Abstract
Primary resistance in Candida albicans to flucytosine (5-FC) was investigated in 25 strains by identifying and sequencing the genes FCA1, FUR1, FCY21, and FCY22, which code for cytosine deaminase, uracil phosphoribosyltransferase (UPRT), and two purine-cytosine permeases, respectively. These proteins are involved in pyrimidine salvage and 5-FC metabolism. An association between a polymorphic nucleotide and resistance to 5-FC was found within FUR1 where the substitution of cytidylate for thymidylate at nucleotide position 301 results in the replacement of arginine with cysteine at amino acid position 101 in UPRT. Isolates that are homozygous for this mutation display increased levels of resistance to 5-FC, whereas heterozygous isolates have reduced susceptibility. Three-dimensional protein modeling of UPRT suggests that the Arg101Cys mutation disturbs the quaternary structure of the enzyme, which is postulated to compromise optimal enzyme activity. A single resistant isolate, lacking the above polymorphism in FUR1, has a homozygous polymorphism in FCA1 that results in a glycine-to-aspartate substitution at position 28 in cytosine deaminase.
Flucytosine (5-FC) was initially synthesized in 1957 as an anticancer drug. Unlike 5-fluorouracil (5-FU), a closely related fluorinated pyrimidine, 5-FC did not exhibit antineoplastic activity but was subsequently found to possess antifungal activity and was used in 1968 to treat human cryptococcosis and candidiasis (25). Flucytosine administered in combination with amphotericin B remains the standard of care for cryptococcal meningitis, and the drug continues to have a role in the treatment of Candida infections which are life threatening or in circumstances where drug penetration may be problematic, such as infections of urine, eyes, and heart valves.
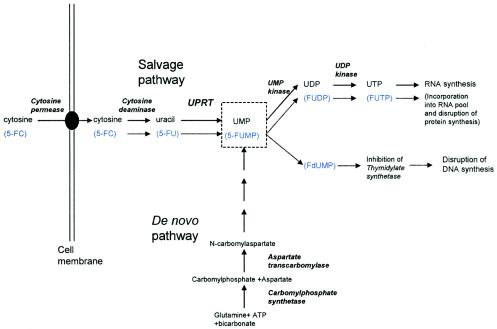
Flucytosine is metabolized via the pyrimidine salvage pathway (Fig. 1), where it acts as a subversive substrate with the subsequent production of toxic nucleotides and disruption of DNA and protein synthesis (18, 26). After being actively transported into the cell by membrane permeases, 5-FC is converted via 5-FU to 5-fluoro-uridylate (synonymous with 5-fluoro-UMP [5-FUMP]) under the action of the enzymes cytosine deaminase and uracil phosphoribosyltransferase (UPRT), respectively. 5-FUMP is in turn phosphorylated by two specific kinases to 5-fluoro-UTP, which is incorporated into RNA. 5-FUMP is also reduced to 5-fluoro-2′-deoxyuridylate, which inhibits the enzyme thymidylate synthetase and thus DNA synthesis by decreasing the available nucleotide pool. Mammalian cells lack the enzyme cytosine deaminase and consequently are not directly subject to the toxic effects of 5-FC.
FIG. 1.
The salvage and de novo pathways for the synthesis of UMP and subsequent incorporation into RNA. The relevant enzymes are shown in italics, and the fluorinated analogues are shown beneath the natural substrates in blue type. Cytosine deaminase is absent from mammalian cells. FUDP, fluoro-UDP; FUTP, fluoro-UTP; FdUMP; fluoro-deoxy-uridylate.
Primary resistance in Candida albicans, the focus of this paper, refers to inherent 5-FC resistance in the absence of prior drug exposure; in recent surveys, this resistance is observed in around 3% of isolates (1, 16). This phenomenon was recognized many years ago to be disproportionately represented in isolates belonging to serogroup B and more recently was found to be confined to a specific clade defined by DNA fingerprinting (19). The possible mechanisms of resistance to 5-FC have been investigated, but most of this work was conducted prior to readily available molecular tools. Initial work employed UV mutagenesis in both C. albicans (4) and Saccharomyces cerevisiae (8), and this work collectively demonstrated that disruption of any of the proteins involved in pyrimidine salvage or their regulation could lead to 5-FC resistance. We therefore considered it opportune to examine the molecular mechanisms of primary resistance in C. albicans by identifying and sequencing the genes coding for the proteins involved in pyrimidine salvage and analyzing the effect of one of these polymorphisms using protein modeling.
The findings in this paper are consistent with a recently published study in which the majority of cases of 5-FC resistance in C. albicans were associated with isolates that were homozygous for a single amino acid substitution, Arg101Cys, in UPRT (2). The present paper explores the relationship of this mutation to an analogous mutation in the UPRT of S. cerevisiae and provides a structural basis by which the mutation induces resistance. It is clear, nevertheless, that this substitution cannot fully account for the level of 5-FC resistance seen in most isolates. This study also implicates an amino acid substitution within cytosine deaminase and further explores the association of substitutions in the purine-cytosine permeases with 5-FC resistance.
(This work was presented in part at the 43rd Interscience Conference on Antimicrobial Agents and Chemotherapy, Chicago, Ill., 14 to 17 September 2003.)
MATERIALS AND METHODS
Isolates.
Twenty-five epidemiologically unrelated clinical isolates of C. albicans of varying susceptibilities to 5-FC were retrieved from our large collection of Candida isolates and maintained on Sabouraud agar (Oxoid, Basingstoke, United Kingdom). Isolates were confirmed to be primarily resistant to 5-FC after clinical case records were reviewed.
Susceptibility testing.
Each isolate was initially tested for 5-FC susceptibility by using the British Society for Mycopathology disk method (7). Briefly, a suspension equivalent to a 0.5 McFarland standard of each isolate was streaked onto one half of a yeast nitrogen base plate supplemented with glucose, and an equivalent suspension of C. kefyr was streaked onto the other half as a control. A 1-μl 5-FC disk was placed in the center of the plate which was then incubated at 37°C for 48 h. Isolates were defined as susceptible if their zone of inhibition was greater than or equal to 80% of that of the control. The MIC for each isolate was then determined twice, according to the EUCAST method (20), by using colonies from around the zone of inhibition in one case and from 48-h-old colonies grown on horse blood agar in the other. A suspension equivalent to a 0.5 McFarland standard was diluted 1:10 in water, 100 μl of which was placed in microtiter plate wells, each containing 100 μl of double-strength RPMI medium with 5-FC to achieve final concentrations of 5-FC ranging from 0.12 to 64 mg/liter. Isolates for which the MICs were <0.12 mg/liter were retested with lower concentrations of 5-FC to enable a definitive MIC to be assigned. The plates were incubated at 37°C for 24 h, and the endpoint, defined as a 50% reduction in growth, was determined spectrophotometrically. The control organism was C. krusei ATCC 6258, for which the 5-FC MIC was 2.0 mg/liter. The classification of the 5-FC MIC results as sensitive, intermediate, or resistant was defined as an MIC of ≤1, 2 to 8, and >8 mg/liter, respectively.
DNA extraction.
Genomic DNA was extracted from yeast cells incubated overnight in Sabouraud broth at 35°C. The suspension was centrifuged at 5,000 × g for 10 min, and the pellet was resuspended in 600 μl of sorbitol buffer (1 M sorbitol, 0.1 M EDTA, 0.014 M β-mercaptoethanol) to which lyticase (200 U; Sigma-Aldrich Co. Ltd., Poole, Dorset, United Kingdom) was added and incubated at 30°C for 30 min. The spheroplasts were pelleted for 10 min at 300 × g. DNA extraction was then achieved by using a DNeasy tissue kit (QIAGEN Ltd., Crawley, West Sussex, United Kingdom) according to the manufacturer's instructions.
Identification of genes and primer design.
The gene FUR1, coding for UPRT, was retrieved from the CandidaDB genome database (accession no. CA2069; http://genolist.pasteur.fr/CandidaDB/), and the gene FCA1, coding for cytosine deaminase, was retrieved from the EMBL DNA database (accession no. U55194) (3). According to CandidaDB, C. albicans possesses four genes encoding putative purine-cytosine permeases, which are FCY21, FCY22, FCY23, and FCY24. The genes FCY22, FCY23, and FCY24 were readily retrieved from the database, but FCY21 is not present as a separate entry; it is, rather, defined as an allele of FCY22. To resolve this situation, the Stanford contig containing the gene for FCY22 (contig 1838) and the corresponding protein sequence were aligned with those of contig 1777 (FCY21), which confirmed that these were definitely different genes and proteins (the DNA sequences of these contigs were obtained from the website of the Stanford Genome Technology Center [http://sequence.stanford.edu/group/candida/index.html]). A BLAST search with Fcy21 against a nonredundant protein database identified related proteins in S. cerevisiae (Fcy2, Fcy21, Fcy22, and Tpn1). These eight protein sequences were aligned by using the program PileUp, which is part of the GCG software package (program manual for the GCG package, version 7; Genetics Computer Group, Madison, Wis.). This alignment was edited manually to remove noninformative residues, and the 421 informative residues were used to calculate evolutionary distance with the Kimura protein correction (11). The program Growtree was used to construct a phylogenetic tree by using neighbor joining, which was viewed using TreeView (15). The genes FCY21 and FCY22 were selected for further analysis. The primer sequences for PCR and sequencing were designed to cover the entire open reading frames of these four genes, and the relevant details of these primers and of the four genes are summarized in Table 1.
TABLE 1.
Pyrimidine salvage pathway genes analyzed and primers used in this studya
| Gene | Gene product | Primers for PCR | Amplicon size (bp) | Primers for sequencing | Size of ORF (bp) | Introns | Intron size (bp) | No. of amino acid residues |
|---|---|---|---|---|---|---|---|---|
| FCY21 | Cytosine permease | For 5′-TATAATTCTCCCATCACCATC-3′ | 1,847 | For 5′-GGTGCCACTTTAGCCATTTG-3′ | 1,545 | 0 | 514 | |
| Rev 5′-ACCGCTGCTTCTCTCTCCAC-3′ | Rev 5′-GTCTTAATCCTAAAGCTGATCC-3′ For 5′-CTAGTAAATCATTTGAAGGTGGTG-3′ | |||||||
| Rev 5′-CAAGCAGCACCCAATATTAAAG-3′ | ||||||||
| FCY22 | Cytosine permease | For 5′-CCCTTCACTCCAACTCTTTCC-3′ | 1,686 | For 5′-CCCTTCACTCCAACTCTTTCC-3′ | 1,581 | 0 | 526 | |
| Rev 3′-CAACATTTAAATAACACGGCAGAATG-3′ | Rev 3′-CAACATTTAAATAACACGGCAGAATG-3′ | |||||||
| For 5′-TCTGGTCCAACTGAAGCTGG-3′ | ||||||||
| Rev 5′-GGTAATGATAACCCGGTGGTC-3′ | ||||||||
| FCA1 | Cytosine deaminase | For 5′-GTCACTTCAAAGTACCAATCTTTA-3′ | 633 | For 5′-GTCACTTCAAAGTACCAATCTTTA-3′ | 523 | 1 | 70 | 150 |
| Rev 3′-GTTTAAGTTTTATATTACACTAATCTG-3′ | Rev 3′-GTTTAAGTTTTATATTACACTAATCTG-3′ | |||||||
| FUR1 | UPRT | For 5′-GTCCCAATTTCTTCATCATG-3′ | 911 | For 5′-GTCCCAATTTCTTCATCATG-3′ | 657 | 0 | 218 | |
| Rev 3′-TCCAGATATAGTACATGTAAAGTG-3′ | Rev 3′-TCCAGATATAGTACATGTAAAGTG-3′ |
ORF, open reading frame.
PCR, sequencing, and sequence analysis.
Reaction volumes of 50 μl were set up, containing approximately 50 ng of genomic DNA, 1.25 U of Taq DNA polymerase, 0.2 mM each deoxynucleoside triphosphate, and 0.1 μM each primer with a standard PCR buffer. The reaction conditions for all primer pairs consisted of 30 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 30 s. Sequencing was performed by using BigDye Terminator version 1.1 (Applied Biosystems, Warrington, United Kingdom) on purified PCR product. A 5-FC-susceptible isolate cultured from a tracheal aspirate (F/10089) was chosen as the reference strain, as none of the polymorphic positions in its four genes were unique to that isolate. Raw sequence data were edited and aligned by using Chromas 2 (Technelysium Pty. Ltd., Gold Coast, Australia) and Gene Lite (BioTools Inc., Edmonton, Canada). The consequences of nucleotide polymorphisms in terms of amino acid change were assessed according to the Blosum62 scoring matrix (5). A radical amino acid change was defined as a matrix score of 0 or lower.
Three-dimensional protein modeling.
The possible effect of the amino acid substitution Arg101Cys was modeled by using the crystal structure of UPRT from Toxoplasma gondii (22, 23). The homology model was generated after sequence alignment using the program LOOK (12) and followed by energy minimization.
Nucleotide sequence accession numbers.
The sequences of the four genes FCY21, FCY22, FCA1, and FUR1 from isolate F/10089 were deposited in GenBank under accession numbers AJ616009, AJ616010, AJ616007, and AJ616008, respectively.
RESULTS
In vitro susceptibility testing.
The 5-FC susceptibility testing results of 25 epidemiologically unrelated clinical isolates of C. albicans are given in Table 2. By using the British Society for Mycopathology disk method (7), five susceptible isolates were identified. The 5-FC MICs were determined twice with the EUCAST method (20) from isolates grown on two different media. The results from these two media were either identical or within 1 dilution of each other. There were 10 susceptible, 3 intermediate, and 12 resistant isolates. Five isolates were resistant to 5-FC by disk testing but deemed susceptible according to the MICs for these isolates, suggesting that the disk method is a sensitive but not necessarily specific method to determine 5-FC susceptibility of C. albicans. The sites of isolation are also summarized in Table 2. The review of case records confirmed that isolates were primarily resistant to 5-FC, although it remains possible that at least some patients acquired C. albicans nosocomially from other patients who may have been exposed to 5-FC.
TABLE 2.
Susceptibility testing of 25 C. albicans clinical isolatesa
| Strain or isolate no. | Lab no. | Site of isolation | % Susceptibleb | Interpretation of disk test | 5-FC MIC (mg/liter)c | Interpretation of 5-FC MIC |
|---|---|---|---|---|---|---|
| Reference | F/10089 | Tracheal aspirate | 92 | S | 0.12 | S |
| 1 | F/10101 | Blood | 88 | S | 0.06 | S |
| 2 | F/10141 | Blood | 88 | S | 0.06 | S |
| 3 | F/10084 | BAL | 91 | S | 0.12 | S |
| 4 | F/10155 | Wound | 84 | S | 0.12 | S |
| 5 | F/7421 | Tracheal aspirate | 0 | R | 0.5 | S |
| 6 | F/7822 | Tracheal aspirate | 0 | R | 0.5 | S |
| 7 | F/6633 | Sputum | 0 | R | 1 | S |
| 8 | F/8894 | BAL | 0 | R | 1 | S |
| 9 | F/9732 | Sputum | 0 | R | 1 | S |
| 10 | F/8750 | BAL | 0 | R | 2 | I |
| 11 | F/9651 | Tracheal aspirate | 0 | R | 4 | I |
| 12 | F/9464 | Urine | 0 | R | 8 | I |
| 13 | F/9406 | Mouth | 0 | R | 16 | R |
| 14 | F/6524 | Catheter tip | 0 | R | >64 | R |
| 15 | F/6563 | BAL | 0 | R | >64 | R |
| 16 | F/6698 | Gastric fluid | 0 | R | >64 | R |
| 17 | F/6997 | Urine | 0 | R | >64 | R |
| 18 | F/7451 | Tracheal aspirate | 0 | R | >64 | R |
| 19 | F/7901 | Vaginal swab | 0 | R | >64 | R |
| 20 | F/8082 | Sputum | 0 | R | >64 | R |
| 21 | F/8166 | Sputum | 0 | R | >64 | R |
| 22 | F/8341 | Surgical wound | 0 | R | >64 | R |
| 23 | F/8556 | CSF | 0 | R | >64 | R |
| 24 | F/9161 | Urine | 0 | R | >64 | R |
| Control | C. krusei ATCC 6258 | 2 | I |
BAL, bronchoalveolar lavage; CSF, cerebrospinal fluid; S, sensitive; I, intermediate; R, resistant.
According to the 5-FC disk method. The percentage value indicates the percent inhibition relative to the control.
MIC data determined from colonies around the zone of inhibition on a yeast nitrogen base plate.
Sequencing of the genes encoding proteins involved in the pyrimidine salvage pathway.
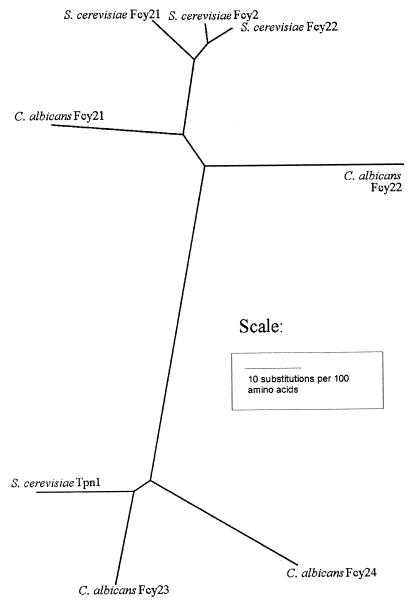
Three proteins are involved in the pyrimidine salvage pathway, and the corresponding C. albicans genes for these proteins were identified in public DNA databases (Table 1). One gene, FCA1, codes for cytosine deaminase, and one gene, FUR1, codes for UPRT. However, four putative purine-cytosine permeases have been identified in C. albicans. A phylogenetic analysis was carried out with these four proteins and four homologous proteins identified in S. cerevisiae (Fig. 2). It can be seen from the tree that Fcy23 and Fcy24 from C. albicans are more closely related to Tpn1 from S. cerevisiae, which has been shown to be a vitamin B6 transporter (24). Fcy21 and Fcy22 from C. albicans cluster with the other three proteins from S. cerevisiae, including Fcy2, which has been shown to transport cytosine and 5-methylcytosine (21, 27). On these grounds, it was decided that only FCY21 and FCY22 should be sequenced, as they are more likely to be primarily involved in the transport of cytosine.
FIG. 2.
Radial phylogenetic tree of the four purine-cytosine permeases in C. albicans and their homologues in S. cerevisiae. The tree was constructed by using neighbor joining from a distance matrix with the Kimura protein correction, and the scale bar indicates 10 substitutions per 100 amino acids.
PCR products spanning the entire open reading frames of the four genes were sequenced from all 25 isolates. These sequences were aligned and positions containing different nucleotides between the isolates were identified. As C. albicans is diploid, the majority of these polymorphic positions had two base calls. For those polymorphic positions falling within the open reading frames, the effect of the different nucleotides on the codons and therefore on the resultant amino acid residue was determined. The majority of the polymorphic nucleotide changes resulted in either no change (synonymous mutation) or a conservative replacement in the respective amino acid residue (Table 3). The nucleotide polymorphisms and resultant amino acid changes between the 25 isolates for FCY21, FCY22, FCA1, and FUR1 and any association with 5-FC resistance are detailed in Tables 4, 5, 6, and 7 respectively.
TABLE 3.
Type and extent of coding changes in genes implicated in 5-FC metabolisma
| Gene | Variable codons/total codon no. (%) | No. of heterozygous loci | No amino acid replacement
|
Amino acid replacements
|
|
|---|---|---|---|---|---|
| No. of synonymous replacements | No. of conservative replacements | No. of radical replacements | |||
| FCY21 | 8/514 (1.6) | 3 | 8 | 0 | 0 |
| FCY22 | 38/526 (7.2) | 37 | 25 | 5 | 8 |
| FCA1 | 11/150 (7.3) | 9 | 7 | 1 | 4 |
| FUR1 | 3/218 (1.4) | 3 | 2 | 0 | 1 |
Data for all 25 isolates.
TABLE 4.
Polymorphic sites within FCY21a
| Strain or isolate no.e | MIC (mg/liter) | Codon position, codons, and amino acidsd
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| 62 | 115 | 212 | 258 | 369 | 456 | 502 | 504 | ||
| Reference | 0.12 | ACT | CTA | GCC | TTC | GTT | TTC | TTT | GTC |
| Thr | Leu | Ala | Phe | Val | Phe | Phe | Val | ||
| 1 | 0.06 | GCT | TTY | GTG | |||||
| 2 | 0.06 | ACC | GTS | ||||||
| 3 | 0.12 | TTT | TTT | GTS | |||||
| 4 | 0.12 | ACY | |||||||
| 5 | 0.5 | ||||||||
| 6 | 0.5 | GTS | |||||||
| 7 | 1 | GTS | |||||||
| 8 | 1 | TTT | TTT | GTG | |||||
| 9 | 1 | GTG | |||||||
| 10 | 2 | ACC | GTC | TTT | GTG | ||||
| 12 | 4 | TTA | GCT | GTS | |||||
| 13 | 8 | GTS | |||||||
| 13 | 16 | GTS | |||||||
| 14 | >64 | ||||||||
| 15 | >64 | GTS | |||||||
| 16 | >64 | TTT | |||||||
| 17 | >64 | GTS | |||||||
| 18 | >64 | GTS | |||||||
| 19 | >64 | ||||||||
| 20 | >64 | GTS | |||||||
| 21 | >64 | GTS | |||||||
| 22 | >64 | GTS | |||||||
| 23 | >64 | GTS | |||||||
| 24 | >64 | GTS | |||||||
| Thrb Thrc | Leuc | Alac | Phec | Valc | Phec | Pheb Phec | Valb Valc | ||
FCY21 is the gene coding for a putative purine-cytosine permease.
Same amino acid and codon as reference strain.
Same amino acid as reference strain, different codon.
Polymorphic nucleotides are underlined. S = G or C; Y = C or T. Blank spaces indicate the codon is the same as in the reference strain.
Amino acids in the bottom row are encoded by the altered codons in the numbered isolates.
TABLE 5.
Polymorphic sites within FCY22a
| Strain or isolate no. | MIC (mg/liter) | Codon position, codons, and amino acids f
|
Codon position, codons, and amino acids f
|
||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2 | 13 | 24 | 26 | 32 | 33 | 34 | 36 | 37 | 38 | 40 | 43 | 45 | 50 | 78 | 80 | 108 | 117 | 142 | 175 | 176 | 203 | 293 | 303 | 335 | 375 | 428 | 460 | 464 | 465 | 474 | 475 | 479 | 496 | 501 | 502 | 512 | 513 | ||
| Reference | 0.12 | AAA | CAG | GTT | ATT | GAT | GAG | CAT | ATT | ACA | TCC | ATT | CCA | ACG | TTA | GAA | CAT | CCC | ACT | AGG | AAT | GCC | GGT | GTC | TTA | ACC | TCT | TCC | CCC | GCC | GGT | TGC | TCT | TCT | AGT | TCT | TTC | GGT | TAC |
| Lys | Gln | Val | Ile | Asp | Glu | His | Ile | Thr | Ser | Ile | Pro | Thr | Leu | Glu | His | Pro | Thr | Arg | Asn | Ala | Gly | Val | Leu | Thr | Ser | Ser | Pro | Ala | Gly | Cys | Ser | Ser | Ser | Ser | Phe | Gly | Tyr | ||
| 1 | 0.06 | ACA | MAG | RTT | MTT | GAY | GAA | GAT | ATC | YCA | ACK | TTR | GGY | YTA | CCY | GGC | TGY | TCY | TCW | ART | TCY | TTY | GGC | TAT | |||||||||||||||
| 2 | 0.06 | ACA | RTT | MTT | GAA | GAT | ATC | YCA | TTG | AGR | YTA | ART | GGY | TAY | |||||||||||||||||||||||||
| 3 | 0.12 | ACA | MAG | RTT | MTT | GAA | GAT | ATC | YCA | TTG | ACY | GTY | YTA | TCY | |||||||||||||||||||||||||
| 4 | 0.12 | ACA | RTT | MTT | GAA | GAT | ATC | YCA | TTG | AGR | YTA | TYT | TCY | ART | GGY | TAY | |||||||||||||||||||||||
| 5 | 0.5 | AMA | RTT | MTT | GAR | SAT | ATY | YCA | TTR | CCY | AAY | GTY | YTA | CCY | GCY | GGY | TCC | TCW | ART | TCY | TTY | GGY | TAY | ||||||||||||||||
| 6 | 0.5 | AMA | RTT | MTT | GAR | SAT | ATY | YCA | TTR | CCY | AAY | GTY | YTA | CCY | GCY | GGY | TGY | TCY | TCW | ART | TCY | TTY | GGY | TAY | |||||||||||||||
| 7 | 1 | AMA | RTT | MTT | GAR | SAT | ATY | YCA | TTR | CCY | AAY | GTY | YTA | CCY | GCY | GGY | TGY | TCY | TCW | ART | TCY | TTY | GGY | TAY | |||||||||||||||
| 8 | 1 | ACA | MAG | RTT | MTT | GAA | GAT | ATC | YCA | TTG | ACY | AGR | GTT | CTA | |||||||||||||||||||||||||
| 9 | 1 | AMA | RTT | MTT | GAR | SAT | ATY | YCA | TTR | CCY | AAY | GTY | YTA | CCY | GCY | GGY | TGY | TCY | TCW | ART | TCY | TTY | GGY | TAY | |||||||||||||||
| 10 | 2 | ACA | MAG | RTT | MTT | GAA | GAT | RTT | ACM | TCM | ATY | YCA | TTR | ACY | GTY | YTA | CCY | GCY | GGY | TGY | TCY | TCW | ART | TCY | TTY | GGY | TAY | ||||||||||||
| 11 | 4 | ACA | ATT | CTT | GAA | GAT | ATC | TCA | TTG | CCT | GCT | GGC | TGT | TCC | TCA | AAT | TCC | TTT | GGC | TAT | |||||||||||||||||||
| 12 | 8 | AMA | RTT | MTT | GAR | SAT | ATY | YCA | TTR | CCY | AAY | GTY | YTA | CCY | GCY | GGY | TGY | TCY | TCW | ART | TCY | TTY | GGY | TAY | |||||||||||||||
| 13 | 16 | AMA | RTT | MTT | GAR | SAT | ATY | YCA | TTR | CCY | AAY | GTY | YTA | CCY | GCY | GGY | TGY | TCY | TCW | ART | TCY | TTY | GGY | TAY | |||||||||||||||
| 14 | >64 | AMA | RTT | MTT | GAR | GAT | ATY | YCA | TTR | CCY | AAY | GTY | YTA | CCY | GCY | GGY | TGY | TCY | TCW | ART | TCY | TTY | GGY | TAY | |||||||||||||||
| 15 | >64 | RTT | MTT | GAR | SAT | ATY | YCA | TTR | CCY | AAY | GTT | YTA | CCY | GCY | GGY | TGY | TCY | TCW | ART | TCY | TTY | GGY | TAY | ||||||||||||||||
| 16 | >64 | ACA | MAG | GAA | GAT | ATC | TCA | TTG | GWA | CRT | GGY | GTY | CTA | AYC | CCY | GCY | GGY | TGY | TCY | TCW | ART | TCY | TTY | GGY | TAY | ||||||||||||||
| 17 | >64 | AMA | RTT | MTT | GAR | SAT | ATY | YCA | TTR | CCY | AAY | GTY | YTA | CCY | GCY | GGY | TGY | TCY | TCW | ART | TCY | TTY | GGY | TAY | |||||||||||||||
| 18 | >64 | AMA | CCT | AAC | GTT | CTA | CCY | GCY | GGY | TGY | TCY | TCW | ART | TCY | TTY | GGY | TAY | ||||||||||||||||||||||
| 19 | >64 | AMA | RTT | MTT | GAR | SAT | ATY | YCA | TTR | CCT | AAY | GTY | YTA | CCY | GGY | TCY | TCW | ART | TCY | TTY | GGY | TAY | |||||||||||||||||
| 20 | >64 | CCT | AAC | GTT | CTA | CCY | GCY | GGY | TGY | TCY | TCW | ART | TCY | TTY | GGY | TAY | |||||||||||||||||||||||
| 21 | >64 | RTT | MTT | GAR | SAT | ATY | YCA | TTG | CCY | AAY | GTT | CTA | CCY | GCY | GGY | TGY | TCY | TCW | ART | TCY | TTY | GGY | TAY | ||||||||||||||||
| 22 | >64 | CCT | AAC | GTT | CTA | CCY | GCY | GGY | TGY | TCY | TCW | ART | TCY | TTY | GGY | TAY | |||||||||||||||||||||||
| 23 | >64 | RTT | MTT | GAR | SAT | ATY | YCA | TTR | CCY | AAY | GGC | YTA | CCY | GCY | GGY | TGY | TCY | TCW | ART | TCY | TTY | GGY | TAY | ||||||||||||||||
| 24 | >64 | RTT | MTT | GAR | SAT | ATY | YCA | TTR | CCY | AAY | GTY | YTA | CCY | GCY | GGY | TGY | TCY | TCW | ART | TCY | TTY | GGY | TAY | ||||||||||||||||
| Lysb Thrd | Glnb Lyse | Valb Ilee | Ileb Leue | Aspb Aspc | Glub Gluc | Hisb Aspd | IlebVale | Thrb Thrc | Serb Serc | Ileb Ilec | Prob Serd | Thrb Thrc | Leub Leuc | GlubVald | HisbArgd | Prob Proc | Thrb Thrc | Argb Argc | Asnb Asnc | Glyd | Glyb Glyc | Valb Valc | Leub Leuc | ThrbIled | Serb Phed | Serb Serc | Prob Proc | Alab Alac | Glyb Glyc | Cysb Cysc | Serb Serc | Serb Serc | Serb Asne | Serb Serc | Pheb Phec | Glyb Glyc | Tyrb Tyrc | ||
FCY22 is the gene coding for a putative purine-cytosine permease.
Same amino acid and codon as reference strain.
Same amino acid as reference strain, different codon.
Radical amino acid replacement.
Conservative replacement.
Polymorphic nucleotides are underlined. R = A or G; Y = C or T; W = A or T; M = A or C; K = G or T; S = G or C. The codon and corresponding amino acid associated with 5-FC resistance are shown in boldface type. The codon and corresponding amino acid associated with intermediate 5-FC resistance are shown in italics. Blank spaces indicate the codon is the same as in the reference strain.
TABLE 6.
Polymorphic sites within FCA1a
| Strain or isolate no. | 5-FC MIC (mg/liter) | Codon position, codon, and amino acidf
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2 | 11 | 28 | 29 | 34 | 36 | 44 | 70 | 73 | 128 | 141 | ||
| Reference | 0.12 | ACG | ATC | GGT | TCA | TCC | GGC | AAC | AAG | AAG | GAA | CCG |
| Thr | Ile | Gly | Ser | Ser | Gly | Asn | Lys | Lys | Glu | Pro | ||
| 1 | 0.06 | GTT | TCT | GAC | AAA | AAA | CCA | |||||
| 2 | 0.06 | RTY | TCT | GRC | AAR | AAA | GWA | CCA | ||||
| 3 | 0.12 | |||||||||||
| 4 | 0.12 | RTY | TCT | AAR | AAA | GWA | CCA | |||||
| 5 | 0.5 | |||||||||||
| 6 | 0.5 | GTT | TCT | GAC | AAA | AAA | CCA | |||||
| 7 | 1 | GTT | TCT | GAC | AAA | AAA | CCA | |||||
| 8 | 1 | AAY | ||||||||||
| 9 | 1 | GTT | TCT | GAC | AAA | AAA | CCA | |||||
| 10 | 2 | TCT | GAC | AAA | AAA | CCA | ||||||
| 11 | 4 | ACA | TTA | CCA | ||||||||
| 12 | 8 | GTT | TCT | GAC | AAA | AAA | CCA | |||||
| 13 | 16 | GTT | TCT | GAC | AAA | AAA | CCA | |||||
| 14 | >64 | GTT | TCT | GAC | AAA | AAA | CCA | |||||
| 15 | >64 | GTT | TCT | AAA | CCR | |||||||
| 16 | >64 | ACA | GAT | CCA | ||||||||
| 17 | >64 | GTY | TCY | GRC | AAR | AAR | ||||||
| 18 | >64 | RTY | TCY | GRC | AAR | AAR | CCR | |||||
| 19 | >64 | RTY | TCY | GRC | AAR | AAR | CCR | |||||
| 20 | >64 | GTT | TCT | GAC | AAA | AAA | CCA | |||||
| 21 | >64 | |||||||||||
| 22 | >64 | GTT | TCT | GAC | AAA | AAA | CCA | |||||
| 23 | >64 | GTT | TCT | GAC | AAA | AAA | CCA | |||||
| 24 | >64 | GTT | TCT | GAC | AAA | AAA | CCA | |||||
| Thrb Thrc | Ileb Vale | Aspd | Leud | Serb Serc | Glyb Aspd | Asnb Asnc | Lysb Lysc | Lysb Lysc | Glub Vald | Prob Proc | ||
FCA1 is the gene for cytosine deaminase.
Same amino acid and codon as reference strain.
Same amino acid as reference strain, different codon.
Radical amino acid replacement.
Conservative replacement.
Polymorphic nucleotides are underlined. R = A or G; Y = C or T, W = A or T. The codon and corresponding amino acid associated with 5-FC resistance are shown in boldface type. The codon and corresponding amino acid associated with intermediate 5-FC resistance are shown in italics. Blank spaces indicate the codon is the same as in the reference strain.
TABLE 7.
Polymorphic sites within FUR1a
| Strain or isolate no. | MIC (mg/liter) | Codon position, codons, and amino acidse
|
||
|---|---|---|---|---|
| 101 | 140 | 147 | ||
| Reference | 0.12 | CGT | GAT | GGR |
| Arg | Asp | Gly | ||
| 1 | 0.06 | |||
| 2 | 0.06 | GGA | ||
| 3 | 0.12 | GGA | ||
| 4 | 0.12 | GGA | ||
| 5 | 0.5 | YGT | GGA | |
| 6 | 0.5 | YGT | GGA | |
| 7 | 1 | YGT | GGA | |
| 8 | 1 | GGA | ||
| 9 | 1 | YGT | GGA | |
| 10 | 2 | GAY | ||
| 11 | 4 | GAY | GGA | |
| 12 | 8 | TGT | GGA | |
| 13 | 16 | TGT | GGA | |
| 14 | >64 | TGT | GGA | |
| 15 | >64 | TGT | GGA | |
| 16 | >64 | GGA | ||
| 17 | >64 | TGT | GGA | |
| 18 | >64 | TGT | GGA | |
| 19 | >64 | TGT | GGA | |
| 20 | >64 | TGT | GGA | |
| 21 | >64 | TGT | GGA | |
| 22 | >64 | TGT | GGA | |
| 23 | >64 | TGT | GGA | |
| 24 | >64 | TGT | GGA | |
| Argb CysdCysdCysd | Aspc Aspb | Glyb | ||
FUR1 is the gene for UPRT.
Same amino acid and codon as reference strain.
Same amino acid as reference strain, different codon.
Radical amino acid replacement.
Polymorphic nucleotides are underlined. R = A or G; Y = C or T. The codon and corresponding amino acid associated with 5-FC resistance are shown in boldface type. The codon and corresponding amino acid associated with intermediate 5-FC resistance are shown in italics. The codon and corresponding amino acid associated with reduced 5-FC susceptibility are shown in boldface italic type. Blank spaces indicate the codon is the same as in the reference strain.
The polymorphic sites found within the open reading frame of FCY21, the gene coding for one of the putative purine-cytosine permeases, are listed in Table 4. Among the resistant isolates, there is only one nonunique synonymous substitution (isolate 16) with no conservative or radical amino acid changes. Interestingly, a microsatellite repeat lies both upstream and downstream of the open reading frame (an ATT repeat 24 bp upstream of the start codon and a TG repeat 36 bp downstream of the stop codon). Not unexpectedly, the number of repeats for both microsatellites varies between isolates.
As can be seen from Tables 3 and 5, there are considerably more polymorphisms relative to FCY21 in FCY22, the gene for the other putative purine-cytosine permease. Even though these genes are of similar size, there are three times as many synonymous mutations in FCY22 as well as eight mutations that result in a radical amino acid substitution and five mutations that result in a conservative amino acid substitution. Among the 12 resistant isolates, all have at least one synonymous mutation (none unique to 5-FC-resistant isolates), all have at least one conservative replacement (none unique), and 10 have at least one radical replacement, four of which are observed uniquely among the 5-FC resistant isolates. Isolate 16 has three out of these four changes (one in each of the codons 78, 80, and 335). However, this isolate has two alleles of this gene, with the other codon at these three positions being the same as that of the reference isolate. Isolate 23 has a GGC triplet rather than a GCC at codon 176, which results in a radical Ala-to-Gly substitution.
Table 6 shows the polymorphisms present in FCA1, the gene coding for cytosine deaminase. This gene has a similar level of polymorphism as that of FCY22 (7.3%), with four radical replacements, one conservative replacement, and six synonymous mutations. Of the 12 resistant isolates, 11 have at least one synonymous mutation (none unique to 5-FC-resistant isolates), 10 have at least one conservative replacement (none unique), and 10 have at least one radical replacement, one of which is unique among 5-FC-resistant isolates. Isolate 16, at position 28, has a GAT codon rather than a GGT codon, which results in a radical amino acid substitution of Gly to Asp. Interestingly, the codon at position 29 has a mutation associated with an isolate of intermediate resistance (isolate 11) and the mutation results in a radical amino acid substitution (TCA [Ser] to TTA [Leu]). In addition to the mutations identified within the open reading frame, a polymorphic site was found 9 bp upstream of the start codon (a G-to-A transition); this mutation is unlikely to have any effect on the regulation or translation of this gene.
Table 7 shows the polymorphisms present in FUR1, the gene coding for UPRT. This gene has a similar level of polymorphism as that of FCY21 (1.4%), with only two synonymous mutations and one that results in a radical amino acid substitution. As with FCA1, there is an association of 5-FC susceptibility with the codon at position 101 being TGT in 10 of the 11 highly resistant isolates (MIC > 64 mg/liter) and isolates 12 and 13 (MIC, 8.0 and 16 mg/liter, respectively). The codon TGT codes for Cys, whereas the CGT codon present in the reference isolate codes for Arg. It is worth noting that the only resistant isolate that does not have this mutation is isolate 16. Moreover, four isolates for which MICs fall between fully resistant and fully susceptible (0.50 and 1.0 mg/liter) are heterozygous at this position and will therefore contain proteins with either an Arg or a Cys at position 101. In addition to the three polymorphic positions within the open reading frame, a variable nucleotide was identified 29 bp downstream of the stop codon (a T-to-C transition).
Protein modeling of the Arg101Cys substitution in UPRT.
In order to provide further insight into the possible significance of the Arg101Cys substitution in UPRT, an alignment of homologous proteins was performed with subsequent modeling of the substitution.
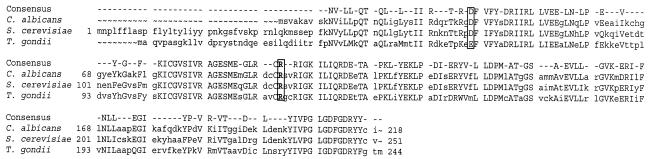
An alignment of the amino acid sequences of UPRT from C. albicans, S. cerevisiae, and Toxoplasma gondii is shown in Fig. 3, which highlights the conservation of Arg at position 101 in C. albicans, position 126 in T. gondii, and position 134 in S. cerevisiae. This residue is replaced with a Cys in all the resistant isolates except isolate 16. The three proteins show a high level of identity to each other, with the C. albicans protein being 72 and 56% identical to S. cerevisiae and T. gondii UPRT, respectively. The crystal structures of UPRT from two bacteria (Thermotoga maritima [structure is available from the Protein Data Bank at http://www.rcsb.org] and Bacillus caldolyticus [6]) and T. gondii (23) were reviewed, which revealed similarities in their protein folds as well as the fact that these enzymes tend to exist in various oligomeric forms. Analysis of the three-dimensional structure of UPRT from T. gondii, the organism bearing the closest phylogenetic relationship to C. albicans for which the crystal structure has been determined, reveals that Arg126 (cognate of Arg101 in C. albicans [Fig. 3]) is engaged in a salt bridge interaction with Glu61 (Asp36 in C. albicans [Fig. 3]). This salt bridge is formed across the dimer interface for both dimers in the tetrameric form, thus contributing to the stability of both quaternary arrangements, and this possible configuration is illustrated schematically in Fig. 4. A close inspection of the structures of UPRT of T. gondii in complex with uracil, α-d-5-phosphoribosyl-1-pyrophosphate (PRPP), and GTP (22) shows that binding of the natural substrate uracil requires only residues contained within one monomer. In contrast, binding of PRPP and GTP involves residues in at least three monomeric subunits within the tetrameric form, suggesting that the tetramer is the configuration required for optimal activity of this enzyme (22). The C. albicans UPRT homology model suggests that substitution of an arginyl side chain with a cysteinyl side chain at residue 101 in C. albicans would eliminate an attractive electrostatic bond with Asp36.
FIG. 3.
Alignment of the amino acid sequences of UPRT from C. albicans, S. cerevisiae, and T. gondii. The alignment was generated by using PileUp, manually edited, and the consensus sequence was created by using the program Pretty, with the threshold for generating the consensus set at 2 by using the Blossum62 scoring matrix. The conserved residues are capitalized and are represented in the consensus sequence. The Asp residues at position 36 in C. albicans and at position 69 in S. cerevisiae, the Glu residue at position 61 in T. gondii, and the Arg residues at position 101 in C. albicans, 134 in S. cerevisiae, and 126 in T. gondii are highlighted within the boxes.
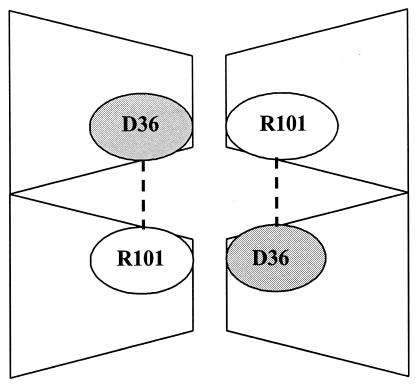
FIG. 4.
Schematic diagram depicting the proposed tetrameric structure of UPRT in C. albicans as a dimer of dimers. There is an attractive electrostatic force between Arg101 and Asp36 at the dimer interface. Mutations involving Arg101 would have the ability to destabilize the oligomer. The illustration is not intended to imply that Asp36 and Arg101 are directly responsible for tetramer formation.
DISCUSSION
Insights into the mechanism of action of 5-FC in yeasts were provided in seminal papers by Jund and Lacroute (8), Kern et al. (9, 10), and Normark and Schonebeck (14). The present work draws upon these studies to investigate the molecular mechanisms of primary resistance to 5-FC in C. albicans.
An understanding of 5-FC metabolism is central to this study. The pathways for de novo synthesis and the salvage of preformed pyrimidines along with the metabolism of 5-FC are illustrated in Fig. 1. Several additional points to those made in the introduction regarding pyrimidine salvage and 5-FC metabolism deserve emphasis. First, UPRT (also referred to as uracil pyrophosphorylase) is the critical regulatory enzyme in pyrimidine salvage, where it converts intracellular uracil to uridylate, thus enabling the recycling of pyrimidine bases. The second important point is the degree of feedback inhibition operating in both the salvage and de novo pathways. It has been shown in S. cerevisiae that in the presence of exogenous uracil, metabolism via the salvage pathway is favored due to the induction of FUR1 transcription and the simultaneous suppression of the de novo pathway (10). In normal circumstances, metabolic end products such as uridylate and UTP (13) exert feedback inhibition of both the salvage and de novo pathways, but the extent to which the respective fluorinated analogues exert the same action is unknown. It is possible that alterations in feedback inhibition could significantly influence the flux of 5-FC through the salvage pathway. This concept was supported by the generation of an S. cerevisiae strain, using UV mutagenesis, which was resistant to 5-FC due to the loss of normal negative feedback inhibition of de novo pyrimidine nucleotide synthesis (8) enabling UMP synthesized by the de novo pathway to compete with the fluorinated analogue 5-FUMP.
Early evidence implicated UPRT in 5-FC resistance in both S. cerevisiae and C. albicans (8-10, 13, 28). While null mutations of the gene coding for UPRT (FUR1) are not lethal because cellular pyrimidine nucleotide requirements can be met by de novo synthesis, they do render yeasts unable to utilize preformed cytosine or uracil (8, 9) and confer resistance to the fluorinated pyrimidines. We have demonstrated an association between the substitution of Arg for Cys at position 101 (Arg101Cys) in UPRT and 5-FC resistance in C. albicans. However, this association does not fully explain 5-FC resistance in this group of isolates, as isolate 12, which is homozygous for the Cys codon at position 101, is an isolate for which 5-FC MICs are only 8.0 mg/liter. Isolates that are heterozygous at this codon demonstrate a reduced susceptibility relative to the homozygous susceptible isolates (at least fourfold less susceptible). This observation is consistent with previous studies (28) in which intermediate levels of UPRT activity and 5-FC resistance in C. albicans were ascribed to a heterozygosity at the putative resistance locus. Transformation studies would be required to establish to what extent this substitution might alter susceptibility to 5-FC.
FUR1 was sequenced in wild-type S. cerevisiae in 1990 (9) and subsequently in a mutant resistant to 5-FU (10). Sequence data from the mutant revealed a point mutation whereby Arg was substituted for Ser at position 134 (Arg134Ser) (10). Figure 3 shows an alignment of the amino acid sequences of UPRT from S. cerevisiae and C. albicans which highlights that the mutated Arg134 residue identified by Kern et al. (10) aligns with Arg101 in C. albicans. Kern et al. (10) provided definitive evidence to associate Arg134Ser with resistance to 5-FU and therefore to 5-FC by using transformation studies but were unable to account for the mechanism by which the mutation induced resistance.
More recently, crystallographic studies of UPRT in both bacteria and eukaryotes have provided insights into the importance of the various quaternary arrangements required for optimal enzyme function (7, 23). The favored quaternary arrangement of UPRT in T. gondii is a tetramer comprised of two tight dimers (23). Dimer formation is facilitated by contacts involving arginine and acidic residues at the respective dimer interfaces. One of the residues particularly important in T. gondii, Arg126, is the cognate of Arg101 in C. albicans and Arg134 in S. cerevisiae. The dimer structure is further reinforced by protruding β arms which, in addition to their important structural role, function to cap the active site of the opposing monomer, thereby shielding bound substrate from bulk solvent (23). The dual role of the β arm and especially its involvement in the catalytic mechanism add credence to the concept that dimer formation is the minimal requirement for physiological activity (23). In addition, however, sedimentation studies suggest that the combination of two dimers to form a tetramer (a dimer of dimers) appears to be the optimal configuration for enzyme activity. There is strong evidence that GTP plays a critical role in this regard (22) since its binding pocket is formed by three monomeric units and GTP binding both stabilizes the tetramer and impacts upon enzyme kinetics by optimally aligning PRPP for catalysis (22). It is possible, therefore, that the effect of Arg101Cys in C. albicans (and Arg134Ser in S. cerevisiae) is due primarily to the disruption of the stability of the dimer and therefore the tetramer because of the interruption of the attractive electrostatic bond ordinarily present between Arg101 and Asp36. Further evidence to support this hypothesis would require sedimentation, three-dimensional structural, and biochemical studies of the mutant forms of UPRT from either yeast species.
Despite the frequent association between the Arg101Cys substitution in UPRT and resistance to 5-FC in both this study and that of Dodgson et al. (2) (where all the resistant isolates were members of one clade), the cumulative data suggest that additional mechanisms must be operational to fully account for primary resistance in C. albicans. For instance, it is not clear whether the Arg101Cys substitution in homozygous isolates can solely account for the MICs for these isolates, which range from 8.0 to >64 mg/liter in both this study and that of Dodgson et al. (3). Moreover, Dodgson et al. (2), described an isolate which was heterozygous at codon 101 for which 5-FC MICs were >128 mg/liter, which is at odds with the conclusion that clade-specific 5-FC resistance is due to the Arg101Cys substitution in UPRT. This data highlights (as acknowledged by Dodgson et al.) that other mechanisms must be present.
The prospect of more than one resistance mechanism is also supported by the early observation that there was a large degree of variability in the incorporation of fluorinated pyrimidine nucleotides into RNA for a group of isolates with otherwise similar levels of susceptibility to 5-FC (17). Finally, in this study, although isolate 16 displays high-level 5-FC resistance and isolates 10 and 11 have intermediate susceptibilities to 5-FC, the three isolates lack the Arg101Cys substitution in UPRT. Isolate 16 is the only one that exhibits the amino acid substitution at Gly28Asp in cytosine deaminase, and a Ser29Leu substitution in this protein is also associated only with isolate 11. It would be of interest, therefore, to study the population genetics of these isolates and explore the possibility, using transformation studies, that the Gly28Asp substitution in cytosine deaminase is directly responsible for 5-FC resistance. The existence of these three isolates (and indeed of isolate 8, for which 5-FC MICs were 1.0 mg/liter), nevertheless, leads to one of two conclusions, either of which extend the work of Pujol et al. (19) and Dodgson et al. (2). Either these isolates are genetically unrelated to those possessing the Arg101Cys substitution, in which case 5-FC resistance is not restricted to one clade, or mechanisms completely distinct and independent of this substitution are operational within isolates belonging to this clade.
This study does not provide strong evidence to implicate substitutions within the purine-cytosine permeases Fcy21 and Fcy22 as a cause of 5-FC resistance, although this remains possible. Indeed, the radical Ala176Gly substitution in Fcy22 was uniquely associated with one resistant isolate. Further work is clearly required to define the other mechanisms that are responsible for 5-FC resistance, which may include changes to the de novo pathway or alterations in gene expression.
Acknowledgments
This work was supported by an unrestricted educational grant from the Fungal Research Trust and from Merck & Co. M.J.A. is supported by the Wellcome Trust.
We thank Sue Howard, Andrew Sharp, and Caroline Moore for help with susceptibility testing and Paul Fullwood, Mark Bond, and Helen Ross for help with sequencing.
REFERENCES
- 1.Barchiesi, F., D. Arzeni, F. Caselli, and G. Scalise. 2000. Primary resistance to flucytosine among clinical isolates of Candida spp. J. Antimicrob. Chemother. 45:408-409. [DOI] [PubMed] [Google Scholar]
- 2.Dodgson, A. R., K. J. Dodgson, C. Pujol, M. A. Pfaller, and D. R. Soll. 2004. Clade-specific flucytosine resistance is due to single nucleotide change in the FUR1 gene of Candida albicans. Antimicrob. Agents Chemother. 48:2223-2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Erbs, P., F. Exinger, and R. Jund. 1997. Characterization of the Saccharomyces cerevisiae FCY1 gene encoding cytosine deaminase and its homologue FCA1 of Candida albicans. Curr. Genet. 31:1-6. [DOI] [PubMed] [Google Scholar]
- 4.Fasoli, M., and D. Kerridge. 1988. Isolation and characterization of fluoropyrimidine-resistant mutants in two Candida species. Ann. N. Y. Acad. Sci. 544:260-263. [DOI] [PubMed] [Google Scholar]
- 5.Henikoff, S., and J. G. Henikoff. 1992. Amino acid substitution matrices from protein blocks. Proc. Natl. Acad. Sci. USA 89:10915-10919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jensen, H. K., N. Mikkelsen, and J. Neuhard. 1997. Recombinant uracil phosphoribosyltransferase from the thermophile Bacillus caldolyticus: expression, purification, and partial characterization. Protein Expr. Purif. 10:356-364. [DOI] [PubMed] [Google Scholar]
- 7.Journal of Antimicrobial Chemotherapy. 1984. Laboratory methods for flucytosine (5-fluorocytosine). Report of a Working Group of the British Society for Mycopathology. J Antimicrob. Chemother. 14:1-8. [PubMed] [Google Scholar]
- 8.Jund, R., and F. Lacroute. 1970. Genetic and physiological aspects of resistance to 5-fluoropyrimidines in Saccharomyces cerevisiae. J. Bacteriol. 102:607-615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kern, L., J. de Montigny, R. Jund, and F. Lacroute. 1990. The FUR1 gene of Saccharomyces cerevisiae: cloning, structure and expression of wild-type and mutant alleles. Gene 88:149-157. [DOI] [PubMed] [Google Scholar]
- 10.Kern, L., J. de Montigny, F. Lacroute, and R. Jund. 1991. Regulation of the pyrimidine salvage pathway by the FUR1 gene product of Saccharomyces cerevisiae. Curr. Genet. 19:333-337. [DOI] [PubMed] [Google Scholar]
- 11.Kimura, M. 1983. The neutral theory of molecular evolution. Cambridge University Press, Cambridge, United Kingdom.
- 12.Lee, C., and K. Perry. 2001. The GeneMine system for genome/proteome annotation and collaborative data mining. IBM Syst. J. 40:592-603. [Google Scholar]
- 13.Natalini, P., S. Ruggieri, I. Santarelli, A. Vita, and G. Magni. 1979. Baker's yeast UMP:pyrophosphate phosphoribosyltransferase. Purification, enzymatic and kinetic properties. J. Biol. Chem. 254:1558-1563. [PubMed] [Google Scholar]
- 14.Normark, S., and J. Schonebeck. 1973. In vitro studies of 5-fluorocytosine resistance in Candida albicans and Torulopsis glabrata. Antimicrob. Agents Chemother. 2:114-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Page, R. D. 1996. TreeView: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12:357-358. [DOI] [PubMed] [Google Scholar]
- 16.Pfaller, M. A., S. A. Messer, L. Boyken, H. Huynh, R. J. Hollis, and D. J. Diekema. 2002. In vitro activities of 5-fluorocytosine against 8,803 clinical isolates of Candida spp.: global assessment of primary resistance using National Committee for Clinical Laboratory Standards susceptibility testing methods. Antimicrob. Agents Chemother. 46:3518-3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Polak, A., and H. J. Scholer. 1975. Mode of action of 5-fluorocytosine and mechanisms of resistance. Chemotherapy 21:113-130. [DOI] [PubMed] [Google Scholar]
- 18.Polak, A., and W. H. Wain. 1977. The influence of 5-fluorocytosine on nucleic acid synthesis in Candida albicans, Cryptococcus neoformans, and Aspergillus fumigatus. Chemotherapy 23:243-259. [DOI] [PubMed] [Google Scholar]
- 19.Pujol, C., M. A. Pfaller, and D. R. Soll. 2004. Flucytosine resistance is restricted to a single genetic clade of Candida albicans. Antimicrob. Agents Chemother. 48:262-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rodriguez-Tudela, J. L., F. Barchiesi, J. Bille, E. Chryssanthou, M. Cuenca-Estrella, D. Denning, J. P. Donnelly, B. Dupont, W. Fegeler, C. Moore, M. Richardson, and P. E. Verweij. 2003. Method for the determination of minimum inhibitory concentration (MIC) by broth dilution of fermentative yeasts. Clin. Microbiol. Infect. 9:i-vii. [Google Scholar]
- 21.Schmidt, R., M. F. Manolson, and M. R. Chevallier. 1984. Photoaffinity labeling and characterization of the cloned purine-cytosine transport system in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 81:6276-6280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schumacher, M. A., C. J. Bashor, M. H. Song, K. Otsu, S. Zhu, R. J. Parry, B. Ullman, and R. G. Brennan. 2002. The structural mechanism of GTP stabilized oligomerization and catalytic activation of the Toxoplasma gondii uracil phosphoribosyltransferase. Proc. Natl. Acad. Sci. USA 99:78-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schumacher, M. A., D. Carter, D. M. Scott, D. S. Roos, B. Ullman, and R. G. Brennan. 1998. Crystal structures of Toxoplasma gondii uracil phosphoribosyltransferase reveal the atomic basis of pyrimidine discrimination and prodrug binding. EMBO J. 17:3219-3232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stolz, J., and M. Vielreicher. 2003. Tpn1p, the plasma membrane vitamin B6 transporter of Saccharomyces cerevisiae. J. Biol. Chem. 278:18990-18996. [DOI] [PubMed] [Google Scholar]
- 25.Vermes, A., H. J. Guchelaar, and J. Dankert. 2000. Flucytosine: a review of its pharmacology, clinical indications, pharmacokinetics, toxicity and drug interactions. J. Antimicrob. Chemother. 46:171-179. [DOI] [PubMed] [Google Scholar]
- 26.Waldorf, A. R., and A. Polak. 1983. Mechanisms of action of 5-fluorocytosine. Antimicrob. Agents Chemother. 23:79-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weber, E., C. Rodriguez, M. R. Chevallier, and R. Jund. 1990. The purine-cytosine permease gene of Saccharomyces cerevisiae: primary structure and deduced protein sequence of the FCY2 gene product. Mol. Microbiol. 4:585-596. [DOI] [PubMed] [Google Scholar]
- 28.Whelan, W. L., and D. Kerridge. 1984. Decreased activity of UMP pyrophosphorylase associated with resistance to 5-fluorocytosine in Candida albicans. Antimicrob. Agents Chemother. 26:570-574. [DOI] [PMC free article] [PubMed] [Google Scholar]