Abstract
Little is known about mechanisms involved in high-level expression of plasmid-associated ampC genes. The sequence for blaMIR-1 has been elucidated, and the gene is not inducible. Although the sequence for the promoter (prA) that drives expression of Enterobacter cloacae chromosomal ampC is present upstream of blaMIR-1, high-level expression from blaMIR-1 is directed from a hybrid promoter (prB) located further upstream of prA. The purpose of this study was to determine the influence of each promoter on blaMIR-1 expression and β-lactam resistance. RNA expression by deletion clones with both promoters was measured and compared to that by clones in which −35 and/or −10 elements of prA and/or prB were altered. Primer extension revealed two start sites for blaMIR-1 transcription. Expression of blaMIR-1 in clones with both promoters was 171-fold higher than that in clones carrying only prA. In addition, blaMIR-1 expression from prA increased 11-fold in the presence of the prB −10 element compared to expression driven from prA alone. Ceftazidime and cefotaxime MICs increased 42- and 64-fold, respectively, for the clone expressing blaMIR-1 from both promoters compared to expression from prA alone. The upstream promoter prB of blaMIR-1 is solely responsible for high-level expression required for cefotaxime and ceftazidime resistance. These data suggest that resistance to extended-spectrum cephalosporins mediated by noninducible plasmid-associated ampC genes requires the formation of novel promoter elements that are capable of increasing ampC expression.
The AmpC or group 1, class C β-lactamase genes are found on the chromosomes of several gram-negative bacteria and are being characterized with increasing frequency on plasmids in cephalosporin-resistant clinical isolates. Genetic evidence suggests that the plasmid-associated ampC genes originated from chromosomal ampC genes that mobilized onto plasmids via mobile genetic elements (30). Several plasmid-associated ampC genes with nucleotide similarity to chromosomal ampC genes from Enterobacter spp., Morganella morganii, Citrobacter freundii, and Hafnia alvei have been identified (1, 8, 9, 17). Recently, sequencing the chromosomal ampC of Aeromonas caviae revealed greater than 96% similarity to the sequences of blaFOX-1-5 (3, 4, 7, 10, 19, 25). The remaining plasmid-associated ampC group with sequences similar to that of blaMOX-1 exhibit ∼70% identity to the genes of Aeromonas spp. (26).
When AmpC is expressed at high levels, AmpC-producing organisms become resistant to almost all β-lactam antibiotics, with the exception of cefepime, cefpirome, and the carbapenems (30). Most chromosomal ampC genes are inducible in the presence of certain agents, such as cefoxitin and imipenem (12). Inducible AmpC expression is regulated by AmpR in the presence of two other gene products, AmpD and AmpG (12, 15, 16, 20). However, AmpR does not regulate expression from the majority of the plasmid-associated ampC genes, and therefore, the mechanisms by which high-level AmpC expression from noninducible plasmid-associated ampC genes occurs remain to be elucidated. The plasmid-associated ampC genes blaDHA-1 and blaDHA-2 of M. morganii origin and blaACT-1 of Enterobacter asburiae origin are inducible, and their induction is by the general mechanism of inducible expression (2, 6, 27, 29). However, a C-to-T transition at the first position of the blaACT-1 −10 promoter element is implicated in increased expression even in the absence of induction (27, 28).
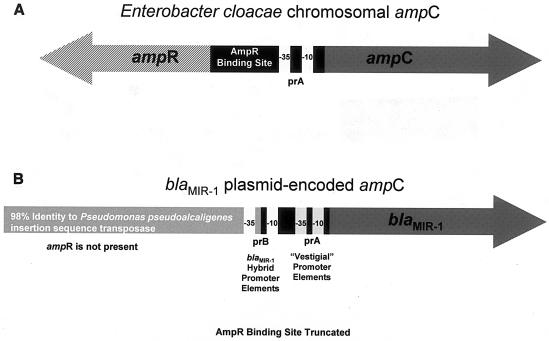
The other plasmid-carried ampC of Enterobacter origin is blaMIR-1 (14, 22). blaMIR-1 expression is not inducible, as the gene lacks the association of an upstream ampR gene and the binding site for AmpR is truncated (14, 28). The 5′ flanking sequence of blaMIR-1 retains the chromosomal ampC −35 and −10 elements (prA) and a portion of the AmpR binding site sequence (Fig. 1B) (14). Upstream of the truncated AmpR binding site is an insertion element with 96% similarity to a transposase gene nucleotide sequence from Pseudomonas pseudoalcaligenes (14).
FIG. 1.
Genetic organization of the chromosomal ampC gene of E. cloacae (A) and the plasmid-carried ampC gene blaMIR-1 (B). The E. cloacae ampC is inducible and is expressed from prA. blaMIR-1 is not inducible; however, prA remains intact, upstream of the structural gene.
A previous study reported that blaMIR-1 is expressed from a putative hybrid promoter (prB) likely formed during mobilization of blaMIR-1 from the chromosome to the plasmid (28). The joining of the upstream insertion element from P. pseudoalcaligenes with the truncated AmpR binding site created prB with the −35 element located in the insertion sequence and the −10 element located 17 bp downstream, within the remnant of the AmpR binding site (Fig. 1B) (28). In this study, the role of expression from the blaMIR-1 promoter prB and the vestigial promoter prA were investigated in relation to β-lactam antibiotic susceptibility.
(A preliminary account of this work has been presented previously [M. D. Reisbig and N. D. Hanson, Abstr. 43rd Intersci. Conf. Antimicrob. Agents Chemother., abstr. C1-676, 2003].)
MATERIALS AND METHODS
Bacterial strains and plasmids.
The bacterial strains and plasmids used in this study are listed in Tables 1 to 3.
TABLE 1.
Clinical isolates used in this study
TABLE 3.
Plasmids used in this study
| Plasmid | Characteristicsa | Size (bp) | Reference or source |
|---|---|---|---|
| pACYC184 | Cm+ Tet+ | 4,245 | 5 |
| pMDR009 | pACYC184 Δ103-515 bp; Cm− Tet+ | 3,833 | This study |
Cm, chloramphenicol; Tet, tetracycline; +, positive for selectable marker; −, negative for selectable marker.
Cloning of blaMIR-1 and deletion clones.
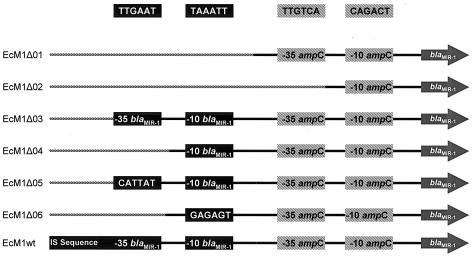
The primers used to amplify by PCR each blaMIR-1 deletion fragment are listed in Table 4. PCR was carried out as previously described to create amplicons containing the entire blaMIR-1 structural gene and various lengths of the 5′ upstream region including different components of prB and prA (Fig. 2). Table 4 identifies the PCR-amplified fragment lengths and the nucleotide positions present in each fragment and the resulting promoter changes. These changes are illustrated in Fig. 2. The seven cDNA amplicons were cloned by using the TOPO-XL cloning kit (Invitrogen). Each fragment was subcloned as an EcoRI fragment into plasmid pMDR009, a pACYC184 derivative that allows for EcoRI insertion without expression interference from the chloramphenicol acetyltransferase promoter (5). Plasmid constructs were transformed into Escherichia coli Top10 (Invitrogen) (11). The promoter inserts were manually sequenced from the plasmid with the Pfu polymerase sequencing kit as described by the manufacturer (Stratagene). The blaMIR-1 structural gene from each clone was amplified by PCR as previously described with Platinum Taq Plus DNA polymerase (28). These amplicons were analyzed by automated sequencing as previously described (23). The constructed plasmids are listed in Tables 2 and 3.
TABLE 4.
Primers used to amplify blaMIR-1 deletion fragments
| Primer | Sequenced (5′-3′) | Product name, size (bp)a | Purposeb | Nucleotidesc | GenBank accession no. | Source or reference(s) |
|---|---|---|---|---|---|---|
| MIRΔUP | GCCATACTTGGCCTCATGCG | M1Δ01, 1,524 | S, C | 786-805 | M37983 | 14 |
| MIR-1R | GCATCAAAAGGCGTGACGACG | 2310-2290 | M37839 | 14 | ||
| MIRΔM35 | CCGTTAATGCTAAATTTAACCG | M1Δ02, 1,490 | S, C | 820-841 | M37983 | 14 |
| MIR-1R | GCATCAAAAGGCGTGACGACG | 2310-2290 | M37839 | 14 | ||
| MIRΔM35M10E35 | CCAACAGACTACAGCGGTCTGACG | M1Δ03, 1,448 | S, C | 862-885 | M37983 | 14 |
| MIR-1R | GCATCAAAAGGCGTGACGACG | 2310-2290 | M37839 | 14 | ||
| MIRΔM35M10 | CGTTTGTCAGCCACAGTCAAATCC | M1Δ04, 1,471 | S, C | 839-863 | M37983 | 14 |
| MIR-1R | GCATCAAAAGGCGTGACGACG | 2310-2290 | M37839 | 14 | ||
| MIRMUTE35 | CCGTTAATGCGAGAGTTAACCCTTTG | M1Δ05, 1,490 | S, C | This study | ||
| MIR-1R | GCATCAAAAGGCGTGACGACG | 2310-2290 | M37839 | 14 | ||
| MIRMUTM35 | CCTCATGCGGCATTATTTCCTATCCG | M1Δ06, 1,513 | S, C | 797-822 | This study | |
| MIR-1R | GCATCAAAAGGCGTGACGACG | 2310-2290 | M37839 | 14 | ||
| MIRUPF | GGGAAGCAAACTGGTGTACC | M1wt, 2,302 | S, C | 008-27 | M37983 | 14 |
| MIR-1R | GCATCAAAAGGCGTGACGACG | 2310-2290 | M37839 | 14 | ||
| ENTB55PE | GGCGAGAGCAGAGCAAGAGATGCC | PE | 80-103 | X07274 | 28 | |
| MIR-1PE | GCGAATGCAGAACTGGCGACG | PE | 966-986 | M37839 | 14, 28 | |
| KpEc16SRNA | CCCAGACATTACTCACCCGTCC | PE | 82-61 | AF390084 | 27, 28 |
Size of the PCR-generated amplicon. For promoter deletions and changes see corresponding name in Fig. 2.
Purpose for the primers. S, sequencing; C, cloning; PE, primer extension.
Nucleotide location of each primer with respect to the cited reference.
Sequences that were mutated are shown in boldface.
FIG. 2.
Diagram of the 5′ flanking regions of each blaMIR-1 construct. Solid lines, wild-type sequence; hatched lines, vector sequence. prA is the vestigial promoter, a remnant of the chromosomal ampC gene of Enterobacter origin. prB is the hybrid promoter, composed of the insertion sequence (IS) and remnant portions of the 5′ flanking region of the chromosomal ampC gene. Deletions of both prA and prB components were generated to assess the contributory effects of these elements on transcription and the susceptibility phenotype. EcM1Δ05 and EcM1Δ06 harbored constructs with mutations that destroyed the −35 and −10 prB elements but kept the original nucleotide spacing. These clones were created to ensure that any changes in the observed in blaMIR-1 transcript levels were not a result of differences in promoter topography. For the specific mutations generated for these clones see Table 4.
TABLE 2.
Other strains used in this studya
| Strain | Relevant blaMIR-1 upstream sequences of the plasmidb |
|---|---|
| EcM1Δ01 | ΔIS, Δ−35prB, Δ−10prB, −35prA+, −10prA+, blaMIR-1+ |
| EcM1Δ02 | ΔIS, Δ−35prB, Δ−10prB, Δ−35prA, −10prA+, blaMIR-1+ |
| EcM1Δ03 | ΔIS, −35prB+, −10prB+, −35prA+, −10prA+, blaMIR-1+ |
| EcM1Δ04 | ΔIS, Δ−35prB, −10prB+, −35prA+, −10prA+, blaMIR-1+ |
| EcM1Δ05 | ΔIS, MUT−35prB, −10prB+, −35prA+, −10prA+, blaMIR-1+ |
| EcM1Δ06 | ΔIS, Δ−35prB, MUT-10prB, −35prA+, −10prA+, blaMIR-1+ |
| EcM1wt | IS+, −35prB+, −10prB+, −35prA+, −10prA+, blaMIR-1+ |
All plasmids were transformed into E. coli Top 10 cells (Invitrogen) [F− mcrA Δ(mrr-hsd RMS-mcrBC) φ80lacZΔM15 ΔlacX74 recA1 araD139 Δ(ara-leu)7697 galU galK rpsL (Strr) endA1 nupG] (11).
Sequences upstream of the blaMIR-1 structural gene cloned into pMDR009. IS, insertion sequence transposase; Δ, deletion; +, present, MUT, mutation inserted by PCR mutagenesis.
Primer extension analysis.
RNA isolation and primer extension analysis were carried out as previously described using 20 μg of RNA for experimental samples and 1 μg for the 16S rRNA control lanes (27). Primers used for primer extension are listed in Table 4. Extension products were visualized, normalized, and quantified as previously described (27).
MICs.
Cefoxitin, cephalothin, ampicillin, cefotaxime, and ceftazidime MICs for each strain were determined by Etest (AB Biodisk North America, Piscataway, N.J.) with a 0.5 McFarland's standard inoculum on 56-ml Mueller-Hinton agar plates incubated in ambient air for 18 h at 37°C.
RESULTS
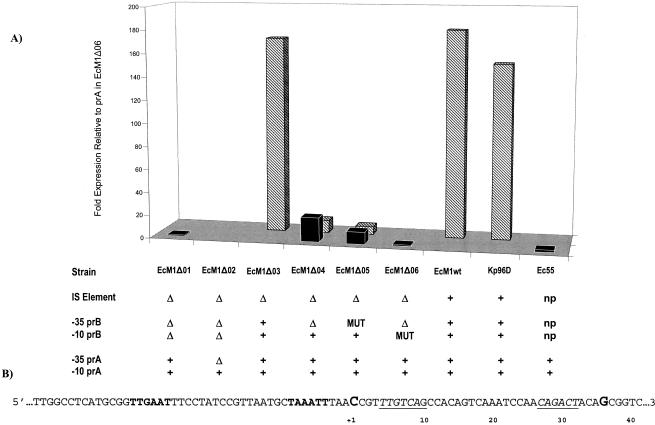
Expression levels of promoter clones were compared to the uninduced wild-type ampC expression from Enterobacter cloacae 55. ampC expression from clone EcM1Δ01 is driven by prA, which includes −10 and −35 elements identical to those found in the wild-type E. cloacae 55 promoter. As shown in Fig. 3, overall expression levels for these two strains were comparable. In addition, the start sites for ampC transcription mapped to position +36 (Fig. 3B) in each clone. As the upstream sequence was extended to include nucleotides which represented the −10 and −35 elements of prA and the −10 element of prB (EcM1Δ04), ampC expression from the prA promoter increased 11-fold compared to expression from EcM1Δ01 and E. cloacae 55 (Fig. 3). In addition, expression from the prB −10 element was seen, as determined by analysis of two ampC transcriptional start sites located at positions +1 for prB and +36 for prA (Fig. 3B). However, the increases in expression from each promoter compared to that from EcM1Δ01 and E. cloacae 55 were almost equivalent (11-fold from prA and 7-fold from prB).
FIG. 3.
(A) Transcription levels expressed relative to the expression observed from prA in EcM1Δ06, as measured by primer extension analysis. Relative expression was calculated by setting the lowest detectable expression level to 1 (prA, EcM1Δ06) after normalization and comparing all expression levels to this value. A value of zero, indicated by the absence of a bar, was given when bands were below the level of detection by primer extension analysis. Solid black bars, expression from prA; striped bars, expression from prB. The key below indicates the presence (+), deletion (Δ), or mutation (MUT) of the promoter elements listed at the left. The insertion sequence (IS) and prB are not present (np) in the E. cloacae 55 clinical isolate. (B) Partial sequence of the upstream region of blaMIR-1. The start sites of transcription are shown in the sequence map below, indicated by G for prA and by C for prB. prB is boldface, and prA is italicized and underlined.
By including the −35 element in prB, represented by clone EcM1Δ03, full blaMIR-1 expression was restored, as expression levels from this clone were comparable to expression levels from the clinical isolate Klebsiella pneumoniae 96D and the full-length clone EcM1wt (Fig. 3). Interestingly, sequence elements other than the −35 promoter element of prB, upstream within the insertion sequence (Fig. 3), did not influence blaMIR-1 expression. To ensure that positional effects due to deletions of sequence within the promoter did not influence the observed levels of expression from clones EcM1Δ01 and EcM1Δ04, clones EcM1Δ06 and EcM1Δ05, which included nonsense sequences in place of the deleted −10 and −35 elements of prB, respectively, were constructed. No difference between levels of blaMIR-1 expression from EcM1Δ01 and EcM1Δ06 (Fig. 3) or EcM1Δ04 and EcM1Δ05 (Fig. 3) was observed. Furthermore, no expression from EcM1Δ02, which contained only the −10 element for prA, was detected (Fig. 3).
The influence of these promoter mutations on the susceptibility phenotype was examined by using MICs of cefotaxime, ceftazidime, ampicillin, cefoxitin, and cephalothin determined by Etest analysis. MICs of the extended-spectrum cephalosporins for all of the clones that had mutations or deletions in any of the prA or prB promoter elements remained in the susceptible category. Etest analysis revealed ceftazidime MICs of 16 and 24 μg/ml for clones EcM1Δ03 and EcM1wt, respectively, which both expressed blaMIR-1 from prB. A significantly lower ceftazidime MIC of 0.75 μg/ml was observed for clones EcM1Δ01 and EcM1Δ06, both of which expressed blaMIR-1 from only the prA promoter. Cefotaxime MICs of 8 μg/ml, observed for clones EcM1Δ03 and EcM1wt, were also significantly higher than MICs for clones EcM1Δ01 and EcM1Δ06, which were 0.125 and 0.094 μg/ml, respectively (Table 5). Even though blaMIR-1 prA expression from clones EcM1Δ04 and EcM1Δ05 showed an ∼11-fold increase over that from EcM1Δ01 and E. cloacae 55, no significant increases in cefotaxime and ceftazidime MICs were observed. However, the cefoxitin MIC for clones EcM1Δ04 and EcM1Δ05 was 24 μg/ml, while ampicillin MICs were 32 and 12 μg/ml, respectively, and cephalothin MICs were >256 and 192 μg/ml, respectively. Cefoxitin, cephalothin, and ampicillin MICs for clones EcM1Δ03 and EcM1wt, both containing the intact prB promoter, were >256 μg/ml. Cephalothin MICs for all clones were above the resistance breakpoint. However, regardless of which β-lactam was tested, the highest MICs correlated to the clones expressing blaMIR-1 at the highest level (Table 5).
TABLE 5.
β-Lactam MICs determined by Etest for clinical isolates and bacterial transformantsa
| β-Lactam antibiotic | MIC (μg/ml) for
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| EcM1Δ01 | EcM1Δ02 | EcM1Δ03 | EcM1Δ04 | EcM1Δ05 | EcM1Δ06 | EcM1wt | 96D | 55 | E. coli Top10pMDR009b | |
| Ceftazidime | 0.75 | 0.125 | 16 | 0.75 | 0.75 | 0.75 | 24 | 32 | 0.38 | 0.125 |
| Cefotaxime | 0.125 | 0.016 | 8 | 0.094 | 0.125 | 0.094 | 8 | 32 | 0.25 | 0.016 |
| Cefoxitin | 6 | 4 | >256 | 24 | 24 | 4 | >256 | >256 | 192 | 3 |
| Cephalothin | 48 | 48 | >256 | >256 | 192 | 32 | >256 | >256 | >256 | 6 |
| Ampicillin | 6 | 6 | >256 | 32 | 12 | 4 | >256 | >256 | 64 | 4 |
DISCUSSION
The events that mobilized blaMIR-1 from the chromosome to the plasmid resulted in the formation of hybrid promoter prB, which is responsible for high-level MIR-1 expression and resistance to all the β-lactam antibiotics tested. In the process, the chromosomal promoter prA was rendered ineffective, as the upstream regulatory elements were not mobilized with the ampC structural gene onto the plasmid. Therefore, prA represents a vestigial promoter associated with blaMIR-1 as a remnant of the chromosomal ampC expression system. Although the level of expression from prA resulted in resistance to cephalothin, this level of expression was not enough to confer resistance to the other β-lactams tested in this study. These data indicate that prB is responsible for high-level expression required for resistance to the extended-spectrum cephalosporins, ceftazidime, and cefotaxime, as well as cefoxitin and ampicillin. Removal of prB from the upstream flanking region of blaMIR-1 did not aid in the ability of prA to express levels of blaMIR-1 sufficient to confer resistance to these drugs. These data suggest that, even in the presence of prA, resistance to the extended-spectrum β-lactams requires the formation of a novel promoter, such as prB, to drive high-level expression.
In the absence of the AmpR regulatory protein, model systems have shown that constitutive ampC expression increased 2.5- to 5.8-fold. However, the data presented from this study show that, in the presence of a stronger promoter, levels of expression of the −35 and −10 elements of the vestigial ampC promoter do not increase above the constitutive level observed from a wild-type strain of E. cloacae. This observation was not predicted by data from promoter gene constructs comparing expression among cloning vectors expressing chromosomal ampC genes with or without the ampR gene (18, 24).
The plasmid-associated ampC genes of Enterobacter origin as a group are distinctive in that the mechanisms driving expression are unique to each gene, blaACT-1 or blaMIR-1. The genetic context of the blaACT-1 promoter is similar to what is observed for the promoter prA of the Enterobacter sp. chromosomal ampC gene, whereas blaMIR-1 is expressed constitutively from the novel hybrid promoter prB. Although prB and prA both exhibit 67% percent identity to the overall E. coli consensus promoter sequence, the observed expression from prB is higher than that observed from prA (13). The difference between prB and prA is in the positions at which these promoters match the consensus sequence. The sequence of prB is more similar than the sequence of prA at positions that have been shown to dramatically increase expression according to studies of the E. coli promoter consensus sequence (13). The prB promoter sequence matched the consensus at position 1 of the −10 element and position 4 of the −35 element compared to that of prA, which did not match the consensus sequence at these positions. Such positional differences would allow the RNA polymerase initiation complex to recognize the prB promoter better than the prA promoter.
It is possible that, because prB expression is 170-fold higher than prA expression, quenching could play a role in promoter usage. Quenching of promoter usage occurs when a strong promoter outcompetes a weak promoter for RNA polymerase recruitment (21). The lack of blaMIR-1 expression in the absence of the upstream promoter, prB, indicated that quenching was not responsible for decreased expression from the vestigial promoter, prA.
Although the formation of new promoters that constitutively express high levels of ampC transcripts is the means by which blaMIR-1 is expressed and is probable in other plasmid-associated ampC promoters, other mechanisms for increased promoter expression cannot be ruled out. The ∼11-fold increase in expression from prA in EcM1Δ04 and EcM1Δ05 compared to expression from prA alone in EcM1Δ06 was likely the result of the prB −10 element attracting the RNA polymerase to the region. Although this increase in expression did not result in a significant change to the observed cefotaxime and ceftazidime MICs, an element with a greater ability to recruit RNA polymerase may enhance the expression of a weak promoter. The expression of blaMIR-1 in EcM1Δ04 and EcM1Δ05 increased cefoxitin and ampicillin MICs to nonsusceptible levels according to breakpoints for resistance set by the National Committee for Clinical Laboratory Standards.
The MICs for EcM1wt and K. pneumoniae 96D were comparable because the copy number of the vector did not result in a difference in relative copy number between the two plasmid-associated systems. blaMIR-1 is carried on plasmids with relative copy numbers of 12 and 11 in K. pneumoniae 96D and EcM1wt, respectively (28).
This study of the blaMIR-1 promoter has provided insight as to how noninducible plasmid-associated ampC genes are expressed. An ampC gene that mobilizes from the chromosome to a plasmid and leaves behind the genetic elements that control inducible expression must evolve some other means of high-level, constitutive expression. As indicated by this study, it is inappropriate to select putative −10 and −35 elements of a sequenced plasmid-associated ampC gene based on the location of the wild-type promoter in the gene of origin. It is likely that the formation of new promoters or the acquisition of strong promoters located within upstream insertion elements (V. L. Herrera and N. D. Hanson, Abstr. 41st Intersci. Conf. Antimicrob. Agents Chemother., abstr. 1488, 2001; M. D. Reisbig and N. D. Hanson, Abstr. 13th Eur. Congr. Clin. Microbiol. Infect. Dis., abstr. P574, 2003) is necessary in order for β-lactam resistance to be observed in an organism that expresses a plasmid-associated ampC gene in the absence of AmpR.
REFERENCES
- 1.Barnaud, G., G. Arlet, C. Danglot, and A. Philippon. 1997. Cloning and sequencing of the gene encoding the AmpC beta-lactamase of Morganella morganii. FEMS Microbiol. Lett. 148:15-20. [DOI] [PubMed] [Google Scholar]
- 2.Barnaud, G., G. Arlet, C. Verdet, O. Gaillot, P. H. Lagrange, and A. Philippon. 1998. Salmonella enteritidis: AmpC plasmid-mediated inducible beta-lactamase (DHA-1) with an ampR gene from Morganella morganii. Antimicrob. Agents Chemother. 42:2352-2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bauernfeind, A., S. Wagner, R. Jungwirth, I. Schneider, and D. Meyer. 1997. A novel class C beta-lactamase (FOX-2) in Escherichia coli conferring resistance to cephamycins. Antimicrob. Agents Chemother. 41:2041-2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bou, G., A. Oliver, M. Ojeda, C. Monzon, and J. Martinez-Beltran. 2000. Molecular characterization of FOX-4, a new AmpC-type plasmid-mediated beta-lactamase from an Escherichia coli strain isolated in Spain. Antimicrob. Agents Chemother. 44:2549-2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang, A. C., and S. N. Cohen. 1978. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J. Bacteriol. 134:1141-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fortineau, N., L. Poirel, and P. Nordmann. 2001. Plasmid-mediated and inducible cephalosporinase DHA-2 from Klebsiella pneumoniae. J. Antimicrob. Chemother. 47:207-210. [DOI] [PubMed] [Google Scholar]
- 7.Fosse, T., C. Giraud-Morin, I. Madinier, and R. Labia. 2003. Sequence analysis and biochemical characterisation of chromosomal CAV-1 (Aeromonas caviae), the parental cephalosporinase of plasmid-mediated AmpC ‘FOX’ cluster. FEMS Microbiol. Lett. 222:93-98. [DOI] [PubMed] [Google Scholar]
- 8.Galleni, M., F. Lindberg, S. Normark, S. Cole, N. Honore, B. Joris, and J. M. Frere. 1988. Sequence and comparative analysis of three Enterobacter cloacae ampC beta-lactamase genes and their products. Biochem. J. 250:753-760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Girlich, D., T. Naas, S. Bellais, L. Poirel, A. Karim, and P. Nordmann. 2000. Biochemical-genetic characterization and regulation of expression of an ACC-1-like chromosome-borne cephalosporinase from Hafnia alvei. Antimicrob. Agents Chemother. 44:1470-1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gonzalez Leiza, M., J. C. Perez-Diaz, J. Ayala, J. M. Casellas, J. Martinez-Beltran, K. Bush, and F. Baquero. 1994. Gene sequence and biochemical characterization of FOX-1 from Klebsiella pneumoniae, a new AmpC-type plasmid-mediated beta-lactamase with two molecular variants. Antimicrob. Agents Chemother. 38:2150-2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grant, S. G., J. Jessee, F. R. Bloom, and D. Hanahan. 1990. Differential plasmid rescue from transgenic mouse DNAs into Escherichia coli methylation-restriction mutants. Proc. Natl. Acad. Sci. USA 87:4645-4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hanson, N. D., and C. C. Sanders. 1999. Regulation of inducible AmpC beta-lactamase expression among Enterobacteriaceae. Curr. Pharm. Des. 5:881-894. [PubMed] [Google Scholar]
- 13.Hawley, D. K., and W. R. McClure. 1983. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res. 11:2237-2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jacoby, G. A., and J. Tran. 1999. Sequence of the MIR-1 beta-lactamase gene. Antimicrob. Agents Chemother. 43:1759-1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Korfmann, G., and C. C. Sanders. 1989. ampG is essential for high-level expression of AmpC beta-lactamase in Enterobacter cloacae. Antimicrob. Agents Chemother. 33:1946-1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lindberg, F., S. Lindquist, and S. Normark. 1987. Inactivation of the ampD gene causes semiconstitutive overproduction of the inducible Citrobacter freundii beta-lactamase. J. Bacteriol. 169:1923-1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lindberg, F., and S. Normark. 1986. Sequence of the Citrobacter freundii OS60 chromosomal ampC beta-lactamase gene. Eur. J. Biochem. 156:441-445. [DOI] [PubMed] [Google Scholar]
- 18.Lindberg, F., L. Westman, and S. Normark. 1985. Regulatory components in Citrobacter freundii ampC beta-lactamase induction. Proc. Natl. Acad. Sci. USA 82:4620-4624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marchese, A., G. Arlet, G. C. Schito, P. H. Lagrange, and A. Philippon. 1998. Characterization of FOX-3, an AmpC-type plasmid-mediated beta-lactamase from an Italian isolate of Klebsiella oxytoca. Antimicrob. Agents Chemother. 42:464-467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Normark, S., S. Lindquist, and F. Lindberg. 1986. Chromosomal beta-lactam resistance in enterobacteria. Scand. J. Infect. Dis. Suppl. 49:38-45. [PubMed] [Google Scholar]
- 21.O'Connor, M. J., S. H. Tan, C. H. Tan, and H. U. Bernard. 1996. YY1 represses human papillomavirus type 16 transcription by quenching AP-1 activity. J. Virol. 70:6529-6539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Papanicolaou, G. A., A. A. Medeiros, and G. A. Jacoby. 1990. Novel plasmid-mediated beta-lactamase (MIR-1) conferring resistance to oxyimino- and alpha-methoxy beta-lactams in clinical isolates of Klebsiella pneumoniae. Antimicrob. Agents Chemother. 34:2200-2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pitout, J. D., K. S. Thomson, N. D. Hanson, A. F. Ehrhardt, E. S. Moland, and C. C. Sanders. 1998. β-Lactamases responsible for resistance to expanded-spectrum cephalosporins in Klebsiella pneumoniae, Escherichia coli, and Proteus mirabilis isolates recovered in South Africa. Antimicrob. Agents Chemother. 42:1350-1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Poirel, L., M. Guibert, D. Girlich, T. Naas, and P. Nordmann. 1999. Cloning, sequence analyses, expression, and distribution of ampC-ampR from Morganella morganii clinical isolates. Antimicrob. Agents Chemother. 43:769-776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Queenan, A. M., S. Jenkins, and K. Bush. 2001. Cloning and biochemical characterization of FOX-5, an AmpC-type plasmid-encoded beta-lactamase from a New York City Klebsiella pneumoniae clinical isolate. Antimicrob. Agents Chemother. 45:3189-3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Raskine, L., I. Borrel, G. Barnaud, S. Boyer, B. Hanau-Bercot, J. Gravisse, R. Labia, G. Arlet, and M. J. Sanson-Le-Pors. 2002. Novel plasmid-encoded class C beta-lactamase (MOX-2) in Klebsiella pneumoniae from Greece. Antimicrob. Agents Chemother. 46:2262-2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reisbig, M. D., and N. D. Hanson. 2002. The ACT-1 plasmid-encoded AmpC beta-lactamase is inducible: detection in a complex beta-lactamase background. J. Antimicrob. Chemother. 49:557-560. [DOI] [PubMed] [Google Scholar]
- 28.Reisbig, M. D., A. Hossain, and N. D. Hanson. 2003. Factors influencing gene expression and resistance for gram-negative organisms expressing plasmid-encoded ampC genes of Enterobacter origin. J. Antimicrob. Chemother. 51:1141-1151. [DOI] [PubMed] [Google Scholar]
- 29.Rottman, M., Y. Benzerara, B. Hanau-Bercot, C. Bizet, A. Philippon, and G. Arlet. 2002. Chromosomal ampC genes in Enterobacter species other than Enterobacter cloacae, and ancestral association of the ACT-1 plasmid-encoded cephalosporinase to Enterobacter asburiae. FEMS Microbiol. Lett. 210:87-92. [DOI] [PubMed] [Google Scholar]
- 30.Thomson, K. S., and E. S. Moland. 2000. Version 2000: the new beta-lactamases of gram-negative bacteria at the dawn of the new millennium. Microbes Infect. 2:1225-1235. [DOI] [PubMed] [Google Scholar]