Abstract
Two methicillin- and vancomycin-resistant Staphylococcus aureus strains, MI-VRSA and PA-VRSA, and Enterococcus faecalis DMC83006B, considered to be the potential donor of glycopeptide resistance to MI-VRSA, were studied. MI-VRSA is highly resistant to both glycopeptides, whereas PA-VRSA displays low-level resistance to vancomycin and reduced susceptibility to teicoplanin. We have analyzed the expression of the vanA operon in the three clinical isolates. Determination of the relative amounts of late peptidoglycan precursors and quantification of the d,d-peptidase activities, in the absence or after induction by glycopeptides, revealed that the resistance genes were expressed at similarly high levels in the three strains. Glycopeptide resistance stability in the three strains was studied by replica plating. Resistance was lost at high frequency, ca. 50%, after overnight growth of PA-VRSA in the absence of antibiotics, whereas it was fully stable in MI-VRSA and E. faecalis DMC83006B. Induction of resistance by vancomycin was significantly delayed in PA-VRSA relative to MI-VRSA. Low-level glycopeptide resistance of S. aureus PA-VRSA is thus likely due to instability of the genetic element, plasmid or transposon, carrying the vanA operon associated with a longer lag phase before growth resumes after induction by vancomycin.
The glycopeptide antibiotics vancomycin and teicoplanin are active against major gram-positive pathogens including methicillin-resistant Staphylococcus aureus (MRSA). They bind to the C-terminal d-alanyl-d-alanine (d-Ala-d-Ala) of pentapeptide peptidoglycan precursors and inhibit the transglycosylation and transpeptidation reactions, preventing incorporation of the precursors into the bacterial cell wall (20). Acquired VanA-type glycopeptide resistance in enterococci is due to the synthesis of modified precursors ending in d-Ala-d-lactate (d-Ala-d-Lac) in place of d-Ala-d-Ala that result in 1,000-fold-lower affinity for glycopeptides (11, 23). VanA-type resistance is characterized by high-level inducible resistance to both vancomycin and teicoplanin and is mediated by Tn1546 or closely related elements (6, 16). Tn1546, which belongs to the Tn3 family of transposons, is composed of seven genes, two of which encode a transposase and a resolvase responsible for the movements of the elements of the vanA operon, and is delineated by imperfect inverted repeats. The vanH, vanA, and vanX genes code for proteins that are necessary for the expression of resistance. VanH is a dehydrogenase that converts pyruvate to d-Lac (4), VanA a ligase that uses d-Lac and a d-Ala residue to synthesize the depsipeptide d-Ala-d-Lac, which is incorporated into the peptidoglycan precursors (11), and VanX is a d,d-dipeptidase that hydrolyzes the dipeptide d-Ala-d-Ala formed by the endogenous Ddl chromosomal d-Ala:D-Ala ligase (22, 29), thus reducing the level of normal peptidoglycan precursors ending in d-Ala-d-Ala. The VanY d,d-carboxypeptidase, not essential for resistance, cleaves the d-Ala C-terminal residue of pentapeptide precursors synthesized by using d-Ala-d-Ala dipeptides that have escaped VanX hydrolysis (8). The vanZ gene is implicated in teicoplanin resistance by an unknown mechanism (2). Expression of the vanA operon is regulated by two genes, vanR and vanS, located upstream from vanH that form a two-component regulatory system (5, 7, 28).
S. aureus is responsible for serious infections and toxinoses in humans in both the hospital environment and the community (17). MRSA harbors the structural mecA gene for penicillin-binding protein 2a (13) that has low affinity for, and thus confers resistance to, all β-lactams. Since MRSA strains are usually also resistant to multiple drug classes, vancomycin is used extensively to treat infections caused by these bacteria.
In 2002, two methicillin- and vancomycin-resistant S. aureus strains, MI-VRSA and PA-VRSA, were isolated from patients in the United States. MI-VRSA is highly resistant to both glycopeptides (24), whereas PA-VRSA (19) is moderately resistant to vancomycin and has reduced susceptibility to teicoplanin (12, 25). The two isolates harbor a plasmid-borne Tn1546 element (26, 27). Vancomycin-resistant Enterococcus faecalis DMC83006B (15, 27) and a susceptible MRSA strain (24, 27) were isolated from the same patient as MI-VRSA and are considered to be the Tn1546 donor and recipient, respectively.
In an attempt to explain the differences in the levels of glycopeptide resistance of the two S. aureus clinical isolates, we studied the expression of the vanA gene cluster in these strains and in the putative Enterococcus donor.
MATERIALS AND METHODS
Bacterial strains.
Methicillin- and vancomycin-resistant S. aureus MI-VRSA (24) and PA-VRSA (19) are clinical isolates and E. faecalis DMC83006B was coisolated with MI-VRSA (15). The vancomycin and teicoplanin MICs were determined by E-test as described by the manufacturer (AB Biodisk, Solna, Sweden). Strains were grown in brain heart infusion (BHI) broth or agar (Difco Laboratories, Detroit, Mich.) at 37°C. Attempts to obtain spontaneous glycopeptide-susceptible derivatives were as follows. MI-VRSA, PA-VRSA, and DMC83006B were streaked onto BHI agar containing vancomycin (6 μg/ml). A single colony of each strain was grown overnight in broth, and the culture was streaked onto BHI agar. One hundred colonies of each plate were replica plated on the same medium without antibiotic or containing vancomycin (6 μg/ml), gentamicin (256 μg/ml), or erythromycin (256 μg/ml) at 37°C for 24 h. Three independent experiments were performed.
Growth rate studies.
Induction of resistance by vancomycin in MI-VRSA and PA-VRSA was studied by determination of growth rates under various conditions. Isolates were grown overnight at 37°C in BHI broth without or with vancomycin (6 μg/ml). The cultures were diluted 1:20 into 20 ml of BHI without or with vancomycin (6 μg/ml) and then grown at 37°C with shaking, and the optical density at 600 nm was monitored.
PCR analysis.
Glycopeptide resistance vanA genotype was confirmed by PCR as described previously (14). Glycopeptide-susceptible derivatives of PA-VRSA and the parental strain were identified as S. aureus by amplification of an internal portion of the thermonuclease nuc gene, with total DNA as a template and specific primers as described previously (10).
Analysis of peptidoglycan precursors.
Extraction and analysis of peptidoglycan precursors were performed as described previously (18). Staphylococci and enterococci were grown in BHI broth at 37°C in the presence (4 μg/ml) or in the absence of vancomycin to an optical density at 600 nm of 1.0 (mid-exponential phase). Ramoplanin (3 μg/ml) was added to inhibit peptidoglycan synthesis, and incubation was continued for 15 min. Bacteria were harvested, and the cytoplasmic precursors were extracted with 8% trichloroacetic acid (15 min. at 4°C), desalted, and analyzed by high-performance liquid chromatography. The results were expressed as the percentages of total late peptidoglycan precursors represented by UDP-Mur-Nac-tetrapeptide, UDP-Mur-Nac-pentapeptide, and UDP-MurNac-pentadepsipeptide that were determined from the integrated peak areas.
D,D-peptidase activities.
The enzymatic activities were assayed as described previously (3, 21). Bacteria grown in BHI broth until the optical density at 600 nm reached 0.7 in the absence or in the presence of glycopeptide (vancomycin or teicoplanin at 4 μg/ml) were harvested and lysed by treatment with lysozyme (2 mg/ml) at 37°C, followed by sonication, and membrane fractions were pelleted (100,000 × g, 45 min). Cytoplasmic and membrane fractions (C100 and S100) were collected and assayed for D,D-dipeptidase (VanX) or D,D-carboxypeptidase (VanY) activities by measuring the d-Ala released from the dipeptide d-Ala-d-Ala (6.56 mM) or from the pentapeptide (l-Ala-γ-d-Glu-l-Lys-d-Ala-d-Ala; 5 mM) by using d-amino acid oxidase, horseradish peroxidase, and O-dianisidine as chromogen. Specific activities were defined as the number of nanomoles of product formed at 37°C per minute per milligram of protein contained in the extracts.
RESULTS AND DISCUSSION
Acquisition of Tn1546 by MI-VRSA.
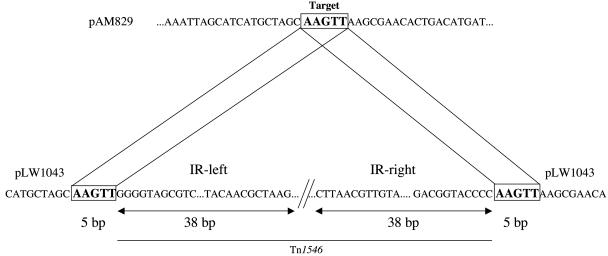
E. faecalis DMC83006B harbors plasmid pAM830, which belongs to the Inc18 family of broad-host-range conjugative plasmids and contains a copy of Tn1546 (15). A vancomycin-susceptible MRSA strain, also coisolated with MI-VRSA, harbors the 47-kb conjugative plasmid pAM829 (27). In MI-VRSA, the Tn1546 element is born by pLW1043, which is identical to pAM829 except for the copy of Tn1546 (27). Therefore, plasmid pAM830 apparently behaves as a suicide Tn1546 delivery vector to pAM829 (27). Analysis of the nucleotide sequences at the junctions of Tn1546 (GenBank accession number M97297) and pLW1043 (accession number AE017171) indicated that the transposon was flanked by an AAGTT duplication of pAM829 target DNA (Fig. 1). Duplications of five base pairs upon insertion are typical of the Tn3 family of transposons (9) to which Tn1546 belongs (6). This observation confirms that Tn1546 had transposed into pAM829.
FIG. 1.
Schematic representation of Tn1546 insertion site. The 5-bp direct repeats of target DNA are indicated in boldface characters and are boxed. The left and right IRL and IRR imperfect terminal 38-bp repeats of Tn1546 are indicated by double arrows. Tn1546 is indicated by a thin line.
Comparative analysis of peptidoglycan precursors.
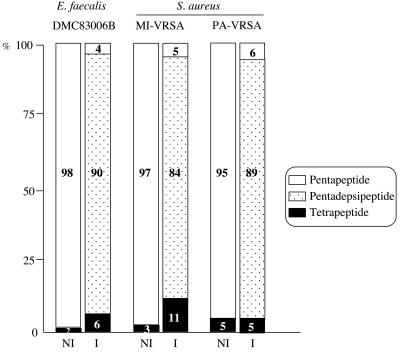
To analyze the cytoplasmic peptidoglycan precursors, strains MI-VRSA, PA-VRSA, and DMC83006B were grown in the presence (4 μg/ml) or in the absence of vancomycin or teicoplanin. Inhibition of cell wall synthesis and consequent accumulation of the precursors was obtained by the addition of ramoplanin (Fig. 2). In the absence of induction, UDP-Mur-Nac-pentapeptide was the main late precursor (95 to 98% of all precursors) synthesized by the three strains. After incubation with vancomycin, UDP-Mur-Nac-tetrapeptide (5 to 11%) and high proportions (84 to 90%) of UDP-Mur-Nac-pentadepsipeptide were detected in the cells, indicating similar high-level expression of the resistance genes in E. faecalis and S. aureus. These data indicate that the three strains were inducibly resistant to vancomycin by production of precursors ending in d-Ala-d-Lac. Although the two S. aureus strains exhibited very different levels of glycopeptide resistance from each other (Table 1), the relative amounts of late peptidoglycan precursors were similar in both strains after induction with vancomycin.
FIG. 2.
Cytoplasmic peptidoglycan precursors in E. faecalis DMC83006B and S. aureus MI-VRSA and PA-VRSA. Peptidoglycan synthesis was inhibited by addition of ramoplanin to the cultures for 15 min. NI, not induced; I, induced by growth in the presence of 4 μg of vancomycin/ml.
TABLE 1.
MICs of glycopeptides for E. faecalis and S. aureus strains
| Strain | MIC (μg/ml)
|
|
|---|---|---|
| Vancomycin | Teicoplanin | |
| E. faecalis DMC83006B | >256 | 128 |
| S. aureus MI-VRSA | >256 | 128 |
| S. aureus PA-VRSAa | 24-64 | 8-16 |
| S. aureus PA-VRSA susceptible derivativesb | 1-2 | 0.5-1 |
Range of five independent determinations.
A total of 24 separate isolates were investigated.
D,D-peptidase activities.
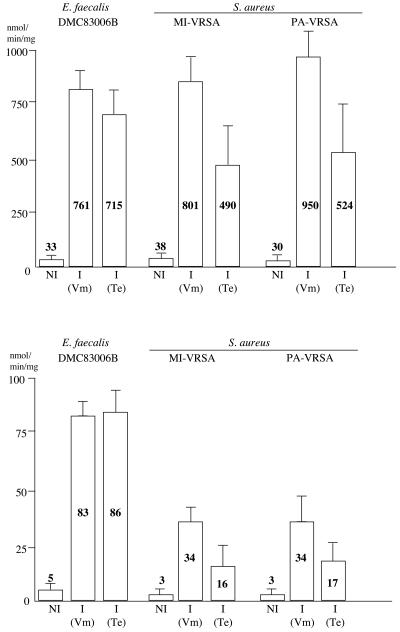
D,D-dipeptidase and D,D-carboxypeptidase activities were analyzed in the three strains. The amount of d-Ala released from hydrolysis of the d-Ala-d-Ala dipeptide (VanX activity) and of the UDP-Mur-Nac-l-Ala-γ-d-Glu-l-Lys-d-Ala-d-Ala pentapeptide (VanY activity) was measured in the cytoplasmic and membrane fractions (Fig. 3). In the absence of induction, very weak VanX and VanY activities were found in the E. faecalis and S. aureus strains. In the presence of vancomycin (4 μg/ml) in the culture medium, high D,D-dipeptidase and D,D-carboxypeptidase activities were found in the three isolates (Fig. 3). As expected for a vanA genotype, teicoplanin (4 μg/ml) was also an inducer, although less efficient than vancomycin for the two S. aureus strains (Fig. 3).
FIG. 3.
D,D-peptidase activities in extracts from E. faecalis DMC83006B and S. aureus MI-VRSA and PA-VRSA. D,D-dipeptidase (VanX) (top) and D,D-carboxypeptidase (VanY) (bottom) activities were measured in the cytoplasmic extracts and in the membrane fractions, respectively. The results are the means from two independent experiments. NI, not induced; I, induced by growth in the presence of 4 μg of vancomycin (Vm) or teicoplanin (Te)/ml.
These results indicate induction at similar levels of expression of the vanA gene clusters by vancomycin and teicoplanin in the three strains. Thus, the difference in glycopeptide resistance between MI-VRSA and PA-VRSA is not due to differences in van gene expression.
Stability of glycopeptide resistance.
We tested the stability of the vanA gene cluster in E. faecalis DMC83006B and in S. aureus MI-VRSA and PA-VRSA by replica plating. In three independent experiments, no glycopeptide susceptible clones of E. faecalis DMC83006B or S. aureus MI-VRSA were obtained. In contrast, ca. 50% (43, 51, and 53%) of the PA-VRSA derivatives obtained after overnight growth in broth in the absence of vancomycin were susceptible to vancomycin (MIC, 2 μg/ml) and teicoplanin (MIC, 1 μg/ml) (Table 1) but remained resistant to gentamicin and erythromycin. PCR amplification with primers specific for the vanA gene (14), which was positive for parental PA-VRSA, was negative for 24 susceptible derivatives (data not shown).
Inducibility of vancomycin resistance in S. aureus.
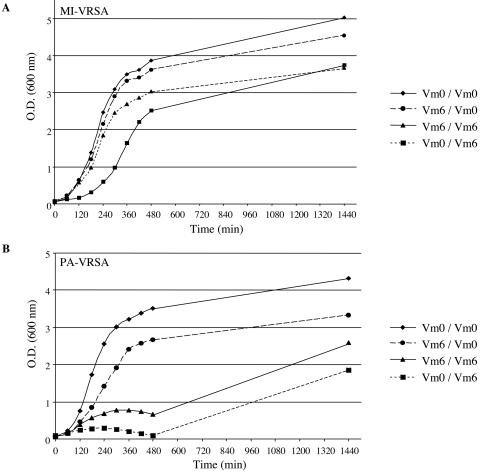
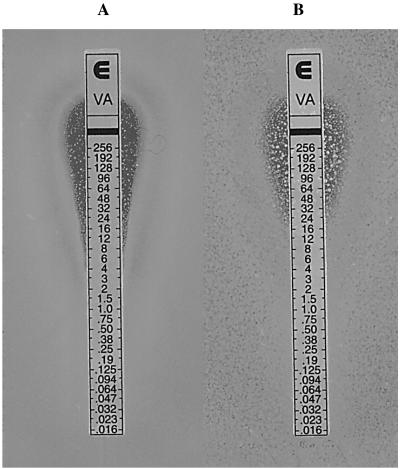
The effect of prior exposure to vancomycin (6 μg/ml) on growth of the MI-VRSA and PA-VRSA isolates was determined in liquid culture (Fig. 4). After overnight incubation of MI-VRSA with vancomycin, no lag phase was observed during the subcultures, whereas overnight culture in the absence of vancomycin resulted in inhibition of growth at the beginning of the subculture in the presence of vancomycin, with growth resuming after a lag phase of ca. 3 h (Fig. 4A). When PA-VRSA was grown overnight in the absence or presence of antibiotic and subcultured with vancomycin, growth was delayed for at least 8 h (Fig. 4B). After overnight growth in the presence of vancomycin and subculture for 3 h in the absence of antibiotic, the D,D-peptidase activities were fourfold weaker than when the subculture contained vancomycin (data not shown), probably reflecting loss and deinduction of resistance. When PA-VRSA was pregrown without vancomycin and then subcultured for 3 h in the presence of vancomycin, weak D,D-peptidase activities were detected (data not shown), indicating that the growing cells possessed a functional vanA gene cluster. After 24 h of incubation in the presence of vancomycin, the vancomycin MIC for PA-VRSA, as determined by E-test, was 32 μg/ml, and numerous large colonies were observed in the inhibition zone up to 256 μg/ml (Fig. 5B), suggesting that the portion of the bacteria that retained resistance was fully induced. When PA-VRSA was not induced by vancomycin, the number and the size of the colonies present in the inhibition zone were significantly lower (Fig. 5A), a finding consistent with higher loss of glycopeptide resistance and slower induction.
FIG. 4.
Effect of overnight growth in the presence of vancomycin (6 μg/ml) on growth of S. aureus MI-VRSA (A) and PA-VRSA (B) in the absence or presence (6 μg/ml) of vancomycin. Vancomycin (Vm) concentration in the overnight culture/in the culture medium. O.D., optical density.
FIG. 5.
Vancomycin MIC determination by E-test of the PA-VRSA isolate after 24 h of incubation in the absence (A) or in the presence (6 μg/ml) (B) of vancomycin.
Taken together, these results suggest that low-level glycopeptide resistance of the PA-VRSA strain could be due to a high rate of emergence of susceptible derivatives associated with a longer delay in induction of resistance. The former could be due to a high rate of spontaneous loss of the glycopeptide resistance determinant or to a low rate of loss of a resistance trait with high biological cost, and the latter could be due to a high rate of appearance of susceptible derivatives or to physiological differences between the two S. aureus strains.
Glycopeptide resistance in S. aureus PA-VRSA is due to the acquisition of a large (∼120-kb) plasmid, probably from enterococcal origin, carrying a copy of Tn1546 (26). Such a large plasmid is likely to be present only in a few copies per cell. Thus, it could take longer than in MI-VRSA, where the vanA gene cluster is present on a plasmid half the size (27), before induction of resistance builds up a sufficient amount of D,D-peptidases and resistant precursors to take over from the susceptible pathway. Under these conditions, vancomycin may trap and immobilize the lipid intermediates and stop growth before resistance can come into play. Consequently, vancomycin MIC is lower than against a strain where induction occurs rapidly.
The genetic instability of the van gene cluster in the new host could be due to inefficient plasmid replication in the new host or to a high rate of excision of the transposon. The former seems more likely since transposons of the Tn3 family (9) and, in particular Tn1546 (1, 6), are very stable genetic elements. Experiments are in progress to discriminate between these two possibilities.
In conclusion, we have demonstrated efficient heterologous expression of the glycopeptide resistance genes in the two S. aureus transconjugants similar to that observed in the E. faecalis donor. Low-level resistance of the PA-VRSA strain is most likely due to loss, at high frequency, of the vanA operon and to a longer lag phase before the induction of resistance.
Acknowledgments
We thank P. E. Reynolds for reading the manuscript.
S. aureus strains were obtained through the Network on Antimicrobial Resistance in Staphylococcus aureus (NARSA) Program supported under NIAID/NIH contract no. N01-AI-95359.
REFERENCES
- 1.Arthur, M., and P. Courvalin. 1993. Genetics and mechanisms of glycopeptide resistance in enterococci. Antimicrob. Agents Chemother. 37:1563-1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arthur, M., F. Depardieu, C. Molinas, P. Reynolds, and P. Courvalin. 1995. The vanZ gene of Tn1546 from Enterococcus faecium BM4147 confers resistance to teicoplanin. Gene 154:87-92. [DOI] [PubMed] [Google Scholar]
- 3.Arthur, M., F. Depardieu, P. Reynolds, and P. Courvalin. 1996. Quantitative analysis of the metabolism of soluble cytoplasmic peptidoglycan precursors of glycopeptide-resistant enterococci. Mol. Microbiol. 21:33-44. [DOI] [PubMed] [Google Scholar]
- 4.Arthur, M., C. Molinas, T. D. H. Bugg, G. D. Wright, C. T. Walsh, and P. Courvalin. 1992. Evidence for in vivo incorporation of d-lactate into peptidoglycan precursors of vancomycin-resistant enterococci. Antimicrob. Agents Chemother. 36:867-869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arthur, M., C. Molinas, and P. Courvalin. 1992. The VanS-VanR two-component regulatory system controls synthesis of depsipeptide peptidoglycan precursors in Enterococcus faecium BM4147. J. Bacteriol. 174:2582-2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arthur, M., C. Molinas, F. Depardieu, and P. Courvalin. 1993. Characterization of Tn1546, a Tn3-related transposon conferring glycopeptide resistance by synthesis of depsipeptide peptidoglycan precursors in Enterococcus faecium BM4147. J. Bacteriol. 175:117-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arthur, M., and R. Quintiliani, Jr. 2001. Regulation of VanA- and VanB-type glycopeptide resistance in enterococci. Antimicrob. Agents Chemother. 45:375-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arthur, M., P. E. Reynolds, and P. Courvalin. 1996. Glycopeptide resistance in enterococci. Trends Microbiol. 4:401-407. [DOI] [PubMed] [Google Scholar]
- 9.Berg, C. M., and D. E. Berg. 1995. Transposable elements as tools for molecular analysis in bacteria, p. 38-68. In D. J. Sherratt (ed.), Mobile genetic elements, 2nd ed. Oxford University Press, Inc., New York, N.Y.
- 10.Brakstad, O. G., K. Aasbakk, and J. A. Maeland. 1992. Detection of Staphylococcus aureus by polymerase chain reaction amplification of the nuc gene. J. Clin. Microbiol. 30:1654-1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bugg, T. D. H., G. D. Wright, S. Dutka-Malen, M. Arthur, P. Courvalin, and C. T. Walsh. 1991. Molecular basis for vancomycin resistance in Enterococcus faecium BM4147: biosynthesis of a depsipeptide peptidoglycan precursor by vancomycin resistance proteins VanH and VanA. Biochemistry 30:10408-10415. [DOI] [PubMed] [Google Scholar]
- 12.Centers for Disease Control and Prevention. 1997. Interim guidelines for prevention and control of staphylococcal infection associated with reduced susceptibility to vancomycin. Morb. Mortal. Wkly. Rep. 46:626-628. [PubMed] [Google Scholar]
- 13.Chambers, H. F. 1997. Methicillin resistance in staphylococci: molecular and biochemical basis and clinical implications. Clin. Microbiol. Rev. 10:781-791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dutka-Malen, S., S. Evers, and P. Courvalin. 1995. Detection of glycopeptide resistance genotypes and identification to the species level of clinically relevant enterococci by PCR. J. Clin. Microbiol. 33:24-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Flannagan, S. E., J. W. Chow, S. M. Donabedian, W. J. Brown, M. B. Perri, M. J. Zervos, Y. Ozawa, and D. B. Clewell. 2003. Plasmid content of a vancomycin-resistant Enterococcus faecalis isolate from a patient also colonized by Staphylococcus aureus with a VanA phenotype. Antimicrob. Agents Chemother. 47:3954-3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Handwerger, S., and J. Skoble. 1995. Identification of chromosomal mobile element conferring high-level vancomycin resistance in Enterococcus faecium. Antimicrob. Agents Chemother. 39:2446-2453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lowy, F. D. 1998. Staphylococcus aureus infections. N. Engl. J. Med. 339:520-532. [DOI] [PubMed] [Google Scholar]
- 18.Messer, J., and P. E. Reynolds. 1992. Modified peptidoglycan precursors produced by glycopeptide-resistant enterococci. FEMS Microbiol. Lett. 73:195-200. [DOI] [PubMed] [Google Scholar]
- 19.Miller, D., V. Urdaneta, A. Weltman, and S. Park. 2002. Vancomycin-resistant Staphylococcus aureus. Morb. Mortal. Wkly. Rep. 51:902.. [PubMed] [Google Scholar]
- 20.Reynolds, P. E. 1989. Structure, biochemistry and mechanism of action of glycopeptide antibiotics. Eur. J. Clin. Microbiol. Infect. Dis. 8:943-950. [DOI] [PubMed] [Google Scholar]
- 21.Reynolds, P. E., O. H. Ambur, B. Casadewall, and P. Courvalin. 2001. The VanYD DD-carboxypeptidase of Enterococcus faecium BM4339 is a penicillin-binding protein. Microbiology 147:2571-2578. [DOI] [PubMed] [Google Scholar]
- 22.Reynolds, P. E., F. Depardieu, S. Dutka-Malen, M. Arthur, and P. Courvalin. 1994. Glycopeptide resistance mediated by enterococcal transposon Tn1546 requires production of VanX for hydrolysis of d-alanyl-d-alanine. Mol. Microbiol. 13:1065-1070. [DOI] [PubMed] [Google Scholar]
- 23.Reynolds, P. E., H. A. Snaith, A. J. Maguire, S. Dutka-Malen, and P. Courvalin. 1994. Analysis of peptidoglycan precursors in vancomycin-resistant Enterococcus gallinarum BM4174. Biochem. J. 301:5-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sievert, D. M., M. L. Boulton, G. Stolman, D. Johnson, M. G. Stobierski, F. P. Downes, P. A. Somsel, J. T. Rudrik, W. Brown, W. Hafeez, T. Lundstrom, E. Flanagan, R. Johnson, J. Mitchell, and S. Chang. 2002. Staphylococcus aureus resistant to vancomycin. Morb. Mortal. Wkly. Rep. 51:565-567. [Google Scholar]
- 25.Tenover, F. C., M. V. Lancaster, B. C. Hill, C. D. Steward, S. A. Stocker, G. A. Hancock, C. M. O'Hara, S. K. McAllister, N. C. Clark, and K. Hiramatsu. 1998. Characterization of staphylococci with reduced susceptibilities to vancomycin and other glycopeptides. J. Clin. Microbiol. 36:1020-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tenover, F. C., L. M. Weigel, P. C. Appelbaum, L. K. McDougal, J. Chaitram, S. McAllister, N. Clark, G. Killgore, C. M. O'Hara, L. Jevitt, J. B. Patel, and B. Bozdogan. 2004. Vancomycin-resistant Staphylococcus aureus isolate from a patient in Pennsylvania. Antimicrob. Agents Chemother. 48:275-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weigel, L. M., D. B. Clewell, S. R. Gill, N. C. Clark, L. K. McDougal, S. E. Flannagan, J. F. Kolonay, J. Shetty, G. E. Killgore, and F. C. Tenover. 2003. Genetic analysis of a high-level vancomycin-resistant isolate of Staphylococcus aureus. Science 302:1569-1571. [DOI] [PubMed] [Google Scholar]
- 28.Wright, G. D., T. R. Holman, and C. T. Walsh. 1993. Purification and characterization of VanR and the cytosolic domain of VanS: a two-component regulatory system required for vancomycin resistance in Enterococcus faecium BM4147. Biochemistry 32:5057-5063. [DOI] [PubMed] [Google Scholar]
- 29.Wu, Z., G. D. Wright, and C. T. Walsh. 1995. Overexpression, purification, and characterization of VanX, a D-,D-dipeptidase which is essential for vancomycin resistance in Enterococcus faecium BM4147. Biochemistry 34:2455-2463. [DOI] [PubMed] [Google Scholar]