Abstract
The sequence of the 45.2-kb multidrug and mercury resistance region of pRMH760, a large plasmid from a clinical isolate of Klebsiella pneumoniae collected in 1997 in Australia, was completed. Most of the modules found in the resistance determinant (r-det), or Tn2670, region of NR1 (also known as R100), isolated from a Shigella flexneri strain in Japan in the late 1950s, were present in pRMH760 but in a different configuration. The location was also different, with the Tn2670-derived region flanked by the transposition module of Tn1696 and a mercury resistance module almost identical to one found in the plasmid pDU1358. This arrangement is consistent with a three-step process. First, the r-det was circularized via homologous recombination between the IS1 elements and reincorporated at a new location, possibly in a different plasmid, via homologous recombination between the 5′-conserved (5′-CS) or 3′-CS of the In34 integron in the r-det and the same region of a second class 1 integron in a Tn1696 relative. Subsequently, resolvase-mediated recombination between the res sites in the r-det and a second mercury resistance transposon removed one end of the Tn1696-like transposon and part of the second transposon. Other events occurring within the r-det-derived portion have also contributed to the formation of the pRMH760 resistance region. Tn2 or a close relative that includes the blaTEM-1b gene had moved into the Tn21 mercury resistance module with subsequent deletion of the adjacent sequence, and all four 38-bp inverted repeats corresponding to Tn21 family transposon termini have been interrupted by an IS4321-like element.
One of the earliest resistance factors to be identified was NR1, subsequently also called R100, a 94.3-kb self-transmissible multiresistance plasmid isolated from a Shigella flexneri strain in Japan in the late 1950s (20). This IncFII plasmid confers resistance to chloramphenicol, streptomycin, spectinomycin, sulfonamides, tetracycline, and mercury, and its complete sequence is now available (GenBank accession no. AP000342). NR1 consists of two components, a resistance transfer factor (RTF) and a resistance determinant region (r-det), separated by two directly oriented copies of the insertion sequence IS1. The RTF contains genes for conjugative transfer (tra) and replication (rep) functions but also includes the transposon Tn10, which contains the tetA(B) tetracycline resistance determinant (30). The r-det includes the other antibiotic resistance markers of NR1 and a mercury resistance (mer) determinant and is essentially equivalent to the transposon Tn2670, which includes both copies of IS1 (13, 16).
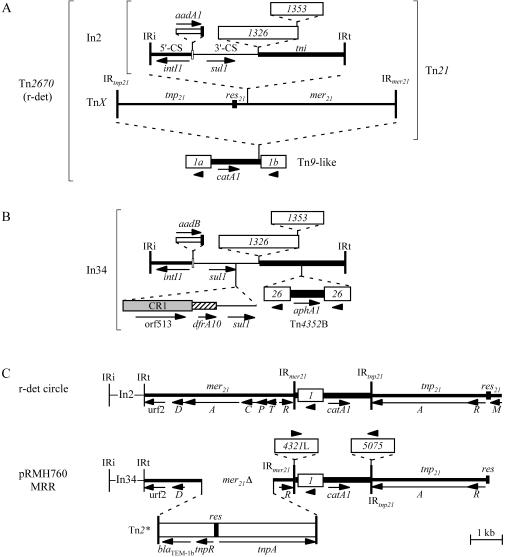
Tn2670 (Fig. 1A) has a complex structure that has arisen via a series of transpositional and other events. It is composed of a transposon closely related to Tn9 (1), with the mercury and antibiotic resistance transposon Tn21 (19) within it. The Tn9-like backbone consists of the two IS1 elements flanking a 1,547-bp central region that contains the catA1 (formerly catI) chloramphenicol resistance gene and is identical to but 445 bp longer at the IS1b end than the central region of Tn9 (GenBank accession no. V00622). Tn21 is inserted 58 bp from the IS1b end and is flanked by a 5-bp duplication of the target sequence, as is characteristic of Tn21 transposition. However, Tn21 is also made up of a transposon within a transposon (2, 19, 23). The class 1 integron In2, a transposon remnant, is situated 340 bp beyond the end of the Tn21 resolution site (res21) and is flanked by a 5-bp target duplication, indicating that it was also acquired by transposition. In2 contains two conserved segments (CS) flanking another mobile element, the aadA1 gene cassette (encoding streptomycin and spectinomycin resistance), as well as a partial transposition (tni) module and two insertion sequences, IS1326 and IS1353 (2). The 5′-CS includes the intI1 gene, the attI1 site, and the Pc promoter typical of class 1 integrons (10, 11), and the 3′-CS contains the sul1 sulfonamide resistance gene.
FIG. 1.
Structure of (A) Tn2670 and (B) In34. IRs at transposon and class 1 integron termini are shown as solid vertical bars. The res sites of Tn21 (res21) and Tn2* (res) are indicated by shorter vertical bars. The names of the various modules are indicated above the lines. Arrows below the lines indicate the positions of relevant genes. Insertion sequences are shown as open boxes containing the IS number, and, where relevant, arrowheads indicate their orientation. Gene cassettes are shown as narrow open boxes with a bar at one end to represent the 59-be, and attI1 is shown as an open vertical bar. CR1 is a shaded box, and the dfrA10 region is a hatched box. The Tn2670 map was constructed from the sequence in GenBank accession no. AP000342 and In34 from GenBank accession no. AY123253.1. (C) Comparison of the circular form of r-det, opened at IRi, with part of the pRMH760 MRR. The structures of the integrons In2 and In34 are as in panels A and B, respectively.
Tn2670 is an active transposon (13, 16), and in NR1, it is flanked by a 9-bp target site duplication characteristic of insertion of IS1. However, RecA-dependent homologous recombination between the IS1a and IS1b elements of Tn2670 can form an extrachromosomal, covalently closed circular molecule containing a single copy of IS1 (3, 4, 29). Although the r-det circle is not a stable replicon, it could effectively act as a mobile element if it was incorporated at other locations by homologous recombination, for example, between the IS1 in the circle and a copy of IS1 located in a new target molecule or between the conserved regions in the integron and a second integron.
We recently reported the structure of an In2-like class 1 integron, In34 (Fig. 1B), from a clinical multidrug resistant Klebsiella pneumoniae strain isolated at a Sydney hospital in 1997 (25). In34 (17.53 kb) includes a backbone identical to that of In2, but the aadB gene cassette (encoding gentamicin, kanamycin, and tobramycin resistance) has replaced the aadA1 cassette, and additional resistance genes have been acquired (Fig. 1B). As part of the tnp module of Tn1696 (tnp1696) (24) was found adjacent to IRi (inverted repeat at intI1 end) and part of the mer21 module was found adjacent to IRt (inverted repeat at tni end) of In34, with precisely the same boundaries between In34 and these modules as in Tn1696 and Tn21, we proposed that this mixed transposon configuration arose by homologous recombination between the 5′-CS or 3′-CS regions of integrons in transposons related to Tn21 and Tn1696 (25). However, two additional antibiotic resistance genes, catA1 (encoding chloramphenicol resistance) and blaTEM (encoding β-lactam resistance) were identified and mapped close to In34, indicating the presence of a larger multiresistance region (MRR) in pRMH760 (25).
As pRMH760 contains several components in common with Tn2670 from NR1 (an In2-like integron, at least part of mer21 and the catA1 gene), it seemed possible that the resistance region of pRMH760 is derived from Tn2670. Also, because NR1 dates from the early days of antibiotic use and pRMH760 is a recent isolate, this derivation would indicate that large, complex MRRs persist as discrete entities but continue to evolve over time. To examine these possibilities, the remainder of the pRMH760 MRR was sequenced to determine its structure and compare it to Tn2670 and other known MRRs.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Escherichia coli DH5α was used to propagate plasmid DNA. pRMH760 was isolated from K. pneumoniae strain 24120497, and subclones were constructed as described previously (25). Bacteria were routinely cultured at 37°C in Luria-Bertani medium or on Luria-Bertani agar. Appropriate antibiotics (Sigma) were added at the following concentrations: ampicillin, 100 μg ml−1; chloramphenicol, 25 μg ml−1; gentamicin, 8 μg ml−1; kanamycin, 15 μg ml−1; and tobramycin, 12.5 μg ml−1.
DNA sequencing.
Plasmid DNA was isolated as described previously (25). PCR was used to amplify fragments of pRMH760 to confirm boundaries between cloned fragments predicted by mapping and to obtain sequences. Amplification reactions were carried out in 50-μl volumes containing 1× PCR Master Mix (Promega), additional MgCl2 to a concentration of 3 mM, 0.4 pmol of each primer, and approximately 20 ng of plasmid template. PCR conditions were generally 94°C for 3 min, followed by 40 cycles of 94°C for 1 min, 57°C for 1 min, and 72°C for 2 min, with a final incubation at 72°C for 5 min. PCR products from Tris-acetate-EDTA-agarose gels were purified for sequencing with an UltraClean DNA purification kit (Mo Bio Laboratories, Inc.). Automated sequencing and DNA sequence assembly were as described previously (25). The sequence of at least one strand of inserts in subclones of pRMH760 was determined for regions for which several sequences are already available in databases (e.g., tnp21, mer21, tnp1696, and IS1). Both strands were sequenced for all novel regions where ambiguities were present or if the sequence differed from published sequences.
Sequence analysis.
GenBank searches were performed with the BLASTN program available through WebANGIS (Australian National Genomic Information Service). Programs in the Genetics Computer Group Wisconsin Package, version 8.1.0, were used via WAG (WebANGIS GCG) to align and analyze the level of identity of DNA sequences and to identify the boundaries of discrete regions.
Nucleotide sequence accession number.
The 45.2-kb sequence of the pRMH760 MRR has been submitted to GenBank under accession no. AY123253.3, which includes 27.5 kb released previously as AY123253.1 (25), AY123253.2, and AY242531-3 (26).
RESULTS
The pRMH760 MRR contains the components of r-det/Tn2670
As the short (250-bp) piece of the mer21 sequence previously found adjacent to IRt of In34 (25) (GenBank accession no. AY123253.1) was identical to that found immediately to the right of In2 in Tn21, the sequence was extended on this side. A region of 15,998 bp determined here (Fig. 1C) contained the components of Tn2670 in the order expected for the circular form of r-det reopened within the integron, but with some additions and deletions. The Tn21 mer module is interrupted by a transposon, here designated Tn2* (see below), that contains the blaTEM-1b ampicillin resistance gene. Part of merA together with merC, merP, and most of merT was missing, presumably due to Tn2*-mediated deletion of the adjacent sequence. Both IRtnp21 and IRmer21 are interrupted by IS4321-like elements (IS5075 and IS4321L) as described previously (26), and the region to the right of res21 that includes the gene known as tnpM is missing. The sequence of the Tn2670-derived region is identical to the corresponding parts of NR1 (AP000342), and the Tn21-derived region is also identical to the corrected version of the Tn21 compilation (AF071413.3). Hence, the pRMH760 MRR backbone appears to be the result of integration of the circular form of a Tn2670-like transposon within a second class 1 integron.
Location of the r-det region.
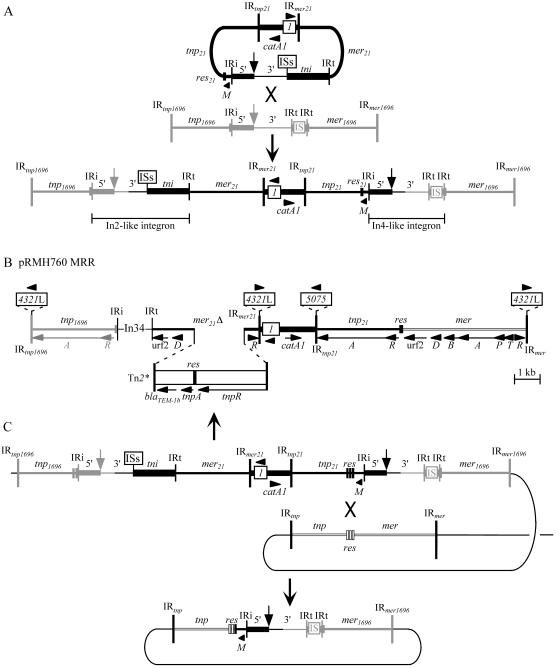
We had also previously found (25) that about 220 bp immediately adjacent to IRi of In34 matched the part of the tnp1696 module immediately adjacent to In4 (GenBank accession no. U12338) (24). This suggested that the tnp1696-In34-mer21 configuration in pRMH760 could have arisen by homologous recombination between the 5′-CS or the 3′-CS of two integrons, one in the same position as In4 in a Tn1696 backbone and the other related to In2 and located in a Tn21 backbone. If the In2-like integron, i.e., In34, was part of a circular r-det molecule, such an event would result in a structure in which two hybrid transposons containing integrons flank the catA1 region and IS1 (Fig. 2A). When the sequence was extended to the left of In34, a complete tnp1696 module was found, but an IS4321L element interrupted IRtnp1696 (Fig. 2B). However, the other end was not as predicted. The segment extending from res21 through the r-det-derived integron to the crossover position, the remainder of the In4-like integron, and the mer1696 module were missing, suggesting that at least one additional step was involved in the formation of the backbone of the pRMH760 MRR. The missing region had been replaced by a mer module different from both mer21 and mer1696 (Fig. 2B).
FIG. 2.
Model for formation of the pRMH760 MRR. (A) Incorporation of a circular r-det by homologous recombination between two integrons, with the crossover within the 3′-CS. (B) The complete pRMH760 MRR. The full structure of In34 is given. (C) Generation of the pRMH760 MRR backbone by resolvase-mediated recombination. Resolvase-mediated excision by crossing over within res would result in the pRMH760 MRR structure shown in panel B plus the excised circle shown below. Features are largely as shown in Fig. 1, except that vertical arrows indicate the positions of gene cassettes, 5′ and 3′ indicate the 5′-CS and 3′-CS, and the boxes labeled ISs and IS represent IS1326 plus IS1353 and IS6100, respectively. Tn1696-derived regions are shown in gray, and the mer region related to that of pDU1358 is shown as a narrow open box. (C) Three subsites of res are shown (not to scale).
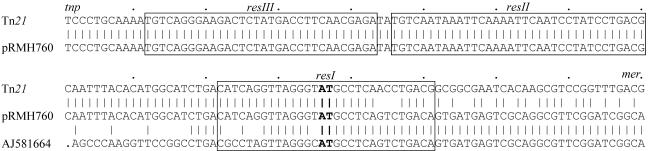
Identity between the sequences of the pRMH760 MRR and Tn21 ends within resI, one of the three subsites of res21, and just after the AT dinucleotide (Fig. 3) at which resolvase-mediated recombinational crossover occurs (28). For 135 bp from this AT dinucleotide, the sequence is identical (Fig. 3) to the region of a partially sequenced transposon (GenBank accession no. AJ581664) immediately preceding a class 1 integron. The sequence identity may continue beyond the other end of that integron, but that sequence is not available. This indicates that a further step in the formation of the pRMH760 MRR backbone was resolvase-mediated recombination between res21 and the res site of a second transposon (Fig. 2C). Resolvase-mediated excision events occur at high frequency only when, as shown in Fig. 2C, the two res sites involved are in the same DNA molecule and in the same orientation (8), and this event would result in formation of the backbone structure found in the pRMH760 MRR plus an excised circular segment.
FIG. 3.
Comparison of res sites. The sequences of the complete res sites of Tn21 (GenBank accession no. AF071413), the pRMH760 MRR, and the partial res site in GenBank accession no. AJ581664 are aligned, with vertical lines indicating identical bases. The first nucleotide shown corresponds to position 3,579 from the start of IRtnp21. The res subsites (12) are boxed, and the AT dinucleotide at which resolvase-mediated recombination takes place is shown in boldface. tnp and mer, regions adjacent to each end of the res site.
The second mer module.
The mer module adjacent to the hybrid res site was also completely sequenced. It is 6.639 kb long and ends with a 38-bp IR identical to IRtnp21 that is interrupted by IS4321L (26). It contains genes corresponding to and 79 to 85% identical to merA, merD, merP, merR, and merT of Tn21, an open reading frame that is 79% identical to urf2M (urf2) of TnX and Tn5060 (18, 19, 24) and a merB gene (Fig. 2B). The last 6.08 kb of this region is nearly identical to the available sequence of the broad-spectrum mer module found in the Serratia marcesens plasmid pDU1358 (obtained by joining sequences from GenBank accession no. M15049, M24940, and AY225348) (7, 21), which has IS4321R in place of IS4321L (26). Identities are 99.6% in the mer module and 97.0% in the IS.
Tn2* and its context.
The 4.95-kb transposon containing the blaTEM1-1b gene located in the mer21 region of the pRMH760 MRR is 98.2% identical to Tn1 from RP1/RP4 (L27758) (22) and 97.4% identical to Tn3 from R1 (V00613) (14). It was designated Tn2* because part of the 4.95-kb sequence is identical to the 1.516 kb, including the blaTEM-1b gene and part of tnpR, that is available for the original Tn2 from RSF1030 (X54607) (5, 6). The remainder may also be identical. Database searches with Tn2* identified three identical, or nearly identical, sequences. A transposon with 22 differences from Tn2*, 8 of which would result in frameshifts, is found in the S. flexneri transposon TnSF1 (AF188331). High identity with the pRMH760 MRR extends on both sides of Tn2*, from the distal end of Tn4352B on the left to IRmer21 (which is uninterrupted in TnSF1) on the right. Some of the differences in Tn2* may be due to sequence errors in TnSF1, as three bases are also missing from the 2.39 kb of the flanking mer module sequence. A partial copy of Tn2* found in the E. coli plasmid p1658/97 (AF550679) is truncated by IS26 and lacks the blaTEM end. The remaining 2.956 kb is identical to Tn2* and is followed by a sequence identical to that of the pRMH760 MRR up to and including IS1, except that IRmer21 is not uninterrupted by an IS4321 or IS5075 element. The third copy, found in a Salmonella enterica serovar Enteritidis strain (AB103092) (17), is identical to Tn2*, but the sequence is incomplete, with 59 bp missing from the blaTEM end. It is located in a context different from that of the other three. The fact that the context of Tn2* is the same in pRMH760, TnSF1, and on one side in p1658/97 suggests that Tn2* may have moved into Tn2670 or a relative prior to its circularization and reincorporation to yield the configuration in the pRMH760 MRR
DISCUSSION
The 45.2-kb MRR of pRMH760 has a complex structure made up of a number of types of mobile elements. However, it appears to have evolved in relatively few steps from other known regions. A 33.5-kb segment that includes In34 (17.53 kb) and all of the antibiotic resistance genes is clearly derived from a transposon related to Tn2670, but a new order of components has been created by circularization of this transposon, followed by incorporation of the circle at a new location via homologous recombination occurring within a region different from that used to form the circle. This part of the MRR has also diverged from Tn2670 by the acquisition of a number of DNA segments containing additional antibiotic resistance genes and of insertion sequences. Two of the additional resistance genes are found within In34 (Fig. 1B), as reported previously (25), and one, the blaTEM-1b gene in Tn2*, is within the region examined here. Two short segments of the Tn2670-derived region have been lost, one that appears to be due to a deletion in the mer21 module adjacent to Tn2* and one that consists of the region of Tn21 found between res21 and the IRi end of In2. While most of these events could have occurred either before or after the r-det excision and integration events, loss of the tnpM segment must have occurred subsequently and is clearly the result of a resolvase-mediated excision event (Fig. 2).
The role of homologous recombination in the dissemination of antibiotic resistance genes is frequently overlooked. However, many small circles can be formed by homologous recombination between pairs of identical sequences in direct orientation, with the circular r-det molecule and the circular form of the common region 1 (CR1)-dfrA10-sul1 region of In34 described previously (25) being just two examples. Integration of these circles either by opening them at a different position, as occurred in the formation of the pRM760 MRR backbone, or at an equivalent position as demonstrated previously in the case of the CR1-containing circle (25), can also involve homologous recombination. Replacement of the aadA1 cassette found in In2 by the aadB cassette in In34 is also likely to have occurred via homologous recombination between the flanking 5′-CS and 3′-CS, and similar exchanges have been demonstrated experimentally (23). These examples clearly illustrate the importance of homologous recombination in evolution of the pRMH760 MRR and other MRRs.
Both the requirement for the two res sites to be on the same molecule and in the same orientation and the phenomenon of transposon exclusion constrain the ways in which resolvase-mediated events can contribute to plasmid evolution. However, the resolution systems that normally complete the transposition process of transposons that form cointegrate intermediates are very efficient. Hence, when two directly oriented res sites are introduced into the same DNA molecule, resolution is likely to occur rapidly. Compatible but nonidentical res sites are also known to recombine efficiently (9). Thus, homologous recombination followed by resolution, as seen here in the formation of the pRMH760 MRR, may be a common theme in the evolution of MRRs, when intact res sites are found in both of the molecules brought together by homologous recombination.
The wide distribution of class 1 integrons provides regions of homology between many different plasmids and has provided the incorporation site for the circular r-det described here. However, the targeting of class 1 integrons to res sites means that they often inactivate the res sites of transposons such as Tn1696 and Tn1403 that contain them (27). Hence, the Tn1696 relative that appears to have been the target of the r-det integration event would not be able to participate in a resolvase-mediated event to remove the region between res1696 and res21 in the initial integration product (Fig. 2A). However, because In2 is a short distance from the res site in Tn21, its resolution system remains active (24) and has interacted with a third transposon only part of which remains in the final MRR. Other combinations of homologous and resolvase-mediated recombination processes can also occur; for example, the circular molecule created by the resolvase-mediated excision step of pRMH760 MRR evolution (Fig. 2C) could be incorporated at new locations by homologous recombination.
Two of the transposons in the pRMH760 MRR, Tn2* and Tn4352B, are found with exactly the same flanking sequences in the Tn21-derived transposon TnSF1 from S. flexneri (GenBank accession no. AF188331). As transposition to precisely the same location is unlikely to occur twice and the changes to the structure surrounding the transposon, such as the deletion adjacent to Tn2*, are also most likely to have arisen only once, this may indicate that TnSF1 and the pRMH760 MRR have a common ancestor. However, other explanations are also possible. For example, these transposons can also potentially be spread by homologous recombination into any region containing the sequence that flanks them. Hence, the two multidrug resistance regions cannot necessarily be assumed to be related to one another by direct lineal decent.
The reason why antibiotic resistance genes are often found clustered within a single large region made up of transposons within other transposons, as seen in Tn2760, the pRMH760 MMR, TnSF1, and other unrelated regions has been the subject of some speculation. However, it may simply reflect the fact that in the compactly organized genomes of plasmids there are few places to go that are without deleterious consequences. Insertion of DNA into the rep region of a plasmid would prevent replication leading to loss of the plasmid, and insertion into a tra region would destroy the ability to transfer into new hosts, restricting its spread by horizontal gene transfer. In contrast, insertion of additional resistance genes into a transposon already in the plasmid would not affect vital plasmid functions. Under the selective pressure of antibiotic use, the additional resistance genes will confer a selective advantage on cells containing the MRR, favoring its retention. The genesis of other clusters of antibiotic resistance genes that have been identified, such as those in the SXT elements of Vibrio cholerae (15), may reflect the same constraints in operation. However, once formed, these complex structures can, as found here, move in a block to a new location and continue to exchange genes with and acquire genes from related regions via homologous recombination. Further examination of the broader context of genes originally found in Tn2670 and its known relatives in both historic and modern-day bacterial isolates should shed further light on the evolutionary pathways involved in creating and spreading derivatives of Tn2670 and how prevalent they are in modern day bacteria.
Acknowledgments
S.R.P. was supported by grant numbers 9936148 and 192108 from the Australian National Health and Medical Research Council.
Part of this work was undertaken at CSIRO Molecular Science, North Ryde, New South Wales, Australia.
REFERENCES
- 1.Alton, N. K., and D. Vapnek. 1979. Nucleotide sequence analysis of the chloramphenicol resistance transposon Tn9. Nature 282:864-869. [DOI] [PubMed] [Google Scholar]
- 2.Brown, H. J., H. W. Stokes, and R. M. Hall. 1996. The integrons In0, In2, and In5 are defective transposon derivatives. J. Bacteriol. 178:4429-4437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chandler, M., B. Allet, E. Gallay, E. Boy de La Tour, and L. Caro. 1977. Involvement of IS1 in the dissociation of the r-determinant and RTF components of the plasmid R100.1. Mol. Gen. Genet. 153:289-295. [DOI] [PubMed] [Google Scholar]
- 4.Chandler, M., L. Silver, D. Lane, and L. Caro. 1979. Properties of an autonomous r-determinant from R100.1. Cold Spring Harb. Symp. Quant. Biol. 43:1223-1231. [DOI] [PubMed] [Google Scholar]
- 5.Chen, S.-T., and R. C. Clowes. 1987. Variations between the nucleotide sequences of Tn1, Tn2, and Tn3 and expression of β-lactamase in Pseudomonas aeruginosa and Escherichia coli. J. Bacteriol. 169:913-916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goussard, S., and P. Courvalin. 1991. Sequence of the genes blaT-IB and blaT-2. Gene 102:71-73. [DOI] [PubMed] [Google Scholar]
- 7.Griffin, H. G., T. J. Foster, S. Silver, and T. K. Misra. 1987. Cloning and DNA sequence of the mercuric- and organomercurial-resistance determinants of plasmid pDU1358. Proc. Natl. Acad. Sci. USA 84:3112-3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grindley, N. D. F. 2002. The movement of Tn3-like elements: transposition and cointegrate resolution, p. 272-302. In N. L. Craig, R. Craigie, M. Gellert, and A. M. Lambowitz (ed.), Mobile DNA II. ASM Press, Washington, D.C.
- 9.Grinsted, J., F. de la Cruz, and R. Schmitt. 1990. The Tn21 subgroup of bacterial transposable elements. Plasmid 2:163-189. [DOI] [PubMed] [Google Scholar]
- 10.Hall, R. M. 2001. Integrons, p. 1041-1045. In S. Brenner and J. H. Miller (ed.), Encyclopedia of genetics. Academic Press, London, United Kingdom.
- 11.Hall, R. M., and C. M. Collis. 1998. Antibiotic resistance in gram-negative bacteria: the role of gene cassettes and integrons. Drug Resist. Updat. 1:109-119. [DOI] [PubMed] [Google Scholar]
- 12.Hall, S. C., and S. E. Halford. 1993. Specificity of DNA recognition in the nucleoprotein complex for site-specific recombination by Tn21 resolvase. Nucleic Acids Res. 21:5712-5719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hänni, C., J. Meyer, S. Iida, and W. Arber. 1982. Occurrence and properties of composite transposon Tn2672: evolution of multiple drug resistance transposons. J. Bacteriol. 150:1266-1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heffron, F., B. J. McCarthy, H. Ohtsubo, and E. Ohtsubo. 1979. DNA sequence analysis of the transposon Tn3: three genes and three sites involved in transposition of Tn3. Cell 18:1153-1163. [DOI] [PubMed] [Google Scholar]
- 15.Hochhut, B., Y. Lotfi, D. Mazel, S. M. Faruque, R. Woodgate, and M. K. Waldor. 2001. Molecular analysis of antibiotic resistance gene clusters in Vibrio cholerae O139 and O1 SXT constins. Antimicrob. Agents Chemother. 45:2991-3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iida, S., C. Hänni, C. Echarti, and W. Arber. 1981. Is the IS1-flanked r-determinant of the R plasmid NR1 a transposon? J. Gen. Microbiol. 126:413-425. [DOI] [PubMed] [Google Scholar]
- 17.Izumiya, H., N. Nojiri, Y. Hashiwata, K. Tamura, J. Terajima, and H. Watanabe. 2003. Salmonella enterica serovar Enteritidis, Japan. Emerg Infect. Dis. 9:1650-1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kholodii, G., S. Mindlin, M. Petrova, and S. Minakhina. 2003. Tn5060 from the Siberian permafrost is most closely related to the ancestor of Tn21 prior to integron acquisition. FEMS Microbiol. Lett. 226:251-255. [DOI] [PubMed] [Google Scholar]
- 19.Liebert, C. A., R. M. Hall, and A. O. Summers. 1999. Transposon Tn21, flagship of the floating genome. Microbiol. Mol. Biol. Rev. 63:507-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakaya, R., A. Nakamura, and Y. Murata. 1960. Resistance transfer factors in Shigella. Biochem. Biophys. Res. Commun. 3:654-659. [DOI] [PubMed] [Google Scholar]
- 21.Nucifora, G., L. Chu, S. Silver, and T. K. Misra. 1989. Mercury operon regulation by the merR gene of the organomercurial resistance system of the plasmid pDU1358. J. Bacteriol. 171:4241-4247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pansegrau, W., E. Lanka, P. T. Barth, D. H. Figurski, D. G. Guiney, D. Haas, D. R. Helinski, H. Schwab, V. Stanisich, and C. M. Thomas. 1994. Complete nucleotide sequence of the Birmingham IncPα plasmids. Compilation and comparative analysis. J. Mol. Biol. 239:623-663. [DOI] [PubMed] [Google Scholar]
- 23.Partridge, S. R., H. J. Brown, and R. M. Hall. 2002. Characterization and movement of the class 1 integron known as Tn2521 and Tn1405. Antimicrob. Agents Chemother. 46:1288-1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Partridge, S. R., H. J. Brown, H. W. Stokes, and R. M. Hall. 2001. Transposons Tn1696 and Tn21 and their integrons In4 and In2 have independent origins. Antimicrob. Agents Chemother. 45:1263-1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Partridge, S. R., and R. M. Hall. 2003. In34, a complex In5 family class 1 integron containing orf513 and dfrA10. Antimicrob. Agents Chemother. 47:342-349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Partridge, S. R., and R. M. Hall. 2003. The IS1111 family members IS4321 and IS5075 have subterminal inverted repeats and target the terminal inverted repeats of Tn21 family transposons. J. Bacteriol. 185:6371-6384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Partridge, S. R., G. D. Recchia, H. W. Stokes, and R. M. Hall. 2001. Family of class 1 integrons related to In4 from Tn1696. Antimicrob. Agents Chemother. 45:3014-3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sherratt, D. 1989. Tn3 and related transposable elements: site-specific recombination and transposition, p. 163-184. In D. E. Berg and M. M. Howe (ed.), Mobile DNA. American Society for Microbiology, Washington, D.C.
- 29.Silver, L., M. Chandler, H. E. D. Lane, and L. Caro. 1980. Production of extrachromosomal r-determinant circles from integrated R100.1: involvement of the E. coli recombination system. Mol. Gen. Genet. 179:565-571. [DOI] [PubMed] [Google Scholar]
- 30.Womble, D. D., and R. H. Rownd. 1988. Genetic and physical map of plasmid NR1: comparison with other IncFII antibiotic resistance plasmids. Microbiol. Rev. 52:433-451. [DOI] [PMC free article] [PubMed] [Google Scholar]