Abstract
Stepwise selection of ciprofloxacin-resistant Haemophilus influenzae mutants produced first-, second-, third-, and fourth-step substitutions in GyrA (S84Y), ParC (S84R), GyrA (D88N), and ParC (E88K), respectively. Successive mutations raised the mutant selection window. The wild-type selection window for garenoxacin, levofloxacin, and moxifloxacin was also measured.
Fluoroquinolone resistance in highly susceptible, gram-negative organisms is thought to arise in a stepwise manner through nucleotide sequence changes occurring largely at mutational hot spots (quinolone resistance-determining regions [QRDRs]) of genes encoding gyrase (gyrA and gyrB) and DNA topoisomerase IV (parC and parE) (7). Mutants at each step are enriched when fluoroquinolone concentrations are within a specific range called the mutant selection window (19). The lower boundary of the window is approximated by MIC(99), the minimal concentration that blocks growth of 99% of cells in a culture. The upper boundary is the MIC of the least susceptible, next-step mutant. Above this concentration two resistance mutations must be acquired concurrently for growth. Since this occurs rarely, the upper boundary of the window is called the mutant prevention concentration (MPC). To characterize stepwise development of fluoroquinolone resistance in vitro, we determined QRDR alterations and changes in fluoroquinolone susceptibility [MIC(99) and MPC] with Haemophilus influenzae.
Strain ATCC 49247 was grown on chocolate II agar (Becton Dickinson and Co., Cockeysville, Md.) or as liquid cultures in Haemophilus test medium broth (HTM broth; Becton Dickinson and Co., Sparks, Md.) at 37°C in 5% CO2. Garenoxacin was obtained from Bristol-Myers Squibb (Wallingford, Conn.), moxifloxacin and ciprofloxacin were from Bayer Corp. (West Haven, Conn.), and levofloxacin was from R. W. Johnson Pharmaceutical Research Institute (Spring House, Pa.). Fluoroquinolone susceptibility [MIC(99)] was determined by counting colonies following plating of serial dilutions on fluoroquinolone-containing agar (Fig. 1A and 2). MPC, which estimates the MIC of resistant mutant subpopulations, was defined as the fluoroquinolone concentration at which no colony was recovered when more than 1010 cells were applied to agar plates (Fig. 1A and 2). Plates were screened for colonies every 24 h during incubation for 96 to 120 h to assure that colony number had stabilized. Colonies obtained at high fluoroquinolone concentration were composed of resistant mutants, as confirmed by regrowth on drug-free agar followed by transfer and growth on agar containing the fluoroquinolone concentration used for selection. Duplicate measurements gave similar results. Nucleotide sequence changes associated with loss of susceptibility were identified from regions of chromosomal DNA amplified by PCR as described previously (12).
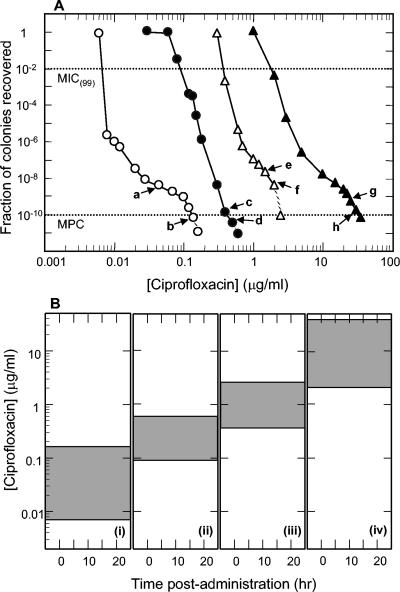
FIG. 1.
Stepwise enrichment of ciprofloxacin-resistant H. influenzae mutants. (A) Effect of ciprofloxacin concentration on recovery of colonies. Wild-type H. influenzae ATCC 49247 (open circles) was applied to ciprofloxacin-containing agar at the indicated concentrations, and colonies were recovered. Arrows indicate ciprofloxacin concentrations used to obtain colonies for which nucleotide sequence information was obtained. Arrows a and b indicate recovery of Ser-84-to-Tyr GyrA variants. Strain KD2308 (arrow b) was used for second-round selection (solid circles). Cells from positions c and d contained an additional parC mutation that changed Ser-84 to Arg. Strain KD2322 (arrow d) was used for a third round (open triangles). Cells from positions e and f contained an additional gyrA mutation that changed Asp-88 to Asn. Strain KD2364 (arrow f) was used for a fourth round (solid triangles). Cells from positions g and h contained an additional parC mutation that changed Glu-88 to Lys. Dashed lines, drug concentrations at which no colony was recovered; dotted lines, guides for determining MIC(99) and MPC, as indicated. (B) Relationship of mutant selection window to serum drug concentration. Shaded areas represent mutant selection window defined by MIC(99) and MPC, which were obtained from data shown in panel A. (i) Wild-type cells; (ii) first-step mutant; (iii) second-step mutant; (iv) third-step mutant.
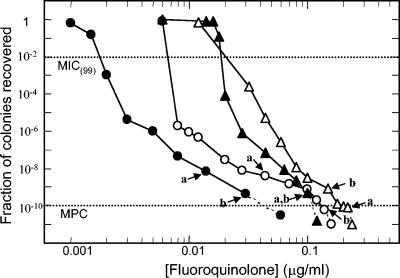
FIG. 2.
Effect of fluoroquinolone concentration on the recovery of first-step, resistant mutants. Wild-type H. influenzae strain ATCC 49247 was applied to agar plates containing the indicated concentrations of garenoxacin (solid circles), ciprofloxacin (open circles), levofloxacin (solid triangles), and moxifloxacin (open triangles). After incubation for 5 to 6 days, colonies were counted, and the fraction recovered relative to the input CFU was determined. Dashed lines, concentrations at which no colony was recovered; dotted lines, MIC(99) and MPC, as indicated. Arrows a and b indicate conditions used to isolate colonies from which the DNA sequence was determined (see Table 1).
Stepwise accumulation of fluoroquinolone resistance mutations was attained by sequential growth of laboratory strain ATCC 49247 on ciprofloxacin-containing agar. Application of wild-type cells to agar (Fig. 1A) allowed selection of a GyrA Ser-84-to-Tyr variant (Fig. 1A). One of these, strain KD2308, served as starter culture for a second round of selection. Colony recovery dropped sharply as ciprofloxacin concentration increased (Fig. 1A). Colonies recovered at concentrations indicated in Fig. 1A contained ParC variants in which Ser-84 changed to Arg. When the second-step gyrA parC mutant, strain KD2322, was applied to ciprofloxacin-containing agar, increasing drug concentration caused colony recovery to drop sharply, pass through an inflection, and drop a second time (Fig. 1A). Mutants recovered at concentrations indicated in Fig. 1A were GyrA variants in which Asp-88 changed to Asn. A third-step mutant, strain KD2364, was then used to select fourth-step mutants at ciprofloxacin concentrations indicated in Fig. 1A. Colonies obtained at the concentrations indicated in Fig. 1A contained a mutation that changed ParC position 88 from Glu to Lys. No mutation was observed in the QRDR of gyrB or parE (not shown). Each successive mutation raised the boundaries of the selection window (Fig. 1B). Each successive mutation also affected the composition of next-step mutant subpopulations, as indicated by the shapes of the recovery curves for the mutant (Fig. 1A).
When strain ATCC 49247 was applied to agar plates containing garenoxacin, levofloxacin, or moxifloxacin, colony recovery dropped with drug concentration as described for ciprofloxacin (Fig. 2). Two resistant mutants, recovered from agar plates containing high concentrations of each compound, had single nucleotide changes causing amino acid substitutions at position 84 or 88 of the GyrA protein (Table 1). Quinolones differed in selection of mutant subpopulations, as indicated by differences in shapes of mutant selection curves (Fig. 2).
TABLE 1.
Selection of resistant mutants by various quinolones
| Quinolone | MIC(99)a (mg/liter) | MPC (mg/liter) | GyrA change in QRDRb for mutant:
|
|
|---|---|---|---|---|
| a | b | |||
| Garenoxacin | 0.0018 | 0.06 | S84 to Y | S84 to Y |
| Levofloxacin | 0.019 | 0.12 | S84 to Y | S84 to Y |
| Ciprofloxacin | 0.007 | 0.16 | S84 to Y | S84 to Y |
| Moxifloxacin | 0.023 | 0.24 | D88 to Y | S84 to Y |
MIC using agar is 1.05 to 1.5 times higher than MIC(99) using agar (Fig. 2); broth MIC is higher than agar MIC by factors of 1, 2, and 3.7 for levofloxacin, ciprofloxacin, and moxifloxacin, respectively (15).
Quinolone concentrations used to obtain mutants are indicated in Fig. 2. Abbreviations: D, aspartic acid; S, serine; Y, tyrosine. First letter represents wild-type amino acid at the position indicated by the number. In each case the mutation changed the amino acid to Y.
GyrA and ParC changes have been observed with clinical isolates of H. influenzae (2-5, 8, 9, 12, 14, 17). GyrA mutants are probably the first to be enriched, as judged from the present in vitro studies (Table 1) and from the recovery of clinical isolates that contain only GyrA variants (2, 8, 9, 14). Since the patterns of topoisomerase changes reported for clinical isolates of H. influenzae (2, 5, 12) often differ from those found with the laboratory strain reported here, clinical isolates can follow a path to resistance different from that followed by the laboratory isolate.
The selection window for ciprofloxacin with wild-type cells was below serum drug concentrations measured at steady state in human volunteers receiving twice-daily doses of 500 mg (10). Comparable statements can be made about the three other compounds examined; this is consistent with the prevalence of fluoroquinolone resistance being low with H. influenzae (3, 4). Resistance may arise largely from sporadic situations in which abnormally low doses (dosing errors) and/or patient characteristics, such as chronic lung disease (16), cause drug concentration to fall into the selection window and allow enrichment of spontaneous gyrA mutants. Since increasing the time that drug concentration is inside the selection window should increase mutant enrichment, the development of resistance is expected to accelerate with each successive mutation. A key to preventing fluoroquinolone resistance in H. influenzae may be to block enrichment of the first gyrA mutation by strictly avoiding low-dose fluoroquinolone use.
The progressive, stepwise acquisition of resistance alleles, which has been likened to hill climbing (1), is likely to be common to many bacterial species. For example, gyrA and parC mutations accumulate in Neisseria gonorrhoeae and Escherichia coli, albeit with the additional occurrence of drug efflux mutations (1, 11, 18). For some species, such as Streptococcus pneumoniae, only two steps appear to be required for resistance (13), and only a single gyrase mutation may be necessary for resistance with Mycobacterium tuberculosis (6). Slowing the development of fluoroquinolone resistance may require adjusting dosing strategies to take into account the number of mutational steps involved in resistance.
Acknowledgments
We thank the following for critical comments on the manuscript: Marila Gennaro, Samuel Kayman, David Perlin, Richard, Pine, and Xilin Zhao.
The work was supported by NIH grant AI 35257, an unrestricted educational grant from Bristol-Myers Squibb, and the BMA Medical Foundation.
REFERENCES
- 1.Belland, R. J., S. G. Morrison, C. Ison, and W. M. Huang. 1994. Neisseria gonorrhoeae acquires mutations in analogous regions of gyrA and parC in fluoroquinolone-resistant isolates. Mol. Microbiol. 14:371-380. [DOI] [PubMed] [Google Scholar]
- 2.Biedenbach, D., and R. Jones. 2003. Five-year analysis of Haemophilus influenza isolates with reduced susceptibility to fluoroquinolones: prevalence results from the SENTRY antimicrobial surveillance program. Diagn. Microbiol. Infect. Dis. 46:55-61. [DOI] [PubMed] [Google Scholar]
- 3.Biedenbach, D., and R. Jones. 2000. Fluoroquinolone-resistant Haemophilus influenzae: frequency of occurrence and analysis of confirmed strains in the SENTRY antimicrobial surveillance program (North and Latin America). Diagn. Microbiol. Infect. Dis. 36:255-259. [DOI] [PubMed] [Google Scholar]
- 4.Bootsma, H., A. Troelstra, A. VanVeen-Rutgers, F. Mool, A. deNeeling, and B. Overbeek. 1997. Isolation and characterization of a ciprofloxacin-resistant isolate of Haemophilus influenzae from The Netherlands. J. Antimicrob. Chemother. 39:292-293. [DOI] [PubMed] [Google Scholar]
- 5.Davies, T., L. Kelly, D. Hoellman, L. Ednie, C. Clark, S. Bajaksouzian, M. Jacobs, and P. Appelbaum. 2000. Activities and postantibiotic effects of gemifloxacin compared to those of 11 other agents against Haemophilus influenzae and Moraxella catarrhalis. Antimicrob. Agents Chemother. 44:633-639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dong, Y., X. Zhao, B. Kreiswirth, and K. Drlica. 2000. Mutant prevention concentration as a measure of antibiotic potency: studies with clinical isolates of Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 44:2581-2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Drlica, K., and M. Malik. 2003. Fluoroquinolones: action and resistance. Curr. Top. Med. Chem. 3:1349-1364. [DOI] [PubMed] [Google Scholar]
- 8.Elliott, E., D. Oosthuizen, M. Johnson, and L. Piddock. 2003. Fluoroquinolone resistance in Haemophilus influenzae. J. Antimicrob. Chemother. 52:734-735. [DOI] [PubMed] [Google Scholar]
- 9.Georgiou, M., R. Munoz, F. Roman, R. Canton, R. Gomez-Lus, J. Campos, and A. G. D. L. Campa. 1996. Ciprofloxacin-resistant Haemophilus influenzae strains possess mutations in analogous positions of gyrA and parC. Antimicrob. Agents Chemother. 40:1741-1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gonzalez, M., F. Uribe, S. Moisen, A. Fuster, A. Selen, P. Welling, and B. Painter. 1984. Multiple-dose pharmacokinetics and safety of ciprofloxacin in normal volunteers. Antimicrob. Agents Chemother. 26:741-744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heisig, P. 1996. Genetic evidence for a role of parC mutations in development of high-level fluoroquinolone resistance in Escherichia coli. Antimicrob. Agents Chemother. 40:879-885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li, X., N. Mariano, J. J. Rahal, C. M. Urban, and K. Drlica. 2004. Quinolone-resistant Haemophilus influenzae in a long-term care facility: nucleotide sequence characterization of alterations in the genes encoding DNA gyrase and DNA topoisomerase IV. Antimicrob. Agents Chemother. 48:3570-3572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li, X., X. Zhao, and K. Drlica. 2002. Selection of Streptococcus pneumoniae mutants having reduced susceptibility to levofloxacin and moxifloxacin. Antimicrob. Agents Chemother. 46:522-524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Perez-Vasquez, M., F. Roman, B. Aracil, R. Canton, and J. Campos. 2003. In vitro activities of garenoxacin (BMS-28756) against Haemophilus influenzae isolates with different fluoroquinolone susceptibilities. Antimicrob. Agents Chemother. 47:3539-3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Perez-Vazquez, M., F. Roman, M. Varela, R. Canton, and J. Campos. 2003. Activities of 13 quinolones by three susceptibility testing methods against a collection of Haemophilus influenzae isolates with different levels of susceptibility to ciprofloxacin: evidence for cross-resistance. J. Antimicrob. Chemother. 51:147-151. [DOI] [PubMed] [Google Scholar]
- 16.Turato, G., R. Zuin, and M. Saetta. 2001. Pathogenesis and pathology of COPD. Respiration 68:117-128. [DOI] [PubMed] [Google Scholar]
- 17.Vila, J., J. Ruiz, F. Sanchez, F. Navarro, B. Mirelis, M. Jimenez DeAnta, and G. Prats. 1999. Increase in quinolone resistance in a Haemophilus influenzae strain isolated from a patient with recurrent respiratory infections treated with ofloxacin. Antimicrob. Agents Chemother. 43:161-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yasuda, M., H. Fukuda, S. Yokoi, S. Ishihara, Y. Kawada, and T. Deguchi. 2000. In vitro selection of fluoroquinolone-resistant Neisseria gonorrhoeae harboring alterations in DNA gyrase and topoisomerase IV. J. Urol. 164:847-851. [DOI] [PubMed] [Google Scholar]
- 19.Zhao, X., and K. Drlica. 2001. Restricting the selection of antibiotic-resistant mutants: a general strategy derived from fluoroquinolone studies. Clin. Infect. Dis. 33(Suppl. 3):S147-S156. [DOI] [PubMed] [Google Scholar]