Abstract
Summary
Genome editing with programmable nucleases has been widely adopted in research and medicine. Next generation sequencing (NGS) platforms are now widely used for measuring the frequencies of mutations induced by CRISPR-Cas9 and other programmable nucleases. Here, we present an online tool, Cas-Analyzer, a JavaScript-based implementation for NGS data analysis. Because Cas-Analyzer is completely used at a client-side web browser on-the-fly, there is no need to upload very large NGS datasets to a server, a time-consuming step in genome editing analysis. Currently, Cas-Analyzer supports various programmable nucleases, including single nucleases and paired nucleases.
Availability and Implementation
Free access at http://www.rgenome.net/cas-analyzer/.
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
Programmable nucleases such as zinc finger nucleases (ZFNs), transcription-activator-like effector nucleases (TALENs), and RNA-guided endonucleases derived from CRISPR-Cas9/Cpf1 systems, which are adaptive immune responses in bacteria and archaea, are widely used for genome editing in many research fields including biology, biotechnology, agriculture, and medical science (Kim and Kim, 2014). The type II Cas9 nuclease from Streptococcus pyogenes (SpCas9) was the first CRISPR nuclease used for genome editing (Cho et al., 2013; Cong et al., 2013; Jinek et al., 2013; Mali et al., 2013); since that time, various orthogonal Cas9 nucleases such as StCas9 (Cong et al., 2013), NmCas9 (Hou et al., 2013) and SaCas9 (Ran et al., 2015) have been developed. Recently, putative type V Cpf1 nucleases from Acidominococcus and Lachnospiraceae were reported to mediate efficient genome editing in human cells (Kim et al., 2016a; Zetsche et al., 2015) and mice (Hur et al., 2016; Kim et al., 2016b). Moreover, dimeric CRISPR nucleases such as RNA-guided nickases (Cho et al., 2014; Ran et al., 2013) and RNA-guided FokI nucleases (Tsai et al., 2014), or biochemical improvement of wild-type SpCas9 (Kleinstiver et al., 2016; Slaymaker et al., 2016) have been developed for genome editing to reduce off-target effects.
Programmable nucleases introduce DNA double-strand breaks at user-defined target sites in the genome, ultimately inducing targeted gene knockout or knock-in via the cell’s own repair systems [error-prone non-homologous end joining or homology-directed repair (HDR) in the presence of a DNA template, respectively]. The induced mutation rates in cells can be estimated in a straightforward manner by using Surveyor nuclease (Perez et al., 2008), the T7 endonuclease I (T7E1) assay (Kim et al., 2009), polyacrylamide gel electrophoresis (Zhu et al., 2014) or droplet digital PCR (Nelson et al., 2016). However, these methods do not allow analysis of mutant sequences and are limited by relatively poor sensitivity. Recently we and other groups have used targeted deep sequencing to detect programmable nuclease-induced mutations with high sensitivity and precision and to analyze mutation patterns (Baek et al., 2016).
However, analysis of next generation sequencing (NGS) data is difficult for many researchers. Although a few web-based tools such as CRISPR-GA (Güell et al., 2014), AGEseq (Xue and Tsai, 2015) and CRISPResso (Pinello et al., 2016) are available, they are inconvenient to use because their web interfaces require a very long time to upload large data files (Supplementary Material S1). AGEseq and CRISPResso also support a command-line interface, but they are not accessible to researchers who are not familiar with bioinformatics. To address this issue, we present a web-based tool, Cas-Analyzer that is constructed with a JavaScript-based algorithm; thus, it wholly runs on the client-side so that large amounts of sequencing data do not need to be uploaded to the server. Thanks to the improvements in the newest JavaScript engines in the most recent web browsers (Supplementary Table S1), this tool works in a reasonable time. Currently, Cas-Analyzer supports a variety of nucleases, including single nucleases (SpCas9, StCas9, NmCas9, SaCas9, CjCas9 and AsCpf1/LbCpf1) and paired nucleases (ZFNs, TALENs, Cas9 nickases and dCas9-FokI nucleases).
2 Implementation
2.1 File loading
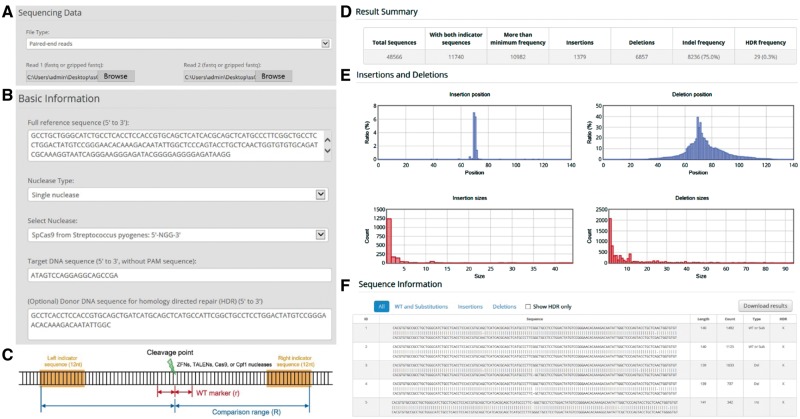
To use Cas-Analyzer, deep sequencing data are needed, which can be obtained by amplifying the target locus of genome edited cells (Supplementary Material S2) followed by NGS. The format of the raw output data is usually Fastq or gzip-compressed, and both data types are acceptable to Cas-Analyzer (Fig. 1A). For the compressed files, we used a JavaScript library ‘pako’ (http://nodeca.github.io/pako/), which is slightly modified to support blocked gzip files. If users upload paired-end sequencing data, Cas-Analyzer first merges paired-end reads by the JavaScript port of Fastq-join, a part of ea-utils (https://code.google.com/archive/p/ea-utils/).
Fig. 1.
Overview of Cas-Analyzer. (A) Uploading NGS data files. Single-end reads, paired-end reads, or already merged sequencing data are allowed. (B) Basic information about the query sequences are required for using Cas-Analyzer. (C) Indicators used in the analysis step. (D) The results are summarized as a table that includes the mutation count and frequency. (E) Insertions and deletions are also visualized as graphs. (F) All filtered sequences from the input data are aligned with the reference sequence
2.2 Data analysis
Cas-Analyzer analyzes the uploaded data and calculates mutation frequencies in three steps (Fig. 1B–D): (i) Cas-Analyzer first finds the cleavage point in the reference sequence for the selected nuclease. Using the given comparison range (R) parameter, Cas-Analyzer defines 12nt of indicator sequences on both sides of the given reference sequence and then selects the valid sequences, which contain both indicators with up to a 1-nt mismatch, from the uploaded data. (ii) For the selected sequences, Cas-Analyzer then counts the recurrent frequency of each sequence and excludes the sequences below the given minimum frequency (n). (iii) Cas-Analyzer finally classifies the filtered sequences into three different groups: ‘insertion’, ‘deletion’ or ‘WT or substitution’ based on comparing the sequence length with the length of the given reference sequence. Optionally, if a WT marker range (r) is given, the short sequence around the cleavage point will be used as the marker of wild-type. If this marker exists in the query sequence, it will always be classified into the ‘WT or substitution’ group regardless of its length. Additionally, if the donor DNA sequence for HDR is given, Cas-Analyzer defines an HDR indicator (>8nt) by comparing the donor sequence with the reference sequence and classifies all query sequences that have the HDR indicator into the ‘HDR’ category.
2.3 Sequence alignment
For user convenience, after data analysis is complete, the results (a relatively small amount of data) are aligned to the reference sequence by using a JavaScript ported EMBOSS Needle (Rice et al., 2000). The aligned results are categorized by mutation type and sorted in descending order by count. In addition, the position and size of insertions or deletions are depicted as interactive graphs on the results web page (Fig. 1E and F).
Funding
This work was supported by a grant of the Korea Healthcare technology R&D Project, Ministry for Health & Welfare Affairs (HI16C1012) to S.B. and Institute for Basic Science (IBS-R021-D1) to J.-S.K.
Conflict of Interest: none declared.
Supplementary Material
References
- Baek K. et al. (2016) DNA-free two-gene knockout in Chlamydomonas reinhardtii via CRISPR-Cas9 ribonucleoproteins. Sci. Rep., 6, 30620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho S.W. et al. (2013) Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat. Biotechnol., 31, 230–232. [DOI] [PubMed] [Google Scholar]
- Cho S.W. et al. (2014) Analysis of off-target effects of CRISPR/Cas-derived RNAguided endonucleases and nickases. Genome Res., 24, 132–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L. et al. (2013) Multiplex Genome Engineering Using CRISPR/Cas Systems. Science, 339, 819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Güell M. et al. (2014) Genome editing assessment using CRISPR Genome Analyzer (CRISPR-GA). Bioinformatics, 30, 2968–2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Z. et al. (2013) Efficient genome engineering in human pluripotent stem cells using Cas9 from Neisseria meningitidis. Proc. Natl. Acad. Sci. USA, 110, 15644–15649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hur J.K. et al. (2016) Targeted mutagenesis in mice by electroporation of Cpf1 ribo nucleoproteins. Nat. Biotechnol., 34, 807–808. [DOI] [PubMed] [Google Scholar]
- Jinek,M. et al. (2013) RNA-programmed genome editing in human cells. eLife, 2, e00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D. et al. (2016a) Genome-wide analysis reveals specificities of Cpf1 endonucle ases in human cells. Nat. Biotechnol., 34, 863–868. [DOI] [PubMed] [Google Scholar]
- Kim H.J. et al. (2009) Targeted genome editing in human cells with zinc finger nucleases constructed via modular assembly. Genome Res., 19, 1279–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H., Kim J.S. (2014) A guide to genome engineering with programmable nucleases. Nat. Rev. Genet., 15, 321–334. [DOI] [PubMed] [Google Scholar]
- Kim Y. et al. (2016b) Generation of knockout mice by Cpf1-mediated gene targeting. Nat. Biotechnol., 34, 808–810. [DOI] [PubMed] [Google Scholar]
- Kleinstiver B.P. et al. (2016) High-fidelity CRISPR–Cas9 nucleases with no detectable genome-wide off-target effects. Nature, 529, 490–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali,P. et al. (2013) RNA-guided human genome engineering via Cas9. Science, 339, 823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson C.E. et al. (2016) In vivo genome editing improves muscle function in a mouse model of Duchenne muscular dystrophy. Science, 351, 403–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez E.E. et al. (2008) Establishment of HIV-1 resistance in CD4+ T cells by genome editing using zinc-finger nucleases. Nat. Biotechnol., 26, 808–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinello L. et al. (2016) Analyzing CRISPR genome-editing experiments with CRISPResso. Nat. Biotechnol., 34, 695–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran F.A. et al. (2013) Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell, 154, 1380–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran F.A. et al. (2015) In vivo genome editing using Staphylococcus aureus Cas9. Nature, 520, 186–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice P. et al. (2000) EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet., 16, 276–277. [DOI] [PubMed] [Google Scholar]
- Slaymaker I.M. et al. (2016) Rationally engineered Cas9 nucleases with improved specificity. Science, 351, 84–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai S.Q. et al. (2014) Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing. Nat. Biotechnol., 32, 569–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue L.J., Tsai C.J. (2015) AGEseq: analysis of genome editing by sequencing. Mol. Plant, 8, 1428–1430. [DOI] [PubMed] [Google Scholar]
- Zetsche B. et al. (2015) Cpf1 Is a Single RNA-Guided Endonuclease of a Class 2 CRISPR-Cas System. Cell, 163, 759–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X. et al. (2014) An efficient genotyping method for genome-modified animals and human cells generated with CRISPR/Cas9 system. Sci. Rep., 4, 6420. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.