolides I and II. J. Antibiot. 33:574-578. [DOI] [PubMed] [Google Scholar]
- 27.He, J., and C. Hertweck. 2003. Iteration as programmed event during polyketide assembly: molecular analysis of the aureothin biosynthesis gene cluster. Chem. Biol. 10:1225-1232. [DOI] [PubMed] [Google Scholar]
- 28.Hutchinson, C. R., and R. McDaniel. 2001. Combinatorial biosynthesis in microorganisms as a route to new antimicrobial, antitumor and neuroregenerative drugs. Curr. Opin. Investig. Drugs 2:1681-1690. [PubMed] [Google Scholar]
- 29.Lancelin, J.-M., and J.-M. Beau. 1989. Complete stereostructure of nystatin A1: a proton study. Tetrahedron Lett. 30:4521-4524. [Google Scholar]
- 30.Marion, D., M. Ikura, R. Tschudin, and A. Bax. 1989. Rapid recordering of 2D NMR spectra without phase cycling. Application to the study of hydrogen exchange in proteins. J. Magn. Reson. 85:393-399. [Google Scholar]
- 31.Mechlinski, W., C. P. Schaffner, P. Ganis, and G. Avitabile. 1970. Structure and absolute configuration of the polyene macrolide antibiotic amphotericin B. Tetrahedron Lett. 44:3873-3876. [Google Scholar]
- 32.Mitrofanova, V. G., I. D. Shenin, O. A. Mirgorodskaia, A. A. Derzhavets, E. R. Matveeva, G. E. Grinberg, I. M. Apter, and L. S. Golubkova. 1991. Determination of nystatin component composition using HPLC and TLC with densitometry. Antibiot. Khimioter. 36:9-11. [PubMed] [Google Scholar]
- 33.Omura, S., and H. Tanaka. 1984. Production, structure, and antifungal activity of polyene macrolides, p. 351-405, In S. Omura (ed.), Macrolide antibiotics: chemistry, biology, and practice. Academic Press, Inc., New York, N.Y.
- 34.Ostrosky-Zeichner, L., S. Bazemore, V. L. Paetznick, J. R. Rodriguez, E. Chen, T. Wallace, P. Cossum, and J. H. Rex. 2001. Differential antifungal activity of isomeric forms of nystatin. Antimicrob. Agents Chemother. 45:2781-2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Palmer, A. G., III, J. Cavanagh, P. E. Wright, and M. Rance. 1991. Sensivity improvement in proton-detected two-dimensional correlation NMR spectroscopy. J. Magn. Reson. 93:151-170. [Google Scholar]
- 36.Pons, J. L., T. E. Malliavin, and M. A. Delsuc. 1996. GIFA V. 4. A complete package for NMR data set processing. J. Biomol. NMR 8:445-452. [DOI] [PubMed] [Google Scholar]
- 37.Prandi, J., and J.-M. Beau. 1989. Stereostructure of nystatin A1: a synthetic assignment of the C1-C10 fragment. Tetrahedron Lett. 30:4517-4520. [Google Scholar]
- 38.Rance, M., O. W. Sorensen, G. Bodenhausen, G. Wagner, R. R. Ernst, and K. Wüthrich. 1983. Improved spectral resolution in COSY 1H NMR spectra of proteins via double quantum filtering. Biochem. Biophys. Res. Commun. 117:479-485. [DOI] [PubMed] [Google Scholar]
- 39.Roush, W. R., and S. M. Hagadorn. 1985. Synthesis of mycarose and epi-axenose from non-carbohydrate precursors. Carbohydr. Res. 136:187-193. [DOI] [PubMed] [Google Scholar]
- 40.Salah-Bey, K., M. Doumith, J.-M. Michel, S. Haydock, J. Cortés, P. F. Leadlay, and M.-C. Raynal. 1998. Targeted gene inactivation for the elucidation of deoxysugar biosynthesis in the erythromycin producer Saccharopolyspora erythraea. Mol. Gen. Genet. 257:542-553. [DOI] [PubMed] [Google Scholar]
- 41.Sekurova, O., H. Sletta, T. E. Ellingsen, S. Valla, and S. B. Zotchev. 1999. Molecular cloning and analysis of a pleiotropic regulatory gene locus from the nystatin producer Streptomyces noursei ATCC 11455. FEMS Microbiol. Lett. 177:297-304. [DOI] [PubMed] [Google Scholar]
- 42.Shimauchi, Y., K. Kubo, K. Osumi, K. Okamura, Y. Fukagawa, and T. Ishikura. 1979. Deltamycins, new macrolide antiobiotics. III. Chemical structures. J. Antibiot. 32:878-883. [DOI] [PubMed] [Google Scholar]
- 43.Sowinski, P., J. Pawlak, E. Borowski, and T. Iwashita. 1985. Structure of amphotericin A. II. The complete structure of the antibiotic. J. Antibiot. 38:175-180. [DOI] [PubMed] [Google Scholar]
- 44.Summers, R. G., S. Donadio, M. J. Staver, E. Wendt-Pienkowski, C. R. Hutchinson, and L. Katz. 1997. Sequencing and mutagenesis of genes from the erythromycin biosynthetic gene cluster of Saccharopolyspora erythraea that are involved in l-mycarose and d-desosamine production. Microbiology 143:3251-3262. [DOI] [PubMed] [Google Scholar]
- 45.Volpon, L., and J.-M. Lancelin. 2002. Solution NMR structure of five representative glycosylated polyene macrolide antibiotics with a sterol-dependent antifungal activity. Eur. J. Biochem. 269:1-9. [DOI] [PubMed] [Google Scholar]
- 46.Wang, L., R. L. White, and L. C. Vining. 1997. Biosynthesis of the dideoxysugar component of jadomycin B: genes in the jad cluster of Streptomyces venezuelae ISP5230 for l-digitoxose assembly and transfer to the angucycline aglycone. Microbiology 14:1091-1103. [DOI] [PubMed] [Google Scholar]
- 47.Wilkinson, B., G. Foster, B. A. Rudd, N. L. Taylor, A. P. Blackaby, P. J. Sidebottom, D. J. Cooper, M. J. Dawson, A. D. Buss, S. Gaisser, I. U. Bohm, C. J. Rowe, J. Cortes, P. F. Leadlay, and J. Staunton. 2000. Novel octaketide macrolides related to 6-deoxyerythronolide B provide evidence for iterative operation of the erythromycin polyketide synthase. Chem. Biol. 7:111-117. [DOI] [PubMed] [Google Scholar]
- 48.Wohlert, S. E., E. Künzel, R. Machinek, C. Méndez, J. A. Salas, and J. Rohr. 1999. The structure of mithramycin reinvestigated. J. Nat. Prod. 62:119-121. [DOI] [PubMed] [Google Scholar]
- 49.Zielinski, J., J. Golik, J. Pawlak, E. Borowski, and L. Falkowski. 1988. The structure of nystatin A3, a component of nystatin complex. J. Antibiot. 41:1289-1291. [DOI] [PubMed] [Google Scholar]
- 50.Zotchev, S. B. 2003. Polyene macrolide antibiotics and their applications in human therapy. Curr. Med. Chem. 10:211-223. [DOI] [PubMed] [Google Scholar]