Abstract
The putative transcriptional corepressor ETO/MTG8 has been extensively studied due to its involvement in a chromosomal translocation causing the t(8;21) form of acute myeloid leukemia. Despite this, the role of ETO in normal physiology has remained obscure. Here we show that ETO is highly expressed in preadipocytes and acts as an inhibitor of C/EBPβ during early adipogenesis, contributing to its characteristically delayed activation. ETO prevents both the transcriptional activation of the C/EBPα promoter by C/EBPβ and its concurrent accumulation in centromeric sites during early adipogenesis. ETO expression rapidly reduces after the initiation of adipogenesis, and this is essential to the normal induction of adipogenic gene expression. These findings define, for the first time, a molecular role for ETO in normal physiology as an inhibitor of C/EBPβ and a novel regulator of early adipogenesis.
Adipose tissue is a key depot for the storage of energy as triglycerides and also plays a dynamic role in the regulation of metabolism (30). Studies of obese and lipodystrophic humans and rodents demonstrate that both increased and decreased adipose tissue mass are associated with insulin resistance and abnormal glucose and lipid metabolism (17, 24, 29). Thus, tight control of adipocyte development, size and insulin-sensitivity appears to be of critical importance in maintaining whole body energy homeostasis. The process of adipogenesis requires highly organized and precisely controlled expression of a cascade of transcription factors within the preadipocyte (25, 32, 35). The rapid and transient induction of the C/CAAT-enhancer binding proteins C/EBPβ and C/EBPδ is one of the earliest steps in this process (35). These transcription factors bind to specific sequences in the promoters of C/EBPα and the nuclear hormone receptor PPARγ, inducing their expression and in turn activating the full adipogenic program of gene expression (11, 34, 47). Although the central involvement of these proteins in adipogenesis has been demonstrated in both cellular systems and knockout animals, important roles for other regulatory molecules in this highly orchestrated transcriptional program are becoming increasingly apparent (25, 35). The C/EBPs are subject to control through heterodimerization with other members of this protein family. Some of these are intrinsically active, such as C/EBPα, C/EBPβ-LAP, and C/EBPδ, whereas others appear inhibitory, including C/EBPβ-LIP, CHOP10, and C/EBPγ (33). Interaction with coactivators such as p300 and corepressors such as histone deacetylase 1 (HDAC1) and Sin3a further modulate function (12, 44). Moreover, C/EBPs are subject to regulation at the levels of transcription and translation, the latter giving rise to alternative forms from the same mRNA as occurs with the LAP and LIP forms of C/EBPβ (4, 10). Posttranslational modification by serine and tyrosine phosphorylation has also been reported for these proteins (33). In addition to regulating target gene expression in a classical fashion C/EBPs may exert nontranscriptionally mediated effects through interaction with cell cycle inhibitors (23). The multifaceted nature of both the control and the function of this family of transcription factors attests to their importance in diverse biological processes and the need for their precise regulation.
In a screen for novel genes regulated by insulin-like growth factor 1 (IGF-1) in 3T3-L1 preadipocytes we identified the transcriptional corepressor ETO/MTG8 as a transcript rapidly repressed by IGF-1 (26). Given the key role of IGF-1 as a stimulus for the conversion of these cells into terminally differentiated adipocytes and the need for tight transcriptional regulation we postulated that ETO might play a role in this process. ETO has been extensively studied in myeloid cells due to its involvement in a chromosomal translocation causing the t(8;21) form of acute myeloid leukemia (19, 46). However, ETO is also clearly detectable in brain, heart, skeletal muscle, and adipose tissue (46), and its presence in metabolically important tissues suggested to us that its hormonal regulation merited further study. ETO is considered to have no inherent DNA-binding activity. Instead, it may form complexes with DNA-bound transcription factors and recruit other corepressors such as Sin3, N-CoR, and HDACs thereby inhibiting transcriptional activity (9). To date, only the transcriptional repressors PLZF, Bcl-6, and Gfi-1 have been identified as ETO targets (5, 9), all of which are involved in hematopoiesis. The function of ETO in vivo remains obscure, although mice lacking this protein have severe abnormalities of midgut development, leading in most cases to embryonic or early neonatal death (3).
We demonstrate here a previously unknown role for ETO in preadipocytes as an inhibitor of C/EBPβ function. By impairing the activity of this key, early modulator of adipogenesis, ETO restrains cells from progressing through the transcriptional program to form mature adipocytes. The rapid disappearance of ETO after exposure of preadipocytes to prodifferentiative hormonal stimuli closely precedes the acquisition of DNA-binding activity by C/EBPβ and the resulting stimulation of transcription from the C/EBPα promoter. This represents not only a novel role for ETO but also defines a new mechanism for its action and reveals its importance in the regulation of adipogenesis.
MATERIALS AND METHODS
RNA isolation, real-time PCR, and Northern blot analyses.
Total RNA was isolated from cultured cells by using an RNeasy kit (Qiagen) or from tissue samples by using RNA STAT-60 (AMS Biotechnology) and quantified by GeneQuant (Amersham Biosciences). Then, 10-μg portions of each sample were analyzed by Northern blotting as described previously (26). Where quantification is shown, blots were reprobed for, and values were normalized to, expression of rRNA. Elsewhere, blots are representative of at least three independent experiments.
Primer Express software (version 1.0; Perkin-Elmer Applied Biosystems) was used to design the probes and primers for real-time quantitative PCR to determine human MTG8 or murine ETO, PPARγ1, PPARγ2, aP2, or Glut4 mRNA expression. RNA was reverse transcribed, and the resulting cDNA was used in 25-μl PCRs, in which 300 nmol of forward and reverse primers/liter and 150 nmol of fluorogenic probe/liter were used. Reactions were carried out in duplicate for each sample on an ABI 7700 sequence detection system (Perkin-Elmer Biosystems) according to the manufacturer's instructions, and target values were normalized to 18S rRNA (reagents from Perkin-Elmer).
Protein analyses and immunoprecipitation.
Protein samples were extracted by scraping in lysis buffer containing 1% NP-40 as described previously (26), followed by sonication. After centrifugation for 10 min at 13,000 × g samples of supernatant containing 30 μg of protein were denatured and analyzed by Western blotting. Green fluorescent protein (GFP)-tagged proteins were immunoprecipitated from lysates containing 150 μg of protein by incubation with agarose conjugated α-GFP antibodies for 3 h at 4°C rotating end over end. Precipitates were washed, denatured, and analyzed by Western blotting essentially as described previously. C/EBPβ or ETO was similarly immunoprecipitated by using an antibody prebound to protein G-Sepharose. All antibodies were from Santa Cruz Biotechnology.
Plasmids and mutagenesis.
Full-length cDNA encoding mouse MTG8/ETO was generated from 3T3-L1 preadipocyte RNA by RT-PCR. This was subsequently cloned in frame downstream of GFP in pEGFP-C1 (Clontech) to generate a construct encoding an N-terminally tagged protein. DNA sequencing confirmed the absence of mutations. GFP-ETO-AA was generated by using a QuikChange site-directed mutagenesis kit (Stratagene). GFP-ETO and GFP-ETO-AA were subsequently subcloned into the SnaB1 site of pBabePuro. The coding regions of ETO and ETO-AA were also subcloned into pcDNA3 (Invitrogen) to produce untagged constructs. The pMT2-C/EBPβ expression vector and the pGL3-C/EBPα promoter reporter construct were generously provided by Q.-Q. Tang and M. D. Lane.
Cell culture and transfection.
3T3-L1 (42), HEK293 (28), and HepG2 (16) cells were cultured as described previously. Preadipocytes were induced to differentiate by transfer to medium containing fetal calf serum and a standard cocktail of insulin, isobutyl methyl xanthine (IBMX), and dexamethasone as previously described (42). HEK293 cells were transiently transfected with Fugene 6 reagent (Roche) according to the manufacturer's protocol. To generate retroviruses, BOSC-293 cells were similarly transfected with 5 μg of pBabePuro vectors encoding either, GFP, GFP-ETO, or GFP-ETO-AA with Fugene 6 reagent (Roche). Supernatants containing virus were collected 48 h later and used to generate stably transfected populations of 3T3-L1 cells essentially as previously described (48). Differentiating 3T3-L1 cells were assessed for lipid content by staining with oil-red O as described previously (44). To assay C/EBPβ activity, HepG2 cells were transfected with 50 ng of pGL3-C/EBPα promoter reporter construct (containing C/EBPα nucleotides −1450 to +125) (37) and 100 ng of pMT2-C/EBPβ (38) in the absence or presence of ETO constructs as indicated with Fugene 6 reagent (Roche). Where indicated, an alternative C/EBPβ responsive promoter, C/EBPwt-LUC was used in which luciferase expression was controlled by two copies of the C/EBPβ binding site of the interleukin-6 promoter cloned upstream of the adenovirus major late promoter as described previously (13). Alternatively, to measure PPARγ activity, cells were transfected with 200 ng of (PPARE)3TKLuc and 100 ng of pcDNA3-PPARγ2 with or without ETO. Activity was assayed 48 h posttransfection by using a dual-luciferase reporter assay system (Promega). Values were normalized to the activity of cotransfected pRL-TK (PPARγ) or pRL-CMV (C/EBPβ) constitutive Renilla luciferase reporter vectors (Promega). For immunofluorescence studies, 3T3-L1 preadipocytes were grown on glass coverslips and transiently transfected with pEGFP or pEGFP-ETO vectors by using Lipofectamine Plus (Invitrogen) according to the manufacturer's instructions.
ChIP assays.
Chromatin immunoprecipitation (ChIP) assays were performed essentially as described earlier (15). Briefly, cells were treated as indicated, rinsed twice in ice-cold phosphate-buffered saline (PBS), and cross-linked by using a 1% solution of formaldehyde for 10 min at room temperature. After two rinses with PBS, cells were scraped in lysis buffer (1% sodium dodecyl sulfate [SDS], 5 mM EDTA, 50 mM Tris-HCl [pH 8.1]), sonicated, and centrifuged at 14,000 × g for 10 min. Samples were diluted 10× in dilution buffer (1% Triton X-100, 2 mM EDTA, 150 mM NaCl, 20 mM Tris-HCl [pH 8.1]) supplemented with protease inhibitors and then incubated with anti-C/EBPβ antibody prebound to protein G-Sepharose at 4°C, with rotation end over end for 4 h. Precipitates were washed once in dilution buffer, once in TSE1 (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl, 150 mM NaCl), once in TSE2 (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl, 500 mM NaCl), and once in buffer 3 (0.25 M LiCl, 1% NP-40, 1% deoxycholate, 1 mM EDTA, 10 mM Tris-HCl). After a final wash in Tris-EDTA buffer, 100 μl of elution buffer (1% SDS, 0.1 M NaHCO3) was added to each pellet or to a 1/10 volume of the corresponding initial lysate sample (input). Samples were incubated at 65°C for 6 h, and DNA was isolated by using a Qiagen PCR cleanup kit according to the manufacturer's instructions. C/EBPα promoter DNA was assayed by using real-time PCR with probe and primers amplifying the C/EBPβ binding site at −190 bp proximal to the transcriptional start site. Values obtained from immunoprecipitated samples were normalized to those from input samples.
Immunofluorescence.
At 3 days posttransfection 3T3-L1 preadipocytes were induced to differentiate for 24 h as described above. Cells were then rinsed in PBS and fixed in 3% paraformaldehyde. Cells were permeabilized with 0.1% Triton X-100 and then incubated with rabbit polyclonal antibodies to C/EBPβ (Santa Cruz Biotechnology). Slides were subsequently probed with Alexa-Fluor 594 goat anti-rabbit secondary antibodies (Molecular Probes, Inc.), mounted, and analyzed by laser scanning confocal microscopy. Similarly, subconfluent 3T3-L1 cells retrovirally transfected with GFP, GFP-ETO, or GFP-ETO-AA were grown on coverslips and fixed, and images were obtained to determine subcellular localization.
Gel shift assays.
ETO and C/EBPβ proteins were synthesized from cDNA templates in pcDNA3.1 by using a TNT quick-coupled transcription/translation system (Promega). Double-stranded oligonucleotide probe (5′-CAGTGGGCGTTGCGCCACGATCTCTCT-3′) was radiolabeled with polynucleotide kinase and [γ-32P]ATP. Protein mixes were preincubated for 1 h at 37°C and then incubated with 0.1 pmol of radiolabeled probe in buffer containing 20 mM HEPES (pH 7.9), 50 mM KCl, 2 mM dithiothreitol, and 10% glycerol for 30 min at room temperature. Analysis of binding complexes was performed by electrophoresis on a 6% polyacrylamide gel in 0.5× Tris-borate-EDTA.
RESULTS
ETO is hormonally regulated, and its expression decreased during adipogenesis.
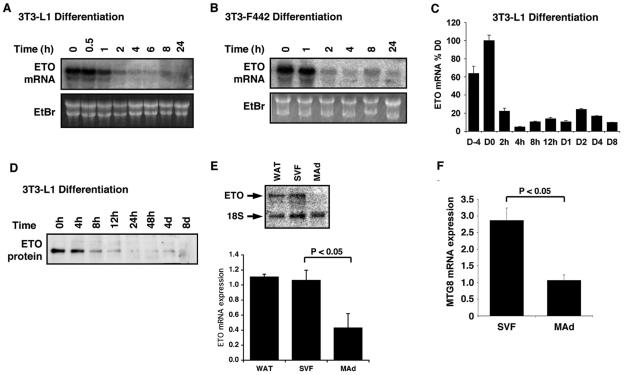
To determine whether ETO was hormonally responsive in 3T3-L1 preadipocytes undergoing differentiation, we treated cells with a standard adipocyte differentiation cocktail containing insulin, IBMX, and dexamethasone. This led to a rapid and sustained decrease in ETO expression, such that the mRNA was significantly decreased within 4 h of treatment (Fig. 1A). Analysis of ETO expression in 3T3-F442 preadipocytes demonstrated that ETO mRNA was also highly expressed in confluent cultures and was again rapidly decreased in response to hormonal induction of adipogenesis in these cells (Fig. 1B). Using real-time quantitative PCR, we examined ETO mRNA expression over a longer time course of differentiation in 3T3-L1 cells. These data confirmed the rapid fall of ETO mRNA expression observed in the Northern blots and demonstrated sustained inhibition of ETO expression even after 8 days (Fig. 1C). Western blot analysis of 3T3-L1 cell lysates during differentiation revealed that ETO protein expression was also rapidly inhibited such that it was almost undetectable 24 h postinduction and again remained suppressed even in the later stages of adipogenesis (Fig. 1D). Having analyzed ETO expression in cultured cells, we sought to assess its potential involvement in vivo. Consistent with previous expression studies (46), we could clearly detect ETO mRNA in rat whole adipose tissue by Northern blotting (Fig. 1E). When this tissue was fractionated, we found that the majority of the ETO mRNA was present in the stromovascular fraction in which the preadipocytes are found. ETO mRNA expression was significantly lower in the mature adipocytes, a finding consistent with our data obtained with fully differentiated 3T3-L1 adipocytes. Identical data were obtained when these samples were assayed by quantitative real-time PCR (data not shown). We further used real-time PCR to quantify the expression of the human ETO homologue, MTG8, in the stromovascular and mature adipocyte fractions of human subcutaneous adipose tissue (Fig. 1F). This gave results almost identical to those seen in the rat with 2.6-fold-higher expression in the preadipocyte-containing than the mature adipocyte fraction. These findings strongly imply that ETO has a role in the function of normal adipose tissue and that decreased expression of ETO is a feature of preadipocyte to adipocyte conversion in vivo.
FIG. 1.
The expression of ETO in preadipocytes is inhibited during adipogenesis. ETO mRNA expression was determined by Northern blotting (A and B) or real-time PCR analysis (C) of RNA samples extracted from 2-day postconfluent 3T3-L1 (A and C) or 3T3-F442 (B) preadipocytes treated for the times indicated with differentiation mixture. (D) Protein samples extracted from 3T3-L1 preadipocytes incubated with differentiation mixture for various times were Western blotted with and anti-ETO polyclonal antibody (Santa Cruz Biotechnology). (E) RNA isolated from rat whole adipose tissue (WAT), cells of the stromovascular fraction (SVF), or mature isolated adipocytes (MAd) was analyzed by Northern blotting to determine ETO expression. Blots were reprobed with a ribosomal probe to control for loading. A representative blot is shown (upper panel) along with mean data ± the SEM from four rats (lower panel). (F) RNA was isolated from cells of the stromovascular fraction (SVF) or mature isolated adipocytes (MAd) from subcutaneous human adipose tissue samples. MTG8 expression was determined by using real-time PCR and normalized to 18S mRNA. EtBr, ethidium bromide.
ETO inhibits adipogenesis in 3T3-L1 cells, whereas dominant-negative ETO augments lipid accumulation.
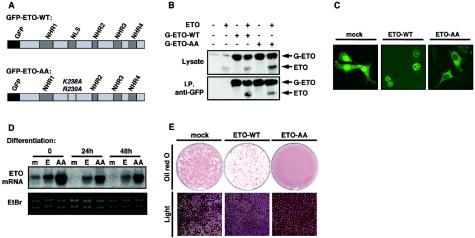
To explore the role of ETO in adipogenesis further, we sought to manipulate the expression of ETO in 3T3-L1 cells. In addition, we attempted to create a dominant-negative form of the molecule. It has been demonstrated that ETO functions as a dimer and is ordinarily localized to the nucleus (21, 28). However, mutation of both lysine 238 and arginine 239 to alanine in the nuclear localization sequence of ETO is known to cause the molecule to be mistargeted to the cytosol (28). We postulated that this mutant form of ETO (hereafter referred to as ETO-AA), if expressed in excess, might dimerize with and cause nuclear exclusion of wild-type ETO, thereby functioning as a dominant negative. Therefore, we generated this mutant by site-directed mutagenesis. Both wild-type ETO and ETO-AA were subsequently tagged with GFP at the N terminus, as represented in Fig. 2A, to allow visualization of the proteins in intact cells and specific identification and isolation of the transfected ETO when expressed in the presence of the endogenous protein.
FIG. 2.
ETO localizes to the nucleus and inhibits adipogenesis, whereas the mutant ETO-AA, in which the nuclear localization signal is disrupted, is targeted to the cytosol and accelerates preadipocyte differentiation. (A) The construction of N terminally GFP-tagged forms of ETO is shown. The nuclear localization sequence (NLS) was disrupted by the introduction of mutations at codons 238 and 239 to generate GFP-tagged ETO-AA. The positions of the NHR domains, involved in protein-protein interactions, are indicated. (B) GFP-ETO (G-ETO-WT) and GFP-ETO-AA were expressed in HepG2 cells in the presence or absence of untagged wild-type ETO (ETO) as indicated. ETO proteins were analyzed by Western blotting in whole-cell lysates (upper panel) or anti-GFP immunoprecipitates obtained by using agarose conjugated anti-GFP polyclonal antibody (lower panel). (C) Subconfluent cultures of 3T3-L1 preadipocytes infected with retroviruses encoding GFP alone (mock) or GFP-tagged ETO (ETO-WT) or ETO-AA were fixed in 4% paraformaldehyde and fluorescent images captured by laser-scanning confocal microscopy. (D) Two-day postconfluent 3T3-L1 preadipocytes retrovirally transfected with GFP (m), GFP-ETO (E), or GFP-ETO-AA (AA) were treated for the times indicated in the absence or presence of differentiation mixture as indicated. RNA was extracted and ETO mRNA expression determined by Northern blotting. E, 3T3-L1 cells expressing GFP, GFP-ETO or GFP-ETO-AA were differentiated for 8 days and lipid accumulation assessed by oil-red O staining (upper panels) or light microscopy (lower panels).
To determine whether these constructs would indeed form heterodimers with endogenous ETO, we coexpressed them with untagged full-length wild-type ETO. Using an antibody directed to GFP, untagged ETO coimmunoprecipitated with GFP-ETO (Fig. 2B). Coprecipitation was equally effective whether wild-type or mutant GFP-ETO was used, demonstrating that both molecules can indeed bind untagged ETO. In addition, the proportion of tagged to untagged ETO seen in the immunoprecipitates was very similar to that in the lysate samples (compare upper and lower panels), suggesting that most of the untagged ETO dimerized with the more highly expressed GFP-tagged forms.
The GFP-tagged ETO constructs were then introduced into 3T3-L1 cells by using retrovirus-mediated gene transfer. As shown in Fig. 2C, GFP-ETO was expressed almost exclusively in the nuclei of the preadipocytes, whereas the GFP-ETO-AA protein was localized instead to the cell cytosol. Mock-transfected cells were also generated expressing GFP alone, which showed a diffuse localization throughout the cell. When induced to differentiate, the cells exhibited a rapid and sustained reduction of mRNA encoding endogenous ETO (Fig. 2D). In contrast, the expression of transfected GFP-ETO or GFP-ETO-AA, which migrated with the endogenous ETO mRNA, was preserved during differentiation. Importantly, the expression levels of GFP-ETO and endogenous ETO were very similar, whereas those of GFP-ETO-AA were much higher. This pattern of expression was mimicked at the protein level (data not shown).
We next determined the effect of constitutive ETO expression on preadipocyte differentiation. As shown in Fig. 2E cells expressing GFP-ETO showed a severely impaired ability to accumulate lipid during differentiation, both when visualized by oil-red O staining (upper panels) and when visualized by light microscopy (lower panels). In contrast, identically treated cells expressing GFP-ETO-AA showed a consistent increase in lipid accumulation compared to untransfected cells.
These data indicate that loss of ETO expression is a key step in the initiation of the full adipogenic program.
ETO inhibits the expression of key proadipogenic genes in intact cells.
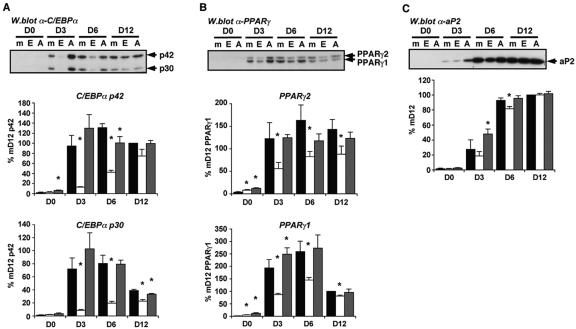
We next examined a number of key markers of adipogenesis to define more clearly the effects of both wild-type and dominant-negative ETO expression on adipocyte differentiation. Figure 3A demonstrates that the induction of C/EBPα protein was significantly impaired in differentiating cells constitutively expressing ETO. The data from four independent time courses are shown, and the quantified data ± the standard error of the mean (SEM) are presented below a representative Western blot. Expression of both the p42 and the p30 isoforms of C/EBPα was almost entirely prevented by expression of ETO 3 days after the induction of differentiation. Conversely, expression of the mutant ETO-AA protein consistently increased C/EBPα expression at this time point. Interestingly, at the later time points, particularly 12 days postinduction, the expression of C/EBPα in ETO-expressing cells appears to reach levels approaching those in control cells. Examination of PPARγ proteins in the same cell extracts revealed that induction of both PPARγ1 and PPARγ2 were also reduced by constitutive ETO expression, again particularly at the earlier time points (Fig. 3B). However, the inhibitory effect of ETO and any stimulatory effect of ETO-AA were weaker than that observed with C/EBPα. The expression of aP2 protein was also examined (Fig. 3C). Again, an overall pattern of expression similar to that seen for C/EBPα and PPARγ was observed in the three cell populations. However, the effects of ETO and ETO-AA expression were much less marked for aP2 than those seen for C/EBPα or PPARγ.
FIG. 3.
ETO inhibits the expression of adipogenic proteins. Cells retrovirally transfected with GFP (▪), GFP-ETO (□) or GFP-ETO-AA (░⃞) were induced to differentiate for 0 to 12 days (D0 to D12) as indicated. Total cell lysates were prepared and proteins analyzed by SDS-PAGE and Western blotting with antibodies to C/EBPα (A), PPARγ (B), or aP2 (C) as appropriate. In each case band intensities were quantified from four independent experiments and the mean data ± the SEM is presented below a representative blot. The data were calculated as the percentage of expression at D12 in the mock transfected cells. Asterisks indicate a statistically significant difference from the expression in mock-transfected cells at the same time point (P < 0.05).
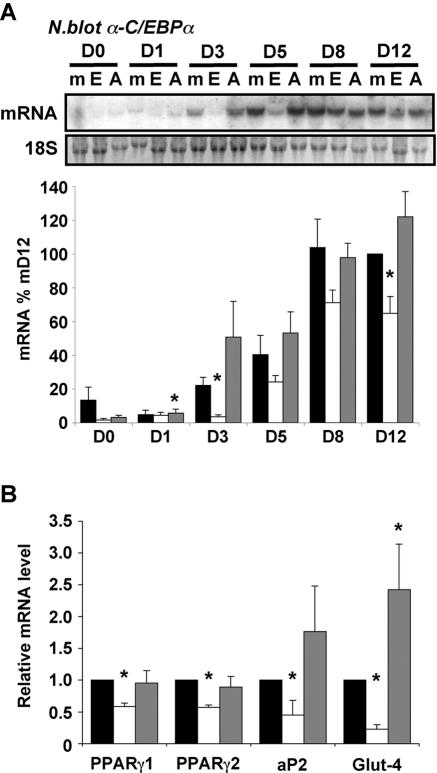
Since C/EBPα expression appeared to be most affected by ETO expression, we examined the induction of its mRNA in ETO- and ETO-AA-expressing cells during differentiation. As shown in Fig. 4A, the effect of constitutive ETO expression on C/EBPα mRNA induction was similar to that seen for its protein and again was most significant at the earlier time points. As with protein expression, the normal induction of C/EBPα mRNA after 3 days was almost entirely prevented by ETO expression, whereas ETO-AA expression led to higher levels of C/EBPα mRNA than those observed in control cells. RNA isolated from cells 3 days postinduction of differentiation were also assayed for aP2, PPARγ2, and Glut4 mRNA expression levels by real-time PCR (Fig. 4B). Again, ETO inhibited the induction of all three mRNAs with the greatest effects observed for Glut4, a well-characterized C/EBPα target that was both significantly lower in cells expressing ETO and higher in cells expressing ETO-AA, reflecting the effects on C/EBPα protein expression, and likely activity, in these cells.
FIG. 4.
ETO inhibits the induction of adipogenic gene expression. RNA was isolated from cells retrovirally transfected with GFP (▪), GFP-ETO (□) or GFP-ETO-AA (░⃞) that had been induced to differentiate for 0 to 12 days (D0 to D8) as indicated. (A) Expression of mRNA encoding C/EBPα was determined by Northern blotting. A representative blot is shown along with 18S rRNA, which was used as a loading control and all values were adjusted accordingly. C/EBPα mRNA expression was quantified in four independent experiments, and the mean ± the SEM is shown in the lower panel. Values were expressed as percentage of those in mock-transfected cells at day 12. Asterisks indicate a statistically significant difference from mock transfected cells at the same time point (P < 0.05). (B) Samples from cells differentiated for 3 days were also assayed for mRNA encoding PPARγ1, PPARγ2, aP2, and Glut-4 by real-time PCR. The data are means ± the SEM of four independent experiments, and values are expressed relative to that in mock-transfected cells. Asterisks indicate a statistically significant difference from mock-transfected cells (P < 0.05).
ETO decreases the expression of C/EBPα by selectively inhibiting the activity of C/EBPβ.
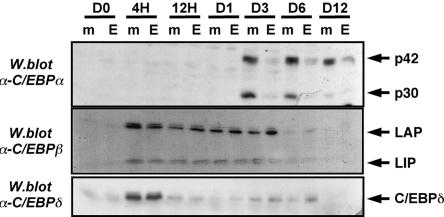
The C/EBP transcription factors C/EBPβ and C/EBPδ play a crucial role in the early induction of genes involved in lipid accumulation by differentiating adipocytes, and their involvement in the induction of C/EBPα and PPARγ expression has been well documented (7, 14, 47). The data obtained thus far suggested that a major effect of ETO was to inhibit C/EBPα mRNA transcription, whereas the extent of the inhibition of this gene in particular suggested that the inhibitory step might be proximal to C/EBPα induction. We therefore examined the possible involvement of C/EBPβ and C/EBPδ in the inhibition of adipogenesis by ETO. Western blotting of lysates from mock-transfected cells or cells constitutively expressing ETO revealed no significant effect of ETO on the induction of either C/EBPβ or C/EBPδ proteins (Fig. 5). In contrast, in the same lysates the expression of C/EBPα was significantly reduced during differentiation in cells expressing ETO as previously observed (see Fig. 3A).
FIG. 5.
ETO impairs the induction of C/EBPα but not C/EBPβ or C/EBPδ during adipogenesis. 3T3-L1 cells expressing GFP or GFP-ETO were differentiated for the times shown. Cell lysates were prepared and analyzed by Western blotting to determine the expression of C/EBPβ, C/EBPδ or C/EBPα as indicated.
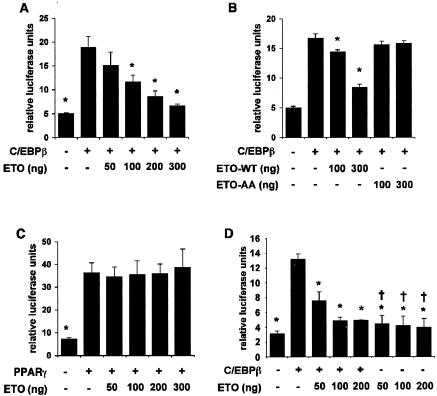
Although C/EBPβ and C/EBPδ are both known to be capable of inducing expression from the C/EBPα promoter (31, 38), the greatest effect of ETO expression on C/EBPα induction was observed at time points up to 3 days after induction of differentiation, when C/EBPβ was also highly expressed (Fig. 5). We therefore tested whether ETO could inhibit the activity of C/EBPβ and so suppress C/EBPα expression. The presence of endogenous ETO in preadipocytes suggested that these cells would be unsuitable for examining the effects of adding exogenous ETO to a C/EBPβ activity assay. Previous studies have demonstrated that ETO expression is undetectable in the liver (46) and, consistent with this, we were unable to detect ETO mRNA in the hepatocyte cell line HepG2 (data not shown). Using these cells, we were able to observe a robust activation of transcription from a luciferase-linked C/EBPα promoter construct in the presence of C/EBPβ. As shown in Fig. 6A, coexpression of ETO led to a repression of C/EBPβ activity in a dose-dependent fashion, demonstrating that ETO can indeed inhibit C/EBPβ activity toward the C/EBPα promoter. In contrast, the mutant ETO-AA was completely unable to inhibit the activity of C/EBPβ in this assay (Fig. 6B), a finding consistent with the inability of this mutant to prevent adipogenesis. To assess the specificity of these effects, we next tested whether ETO affected the transcriptional activity of PPARγ which, although also critical in the differentiation process, belongs to a different family of transcription factors. In contrast to C/EBPβ, the activity of PPARγ was completely unaffected by ETO expression at levels that almost entirely inhibited C/EBPβ activity (Fig. 6C). To test further whether ETO acted directly to inhibit C/EBPβ activity or acted more generally to inhibit the activity of the C/EBPα promoter, we repeated our assays with an alternative minimal C/EBPβ responsive reporter construct. Using this construct, we observed almost identical inhibition of C/EBPβ activity by ETO to that seen with the C/EBPα promoter construct (Fig. 6D). In addition, no significant reduction in the basal activity of the promoter was observed when ETO was present in the absence of C/EBPβ. Since the effect of ETO is similar with two different C/EBPβ target promoters, these data strongly suggest a direct effect of ETO on the activity of this transcription factor. In addition, the lack of effect on the transactivating capacity of PPARγ argues strongly against ETO affecting the basal transcriptional machinery in these assays.
FIG. 6.
ETO but not ETO-AA selectively inhibits C/EBPβ transcriptional activity. HepG2 cells were transfected with either a C/EBPα promoter-luciferase reporter construct (A and B) or a minimal C/EBP responsive reporter construct C/EBPwt-LUC (D) with or without C/EBPβ in the absence or presence of increasing quantities of ETO (A, B, and D) or ETO-AA (B) as indicated. (C) Cells were transfected with a PPRE-luciferase reporter construct alone or in combination with PPARγ and increasing quantities of ETO. In each case data shown are means ± the SEM of four independent experiments. Asterisks indicate statistically significant difference from activity in the presence of C/EBPβ (A, B, and D) or PPARγ (C) alone. In panel D, a dagger (†) indicates no significant difference from activity in cells transfected with neither C/EBPβ nor ETO.
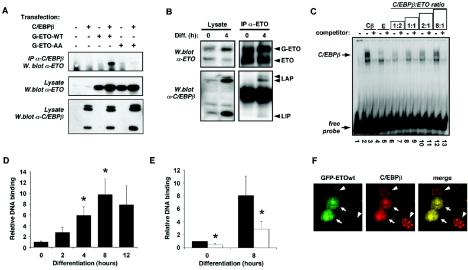
We next sought to examine more closely the mechanism of ETO’s actions on C/EBPβ. To assess whether the inhibition of C/EBPβ activity by ETO involved a direct physical interaction, we coexpressed C/EBPβ with GFP-ETO or GFP-ETO-AA. As shown in Fig. 7A, we were able to immunoprecipitate GFP-ETO with an antibody specific to C/EBPβ but only when C/EBPβ was also present in the cells. However, no GFP-ETO-AA could be detected in C/EBPβ immunoprecipitates when these two proteins were coexpressed. This was not surprising given that C/EBPβ displays an almost exclusively nuclear localization, whereas GFP-ETO-AA is excluded from this cellular compartment. We also examined the interaction of these proteins in differentiating 3T3-L1 preadipocytes. ETO was immunoprecipitated from 3T3-L1 preadipocytes constitutively expressing ETO that had been induced to differentiate for 4 h (Fig. 7B). Western blotting revealed the interaction of ETO with endogenous C/EBPβ; indeed, both the LAP and the LIP isoforms were effectively coimmunoprecipitated with ETO, not only suggesting that both are susceptible to regulation by ETO but also demonstrating that the interaction between the two proteins involves the C-terminal portion of C/EBPβ that is present in all isoforms.
FIG. 7.
ETO interacts directly with C/EBPβ inhibiting its DNA-binding activity toward the C/EBPα promoter and preventing centromeric localization during adipogenesis. (A) HEK293 cells were transfected with control vector, GFP-ETO (G-ETO-WT), or GFP-ETO-AA in the absence or presence of C/EBPβ as indicated. Anti-C/EBPβ immunoprecipitates were analyzed for associated ETO protein (upper panel), whereas corresponding cell lysates were probed for ETO (middle panel) or C/EBPβ (lower panel) by Western blotting. (B) 3T3-L1 preadipocytes expressing GFP-ETO were treated for 4 h in the absence or presence of differentiation cocktail as indicated prior to lysis. Cell lysates (left panels) or immunoprecipitates prepared by using an anti-ETO antibody (Santa Cruz Biotechnology) (right panels) were analyzed by Western blotting to detect ETO (upper panels) or C/EBPβ isoforms (lower panels). (C) In vitro transcribed/translated C/EBPβ (Cβ) and/or ETO (E) were incubated with radiolabeled DNA probe corresponding to the proximal C/EBPα binding site of the C/EBPα promoter in a gel shift assay. Various ratios of C/EBPβ to ETO were achieved by adjusting the quantity of ETO. In all lanes total protein input was kept constant by appropriate addition of rabbit reticulocyte lysate, except in lane 1, where free labeled probe was run alone. (D) 3T3-L1 preadipocytes were induced to differentiate for the times indicated and ChIP assays performed by using an anti-C/EBPβ antibody to isolate C/EBPβ-associated DNA. DNA from these immunoprecipitates corresponding to the C/EBPβ binding site in the C/EBPα promoter was quantified by using real-time PCR and normalized to DNA from a 10% sample of corresponding input lysate. The data are means ± the SEM from three independent experiments. Asterisks indicate a statistically significant difference (P < 0.05) from values obtained at time zero. (E) 3T3-L1 preadipocytes expressing GFP (▪) or GFP-ETO (□) were differentiated for 8 h as indicated and ChIP assays performed to determine occupancy of the C/EBPα promoter by C/EBPβ as in panel D. The data are means ± the SEM from five independent experiments. Asterisks indicate a statistically significant difference (P < 0.05) from values obtained in GFP-transfected cells at the corresponding time point. (F) Confocal microscope images of 3T3-L1 cells transfected with GFP-ETO, grown to confluence, and differentiated for 24 h. Cells were fixed in 4% paraformaldehyde, and C/EBPβ was visualized by using anti-C/EBPβ antibody (Santa Cruz Biotechnology) and an anti-rabbit Alexa-Fluor 594 secondary antibody. Arrows indicate the nuclei of cells transfected with GFP-ETO. Arrowheads indicate cells not expressing ETO but immunostaining for endogenous C/EBPβ.
We next examined whether ETO affects the DNA-binding activity of C/EBPβ. The promoter of C/EBPα contains a well-characterized C/EBPβ consensus binding site 190 bp proximal to the transcriptional start site (6), and therefore we performed gel shift assays with an oligonucleotide probe corresponding to this sequence. As shown in Fig. 7C, in vitro-translated C/EBPβ formed a stable complex with radiolabeled DNA probe, the binding of which could be competed away by using a 100-fold excess of unlabeled probe. ETO itself formed no stable complex with the DNA and, when incubated at a 2:1 ratio with C/EBPβ, almost completely prevented the latter from binding to the probe. As the ratio of ETO to C/EBPβ was decreased, the DNA-binding activity was restored in a dose-dependent fashion. These data strongly suggested that the mechanism by which ETO inhibited C/EBPβ activity involved a direct association between ETO and C/EBPβ, causing the latter to lose affinity for its target DNA sequence. To examine this effect further in intact cells, we performed chromatin immunoprecipitation (ChIP) assays in which C/EBPβ was immunoprecipitated and its associated DNA was isolated. In a novel modification of the commonly used methods in which limited cycle PCR is used to determine the relative levels of associated DNA sequences in different samples, we instead used real-time PCR to allow more accurate quantification of the extent of DNA binding. The total genomic DNA input was similarly quantified for each sample, and results from immunoprecipitated samples were adjusted accordingly. Examination of C/EBPβ binding to DNA over the first 12 h of differentiation in 3T3-L1 preadipocytes revealed that maximum binding was achieved some 8 h after induction of differentiation (Fig. 7D). We believe that this is the first assessment of these early time points of differentiation by using this method. However, the data agree well with previously published time courses assessed by gel shift analysis with nuclear extracts from 3T3-L1 preadipocytes (8), which has demonstrated strong binding of C/EBPβ to the C/EBPα promoter within 12 h of differentiation. Subsequent analysis of C/EBPβ binding to the C/EBPα promoter in cells constitutively expressing ETO revealed that C/EBPβ binding activity was significantly reduced in these cells compared to mock-transfected cells (Fig. 7E). These data demonstrate that ETO does indeed inhibit association of C/EBPβ with the C/EBPα promoter in intact cells.
In differentiating 3T3-L1 preadipocytes it is known that, although C/EBPβ is rapidly induced within 4 h, it acquires DNA-binding activity only after 12 to 24 h (39). Tang and Lane demonstrated that concomitant with the acquisition of DNA-binding activity to both the C/EBPα promoter and centromeric DNA sequences, C/EBPβ shifts from a diffuse to a punctate nuclear localization pattern, which can be clearly detected by immunohistochemistry (39). Having demonstrated that ETO could inhibit the DNA-binding activity of C/EBPβ, we next tested whether its centromeric localization during adipogenesis might be affected by ETO expression. To address this, 3T3-L1 preadipocytes were transiently transfected with GFP-ETO, grown to confluence, and then induced to differentiate for 24 h to bring about the expression and subsequent centromeric localization of endogenous C/EBPβ. Cells were then fixed and stained with antibodies to C/EBPβ, and the localization of both C/EBPβ and GFP-ETO was determined by confocal microscopy. Cells expressing GFP-ETO showed a diffuse pattern of fluorescence for this protein within the nucleus (Fig. 7F). As expected with transient transfection, some cells showed high levels of GFP-ETO expression, whereas others did not. Cells that had not been transfected with ETO showed the expected punctate centromeric pattern of C/EBPβ expression. In marked contrast, in all cells expressing GFP-ETO, C/EBPβ immunostaining was diffusely distributed in the nucleus. Of particular note was the fact that inhomogeneities in this diffuse nuclear staining pattern precisely mimicked the pattern seen with ETO-GFP, strongly suggesting an intimate colocalization of these two proteins.
These data strongly suggest that ETO associates with C/EBPβ in intact cells and is likely to contribute significantly to the delayed acquisition of DNA-binding activity of this transcription factor.
DISCUSSION
The results presented here demonstrate that ETO has an important role in the early stages of adipogenesis as an inhibitor of C/EBPβ-driven transcription. We show that ETO can directly inhibit C/EBPβ-induced transcription from the C/EBPα promoter and that this results in decreased expression of C/EBPα in intact 3T3-L1 preadipocytes. Gel shift assays, ChIP assays, and immunohistochemical data indicate that ETO causes loss of C/EBPβ binding to target DNA sequences, including the proximal C/EBP consensus site in the C/EBPα promoter. As a result the early induction of C/EBPα, normally visible within 3 days of the induction of differentiation, is entirely inhibited in cells constitutively expressing ETO, whereas a more rapid and robust increase in C/EBPα expression can be observed in cells expressing a putative dominant-negative ETO. The induction of PPARγ1 and PPARγ2 is also decreased in differentiating cells in which ETO expression cannot be appropriately inhibited. However, it is noteworthy that the effect is less marked that that seen for C/EBPα and that this is also true for aP2. We propose that this reflects differences in the relative individual contributions of different transcription factors to the induction of these genes. Although C/EBPβ and C/EBPδ have been identified as important regulators of C/EBPα, PPARγ, and aP2 induction during adipogenesis (36, 47), their regulation in vivo is likely to involve an array of transcriptional regulators. The data presented here are consistent with a critical role for C/EBPβ in early C/EBPα induction and an important but less essential role in the early induction of PPARγ and aP2. Pertinent to this, examination of the proximal PPARγ2 promoter sequence has demonstrated that this may be effectively activated by C/EBPδ and C/EBPα but that C/EBPβ alone lacks the ability to directly stimulate transcription from this region of the promoter (11). Thus, although a role for C/EBPβ in PPAR2 induction has been demonstrated in other studies (36, 47), this is probably due to an indirect effect or is mediated by more distal regions of the promoter.
It is not yet clear whether C/EBPβ represents the sole target of ETO in preadipocytes. Most of our observations in 3T3-L1 cells constitutively expressing ETO can be accounted for by inhibition of known C/EBPβ functions. However, it is possible that, given that other ETO targets, such as the zinc-finger repressor PLZF and Bcl-6, have been described (5, 9), ETO may also exert its actions through other transcription factors present in preadipocytes.
Whether or not ETO also targets other transcription factors, several pieces of evidence demonstrate that ETO does not function nonspecifically to inhibit differentiation. First, it has no effect on the rapid expression of C/EBPβ or C/EBPδ upon induction of adipogenesis. In addition, the coexpression of ETO had no effect on the transactivating capacity of PPARγ in luciferase reporter assays or on the binding of PPARγ to target DNA probe in gel shift assays (data not shown). Finally, cells expressing ETO constitutively appear to be capable of at least partially restoring the expression of adipogenic genes such as PPARγ, C/EBPα, and aP2 at later time points in this process. Since C/EBPβ expression has by this stage subsided, we propose that it makes a less critical contribution to the expression of these genes and so the effect of ETO is attenuated. It is noteworthy that mock-transfected 3T3-L1 preadipocytes, being competent to fully differentiate, eventually accumulate equivalent levels of lipid to the more rapidly differentiating ETO-AA-expressing cells. However, cells constitutively expressing ETO fail to do so fully even when differentiated for extended periods of up to 16 days. At present, it is unclear whether this results from ETO affecting critical early C/EBPβ-mediated events, which cannot subsequently be overcome, or from modulation of other targets by ETO in mature cells. However, we have observed that Glut4 mRNA expression remains suppressed longer than other genes tested and, since glucose may represent an important substrate for lipid production in these cultured adipocytes, it is possible that this explains the lack of lipid accumulation despite almost normal levels of aP2, PPARγ, and C/EBPα.
Although ETO mRNA levels fell acutely in response to proadipogenic signals, the disappearance of ETO protein within the cells was less rapid and was absent only after 8 to 24 h of differentiation. This coincides well with the previously reported acquisition of DNA-binding activity by C/EBPβ, which lags considerably behind C/EBPβ expression (39), which is apparent within 2 h of induction of differentiation. Our data from gel shift assays, ChIP assays, and immunostaining of C/EBPβ subnuclear localization strongly suggest that not only does ETO inhibit the binding activity of C/EBPβ but that this occurs in intact cells, thus explaining the loss of induction of C/EBPβ target genes in differentiating preadipocytes constitutively expressing ETO. A similar mechanism has been described for the C/EBP homologous protein CHOP-10, a dominant-negative member of the C/EBP family of transcription factors, which heterodimerizes with C/EBPβ via leucine zipper domains (40). However, because ETO is not a member of the C/EBP family its control of C/EBPβ activity in this way represents a novel mechanism for its regulation. Since ETO has no conventional C/EBP binding domains, it is not clear which regions of the two proteins are involved in their interaction, and we are currently addressing this question. Given the colocalization of ETO and C/EBPβ that we have observed in preadipocytes, it is intriguing that ETO has been reported to reside in transcriptionally inert regions of the nucleus (2). Thus, in addition to inhibiting the DNA-binding activity of C/EBPβ per se, ETO may also actively sequester it within these regions.
Wiper-Bergeron et al. have demonstrated that, once capable of binding DNA, C/EBPβ forms a complex on the C/EBPα promoter that is inactive due to the presence of the transcriptional corepressors Sin3a and HDAC1 (44). Only once HDAC1 is degraded is transcription activated, and this may explain the additional lag between the reported binding of C/EBPβ to the C/EBPα promoter and the appearance of C/EBPα mRNA (39). Although our data demonstrate that the effect of ETO is to inhibit binding of C/EBPβ to the C/EBPα promoter, several studies have reported the binding of Sin3a and HDAC1 to ETO (1). This raises the possibility that ETO may assist in the assembly of this complex before the disappearance of ETO allows its association with the C/EBPα promoter.
ETO is selectively expressed at much higher levels in the preadipocyte than the mature adipocyte fraction of rat adipose tissue, mimicking the situation in the 3T3-L1 cells differentiated in culture. It therefore appears likely that the mechanism we have described is operative in adipogenesis in vivo. The preponderance of ETO in the preadipocyte fraction of human fat further suggests that our data are relevant to human adipose tissue development, raising the possibility that decreased ETO expression or activity may play a role in the development of obesity. Indeed, an association between obesity and a polymorphism in the 3′-untranslated region of ETO has been reported in male Pima Indians (45). Although that study failed to find any mutations in the coding sequence of the gene itself, in light of our present data one would postulate that a mutation affecting RNA stability or the expression of ETO might also predispose to obesity. Although an ETO knockout mouse model has been generated, the few mice surviving past the neonatal stage exhibit severe abnormalities in midgut development (3). The resulting nutrient malabsorption makes them unsuitable for assessing any effects on obesity. Evidently, just as decreased ETO expression may contribute to obesity, the potential involvement of ETO overexpression in syndromes of lipodystrophy also warrants examination. Thus, ETO-overexpressing mice or tissue-specific manipulation of this gene may provide insights in the future.
In addition to its role in adipogenesis C/EBPβ has been implicated in the regulation of diverse processes, including tumor development, neuronal survival, memory consolidation in the hippocampus, and myeloid differentiation (18, 22, 27, 41). Interestingly, C/EBPβ but not C/EBPα is capable of inducing myeloid differentiation of pluripotent hematopoietic progenitor cells (27). We have not investigated the effect of the leukemogenic AML1-ETO fusion protein on C/EBPβ activity. However, if it were similarly inhibitory, this may contribute to the development of AML in subjects bearing the AML1-ETO t(8;21) translocation via this mechanism. From a metabolic perspective analysis of knockout mice has revealed that loss of C/EBPβ protein throughout the body results in decreased epididymal fat mass, decreased gluconeogenesis and lipolysis during fasting, and diabetes and increased skeletal muscle insulin sensitivity (20, 36, 43). Having demonstrated its ability to inhibit C/EBPβ activity, the involvement of ETO in these fundamental C/EBPβ-regulated processes in both normal and disease states merits examination. We believe that this is likely to reveal further important physiological roles for ETO and may also suggest opportunities for therapeutic manipulation.
In summary, ETO acts in preadipocytes to inhibit C/EBPβ activity and promote the maintenance of the undifferentiated state. Its rapid downregulation by hormonal stimuli plays an essential role in coordinating the differentiation process. These findings define, for the first time, a precise function for ETO in normal cellular physiology, revealing its novel and important role in the regulation of adipogenesis.
Acknowledgments
This study was supported by the Wellcome Trust (J.J.R., R.K.S., K.B.B., D.H., M.M., and S.O.R.), the Deutsche Forschungsgemeinschaft (M.L. and S.S.), the UK MRC (C.C., C.J.L., and C.M.), and the Raymond and Beverly Sackler Foundation (R.K.S.). J.K.S. is a BBSRC David Phillips Fellow.
We are particularly grateful to the members of the O'Rahilly and Siddle labs for many helpful discussions. Q.-Q. Tang and M. D. Lane generously provided C/EBPα promoter and C/EBPβ expression constructs. The C/EBPwt-LUC construct was kindly provided by S. Smola-Hess.
REFERENCES
- 1.Amann, J. M., J. Nip, D. K. Strom, B. Lutterbach, H. Harada, N. Lenny, J. R. Downing, S. Meyers, and S. W. Hiebert. 2001. ETO, a target of t(8;21) in acute leukemia, makes distinct contacts with multiple histone deacetylases, and binds mSin3A through its oligomerization domain. Mol. Cell. Biol. 21:6470-6483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barseguian, K., B. Lutterbach, S. W. Hiebert, J. Nickerson, J. B. Lian, J. L. Stein, A. J. van Wijnen, and G. S. Stein. 2002. Multiple subnuclear targeting signals of the leukemia-related AML1/ETO and ETO repressor proteins. Proc. Natl. Acad. Sci. USA 99:15434-15439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Calabi, F., R. Pannell, and G. Pavloska. 2001. Gene targeting reveals a crucial role for MTG8 in the gut. Mol. Cell. Biol. 21:5658-5666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Calkhoven, C. F., C. Muller, and A. Leutz. 2000. Translational control of C/EBPα and C/EBPβ isoform expression. Genes Dev. 14:1920-1932. [PMC free article] [PubMed] [Google Scholar]
- 5.Chevallier, N., C. M. Corcoran, C. Lennon, E. Haijek, A. Chadburn, V. J. Bardwell, J. D. Licht, and A. Melnick. 2003. The ETO protein of t(8;21) AML is a corepressor for the Bcl-6 B-cell lymphoma oncoprotein. Blood. [DOI] [PubMed]
- 6.Christy, R. J., K. H. Kaestner, D. E. Geiman, and M. D. Lane. 1991. CCAAT/enhancer binding protein gene promoter: binding of nuclear factors during differentiation of 3T3-L1 preadipocytes. Proc. Natl. Acad. Sci. USA 88:2593-2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clarke, S. L., C. E. Robinson, and J. M. Gimble. 1997. CAAT/enhancer binding proteins directly modulate transcription from the peroxisome proliferator-activated receptor gamma 2 promoter. Biochem. Biophys. Res. Commun. 240:99-103. [DOI] [PubMed] [Google Scholar]
- 8.Cole, K. A., A. W. Harmon, J. B. Harp, and Y. M. Patel. 2004. Rb regulates C/EBPβ-DNA-binding activity during 3T3-L1 adipogenesis. Am. J. Physiol. Cell Physiol. 286:C349-C354. [DOI] [PubMed] [Google Scholar]
- 9.Davis, J. N., L. McGhee, and S. Meyers. 2003. The ETO (MTG8) gene family. Gene 303:1-10. [DOI] [PubMed] [Google Scholar]
- 10.Descombes, P., and U. Schibler. 1991. A liver-enriched transcriptional activator protein, LAP, and a transcriptional inhibitory protein, LIP, are translated from the same mRNA. Cell 67:569-579. [DOI] [PubMed] [Google Scholar]
- 11.Elberg, G., J. M. Gimble, and S. Y. Tsai. 2000. Modulation of the murine peroxisome proliferator-activated receptor gamma 2 promoter activity by CCAAT/enhancer-binding proteins. J. Biol. Chem. 275:27815-27822. [DOI] [PubMed] [Google Scholar]
- 12.Erickson, R. L., N. Hemati, S. E. Ross, and O. A. MacDougald. 2001. p300 coactivates the adipogenic transcription factor CCAAT/enhancer-binding protein alpha. J. Biol. Chem. 276:16348-16355. [DOI] [PubMed] [Google Scholar]
- 13.Hadaschik, D., K. Hinterkeuser, M. Oldak, H. J. Pfister, and S. Smola-Hess. 2003. The papillomavirus E2 protein binds to and synergizes with C/EBP factors involved in keratinocyte differentiation. J. Virol. 77:5253-5265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hamm, J. K., B. H. Park, and S. R. Farmer. 2001. A role for C/EBPβ in regulating peroxisome proliferator-activated receptor gamma activity during adipogenesis in 3T3-L1 preadipocytes. J. Biol. Chem. 276:18464-18471. [DOI] [PubMed] [Google Scholar]
- 15.Hartman, H. B., X. Hu, K. X. Tyler, C. K. Dalal, and M. A. Lazar. 2002. Mechanisms regulating adipocyte expression of resistin. J. Biol. Chem. 277:19754-19761. [DOI] [PubMed] [Google Scholar]
- 16.Herzig, S., F. Long, U. S. Jhala, S. Hedrick, R. Quinn, A. Bauer, D. Rudolph, G. Schutz, C. Yoon, P. Puigserver, B. Spiegelman, and M. Montminy. 2001. CREB regulates hepatic gluconeogenesis through the coactivator PGC-1. Nature 413:179-183. [DOI] [PubMed] [Google Scholar]
- 17.Kahn, B. B., and J. S. Flier. 2000. Obesity and insulin resistance. J. Clin. Investig. 106:473-481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lamb, J., S. Ramaswamy, H. L. Ford, B. Contreras, R. V. Martinez, F. S. Kittrell, C. A. Zahnow, N. Patterson, T. R. Golub, and M. E. Ewen. 2003. A mechanism of cyclin D1 action encoded in the patterns of gene expression in human cancer. Cell 114:323-334. [DOI] [PubMed] [Google Scholar]
- 19.Licht, J. D. 2001. AML1 and the AML1-ETO fusion protein in the pathogenesis of t(8;21) AML. Oncogene 20:5660-5679. [DOI] [PubMed] [Google Scholar]
- 20.Liu, S., C. Croniger, C. Arizmendi, M. Harada-Shiba, J. Ren, V. Poli, R. W. Hanson, and J. E. Friedman. 1999. Hypoglycemia and impaired hepatic glucose production in mice with a deletion of the C/EBPβ gene. J. Clin. Investig. 103:207-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lutterbach, B., D. Sun, J. Schuetz, and S. W. Hiebert. 1998. The MYND motif is required for repression of basal transcription from the multidrug resistance 1 promoter by the t(8;21) fusion protein. Mol. Cell. Biol. 18:3604-3611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marshall, J., B. M. Dolan, E. P. Garcia, S. Sathe, X. Tang, Z. Mao, and L. A. Blair. 2003. Calcium channel and NMDA receptor activities differentially regulate nuclear C/EBPβ levels to control neuronal survival. Neuron 39:625-639. [DOI] [PubMed] [Google Scholar]
- 23.McKnight, S. L. 2001. McBindall: a better name for CCAAT/enhancer binding proteins? Cell 107:259-261. [DOI] [PubMed] [Google Scholar]
- 24.Moitra, J., M. M. Mason, M. Olive, D. Krylov, O. Gavrilova, B. Marcus-Samuels, L. Feigenbaum, E. Lee, T. Aoyama, M. Eckhaus, M. L. Reitman, and C. Vinson. 1998. Life without white fat: a transgenic mouse. Genes Dev. 12:3168-3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morrison, R. F., and S. R. Farmer. 1999. Insights into the transcriptional control of adipocyte differentiation. J. Cell Biochem. Suppl. 32-33:59-67. [DOI] [PubMed] [Google Scholar]
- 26.Mulligan, C., J. Rochford, G. Denyer, R. Stephens, G. Yeo, T. Freeman, K. Siddle, and S. O'Rahilly. 2002. Microarray analysis of insulin and IGF-1 receptor signalling reveals the selective upregulation of the mitogen HB-EGF by IGF-1. J. Biol. Chem. 277:42480-42487. [DOI] [PubMed] [Google Scholar]
- 27.Nerlov, C., K. M. McNagny, G. Doderlein, E. Kowenz-Leutz, and T. Graf. 1998. Distinct C/EBP functions are required for eosinophil lineage commitment and maturation. Genes Dev. 12:2413-2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Odaka, Y., A. Mally, L. T. Elliott, and S. Meyers. 2000. Nuclear import and subnuclear localization of the proto-oncoprotein ETO (MTG8). Oncogene 19:3584-3597. [DOI] [PubMed] [Google Scholar]
- 29.Petersen, K. F., E. A. Oral, S. Dufour, D. Befroy, C. Ariyan, C. Yu, G. W. Cline, A. M. DePaoli, S. I. Taylor, P. Gorden, and G. I. Shulman. 2002. Leptin reverses insulin resistance and hepatic steatosis in patients with severe lipodystrophy. J. Clin. Investig. 109:1345-1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rajala, M. W., and P. E. Scherer. 2003. Minireview: the adipocyte: at the crossroads of energy homeostasis, inflammation, and atherosclerosis. Endocrinology 144:3765-3773. [DOI] [PubMed] [Google Scholar]
- 31.Rana, B., Y. Xie, D. Mischoulon, N. L. Bucher, and S. R. Farmer. 1995. The DNA binding activity of C/EBP transcription factor is regulated in the G1 phase of the hepatocyte cell cycle. J. Biol. Chem. 270:18123-18132. [DOI] [PubMed] [Google Scholar]
- 32.Rangwala, S. M., and M. A. Lazar. 2000. Transcriptional control of adipogenesis. Annu. Rev. Nutr. 20:535-559. [DOI] [PubMed] [Google Scholar]
- 33.Roesler, W. J. 2001. The role of C/EBP in nutrient and hormonal regulation of gene expression. Annu. Rev. Nutr. 21:141-165. [DOI] [PubMed] [Google Scholar]
- 34.Rosen, E. D., C. H. Hsu, X. Wang, S. Sakai, M. W. Freeman, F. J. Gonzalez, and B. M. Spiegelman. 2002. C/EBPα induces adipogenesis through PPARγ: a unified pathway. Genes Dev. 16:22-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rosen, E. D., and B. M. Spiegelman. 2000. Molecular regulation of adipogenesis. Annu. Rev. Cell Dev. Biol. 16:145-171. [DOI] [PubMed] [Google Scholar]
- 36.Tanaka, T., N. Yoshida, T. Kishimoto, and S. Akira. 1997. Defective adipocyte differentiation in mice lacking the C/EBPβ and/or C/EBPδ gene. EMBO J. 16:7432-7443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tang, Q. Q., M. S. Jiang, and M. D. Lane. 1997. Repression of transcription mediated by dual elements in the CCAAT/enhancer binding protein alpha gene. Proc. Natl. Acad. Sci. USA 94:13571-13575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tang, Q. Q., M. S. Jiang, and M. D. Lane. 1999. Repressive effect of Sp1 on the C/EBPα gene promoter: role in adipocyte differentiation. Mol. Cell. Biol. 19:4855-4865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tang, Q. Q., and M. D. Lane. 1999. Activation and centromeric localization of CCAAT/enhancer-binding proteins during the mitotic clonal expansion of adipocyte differentiation. Genes Dev. 13:2231-2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tang, Q. Q., and M. D. Lane. 2000. Role of C/EBP homologous protein (CHOP-10) in the programmed activation of CCAAT/enhancer-binding protein-beta during adipogenesis. Proc. Natl. Acad. Sci. USA 97:12446-12450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Taubenfeld, S. M., M. H. Milekic, B. Monti, and C. M. Alberini. 2001. The consolidation of new but not reactivated memory requires hippocampal C/EBPβ. Nat. Neurosci. 4:813-818. [DOI] [PubMed] [Google Scholar]
- 42.Urso, B., D. L. Cope, H. E. Kalloo-Hosein, A. C. Hayward, J. P. Whitehead, S. O'Rahilly, and K. Siddle. 1999. Differences in signaling properties of the cytoplasmic domains of the insulin receptor and insulin-like growth factor receptor in 3T3-L1 adipocytes. J. Biol. Chem. 274:30864-30873. [DOI] [PubMed] [Google Scholar]
- 43.Wang, L., J. Shao, P. Muhlenkamp, S. Liu, P. Klepcyk, J. Ren, and J. E. Friedman. 2000. Increased insulin receptor substrate-1 and enhanced skeletal muscle insulin sensitivity in mice lacking CCAAT/enhancer-binding protein beta. J. Biol. Chem. 275:14173-14181. [DOI] [PubMed] [Google Scholar]
- 44.Wiper-Bergeron, N., D. Wu, L. Pope, C. Schild-Poulter, and R. J. Hache. 2003. Stimulation of preadipocyte differentiation by steroid through targeting of an HDAC1 complex. EMBO J. 22:2135-2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wolford, J. K., C. Bogardus, and M. Prochazka. 1998. Polymorphism in the 3′ untranslated region of MTG8 is associated with obesity in Pima Indian males. Biochem. Biophys. Res. Commun. 246:624-626. [DOI] [PubMed] [Google Scholar]
- 46.Wolford, J. K., and M. Prochazka. 1998. Structure and expression of the human MTG8/ETO gene. Gene 212:103-109. [DOI] [PubMed] [Google Scholar]
- 47.Wu, Z., N. L. Bucher, and S. R. Farmer. 1996. Induction of peroxisome proliferator-activated receptor gamma during the conversion of 3T3 fibroblasts into adipocytes is mediated by C/EBPβ, C/EBPδ, and glucocorticoids. Mol. Cell. Biol. 16:4128-4136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu, Z., E. D. Rosen, R. Brun, S. Hauser, G. Adelmant, A. E. Troy, C. McKeon, G. J. Darlington, and B. M. Spiegelman. 1999. Cross-regulation of C/EBPα and PPARγ controls the transcriptional pathway of adipogenesis and insulin sensitivity. Mol. Cell 3:151-158. [DOI] [PubMed] [Google Scholar]