Abstract
Whether or not resistant mutants will be present before the start of antibiotic treatment of an initially susceptible population of bacteria depends on the size of the infecting population, the rate of mutation to resistance, and the amount of time that the population has been maintained. In the present investigation, we argue that for the treatment of chronic infections caused by hypermutable Pseudomonas aeruginosa of the sort frequently found in cystic fibrosis patients, mutants resistant to all single antipseudomonal drugs will almost invariably be present in a high proportion at the onset of treatment, and consequently, these strains should be considered resistant to all agents when they are used as monotherapy. Using a construct of P. aeruginosa strain PAO1 with a mutS deletion (strain PAOΔmutS), we show that when in vitro populations of less than 5 × 104 seemingly susceptible hypermutable bacteria are confronted with any of 11 antipseudomonal agents, mutants for which the MICs and the minimum bactericidal concentrations are in the range of clinical resistance will almost invariably ascend to dominance within 24 to 36 h. This does not occur for PAO1 without the mutS deletion. The results of our detailed analysis of this evolution of acquired resistance to two of these antibiotics, imipenem and ciprofloxacin, indicate that although the rates of mutation to resistance in PAOΔmutS are on the order of 1 × 10−6 per generation, resistant mutants are very likely to either be present in cultures of between 2 × 104 and 4 × 104 bacteria or arise after the bacterial populations are confronted with antibiotics. We also demonstrate with in vitro experiments that the problem of acquired resistance to treatment with single antibiotics can be thwarted by combination therapy with pairs of antibiotics of different classes with synergistic activities. We discuss the clinical implications of our analysis of these observations.
Chronic lung infection with Pseudomonas aeruginosa is the major cause of morbidity and mortality in cystic fibrosis (CF) patients (8, 10, 22). While long-term treatment with antipseudomonal agents is necessary to avoid a fast decline in the respiratory functions of the infected patients, mutants resistant to multiple antimicrobials almost invariably evolve and lead to treatment failure. P. aeruginosa, unlike many other bacteria, can generate mutants that are resistant to the clinical concentrations of all antimicrobial agents used for therapy by making changes in single genes (9, 11, 12, 16, 20, 43, 45).
On first consideration it would seem that this problem of acquired resistance due to chromosomal gene mutations in P. aeruginosa strains causing infections in CF patients is further exacerbated by the anticipated presence of strains with elevated mutation rates at high frequencies. The presence of hypermutable (mutator) P. aeruginosa strains in the lungs of CF patients at high frequencies has been reported previously (29). Mutators represented 20% of the total P. aeruginosa isolates in CF patients and were found in 37% of the patients. In those patients, the overall proportion of mutator isolates was 43%. In that study, it was also found that for all antibiotics the resistance rates of mutator isolates were substantially higher than those of nonmutator isolates. The majority of the mutator P. aeruginosa strains isolated from these CF patients are deficient in the mismatch repair system, with mutS being the gene most frequently involved (30).
Are these hypermutable strains responsible for the generation of the mutants responsible for acquired resistance in the P. aeruginosa strains infecting CF patients, or are these mutator strains the product of antibiotic-mediated selection in treated CF patients? In theory at least, when populations of bacteria are challenged with novel environmental stresses, like antibiotic treatment, selection can favor the evolution of mutator genes (23, 40, 41). If these two products of antibiotic-mediated selection, acquired resistance and the evolution of hypermutation, are broadly applicable, then it is likely that they will act synergistically. Antibiotic-mediated selection leads to the evolution of mutator genes as well as to the ascent of resistance, and once they are present, strains with mutator genes augment the likelihood of the subsequent generation of mutants resistant to additional antibiotics.
In the investigation described here we explored the effects of hypermutation on the likelihood of acquired resistance in P. aeruginosa causing chronic infections in antibiotic-treated hosts. We show that in vitro, mutants resistant to all single antipseudomonal drugs are likely to be present in even relatively small populations of hypermutable P. aeruginosa or are likely to evolve after these bacteria are confronted with antibiotics. We also show that this effective resistance to treatment of seemingly susceptible populations of mutator P. aeruginosa can be overcome by therapy with multiple antibiotics that act synergistically. We discuss the clinical implications of these results and recommend that as a consequence of the anticipated presence of hypermutable strains, first-line treatment for chronic P. aeruginosa infections be with combinations of drugs rather than single drugs.
MATERIALS AND METHODS
Construction of PAOΔmutS strain.
A 2.6-kb BamHI fragment containing the wild-type mutS gene from P. aeruginosa strain PAO1 obtained from plasmid pMBMS (30) was ligated to plasmid pJQ200mp18 (34) digested with the same enzyme to obtain plasmid pJQMS, which was transformed into Escherichia coli XL1Blue made competent by treatment with CaCl2. Transformants were selected on Luria-Bertani (LB) agar plates with 30 μg of gentamicin per ml. Plasmid pJQ200mp18 acts as a suicide vector in Pseudomonas and contains the RP4 transfer origin; the sacB gene, which has a lethal effect when 5% sucrose is present; and a gentamicin resistance (Gmr) marker [AAC(3)-I]. Plasmid pJQMS was digested with NotI, which liberates a 1.5-kb internal fragment of the mutS gene, and then ligated with a 1.2-kb kanamycin resistance cassette digested with the same enzyme to obtain plasmid pJQMSKm, which was subsequently transformed into E. coli XL1Blue. Transformants were selected on LB agar plates containing 50 μg of kanamycin per ml and 30 μg of gentamicin per ml. The kanamycin resistance (Kmr) cassette was obtained by PCR amplification of the Kmr gene [APH(3′)I] from plasmid pBGS18− (37) with specific primers containing NotI restriction sites (underlined): 5′-AAGGAAAAAAGCGGCCGCGTTGTAGGTGGACCAGTTGGTG-3′ and 5′-AAGGAAAAAAGCGGCCGCGAAGTCAGCGTAATGCTCTGCC-3′. Plasmid pJQMSKm was transformed into E. coli S17λ-pir (5), from which it was mobilized by conjugation into P. aeruginosa strain PAO1. The recombination of the ΔmutS::Km region of plasmid pJQMSKm into the PAO1 chromosome was performed in two steps. First, recombinants with a single crossing over (with the whole plasmid recombined) were selected after mating experiments by using S17λ-pir(pJQMSKm) as the donor and PAO1 as the recipient on LB agar plates containing 100 μg of kanamycin per ml, 50 μg of ampicillin per ml, and 30 μg of gentamicin per ml. After overnight incubation in LB broth, double-crossing-over recombinants were then selected on LB agar plates containing 100 μg of kanamycin per ml and 5% sucrose. Growing colonies were checked for gentamicin susceptibility and tested by PCR amplification with combinations of mutS-specific internal and external primers.
Complementation of PAOΔmutS with PAO1 wild-type gene.
The 2.6-kb BamHI fragment containing the wild-type mutS gene from P. aeruginosa strain PAO1 was ligated to plasmid pUCP24 (42), which had been digested with the same enzyme, to obtain plasmid pUCPMS, which was transformed into E. coli XL1Blue. Transformants were selected on LB agar plates containing 30 μg of gentamicin per ml. pUCPMS was then electroporated into PAOΔmutS, as described previously (36). PAOΔmutS(pUCPMS) transformants were selected on LB agar plates containing 50 μg of gentamicin per ml. Complementation of mutS deficiency was demonstrated by the reversion of the increased rate of mutation for rifampin resistance. PAOΔmutS(pUCPMS) was used as a control to demonstrate that the observed phenotype of PAOΔmutS was a consequence of the mutS deletion and not of secondary mutations that may occur during gene replacement (13).
Determination of MICs, MBCs, and killing kinetics.
MICs, minimal bactericidal concentrations (MBCs), and killing kinetics were determined by the broth microdilution method according to the recommendations of NCCLS (26, 27). The bacterial inoculum was prepared by the same procedure; approximately 103 cells from overnight cultures were inoculated into tubes containing 10 ml of Mueller-Hinton broth, and the tubes were incubated at 37°C with strong agitation until the mid-log phase of growth (approximately 108 cells/ml). Then, 2 × 104 to 4 × 104 cells from these cultures were inoculated into each microdilution well (1 × 105 to 2 × 105 CFU/ml). For MBC determination, 100-μl aliquots, as well as serial 1/10 dilutions, from all wells with no visible growth after 16 h of incubation were plated onto Mueller-Hinton agar (MHA) plates, which were incubated for 24 h. The MBC was defined as the lowest concentration that brought about a >99.9% reduction in the size of the initial inoculum. Killing kinetics studies with imipenem (4 and 8 μg/ml) and ciprofloxacin (0.5 and 1 μg/ml) were performed as for determination of MBCs, but aliquots (and serial 1/10 dilutions) were plated on MHA plates at 1, 2, 3, 4, 5, 6, 12, and 24 h. Final results represent the median values of six independent experiments. For synergy studies the antibiotics were combined in a ratio proportional to their respective MICs, and MICs and MBCs were determined as described above.
Estimation of mutation rates.
Approximately 102 cells from overnight cultures were inoculated into 10 tubes, each of which contained 10 ml of Mueller-Hinton broth, and the tubes were incubated at 37°C with strong agitation for 24 h. Aliquots from successive dilutions were plated onto MHA plates with and without the different antibiotics. The numbers of colonies growing after 24 h of incubation were counted, and the mutation rate (number of mutants per cell per division) was estimated by a method described elsewhere (3). The final result represents the mean value of two independent experiments. For the definition of the mutation rates, MHA plates containing 300 μg of rifampin per ml were used as described previously (29, 30). Additionally, rates of mutation to imipenem (4 and 8 μg/ml) and ciprofloxacin (0.5 and 1 μg/ml) resistance were also estimated by the same procedure.
Characterization of antibiotic-resistant PAOΔmutS mutants.
For the characterization of β-lactam-resistant mutants, the chromosomal β-lactamase activity was quantified by measuring the nitrocefin hydrolysis rates (micromoles of substrate hydrolyzed per minute and per milligram of protein), as described previously (31). For the characterization of imipenem-resistant mutants, oprD was amplified by PCR with primers OprDF (5′-CGCCGACAAGAAGAACTAG-3′) and OprDR (5′-GTCGATTACAGGATCGACAG-3′), designed to amplify a 1.4-kb DNA fragment containing oprD from the published genome sequence of PAO1 (39). For the characterization of ciprofloxacin-resistant mutants, the quinolone resistance-determining regions of gyrA and parC were amplified by PCR as described previously (9). In all cases, two independent PCR products were sequenced from both strands. The BigDye Terminator kit (PE-Applied Biosystems, Foster City, Calif.) was used for the sequencing reactions, and the sequences were analyzed with an ABI Prism 3100 DNA sequencer (PE-Applied Biosystems).
RESULTS
Strain PAOΔmutS was constructed as described in Materials and Methods. As expected, the deletion of mutS produced an increase in the spontaneous mutation rate of more than 2 orders of magnitude. As estimated from fluctuation test experiments, the rates of mutation to rifampin (300 μg/ml) resistance were 4.9 × 10−9 for PAO1 and 1.0 × 10−6 for PAOΔmutS. Complementation of PAOΔmutS with the cloned wild-type gene restored the mutation rate back to the levels for PAO1 [mutation rate of PAOΔmutS(pUCPMS), 7.4 × 10−9].
Mismatch repair inactivation immediately increases the MICs and MBCs of antipseudomonal agents for P. aeruginosa.
The MICs and MBCs of 11 antipseudomonal agents used to treat lung infections in CF patients were determined for strains PAO1 and PAOΔmutS. When the MICs were estimated at 16 h, they were essentially identical for both strains. On the other hand, while all agents tested were highly bactericidal for PAO1 (the MBCs were within 1 dilution of the MICs), for all the replicates a significant increase in the MBCs (median increases in the MBCs of the different antibiotics, 4- to 16-fold) was observed for strain PAOΔmutS (Table 1). Additionally, when the turbidities of these cultures were examined after prolonged incubation (36 h), significant increases in the MICs of all the antibiotics were observed for PAOΔmutS but not for PAO1 (Table 1). The increased resistance was demonstrated to be dependent on mutS inactivation, since when PAOΔmutS harbored a plasmid (pUCPMS) with the wild-type gene, the MBCs and the MICs for the strain at 36 h were found to be essentially identical to those for PAO1.
TABLE 1.
MICs and MBCs for PAO1 and PAOΔmutS strains
| Antibiotic | MIC (μg/ml)
|
MBC (μg/ml)
|
MBC ratio (PAOΔmutS/PAO1) | NCCLS breakpoint (S-R)c | |||
|---|---|---|---|---|---|---|---|
| PAO1 (16 h)a,b | PAOΔmutS (16 h)b | PAOΔmutS (36 h)b | PAO1b | PAOΔmutSb | |||
| Ceftazidime | 1 | 1 | 16 | 2 | 32 | 16 | ≤8-≥32 |
| Cefepime | 1 | 1 | 4 | 1 | 8 | 8 | ≤8-≥32 |
| Imipenem | 1 | 1 | 16 | 2 | 16 | 8 | ≤4-≥16 |
| Meropenem | 0.12 | 0.25 | 2 | 0.12 | 2 | 16 | ≤4-≥16 |
| Ticarcillin | 16 | 16 | 64 | 16 | 128 | 8 | ≤64-≥128 |
| Piperacillin | 8 | 8 | 64 | 16 | 128 | 8 | ≤64-≥128 |
| Tobramycin | 0.25 | 0.5 | 4 | 0.5 | 4 | 8 | ≤4-≥16 |
| Gentamicin | 0.25 | 0.5 | 8 | 1 | 16 | 16 | ≤4-≥16 |
| Amikacin | 1 | 2 | 32 | 2 | 32 | 16 | ≤16-≥64 |
| Colistin | 0.25 | 0.25 | 0.5 | 0.25 | 1 | 4 | NDd |
| Ciprofloxacin | 0.06 | 0.06 | 1 | 0.12 | 2 | 16 | ≤1-≥4 |
No changes in the MICs for PAO1 were observed after 36 h of incubation.
The values are medians from six different experiments. The estimated MICs at 36 h and the MBCs for PAOΔmutS from different replicates varied within 1 dilution.
S-R, standard for nonsusceptibility and resistance breakpoints, respectively.
ND, not defined.
Genetic rather than physiological resistance.
To ascertain whether the higher MBCs and the higher estimates of the MICs for the mutator strain at 36 h were due to a physiological response or inherited resistance, these parameters were estimated for 10 surviving colonies (from each of three independent experiments) isolated from the microdilution wells with the breakpoint MBCs of the 11 antipseudomonal agents. After subculture on antibiotic-free medium, the MICs (16 h) for the selected colonies were equal to or greater than the antibiotic concentration in the microdilution wells from which they were isolated. Thus, it is reasonable to interpret these findings as indicating that the increases in the MICs and the MBCs for PAOΔmutS shown in Table 1 were a consequence of the selection of resistant mutants rather than a physiological response.
The level of resistance of PAOΔmutS in the MBC experiments and the MIC experiments at 36 h reached or surpassed the NCCLS breakpoints for nonsusceptibility to 7 of the 11 antibiotics used to challenge the strains (ticarcillin, piperacillin, ceftazidime, imipenem, gentamicin, amikacin, and ciprofloxacin) and remained just 1 dilution below the breakpoints for two of these antibiotics (tobramycin and cefepime). Although the meropenem MICs for the PAOΔmutS mutants isolated from these cultures were 16-fold greater than those for their susceptible ancestors, the MICs for these mutants were 2 dilutions below the NCCLS breakpoints for nonsusceptibility (Table 1). Finally, the MIC of colistin for the PAOΔmutS mutants isolated was only fourfold greater than that for the ancestors. At this time, NCCLS has not recommended breakpoints for nonsusceptibility to colistin.
To determine if the nature of the resistant mutants selected during the MBC experiments and the MIC experiments at 36 h was consistent with that expected from our knowledge of mutational antibiotic resistance mechanisms in P. aeruginosa, a number of isolates were further characterized. Five colonies (from five independent experiments) recovered from microdilution wells with 16 μg of ceftazidime (as a representative of β-lactams) per ml, 4 μg of imipenem (as a representative of carbapenems) per ml, and 0.5 μg of ciprofloxacin (as a representative of fluoroquinolones) per ml were characterized after passage in antibiotic-free medium. The MICs (16 h) of ceftazidime for the selected colonies ranged from 64 to 128 μg/ml, and the colonies showed cross-resistance to ticarcillin, piperacillin, and cefepime, a phenotype consistent with AmpC hyperproduction, which is the most frequent mutational mechanism of β-lactam resistance in P. aeruginosa (9). Quantification of the chromosomal β-lactamase activity revealed that all the mutants were in fact AmpC hyperproducers. The β-lactamase activities of the five selected mutants were 81, 40, 22, 272, and 114 times higher, respectively, than that of the parent strain, PAOΔmutS. The MICs (16 h) of imipenem for the selected colonies ranged from 16 to 32 μg/ml, whereas those of meropenem ranged from 4 to 8 μg/ml, a phenotype consistent with OprD inactivation, the most frequent imipenem resistance mechanism in P. aeruginosa (32). Sequencing of oprD revealed that three of the five mutants contained mutations (in boldface) that lead to the creation of a premature stop codon: W339X (TGG to TGA at nucleotide 1017), W277X (TGG to TAG at nucleotide 830), and W277X (TGG to TGA at nucleotide 831). A fourth mutant contained the missense mutation S278P (TCC to CCC at nucleotide 832), which, to the best of our knowledge, has not been described previously. Finally, the last mutant did not contain any mutation in oprD, suggesting that mutations in regulatory systems affecting oprD expression could be involved, as has been noted previously for a few clinical strains (32). The MICs (16 h) of ciprofloxacin for the selected colonies ranged from 1 to 2 μg/ml. Sequencing of the quinolone resistance-determining regions of gyrA and parC revealed no mutations in any of the five selected mutants, ruling out the possibility that mutation in the topoisomerases is a fluoroquinolone resistance mechanism. On the other hand, analysis of the resistance patterns of all five mutants revealed no modifications in the MICs of ceftazidime, imipenem, meropenem, and gentamicin (MICs were within 1 dilution), whereas a four- to eightfold increase in the MIC of cefepime was observed. Extended antibiotic susceptibility testing revealed no modifications of the tetracycline MICs, whereas a significant (more than twofold dilution) increase in chloramphenicol MICs was noted. This resistance pattern is typical of nfxB-type mutants and is known to be produced by hyperexpression of the MexCD-OprJ efflux system, which has been found to be selected for by quinolones both in vitro and in vivo (24, 25).
Dynamics of selection process.
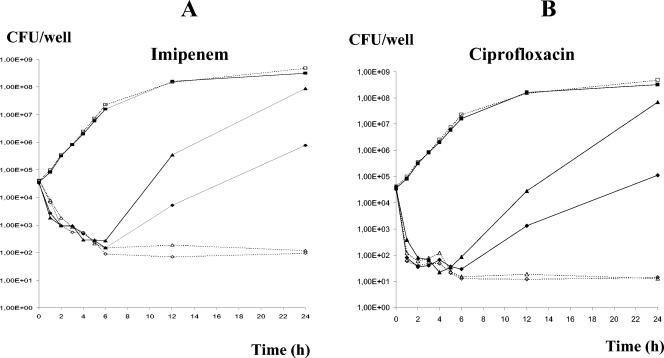
To explore in more detail the selection for resistance in the MBC assays and the assays of the MICs at 36 h, killing kinetics experiments were conducted with PAO1 and PAOΔmutS and two representative antipseudomonal agents, imipenem and ciprofloxacin. The two highest concentrations of antibiotics at which mutants were selected in the MIC and MBC experiments were used: 4 and 8 μg/ml for imipenem and 0.5 and 1 μg/ml for ciprofloxacin. As can be seen in Fig. 1A and B, the initial killing kinetics were very similar for strains PAO1 and PAOΔmutS, with these antibiotics having marked bactericidal effects in the first few hours. Starting at approximately 6 h, the number of CFU of the PAOΔmutS mutant but not of the wild-type strain increased in the antibiotic-containing cultures. As was the case in the original MBC experiments and MIC experiments at 36 h, the colonies isolated after the recovery of PAOΔmutS were resistant to clinically achievable concentrations of these antibiotics. Moreover, as observed in the MBC experiments and the MIC experiments at 36 h, the killing kinetics for strain PAOΔmutS harboring plasmid pUCPMS were nearly identical to those for PAO1.
FIG. 1.
Killing kinetics for PAO1 (dashed lines and open symbols) and PAOΔmutS (solid lines and solid symbols). (A) Imipenem killing kinetics. Squares, control with no antibiotic; triangles, imipenem at 4 μg/ml; diamonds, imipenem at 8 μg/ml. (B) Ciprofloxacin killing kinetics. Squares, control with no antibiotic; triangles, ciprofloxacin at 0.5 μg/ml; diamonds; ciprofloxacin at 1 μg/ml.
Likelihood of preexisting mutations.
Can the ascent of resistant Pseudomonas strains in the MIC, MBC, and killing kinetics experiments described above with PAOΔmutS but not with PAO1 be attributed to preexisting resistant mutants among the 2 × 104 to 4 × 104 bacteria used to initiate these experiments? To address this question, we estimated the rate of spontaneous mutation of PAO1 and PAOΔmutS to imipenem (4 and 8 μg/ml) and ciprofloxacin (0.5 and 1 μg/ml) resistance by fluctuation test experiments and by the method described in the Appendix. With these estimated mutation rates and the method described in the Appendix, we calculated the probability that one or more resistant mutants were present in these initial inocula.
As would be anticipated, the rate of mutation to resistance for the mutator strain was substantially greater than that for the nonmutator strain, with the increase ranging from 2 to 3 orders of magnitude (Table 2). On the basis of these mutation rates and the calculations presented in the Appendix, the probability that one or more ciprofloxacin- or imipenem-resistant mutants were present among the 4 × 104 PAO1 cells used in the MBC experiments, the MIC experiments at 36 h, and the killing kinetic experiments was less than 0.001 for both of these antibiotics at both concentrations. The corresponding probabilities that one or more resistant mutants were present in the cultures initiated with 4 × 104 PAOΔmutS bacteria are 0.99, 0.36, 0.52, and 0.33 for imipenem at 4 μg/ml, imipenem at 8 μg/ml, ciprofloxacin at 0.5 μg/ml, and ciprofloxacin at 1 μg/ml, respectively.
TABLE 2.
Estimates of rates of mutation to ciprofloxacin and imipenem resistance for PAO1 and PAOΔmutS
| Antibiotic and concn | Mutation rate (no. of mutants per cell per division)
|
|
|---|---|---|
| PAO1 | PAOΔmutS | |
| Ciprofloxacin | ||
| 1 μg/ml | 1.2 × 10−8 | 1.1 × 10−6 |
| 0.5 μg/ml | 2.2 × 10−8 | 1.9 × 10−6 |
| Imipenem | ||
| 8 μg/ml | 6.5 × 10−9 | 1.1 × 10−6 |
| 4 μg/ml | 1.3 × 10−8 | 1.2 × 10−5 |
To ascertain whether the observed frequencies of PAOΔmutScultures with one or more imipenem- or ciprofloxacin-resistant mutants were consistent with those expected from these calculations, we set up 40 microtiter well cultures with between 2 × 104 and 4 ×104 bacteria and either 0.5 or 1 μg of ciprofloxacin per ml or 4 or 8 μg of imipenem per ml. To minimize the probability that this hypothesis would be rejected due to mutant jackpots (21) in the original inoculum, the bacteria in these wells were derived from 10 independent overnight cultures, each initiated with a different PAOΔmutS colony, that were used to inoculate four of the cultures containing ciprofloxacin and imipenem. Of the 40 cultures used for each treatment, the numbers that were turbid at 36 h were 40, 15, 35, and 17 for imipenem at 4 μg/ml, imipenem at 8 μg/ml, ciprofloxacin at 0.5 μg/ml, and ciprofloxacin at 1 μg/ml, respectively. Assuming that these cultures were initiated with 4 × 104 cells and with the above estimates of the expected frequencies of cultures with one or more resistant bacteria, based on a chi-square goodness-of-fit test (with 1 degree of freedom with Yates' correction), there was only one statistically significant deviation from the expected number of cultures with one or more resistant cells. There were too many cultures with one or more mutants resistant to 0.5 μg of ciprofloxacin per ml (P < 0.001). There is evidence that in E. coli ciprofloxacin may induce resistance mutations among the surviving exposed cells (35). At this time, we are exploring some form of adaptive mutation mechanism as the explanation for this greater than expected number of resistant mutants in the cultures with 0.5 μg of ciprofloxacin per ml.
Use of combinations of antipseudomonal agents to minimize the selection of hypermutable antibiotic-resistant mutants.
When the activities of the antipseudomonal agents against strain PAOΔmutS are considered, it seems reasonable that no antimicrobial agent should be used alone for the treatment of lung infections in patients with CF, in whom the prevalence of Pseudomonas mutator variants is high, due to the inevitable selection of resistant mutants and the immediate dominance of the resulting resistant population. To explore the potential activities of combinations of antipseudomonal agents against mutator P. aeruginosa strains, synergy studies were conducted with strains PAO1 and PAOΔmutS. The MICs and MBCs of several combinations of two antimicrobial agents are shown in Table 3. No significant modifications of the MICs or the MBCs for PAO1 were observed with any of the combinations compared with those obtained with the individual antimicrobial agents (Table 3). On the other hand, the increases in the MBCs of the individual antibiotics previously found for PAOΔmutS almost disappeared with most of the combinations of two bactericidal antipseudomonal agents. As shown in Table 3, the MBCs of the antibiotic combinations were 2- to 16-fold lower than those of the individual agents for strain PAOΔmutS. Nevertheless, a twofold-higher MBC of most combinations was observed when those for PAOΔmutS were compared with those for PAO1, suggesting that very low (peri-inhibitory) concentrations of two antibiotics could select for phenotypes of low-level multidrug resistance (28).
TABLE 3.
MICs and MBCs of selected antibiotic combinations for PAO1 and PAOΔmutS strains
| Antibiotic combination | MIC (μg/ml)
|
MBC (μg/ml)
|
MBC ratio for PAOΔmutS/PAO1a | MBC ratio for PAOΔmutS (individual/combination)b | ||
|---|---|---|---|---|---|---|
| PAO1 (16 h) | PAOΔmutS (16 h) | PAO1 | PAOΔmutS | |||
| Ceftazidime with: | ||||||
| Ciprofloxacin | 1/0.06 | 1/0.06 | 2/0.12 | 4/0.25 | 2 | 8 |
| Tobramycin | 1/0.25 | 1/0.25 | 1/0.25 | 2/0.5 | 2 | 16 |
| Imipenem with: | ||||||
| Ciprofloxacin | 1/0.06 | 1/0.06 | 1/0.12 | 2/0.25 | 2 | 8 |
| Tobramycin | 2/0.5 | 2/0.5 | 2/0.5 | 4/1 | 2 | 4 |
| Meropenem with: | ||||||
| Ciprofloxacin | 0.12/0.06 | 0.12/0.06 | 0.25/0.12 | 0.5/0.25 | 2 | 4 |
| Tobramycin | 0.12/0.25 | 0.25/0.5 | 0.25/0.5 | 1/2 | 4 | 2 |
| Ciprofloxacin and tobramycin | 0.12/0.5 | 0.12/0.5 | 0.12/0.5 | 0.25/1 | 2 | 4 |
Ratio of MBC for strain PAOΔmutS and MBC for strain PAO1 for the selected antibiotic combinations.
Ratio of the MBC of each agent alone and the MBC of the antibiotic combination for strain PAOΔmutS.
DISCUSSION
In theory at least, if resistant mutants are not initially present, treatment failure due to the ascent of resistant mutants can be avoided by aggressive treatment with even single antibiotics (6, 18). This “hit-them-hard” approach to antimicrobial chemotherapy, which Paul Ehrlich already recommended early last century (7), is less likely to be effective if resistant mutants are already present in the treated population of microbes. The results of this study, together with those presented elsewhere (29), suggest that in patients with chronic P. aeruginosa infections, such as those in CF patients, mutants resistant to virtually all single antipseudomonal agents should be anticipated to be already present in a high proportion prior to treatment.
There are several reasons to anticipate the existence of substantial numbers of antibiotic-resistant mutants in chronic P. aeruginosa infections prior to treatment with the selecting antibiotics. The first is the large numbers of P. aeruginosa bacteria in infecting populations, typically greater than 107 CFU/g of sputum (44). The second is the likely presence of hypermutable strains and, thus, elevated rates of mutation in the bacterial population. The third is the impressive capacity of P. aeruginosa to adapt to environmental changes by mutation and selection. Perhaps because of its exceptionally large genome (6.2 Mb) (39), P. aeruginosa can become resistant to the clinically achievable concentrations of virtually all agents used for therapy as a result of mutations of single chromosomal genes, and this often occurs in treated hosts (20). Among the mechanisms predisposing P. aeruginosa to inherited resistance to antimicrobial agents are the impressive number of efflux systems encoded in its genome. The complex of efflux machinery provides P. aeruginosa with intrinsic resistance to high concentrations of several antibiotics and low-level resistance to many others. As a consequence of regulatory and other mutations leading to the hyperexpression of these efflux mechanisms, these intrinsic low-level resistance mechanisms can result in inherited resistance to high concentrations of other antibiotics (25, 33). Another example could be AmpC hyperproduction, at least compared with the level of production by E. coli. Whereas AmpC hyperproduction in E. coli depends exclusively on a few specific point mutations in the β-lactamase promoter region, P. aeruginosa has many targets that can be mutated for this purpose, including several mutations in ampR (and the ampR-ampC intergenic region), any inactivating mutation in ampD, and mutations in other genes that have not yet been characterized, since many AmpC-hyperproducing strains contain no mutations in these two genes (2).
Finally, the bacteria in a chronic infection replicate as well as die, and in the course of this birth-death process they produce antibiotic resistance and other mutations. Save for occasional purging by selective sweeps, or “periodic selection” (1, 15, 17), resistant mutants will continue to accumulate even in the absence of the antibiotics selecting for their ascent. Equilibrium will be reached when the rate of production of new mutants is exactly equal to their rate of loss due to mutation to the wild type and selection against them. For example, if the rate of mutation to resistance to a particular antibiotic is 10−6 and resistance engenders a 1% fitness cost, then at equilibrium the relative frequency of resistant mutants would be approximately 10−4 (4). Stated another way, in a population of 109 bacteria, 105 would be resistant to that antibiotic prior to its use.
While the present experiments do not directly simulate the accumulation of mutants in a chronic infection, they do account for the anticipated increase in the number of mutants in the course of replication. The reasons for this are beautifully described in Luria and Delbruck's classical paper (21) and can also be seen with a numerical example. The 2 × 104 to 4 × 104 cells used for the MIC, MBC, and killing rate experiments were taken from cultures of 108 cells. If the rate of mutation to resistance was 10−6, then, on average, the first mutant would occur when the culture was 106 cells, and in each successive generation the existing mutants would produce two cells. Moreover, in each generation new mutants would be produced from the ancestral cells that dominate the population. Thus, after one division the population of 106 bacteria with one mutant would be 2 × 106 total cells, of which two would be mutants derived by replication from the original mutant and two would be produced in the course of cell division by the ancestral population, for a total of four mutants. In the next generation, the total population size will be 4 × 104, the 4 mutants present in the previous generation would become 8 by cell division, and approximately 4 additional mutants will be produced from the ancestral population, for a total of 12. This process continues, and by the time the total number of bacteria in the population reaches 108 there will be approximately 700 mutants, for a frequency of mutants of 7 × 10−6. Thus, in a sample of 2 × 104 cells there will be, on average, 0.14 mutant and in a sample of 4 × 104 cells there will be, on average, 0.28 mutant. If we neglect this accumulation, the expected number of mutants in these cultures would be the product of the mutation rate and the total number of cells, or 0.02 and 0.04 for samples with 2 × 104 and 4 × 104 bacteria, respectively. In the Appendix we consider the contribution of this accumulation to the expected number of mutants in a population as well as the stochastic nature of the mutation process.
From a clinical perspective, these results and the considerations outlined above suggest that the treatment of CF or other patients with long-standing chronic Pseudomonas infections with single antibiotics, or monotherapy, is likely to fail because of the preexistence and subsequent ascent of resistant mutants. What about multidrug therapy, that is, treatment with drugs with synergistic activities, resistance to which requires separate and independent mutations? Are bacteria with mutations for resistance to clinically achievable concentrations of multiple drugs likely to be present in patients with long-standing chronic infection with P. aeruginosa? Although this possibility cannot be completely excluded due to the large numbers of bacteria in the P. aeruginosa populations frequently found in these patients, treatment with combinations of antibiotics may help, as found in our synergy testing, minimize the selection and ascent to dominance of antibiotic-resistant mutants.
In designing the treatment protocols for combination therapy, it is critical to consider the fact that although mutants with simultaneous high-level resistance to multiple antibiotics may not be present prior to treatment, they may rapidly evolve during the course of multidrug treatment. To minimize the likelihood of that occurring whenever possible, the treatment protocol should preclude situations in which the antibiotic concentrations decline to levels at which only one of the two antibiotics is limiting the growth of the bacterial population. Under these conditions, mutants resistant solely to the limiting antibiotic would be favored and may well increase in sufficient numbers that mutants that are also resistant to the second antibiotic are generated (18, 19). Another critical point to consider in the design of multidrug therapy protocols is the fact that low (peri-inhibitory) concentrations of multiple drugs could provide conditions for the selection for high-level resistance to one or more of the drugs used. Thus, in the design of multidrug treatment protocols, dosing intervals should be adjusted to minimize and, ideally, prevent the antibiotic concentrations from falling to levels below the intrinsic level of resistance to these agents. Finally, our results suggest that multidrug therapy for lung infections in patients with CF should probably be applied at any stage of the infective process, including any stage in which a low bacterial load is expected to occur, and particularly if there is suspicion that the founder population originated in another patient, in whom resistant mutants may have accumulated during chronic infection.
Acknowledgments
We thank J. L. Pérez, chief of the Microbiology Department, and J. A. Bengoechea and S. Alberti, of the Research Unit, Hospital Son Dureta, for continuous support of this project. We are grateful to C. Vidal and C. Santos of the DNA Sequencing Unit of Hospital Son Dureta for inestimable participation in the sequencing experiments and S. Martín for excellent technical assistance. We thank Arjan deVisser and three anonymous reviewers for useful comments and suggestions on an earlier version of this report. Plasmids pUCP24 and pJQ200mp18 were kindly provided by H. P. Schweizer and M. F. Hynes, respectively.
This work was partially financed by grant 01/0020-02 from the Ministerio de Sanidad y Consumo (FIS) to J.B. and grants BMC 2001-0012 and SAF2003-02851 from the Ministerio de Ciencia y Tecnología to J.B. and A.O., respectively. It was also partially financed by U.S. National Institutes of Health grants GM 33782 and AI40662 and grants from The British National Trust IPRAVE Project to B.R.L. It was also supported in part by “Red Española de Investigación en Patología Infecciosa (REIPI)” and Red Española de Investigación en Patología Respiratoria (RESPIRA).
APPENDIX
Expected number of mutants in a sample. As noted in the Discussion of this report and much earlier by Luria and Delbruck (21), as a consequence of the replication of existing mutants and the production of new ones, the expected number of mutants in a culture of N bacteria or a sample of Z cells from that culture (Z < N) will not be equal to the product of the mutation rate (μ) and the number of cells in the culture or the sample. If the mutation rate is high and N is large, the expected number of mutants in a culture or a sample can be substantially greater than Nμ or Zμ. Moreover, there will be considerable variation in the number of mutants produced among cultures that have the same total number of cells but that started independently with relatively few cells (21). This stochasticity is a consequence of the variation in the number of mutational events occurring in these cultures and the time during the course of population growth that these mutants were generated.
The accumulation of existing mutants, the production of new ones, and the stochasticity of the mutation process are taken into account in the algorithms used to estimate the mutation rates from fluctuation test data (14, 21, 38). The expected number of mutants per culture in an array of c cultures of N bacteria can be calculated from the estimated mutation rate by using the transcendental equation in Luria and Delbruck’s 1943 article (21). To our knowledge, however, there is no formula for estimation of the expected number of mutants in a culture that takes into account the stochastic nature of the accumulation of existing mutants, the production of new ones, and, if appropriate, differences in the growth rates (relative fitness) of the mutants and their ancestors, as well as other inconveniences of reality. One can, however, obtain realistic estimates of the expected number of mutants and the variation in those numbers with Monte Carlo simulations of fluctuation experiments. This is how we determined the expected frequencies of cultures with one or more mutations that we used for the chi-square test described above. Our fluctuation test simulation was programmed with the Berkeley Madonna software package and can be downloaded from www.eclf.net.
As input for these simulations we used the estimated mutation rate for resistance presented in Table 2 and set the parameters (resource concentration and conversion efficiency) so that when the cultures stop growing due to resource limitations there are 109 bacteria. Because we had no information to the contrary, we assumed that mutants and their ancestors were equally fit. For each mutation rate, we ran two 20-culture fluctuation test simulations and for each simulation estimated the expected number of mutations from the median number of mutants in the 20 cultures. We used the mean of two estimated medians of the total number of mutants in the simulated cultures, M, to calculate the relative frequency of resistant cells (F = M/10−9), and from that frequency we calculated the expected number of mutants, X, in samples of 4 × 104 bacteria [X = F · (4 × 104)]. To calculate the probability that a culture of 4 × 104 cells would have at least one resistant mutant, y > 0, we assumed a Poisson distribution so that P(y > 0) = 1 − ex.
To illustrate how we used this method, we consider the expected number of PAOΔmutS cultures with one or more mutants with mutations for resistance to imipenem at 8 μg/ml. From the fluctuation test, we estimated the rate of mutation for resistance to this concentration of imipenem to be 1.1 × 10−6 per cell per generation (Table 2). The mutation rates calculated from the two simulations with this empirically estimated mutation rate and 109 bacteria per culture were 1.2 × 10−6 and 1.1 × 10−6, respectively. The relative frequencies of mutants in these simulated cultures, F, were substantially greater than that expected from the product of the mutation rate and the total number of bacteria, 1.2 × 10−5 and 1.0 × 10−5, for an average of 1.1 × 10−5. Thus, in a sample of 4 × 104 PAOΔmutS cells, the expected number of mutants resistant to imipenem at 8 μg/ml would be (1.1 × 10−5) × (4.0 × 104), or X = 0.44, and the expected frequency of cultures with one or more resistant mutants would be 1 − e−0.44 = 0.36. Therefore, of 40 independent cultures, we would anticipate that 14.4 (0.36 × 40) would have one or more resistant mutants.
REFERENCES
- 1.Atwood, K. C., L. K. Scheider, and F. J. Ryan. 1951. Periodic selection in Escherichia coli. Proc. Natl. Acad. Sci. USA 37:146-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bagge, N., O. Ciofu, M. Hentzer, J. I. Campbell, M. Givskov, and N. Hoiby. 2002. Constitutive high expression of chromosomal β-lactamase in Pseudomonas aeruginosa caused by a new insertion sequence (IS1669) located in ampD. Antimicrob. Agents Chemother. 46:3406-3411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Crane, G. J., S. M. Thomas, and M. E. Jones. 1996. A modified Luria-Delbruck fluctuation assay for estimating and comparing mutation rates. Mutat. Res. 354:171-182. [DOI] [PubMed] [Google Scholar]
- 4.Crow, J. F., and M. Kimura. 1971. An introduction to population genetics theory. Harper & Row, New York, N.Y.
- 5.De Lorenzo, V., S. Fernández, M. Herrero, U. Jakubzik, and K. Timmis. 1993. Engineering of alkyl- and haloaromatic-responsive gene expression with mini-transposons containing regulated promoters of biodegradative pathways of Pseudomonas. Gene 130:41-46. [DOI] [PubMed] [Google Scholar]
- 6.Drlica, K. 2001. Antibiotic resistance: can we beat the bugs? Drug Discov. Today 6:714-715. [DOI] [PubMed] [Google Scholar]
- 7.Ehrlich, P. 1913. Chemotherapeutics: scientific principles, methods and results. Lancet ii:445-451. [Google Scholar]
- 8.Gibson, R. L., J. L. Burns, and B. W. Rammsey. 2003. Pathophysiology and management of pulmonary infections in cystic fibrosis. Am. J. Respir. Crit. Care Med. 168:918-951. [DOI] [PubMed] [Google Scholar]
- 9.Giwercman, B., P. Lambert, V. T. Rosdahl, G. H. Shand, and N. Hoiby. 1990. Rapid emergence of resistance in Pseudomonas aeruginosa in cystic fibrosis patients due to in-vivo selection of stable partially derepressed β-lactamase producing strains. J. Antimicrob. Chemother. 26:247-259. [DOI] [PubMed] [Google Scholar]
- 10.Govan, J. R., and V. Deretic. 1996. Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol. Rev. 60:539-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hocquet, D., X. Bertrand, T. Köhler, D. Talon, and P. Plésiat. 2003. Genetic and phenotypic variations of a resistant Pseudomonas aeruginosa epidemic clone. Antimicrob. Agents Chemother. 47:1887-1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jalal, S., O. Ciofu, N. Hoiby, N. Gotoh, and B. Wretlind. 2000. Molecular mechanisms of fluoroquinolone resistance in Pseudomonas aeruginosa isolates from cystic fibrosis patients. Antimicrob. Agents Chemother. 44:710-712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnson, J. R., H. A. Lockman, K. Owens, S. Jelacic, and P. I. Tarr. 2003. High-frequency secondary mutations after suicide-driven allelic exchange mutagenesis in extraintestinal pathogenic Escherichia coli. J. Bacteriol. 185:5301-5305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jones, M. E., S. M. Thomas, and A. Rogers. 1994. Luria-Delbruck fluctuation experiments: design and analysis. Genetics 136:1209-1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koch, A. L. 1974. The pertinence of the periodic selection phenomenon to prokaryote evolution. Genetics 77:127-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kohler, T., M. Michea-Hamzehpour, S. F. Epp, and J. C. Pechere. 1999. Carbapenem activities against Pseudomonas aeruginosa: respective contributions of OprD and efflux systems. Antimicrob. Agents Chemother. 43:424-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Levin, B. R. 1981. Periodic selection, infectious gene exchange and the genetic structure of E. coli populations. Genetics 99:1-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lipsitch, M., and B. R. Levin. 1997. The population dynamics of antimicrobial chemotherapy. Antimicrob. Agents Chemother. 41:363-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lipsitch, M., and B. R. Levin. 1998. Population dynamics of tuberculosis treatment: mathematical models of the roles of non-compliance and bacterial heterogeneity in the evolution of drug resistance. Int. J. Tuberc. Lung Dis. 2:187-199. [PubMed] [Google Scholar]
- 20.Livermore, D. M. 2002. Multiple mechanisms of antimicrobial resistance in Pseudomonas aeruginosa: our worst nightmare? Clin. Infect. Dis. 34:634-640. [DOI] [PubMed] [Google Scholar]
- 21.Luria, S., and M. Delbruck. 1943. Mutations of bacteria from virus sensitivity to virus resistance. Genetics 28:491-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lyczak, J. B., C. L. Cannon, and G. B. Pier. 2002. Lung infections associated with cystic fibrois. Clin. Microbiol. Rev. 15:194-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mao, E. F., L. Lane, J. Lee, and J. H. Miller. 1997. Proliferation of mutators in a cell population. J. Bacteriol. 179:417-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Masuda, N., E. Sakagawa, and S. Ohya. 1995. Outer membrane proteins responsible for multiple drug resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 39:645-647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Masuda, N., E. Sakagawa, S. Ohya, N. Gotoh, H. Tsujimoto, and T. Nishino. 2000. Substrate specificities of MexAB-OprM, MexCD-OprJ, and MexXY-OprM efflux pumps in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 44:3322-3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.National Committee for Clinical Laboratory Standards. 1999. Methods for determining bactericidal activity of antimicrobial agents. Approved standard M21-A. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 27.National Committee for Clinical Laboratory Standards. 2000. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard M7-A5. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 28.Negri, M. C., M. Lipsitch, J. Blazquez, B. R. Levin, and F. Baquero. 2000. Concentration-dependent selection of small phenotypic differences in TEM β-lactamase-mediated antibiotic resistance. Antimicrob. Agents Chemother. 44:2485-2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oliver, A., R. Cantón, P. Campo, F. Baquero, and J. Blázquez. 2000. High frequency of hypermutable Pseudomonas aeruginosa in cystic fibrosis lung infection. Science 288:1251-1254. [DOI] [PubMed] [Google Scholar]
- 30.Oliver, A., F. Baquero, and J. Blazquez. 2002. The mismatch repair system (mutS, mutL and uvrD genes) in Pseudomonas aeruginosa: molecular characterization of naturally occurring mutants. Mol. Microbiol. 43:1641-1650. [DOI] [PubMed] [Google Scholar]
- 31.Oliver, A., L. M. Weigel, J. K. Rasheed, J. E. McGowan, P. Raney, and F. C. Tenover. 2002. Mechanisms of decreased susceptibility to cefpodoxime in Escherichia coli. Antimicrob. Agents Chemother. 46:3829-3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pirnay, J. P., D. De Vos, D. Mossialos, A. Vanderkelen, P. Cornells, and Z. Martin. 2002. Analysis of the Pseudomonas aeruginosa oprD gene from clinical and environmental isolates. Environ. Microbiol. 4:872-882. [DOI] [PubMed] [Google Scholar]
- 33.Poole, K. 2001. Multidrug efflux pumps and antimicrobial resistance in Pseudomonas aeruginosa and related organisms. J. Mol. Microbiol. Biotechnol. 3:255-264. [PubMed] [Google Scholar]
- 34.Quandt, J., and M. F. Hynes. 1993. Versatile suicide vectors which allow direct selection for gene replacement in gram-negative bacteria. Gene 127:15-21. [DOI] [PubMed] [Google Scholar]
- 35.Riesenfeld, C., M. Everett, L. J. Piddock, and B. G. Hall. 1997. Adaptive mutations produce resistance to ciprofloxacin. Antimicrob. Agents Chemother. 41:2059-2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith, A. W., and B. H. Iglewski. 1989. Transformation of Pseudomonas aeruginosa by electroporation. Nucleic Acids Res. 17:10509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Spratt, B. G., P. J. Hedge, S. T. Heesen, A. Edelman, and J. K. Broome-Smith. 1986. Kanamycin-resistant vectors that are analogues of plasmids pUC8, pUC9, pEMBL8, and pEMBL9. Gene 41:337-342. [DOI] [PubMed] [Google Scholar]
- 38.Stewart, F. M., D. M. Gordon, and B. R. Levin. 1990. Fluctuation analysis: the probability distribution of the number of mutants under different conditions. Genetics 124:175-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stover, C. K., X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. Brinkman, W. O. Hufnagle, D. J. Kowalik, M. Lagrou, et al. 2000. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406:959-964. [DOI] [PubMed] [Google Scholar]
- 40.Taddei, F., M. Radman, J. Maynard-Smith, B. Toupance, P. H. Gouyon, and B. Godellete. 1997. Role of mutator alleles in adaptive evolution. Nature 387:700-702. [DOI] [PubMed] [Google Scholar]
- 41.Tanaka, M. M., C. T. Bergstrom, and B. R. Levin. 2003. The evolution of mutator genes in bacterial populations: the roles of environmental change and timing. Genetics 164:843-854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.West, S. E., H. P. Schweizer, C. Dall, A. K. Sample, and L. J. Runyen-Janecky. 1994. Construction of improved Escherichia-Pseudomonas shuttle vectors derived from pUC18/19 and sequence of the region required for their replication in Pseudomonas aeruginosa. Gene 148:81-86. [DOI] [PubMed] [Google Scholar]
- 43.Westbrock-Wadman, S., D. R. Sherman, M. J. Hickey, S. N. Coulter, Y. Q. Zhu, P. Warrener, L. Y. Nguyen, R. M. Shawar, K. R. Folger, and C. K. Stover. 1999. Characterization of a Pseudomonas aeruginosa efflux pump contributing to aminoglycoside impermeability. Antimicrob. Agents Chemother. 43:2975-2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wong, K., M. C. Roberts, L. Owens, M. Fife, and A. L. Smith. 1984. Selective media for the quantitation of bacteria in cystic fibrosis sputum. J. Med. Microbiol. 17:113-119. [DOI] [PubMed] [Google Scholar]
- 45.Ziha-Zarifi, I., C. Llanes, T. Köhler, J. C. Pecher, and P. Plesiat. 1999. In vivo emergence of multidrug-resistant mutants of Pseudomonas aeruginosa overexpressing the active efflux system MexA-MexB-OprM. Antimicrob. Agents Chemother. 43:287-291. [DOI] [PMC free article] [PubMed] [Google Scholar]