Abstract
Rapid detection of resistance in Mycobacterium tuberculosis can optimize the efficacy of antituberculous therapy and control the transmission of resistant M. tuberculosis strains. Real-time PCR has minimized the time required to obtain the susceptibility pattern of M. tuberculosis strains, but little effort has been made to adapt this rapid technique to the direct detection of resistance from clinical samples. In this study, we adapted and evaluated a real-time PCR design for direct detection of resistance mutations in clinical respiratory samples. The real-time PCR was evaluated with (i) 11 clinical respiratory samples harboring bacilli resistant to isoniazid (INH) and/or rifampin (RIF), (ii) 10 culture-negative sputa spiked with a set of strains encoding 14 different resistance mutations in 10 independent codons, and (iii) 16 sputa harboring susceptible strains. The results obtained with this real-time PCR design completely agreed with DNA sequencing data. In all sputa harboring resistant M. tuberculosis strains, the mutation encoding resistance was successfully detected. No mutation was detected in any of the susceptible sputa. The test was applied only to smear-positive specimens and succeeded in detecting a bacterial load equivalent to 103 CFU/ml in sputum samples (10 acid-fast bacilli/line). The analytical specificity of this method was proved with a set of 14 different non-M. tuberculosis bacteria. This real-time PCR design is an adequate method for the specific and rapid detection of RIF and INH resistance in smear-positive clinical respiratory samples.
Tuberculosis has been treated effectively for many years. Nevertheless, it is still the second-leading cause of death from an infectious disease worldwide, and it has been reported that resistance to antituberculous drugs is increasing dramatically (28). One of the most important challenges in the control of tuberculosis is a rapid diagnosis of cases and the optimization of antituberculous treatment, mainly to prevent the development of resistance and the dissemination of resistant strains.
Isoniazid (INH) and rifampin (RIF) are considered the mainstays of antituberculous treatment (1, 29). The knowledge of the genetic basis of resistance to both drugs has provided molecular tools to rapidly detect the principal mutations conferring resistance to INH and RIF (2, 14, 16, 19). Ninety-five percent of RIF resistance mutations in Mycobacterium tuberculosis are located in an 81-bp region of the rpoB gene (20). The molecular basis of INH resistance is less well characterized, and mutations in several genes have been associated with it, although mutations in codon 315 of the katG gene are considered to be the most prevalent mutations encoding higher levels of resistance to INH (26).
Despite the use of new liquid medium cultures, the isolation of M. tuberculosis is still time consuming and leads to delays in obtaining susceptibility patterns. Rapid methods to detect resistance are necessary to optimize antituberculous treatment and avoid the transmission of resistant strains. In the last few years, several molecular methods to detect resistance mutations in M. tuberculosis have been described (2, 6, 14); however, they are labor intensive and frequently require well-grown cultures. Recently, real-time PCR has made the rapid detection of resistance mutations from M. tuberculosis isolates possible (4, 5, 7, 22). Nevertheless, little effort has been made to adapt these rapid methods to the detection of resistance directly from clinical samples (8, 10, 11, 13, 24, 27); therefore, genotypic susceptibility patterns still depend on culture.
Our aim is to adapt a real-time PCR design, recently developed in our laboratory (7), to assess resistance against RIF and INH in M. tuberculosis directly from clinical respiratory samples.
(This study was partially presented at the 13th European Congress of Clinical Microbiology and Infectious Diseases, Glasgow, United Kingdom, 10 to 13 May 2003 [P-636].)
MATERIALS AND METHODS
Clinical samples.
Fifteen auramine-positive clinical respiratory samples whose cultures were positive for drug-resistant M. tuberculosis were recovered from our laboratory collection. Sixteen stain-positive respiratory samples harboring susceptible M. tuberculosis isolates were used as controls.
The specimens were decontaminated by the N-acetyl-l-cysteine NaOH method (9) and conserved frozen at −70°C until use.
Spiked control specimens.
An M. tuberculosis-negative sputum sample spiked with 104 CFU of M. tuberculosis H37Rv/ml was used as a susceptible control.
To increase the variety of resistance mutations assayed, we obtained a set of 10 resistant controls after spiking a pool of M. tuberculosis-negative sputa with 104 CFU of 10 different M. tuberculosis strains/ml encoding 11 different resistance mutations in rpoB (codons 511, 513, 514, 515, 516, 517, 526, 531, and 533) and three different substitutions in codon 315 of katG (see Table 2). The resistance mutations assayed for the rpoB gene represent the different mutations detected in Spain up to the year 2002 because they are a selection of resistant strains compiled nationwide (provided by the Mycobacterium Reference Laboratory, Madrid, Spain).
TABLE 2.
Genotypic and phenotypic characteristics of resistant bacilli spiked in negative sputum samples and Tms obtained with the real-time PCR assay
| Mutant control strain | Resistance phenotypea
|
Mutant codon | Nucleotidic substitution | Amino acid substitution |
Tm (°C) (deviation from wild type)b
|
|||
|---|---|---|---|---|---|---|---|---|
| RIF | INH | RPO1 | RPO2 | KAT | ||||
| 1C | R | S | rpo deletion 515-516 | Deletion TGG | 57.62 (−9.38) | |||
| 2C | R | R | rpo531 | TCG→TGG | Ser→Trp | 59.12 (−5.76) | ||
| kat, no mutant | ||||||||
| 3C | R | R | rpo516 | GAC→GTC | Asp→Val | 64.14 (−2.86) | 70.81 (1.93) | |
| kat315 mutant | AGC→AAC | Ser→Asn | ||||||
| 4C | R | R | rpo531 | TCG→TTG | Ser→Leu | 58.82 (−6.06) | 70.85 (1.97) | |
| kat315 mutant | AGC→ACA | Ser→Thr | ||||||
| 5C | R | S | rpo533 | CTG→CCG | Leu→Pro | 63.86 (−1.02) | ||
| 6C | R | S | rpo526 | CAC→CTC | His→Leu | 64.38 (−0.50) | ||
| 7C | R | R | rpo511 | CTG→CCG | Leu→Pro | 60.78 (−6.22) | 70.84 (1.96) | |
| rpo516 | GAC→GGC | Asp→Gly | ||||||
| kat315 mutant | AGC→ACC | Ser→Thr | ||||||
| 8C | R | R | rpo deletion 516-517 | Deletion GAC-CAG | Ser→Thr | 60.92 (−6.08) | 70.86 (1.98) | |
| kat315 mutant | AGC→ACC | |||||||
| 9C | R | R | rpo526 | CAC→TAC | His→Tyr | 64.38 (−0.50) | 70.91 (2.03) | |
| kat315 mutant | AGC→ACC | Ser→Thr | ||||||
| 10C | R | S | rpo513 | CAA→CGA | Gln→Arg | 61.66 (−5.34) | ||
R, resistant; S, susceptible.
Wild-type Tms for RPO1, RPO2, and KAT are 67.0, 64.88, and 68.88°C, respectively.
As specificity controls, we spiked a pool of M. tuberculosis-negative sputa with six mycobacteria (final concentration, 104 CFU/ml) (M. bovis, M. avium, M. kansasii, M. gordonae, M. fortuitum, and M. chelonae) and eight nonmycobacterial species (Streptococcus pneumoniae, Staphylococcus aureus, Enterococcus faecalis, Escherichia coli, Pseudomonas aeruginosa, Nocardia asteroides, Haemophilus influenzae, and Klebsiella pneumoniae).
Antibiotic susceptibility testing of isolates.
The phenotypic susceptibility to INH and RIF was assayed by means of the SIRE method (Becton-Dickinson, Sparks, Md.), which is based on the agar proportion method.
DNA sequencing.
In all cases, resistance mutations in rpoB and katG were determined by DNA sequencing with the same primers as those used for the real-time PCR. The amplicons obtained were purified by a commercial system (GFX PCR purification kit; Amersham Biosciences GmbH). They were then sequenced by a PCR-based reaction with the Big Dye Terminator method (Applied Biosystems Inc.) according to the manufacturer's instructions and detected with an AbiPrism 3100 automatic DNA sequencer (Applied Biosystems Inc.).
DNA extraction.
Five hundred microliters of each decontaminated sample was centrifuged at 13,000 rpm with a Hettich Milkro 20 centrifuge for 15 min, and the supernatant was discarded. The pellet was resuspended in 50 μl of lysis solution (reagents 1 and 2 from an Accuprobe culture identification reagent kit; Gene Probe Inc., San Diego, Calif.) diluted 1:16 in fresh MGIT medium (Becton-Dickinson) and boiled for 5 min. Twenty-five microliters of sterile 106-μm glass beads (Sigma) was added, and the mixture was sonicated in a bath for 5 min. For sequencing purposes, the DNA was extracted from M. tuberculosis isolates in the same way.
Real-time PCR.
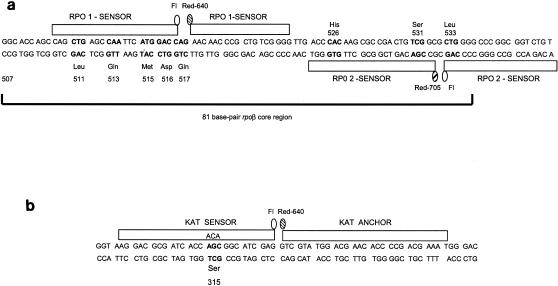
The real-time PCR general design corresponded to that described previously (7). Briefly, two pairs of fluorescence resonance energy transfer (FRET) probes (RPO1 and RPO2) labeled with different fluorophores were used to cover the rpoB core region (Fig. 1a). RPO1 consisted of one probe labeled with fluorescein and the other labeled with LC Red 640. RPO2 probes were labeled with fluorescein and LC Red 705. Both probes were homologous to the wild-type sequence. Probes were detected by using two independent LC fluorescence channels (F2 for Red 640 and F3 for Red 705). To detect mutations in katG, a pair of FRET probes (KAT) labeled with fluorescein and LC Red 640 was used (Fig. 1b). The probes were homologous with the mutant sequence in codon 315 and detected in channel F2. Each PCR run included a water negative control and a sputum sample spiked with the wild-type H37Rv control.
FIG. 1.
Schematic representation of (a) the rpoB core region and RPO1 and RPO2 FRET probes and (b) the katG region and KAT probes. In both figures, the most frequent mutant codons are shown in boldface type.
To adapt the real-time PCR to detect mutations directly on clinical samples, two major changes were included. First, the PCR was performed with three independent reactions to detect mutations in (i) rpoB with the RPO1 probes, (ii) rpoB with the RPO2 probes (Fig. 1a), and (iii) katG with KAT probes (Fig. 1b). Second, an asymmetric PCR format was designed to increase the amount of target DNA which is complementary to the FRET probes.
(i) Primers and probes.
The primers used were TR8 and TR9 (21) for the rpoB region and TB86 (21) and KatGR (5′-CTCCCACTCGTAGCCGTACA-3′) for the katG gene. Probes were designed by TibMolBiol (DNA synthesis service; Roche Diagnostics. Berlin, Germany), and their sequences were as follows: for RPO1, 5′-CAGCTGAGCCAATTCATGGACC-fluorescein-3′ and 5′-LC Red 640-AACACCCCGCTGTCGGG-phosphate-3′; for RPO2, 5′-ACAGACCGCCGGGCCCCAG-fluorescein-3′ and 5′-LC Red 705-CGACAGTCGGCGCTTGTGGGT-phosphate-3′; and for katG probes, 5′-GACGCGATCACCACAGGCATCGAGG-fluorescein-3′ and 5′-LC Red 640-CGTATGGACGAACACCCCGACGAAATGG-phosphate-3′.
(ii) Master mix.
Two microliters of a 1:10 dilution of the DNA extract was used as a template for the PCR. The master mix was composed of 2 μl of LC FastStart plus deoxynucleotide mix (Roche Diagnostics), 10 pmol of each of the primers amplifying the strand which is complementary to each probe used (TR8 with RPO1, TR9 with RPO2, and KatGR with KAT), 2 pmol of the remaining primers, MgCl2 (5 mM for rpoB and 4 mM for katG), 0.2 μM each Red-labeled probe, and 0.1 μM each fluorescein-labeled probe. The final reaction volume was 20 μl.
(iii) PCR conditions.
Three independent reactions (with RPO1, RPO2, and KAT probes) were performed for each sample, and all of them shared the same PCR profile. Prior to PCR, a preincubation step of 95°C for 7 min was performed to activate the FastStart enzyme. The PCR consisted of 40 cycles with the following thermal sequence: 95°C for 10 s, 55°C for 8 s, and 72°C for 20 s. PCRs were performed in capillary tubes with a Light-Cycler real-time PCR instrument (Roche Diagnostics).
(iv) Post-PCR analysis.
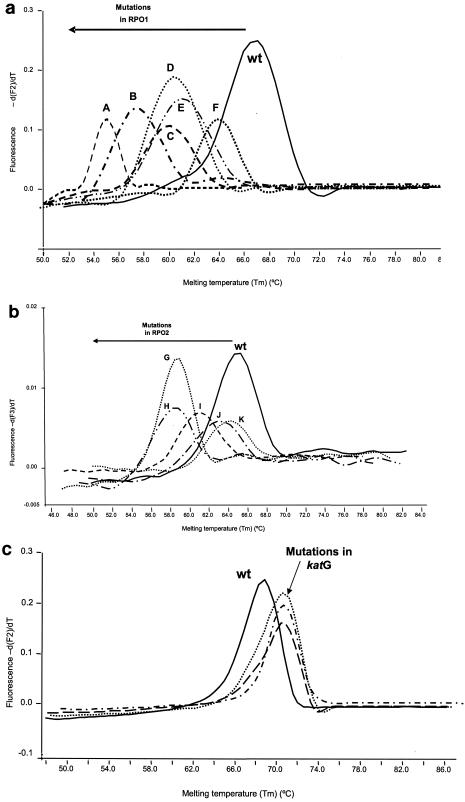
PCR was followed by a melting step involving two sequential melting ramps, and the measurements were taken during the second step because it offered better definition in the melting curves (Fig. 2). The first melting step consisted of 95°C for 5 s, 65°C (annealing temperature) for 30 s, 40°C for 0 s, and 95° for 0 s. This pattern was followed by 95°C for 5 s, 60°C for 30 s, 40°C for 0 s, and 95°C for 0 s. The rate of temperature increase was 0.2°C/s, and fluorescence was continuously acquired. Interpretation of melting data was done as previously described (7). A mutation was suspected in the region covered by the probes when the deviations from the reference melting temperature (Tm) were higher than two times the standard deviation. Mutations in rpoB led to reductions in the reference Tm, and mutations in katG led to an increase in the reference Tm.
FIG. 2.
Melting curves of wild-type (wt) strains and some representatives of mutant strains. (a) Melting curves obtained with RPO1 probes. A, deletion at codons 514 and 515; B, deletion at codons 515 and 516; C, double mutant in codons 511 and 516; D, deletion at codons 516 and 517; E, mutant in codon 513 (CAA→CGA); F, mutant in codon 516 (GAC→GTC). (b) Melting curves obtained with RPO2 probes. G, mutant in codon 531 (TCG→TGG); H, mutant in codon 531 (TCG→TTG); I, mutant in codon 526 (CAC→GAC); J, mutant in codon 533; K, mutant in codon 526 (CAC→TAC). (c) Melting curves obtained with KAT probes and mutations at codon 315 (AGC→ACC, AGC→ACA, and AGC→AAC).
No result is given if a melting peak is not detected, which may be due to either amplification inhibition or undetectable amplification of an insufficient amount of template.
Analytical sensitivity.
In order to assess the sensitivity of this method, 1 ml of a pool of culture-negative sputa was spiked with 10-fold dilutions of (i) M. tuberculosis H37Rv, (ii) rpoB mutants in codons 515 and 516 and 531 (TCG→TTG), and (iii) a katG315 ACG→ACC mutant. Spiked sputa were cultured quantitatively in 7H11 Middlebrook agar. The amount of bacilli in each dilution was defined by determining the number of CFU in culture and by microscopic quantification of the number of bacilli by auramine staining performed with 50 μl of these controls. The spiked sputa were analyzed with a Light-Cycler instrument as described above for clinical samples.
RESULTS
From 2000 to 2002, 30 INH- and/or RIF-resistant M. tuberculosis isolates were cultured from respiratory samples in our laboratory. Of these samples, 18 were stain positive and 15 (83%) were available for study from our collection.
DNA sequencing.
The isolates which had been cultured from the specimens in analysis were sequenced. All samples containing RIF-resistant bacilli encoded mutations in rpoB (codons 514 and 515, 515 and 516, 526, and 531), and five samples harboring INH-resistant bacilli encoded mutations in codon 315 of katG (Table 1). Four samples harboring INH-resistant bacilli were excluded from the study due to the lack of mutations in katG and thus, resistance in these strains could not be detected genotypically.
TABLE 1.
Genotypic and phenotypic characteristics of M. tuberculosis isolates cultured from the clinical samples studied and Tms obtained with the real-time PCR assay
| Clinical sample no. (type) | Acid-fast auramine stain resultb | Resistance phenotypec
|
Mutant codon | Nucleotidic substitution | Amino acid substitution |
Tm (°C) (deviation from wild type)a
|
|||
|---|---|---|---|---|---|---|---|---|---|
| RIF | INH | RPO1 | RPO2 | KAT | |||||
| 1 (sputum) | + (10/line) | R | S | rpo deletion 515-516 | Deletion TGG | 57.20 (−9.8) | |||
| 2 (bronchial aspirate) | + (>50/field) | R | S | rpo deletion 514-515 | Deletion TCA-TGG | 55.0 (−12) | |||
| 3 (sputum) | + (5/field) | R | R | rpo531 | TCG→TTG | Ser→Leu | 58.72 (−6.16) | 71.15 (+2.27) | |
| kat315 mutant | AGC→ACC | Ser→Thr | |||||||
| 4 (sputum) | + (>50/field) | R | R | rpo531 | TCG→TTG | Ser→Leu | 59.05 (−5.83) | ||
| kat, no mutant | |||||||||
| 5 (sputum) | + (10/field) | R | I | rpo526 | CAC→GAC | His→Asp | 61.24 (−3.64) | ||
| kat, no mutant | |||||||||
| 6 (sputum) | + (>50/field) | S | R | kat315 mutant | AGC→ACC | Ser→Thr | 71.32 (+2.44) | ||
| 7 (sputum) | + (10/line) | R | S | rpo526 | CAC→GAC | His→Asp | 61.48 (−3.4) | ||
| 8 (sputum) | + (20/line) | R | S | rpo531 | TCG→TTG | Ser→Leu | 58.54 (−6.34) | ||
| 9 (sputum) | + (10/line) | R | R | rpo531 | TCG→TTG | Ser→Leu | 59.83 (−5.05) | 71.32 (+2.44) | |
| kat315 mutant | AGC→ACC | Ser→Thr | |||||||
| 10 (sputum) | + (20/field) | S | R | kat315 mutant | AGC→ACC | Ser→Thr | 71.62 (+2.74) | ||
| 11 (sputum) | + (>50/field) | R | R | rpo531 | TCG→TTG | Ser→Leu | 58.77 (−6.11) | 71.46 (+2.58) | |
| kat315 mutant | AGC→ACC | Ser→Thr | |||||||
Wild-type Tms for RPO1, RPO2, and KAT are 67.0, 64.88, and 68.88°C, respectively.
Obtained at a magnification of ×250. +, positive stain.
R, resistant; S, susceptible; I, intermediate.
Real-time PCR detection of resistance in clinical samples.
The reference Tm values for wild-type sequences were obtained by 10 repetitions of the assay using sputa spiked with the reference susceptibility control (M. tuberculosis H37Rv). The average reference values for wild-type sequences were as follows: 67°C for RPO1, 64.88°C for RPO2, and 68.88°C for KAT (standard deviations, 0.1, 0.17, and 0.22°C, respectively).
All clinical samples containing M. tuberculosis strains with a mutation in rpoB or katG were efficiently detected by this real-time PCR design. In all cases, Tm deviations (more than two times standard deviation) were detected with respect to the susceptible reference Tm (Table 1).
All clinical samples containing RIF- or INH-susceptible strains showed a Tm within two times the standard deviation of the reference susceptible Tm.
Real-time PCR detection of resistance in spiked control specimens.
In order to evaluate this real-time PCR design with a broader spectrum of resistance mutations, we tested 10 control specimens obtained by spiking a pool of negative sputa with different M. tuberculosis strains with known resistant mutations: for rpoB, nine different substitutions in eight independent codons, one deletion, and one double mutant in two codons were found; and for katG, three different substitutions in codon 315 (Table 2).
Again, all samples harboring resistant strains were correctly detected by significant deviations from the reference wild-type Tm values (Table 2).
Analytical sensitivity and specificity of real-time PCR.
In order to assess the analytical sensitivity of the method, culture-negative sputa were spiked with serial dilutions of M. tuberculosis H37Rv and three different resistant strains. The amount of bacilli in each dilution was defined by determining the number of CFU in culture and by microscopic quantification of the number of bacilli. The test succeeded in detecting a bacterial load equivalent to 103 CFU/ml (10 acid-fast bacilli/line).
To determine whether the assay was specific for M. tuberculosis, we tested a set of negative sputa spiked with 14 nontuberculous mycobacteria and other bacterial genera. Only M. bovis (belonging to the M. tuberculosis complex) and M. avium (only with RPO1) led to a post-PCR melting fluorescent signal. Nevertheless, the Tm value obtained with M. avium was completely different from that obtained with M. tuberculosis (Tm = 53.7°C).
DISCUSSION
A rapid assessment of antituberculous drug resistance is essential for the clinical management of patients with tuberculosis and to avoid the transmission of resistant strains. At present, the quickest way of analyzing the susceptibility of M. tuberculosis isolates is by real-time PCR. In recent years, we and other authors have developed real-time PCR designs specialized for detecting mutations encoding resistance to RIF and INH (3-5, 7, 22, 25). Nevertheless, most of these assays have been applied only to cultured isolates and therefore lead to delays in assessing the susceptibility patterns, as the methods still require culture. Furthermore, the few studies which have applied real-time PCR for detecting resistance directly from clinical samples were able to detect only the most prevalent resistance mutations (4, 5, 25).
In the present study, we tried to resolve this dual requirement of rapidity and polyvalent detection of a wide variety of resistance mutations in M. tuberculosis. A real-time PCR assay based on rapid cycle technology, recently developed by García de Viedma et al. (7) to detect multiple resistance mutations in a single reaction tube, was adapted to analyze M. tuberculosis resistance directly from clinical samples. This adaptation required (i) the real-time PCR design to be converted to an asymmetric format in order to preferentially amplify the DNA strands complementary to the probes and (ii) the three PCRs (with RPO1, RPO2, and KAT probes) to be performed in three independent reaction tubes.
In this sense, this design is methodologically similar to the design recently published by Ruiz et al. (17), which used FRET probes and rapid-cycle PCR to detect resistance to RIF and INH directly from clinical samples. Ruiz et al. (17) succeeded in detecting mutations from four patients with RIF-resistant isolates and seven patients with INH-resistant isolates. The present study is different in several ways. First, we evaluated a much wider variety of mutations encoding resistance (14 different mutations in 10 independent codons). Secondly, in the study by Ruiz et al. (17), sequencing data were not available; therefore, neither the correlation of real-time PCR data with the genotypic “gold standard ” nor the efficiency in detecting certain nucleotidic substitutions in different codons can be assessed. In this study, sequencing data are available for all 46 specimens, and thus, we can assure (i) 100% correlation with DNA sequencing and (ii) the efficiency of this system for detecting not only some specific prevalent mutations but also a huge variety of different mutations, some of them very rare. This result makes our system suitable for different geographic settings, as has been proved in a study done in Poland (18) in which part of this design has been applied, allowing the detection of mutations different from those tested here. In addition, in our method, the search of all rpoB and katG regions shares PCR conditions and allows all mutations to be detected in a single run, which makes for earlier availability of results. As a potential methodological limitation, it could be argued that this design does not detect INH-resistant mutations out of katG315. Resistance to INH is less frequently found to be associated with mutations out of katG, and it is important to specify that isolates with mutations in katG315 show high-level resistance to INH, whereas isolates with mutations in loci other than katG are associated with low or intermediate levels of resistance to INH. In this context (7), in 92% of the resistant strains with a mutation in katG315, high MICs of INH (>3 μg/ml) were obtained, whereas most of the resistant strains with mutations other than those at katG315 had low MICs (<1 μg/ml). Other authors (12; M. E. Verdú et al., Abstr. 12th Eur. Cong. Clin. Microbiol. Infect. Dis. 2002, abstr. P528) have found similar values, with most of the isolates encoding a katG315 mutation showing high levels of resistance to INH. It has recently been shown that among all the INH-resistant M. tuberculosis strains from Equatorial Guinea, none had mutations in katG and 80.5% had mutations in inhA (23). All these resistant strains, with mutations mapping out of katG315, showed low levels of resistance to INH, and the patients responded to standard therapy. Additionally, katG315 mutations have also been found to be a marker for multidrug-resistant tuberculosis, and they are successfully transmitted within the population (26). Taking all these data together, we believe that the inability of this real-time PCR design to detect INH-resistant mutations other than katG315 should not be cause for concern.
When searching for resistant mutations directly from clinical samples, the two main requirements an assay should be its ability to (i) guarantee specificity in the detection of mutations, considering that respiratory samples frequently include additional bacteria, and (ii) obtain high analytical sensitivity values.
We first tested whether amplification occurred when different respiratory pathogens were used as PCR templates. No amplification signal was found with 14 different non-M. tuberculosis pathogens. The only pathogens which led to an amplification signal were M. bovis and M. avium. It is not unexpected that M. bovis would lead to an amplification signal because it is included in the M. tuberculosis complex. With regard to M. avium, only the RPO1 probe produced a hybridization signal, and the Tm of the probe was easily differentiated from that obtained from M. tuberculosis (53.7 versus 67°C). This finding rules out the possibility of a misinterpretation of the results.
Second, the analytical sensitivity of the assay was calculated to be 103 CFU/ml (10 acid-fast bacilli/line) for the three regions assayed (RPO1, RPO2, and KAT), for both wild-type and mutant strains. In our laboratory, of the stain-positive samples obtained over 3 years, only three (17%) had a lower bacterial load. This finding means that this method would succeed in analyzing the susceptibility patterns of the majority of the stain-positive respiratory samples in this context. This method could be criticized for having used only stain-positive sputum samples. How useful this design is going to be in smear-negative specimens remains to be established. Few studies have data from stain-negative samples (10, 15, 27), and those studies are all based on traditional PCR and detect either rifampin or isoniazid resistance only, but never both. In our opinion, this limitation of analytical sensitivity should not be considered a weakness of this real-time PCR approach, especially if we consider the diagnostic context in which the susceptibility pattern of M. tuberculosis is usually requested. Generally, clinicians demand a rapid assessment of susceptibility when they receive the first laboratory evidence of M. tuberculosis infection, that is, microscopic observation of bacilli. Additionally, patients with stain-positive samples have a higher risk of causing transmission; therefore, the need for susceptibility patterns is particularly important in these cases. In all these situations, real-time PCR works properly, and requests for further assessment can be met satisfactorily. For stain-negative samples, it generally makes no sense to apply the real-time PCR, considering the low rate of positivity for M. tuberculosis in most mycobacterial laboratories in this context. In this case, M. tuberculosis was isolated in only 5.7% of all samples received from requests for mycobacteria culture.
The method described here combines an easy and short DNA extraction from clinical samples with a rapid, sensitive, and specific real-time PCR design. The complete assay, including analysis, enables us to obtain resistance results in 1[1/2] hours. To our knowledge, this assay is the first one based on real-time PCR that is able to detect, simultaneously and directly from clinical respiratory samples, numerous different mutations conferring resistance to RIF and those most frequently associated with high resistance to INH by using several FRET probes simultaneously in a single PCR run. The flexibility of real-time PCR assays to detect resistance mutations in different geographic settings cannot yet be answered, and international panels should be compiled and tested to answer this question.
The results obtained in this study lead us to propose this real-time PCR design as an adequate tool for the direct analysis of M. tuberculosis resistance in clinical samples. We are conscious that the introduction of real-time tools in the diagnostic laboratory requires specific installations and personnel to guarantee that the assay can be run upon receipt of the sample while traditional approaches to susceptibility tests are maintained, although this may not be the reality with regard to many laboratories. Efforts should be made, at least in specific reference laboratories, to adapt these new molecular procedures which allow the susceptibility pattern of M. tuberculosis to be obtained quickly, thus considerably reducing the time between diagnosis and initiation of individualized therapy.
Acknowledgments
We thank José Luis Jiménez and Milagros González from the central sequencing service of the Hospital G. U. Gregorio Marañón for their technical assistance in obtaining DNA sequencing data.
The sequencer of Hospital Gregorio Marañon was acquired with a grant from Fondo de Investigaciones Sanitarias (01/3624). This study was partially financed by grants from Comunidad de Madrid (08.2/0029.1/2001) and Fondo de Investigaciones Sanitarias (020882). M. Marín has a contract from the Fondo de Investigaciones Sanitarias (FO1/48).
We are indebted to Thomas O'Boyle for his revision of the English in the manuscript.
REFERENCES
- 1.American Thoracic Society, Centers for Disease Control and Prevention, and Infectious Diseases Society of America. 2003. Treatment of tuberculosis. Am. J. Respir. Crit. Care Med. 167:603-662. [DOI] [PubMed] [Google Scholar]
- 2.Caws, M., and F. A. Drobniewski. 2001. Molecular techniques in the diagnosis of Mycobacterium tuberculosis and the detection of drug resistance. Ann. N. Y. Acad. Sci. 953:138-145. [DOI] [PubMed] [Google Scholar]
- 3.Edwards, K. J., L. A. Metherell, M. Yates, and N. A. Saunders. 2001. Detection of rpoB mutations in Mycobacterium tuberculosis by biprobe analysis. J. Clin. Microbiol. 39:3350-3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.El-Hajj, H. H., S. A. Marras, S. Tyagi, F. R. Kramer, and D. Alland. 2001. Detection of rifampin resistance in Mycobacterium tuberculosis in a single tube with molecular beacons. J. Clin. Microbiol. 39:4131-4137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Espasa, M. A., J. M. Gonzalez, F. Alcaide, J. Lonca, X. M. Manterola, E. Verdu, and P. Coll. 2002. Use of real time PCR for direct detection in clinical samples of genetic polymorphisms causing resistance to isoniazid and rifampin in Mycobacterium tuberculosis. Clin. Microbiol. Infect. 8:O131. [Google Scholar]
- 6.Garcia de Viedma, D. 2003. Rapid detection of resistance in Mycobacterium tuberculosis: a review discussing molecular approaches. Clin. Microbiol. Infect. 9:349-359. [DOI] [PubMed] [Google Scholar]
- 7.García de Viedma, D., M. del Sol Díaz Infantes, F. Lasala, F. Chaves, L. Alcalá, and E. Bouza. 2002. New real-time PCR able to detect in a single tube multiple rifampin resistance mutations and high-level isoniazid resistance mutations in Mycobacterium tuberculosis. J. Clin. Microbiol. 40:988-995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johansen, I. S., B. Lundgren, A. Sosnovskaja, and V. O. Thomsen. 2003. Direct detection of multidrug-resistant M. tuberculosis in clinical specimens in low- and high-incidence countries by line probe assay. J. Clin. Microbiol. 41:4454-4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kubica, G. P., W. E. Dye, M. L. Cohn, and G. Middlebrook. 1963. Sputum digestion and decontamination with N-acetyl-L-cysteine-sodium hydroxide for culture of mycobacteria. Am. Rev. Respir. Dis. 87:775-779. [DOI] [PubMed] [Google Scholar]
- 10.Leung, E. T.-Y., K.-M. Kam, A. Chiu, P.-L. Ho, W.-H. Seto, K.-Y. Yuen, and W.-C. Yam. 2003. Detection of katG Ser315Thr substitution in respiratory specimens from patients with isoniazid-resistant M. tuberculosis using PCR-RFLP. J. Med. Microbiol. 52:999-1003. [DOI] [PubMed] [Google Scholar]
- 11.Marttila, H. J., H. Soini, E. Vyshnevskaya, B. I. Vyshnevskiy, T. F. Otten, A. V. Vasilyef, et al. 1999. Line probe assay in the rapid detection of rifampin-resistant Mycobacterium tuberculosis directly from clinical specimens. Scand. J. Infect. Dis. 31:269-273. [DOI] [PubMed] [Google Scholar]
- 12.Marttila, H. J., H. Soini, E. Eerola, E. Vyshnevskaya, B. I. Vyshnevskiy, T. F. Otten, A. V. Vasilyef, and M. K. Viljanen. 1998. A Ser315Thr substitution in KatG is predominant in genetically heterogeneous multidrug-resistant Mycobacterium tuberculosis isolates originating from the St. Petersburg area in Russia. Antimicrob. Agents Chemother. 42:2443-2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mokrousov, I., T. Otten, M. Filipenko, A. Vyazovaya, E. Chrapov, E. Limeschenko, et al. 2002. Detection of isoniazid-resistant Mycobacterium tuberculosis strains by a multiplex allele-specific PCR assay targeting katG codon 315 variation. J. Clin. Microbiol. 40:2509-2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Musser, J. M. 1995. Antimicrobial agent resistance in mycobacteria: molecular genetic insights. Clin. Microbiol. Rev. 8:496-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Patnaik, M., K. Liegmann, and J. B. Peter. 2001. Rapid detection of smear-negative Mycobacterium tuberculosis by PCR and sequencing for rifampin resistance with DNA extracted directly from slides. J. Clin. Microbiol. 39:51-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rattan, A., A. Kalia, and N. Ahmad. 1998. Multidrug-resistant Mycobacterium tuberculosis: molecular perspectives. Emerg. Infect. Dis. 4:195-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ruiz, M. T., M. J. Torrez, A. C. Llanos, A. Arroyo, J. C. Palomares, and J. Aznar. 2004. Direct detection of rifampin- and isoniazid-resistant Mycobacterium tuberculosis in auramine-rhodamine-positive sputum specimens by real-time PCR. J. Clin. Microbiol. 42:1585-1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sajduda, A., A. Brzostek, M. Poplawska, E. Augustynowicz-Kopee, Z. Zwolska, S. Nieman, J. Dziadek, and D. Hillemann. 2004. Molecular characterization of rifampin- and isoniazid-resistant Mycobacterium tuberculosis strains isolated in Poland. J. Clin. Microbiol. 42:2425-2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Telenti, A. 1997. Genetics of drug resistance in tuberculosis. Clin. Chest Med. 18:55-64. [DOI] [PubMed] [Google Scholar]
- 20.Telenti, A., P. Imboden, F. Marchesi, D. Lowrie, S. Cole, M. J. Colston, et al. 1993. Detection of rifampicin-resistance mutations in Mycobacterium tuberculosis. Lancet 341:647-650. [DOI] [PubMed] [Google Scholar]
- 21.Telenti, A., N. Honore, C. Bernasconi, J. March, A. Ortega, B. Heym, et al. 1997. Genotypic assessment of isoniazid and rifampin resistance in Mycobacterium tuberculosis: a blind study at reference laboratory level. J. Clin. Microbiol. 35:719-723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Torres, M. J., A. Criado, J. C. Palomares, and J. Aznar. 2000. Use of real-time PCR and fluorimetry for rapid detection of rifampin and isoniazid resistance-associated mutations in Mycobacterium tuberculosis. J. Clin. Microbiol. 38:3194-3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tudó, G, J. González, R. Obama, J. M. Rodriguez, J. R. Franco, M. Espasa, P. P. Simarro, G. Escaramis, C. Ascaso, A. Garcia, and M. T. Jimenez de Anta. 2004. Study of resistance to anti-tuberculous drugs in five districts of Equatorial Guinea: rates, risk factors, genotyping of gene mutations and molecular epidemiology. Int. J. Tuberc. Lung Dis. 8:15-22. [PubMed] [Google Scholar]
- 24.Van Der Zanden, A. G., E. M. Te Koppele-Vije, N. Vijaya Bhanu, D. Van Soolingen, and L. M. Schouls. 2003. Use of DNA extracts from Ziehl-Neelsen-stained slides for molecular detection of rifampin resistance and spoligotyping of Mycobacterium tuberculosis. J. Clin. Microbiol. 41:1101-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van Doorn, H. R., E. C. J. Claas, K. E. Templeton, A. G. M. van der Zanden, A. K. Vije, M. D. Jong, J. Dankert, and E. J. Kuijper. 2003. Detection of a point mutation associated with high-level isoniazid resistance in Mycobacterium tuberculosis by using real-time PCR technology with 3′-minor groove binder-DNA probes. J. Clin. Microbiol. 41:4630-4635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van Soolingen, D., P. E. de Haas, H. R. van Doorn, E. Kuijper, H. Rinder, and M. W. Borgdorff, M. W. 2000. Mutations at amino acid position 315 of the katG gene are associated with high-level resistance to isoniazid, other drug resistance, and successful transmission of Mycobacterium tuberculosis in The Netherlands. J. Infect. Dis. 182:1788-1790. [DOI] [PubMed] [Google Scholar]
- 27.Whelen, A. C., T. A. Felmlee, J. M. Hunt, D. L. Williams, G. D. Roberts, L. Stockman, et al. 1995. Direct genotypic detection of Mycobacterium tuberculosis rifampin resistance in clinical specimens by using single-tube heminested PCR. J. Clin. Microbiol. 33:556-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.World Health Organization. 2000. Anti-tuberculosis drug resistance in the world. Prevalence and trends. Report no. 2. Publication W. H. O./CDS/TB/2000.278. World Health Organization, Geneva, Switzerland.
- 29.World Health Organization. 2003. Global Tuberculosis Programme. Treatment of tuberculosis: guidelines for national programmes. Publication W. H. O./CDS/TB/2003.13. World Health Organization, Geneva, Switzerland.