Abstract
Cell cycle arrest by FoxO transcription factors involves transcriptional repression of cyclin D, although the exact mechanism remains unclear. In this study, we used the BCR-ABL-expressing cell line BV173 as a model system to investigate the mechanisms whereby FoxO3a regulates cyclin D2 expression. Inhibition of BCR-ABL by STI571 results in down-regulation of cyclin D2 expression, activation of FoxO3a activity, and up-regulation of BCL6 expression. Using reporter gene assays, we demonstrate that STI571, FoxO3a, and BCL6 can repress cyclin D2 transcription through a STAT5/BCL6 site located within the cyclin D2 promoter. We propose that BCR-ABL inhibition leads to FoxO3a activation, which in turn induces the expression of BCL6, culminating in the repression of cyclin D2 transcription through this STAT5/BCL6 site. This process was verified by mobility shift and chromatin immunoprecipitation analyses. We find that conditional activation of FoxO3a leads to accumulation of BCL6 and down-regulation of cyclin D2 at protein and mRNA levels. Furthermore, silencing of FoxO3a and BCL6 in BCR-ABL-expressing cells abolishes STI571-mediated effects on cyclin D2. This report establishes the signaling events whereby BCR-ABL signals are relayed to cyclin D2 to mediate cell cycle progression and defines a potential mechanism by which FoxO proteins regulate cyclin D2 expression.
In mammalian cells, the commitment to divide is made in the G1 phase of the cell cycle in response to various stimuli, including growth factors. After passing the restriction point at mid- to late G1, cells become refractory to growth inhibition signals or do not require growth factors to progress into S phase (37). Progression of eukaryotic cells through the cell cycle is controlled by the two families of G1 cyclins: (i) D-type cyclins (cyclins D1, D2, and D3) and cyclin E (cyclins E1 and E2) (29, 44) and (ii) the cyclin-dependent kinases (cdk's), their catalytic counterparts. The primary targets of the G1 cyclin-cdk complexes are the retinoblastoma protein (pRb) family of pocket proteins, consisting of pRb, p107, and p130 (20, 27, 34). The phosphorylation state of pRb regulates the activity of the E2F family of transcription factors; in their hypophosphorylated forms, the pRb-related pocket proteins associate with members of the E2F family, negatively regulating transcription activity of E2F-regulated genes that are required for entry into the S phase of the cell cycle (15, 35, 40).
In mammals, the phosphatidylinositol 3-kinase/protein kinase B (PI3-K/PKB) pathway is stimulated by a variety of growth factors and cytokines and by cell-matrix interactions, and it controls many biological functions, including cell proliferation, cell survival, and insulin responses (30). Importantly, constitutive activation of the PI3-K pathway facilitates tumor formation by two different mechanisms: it supports S-phase entry, and it confers resistance to apoptotic signals which normally restrict uncontrolled cell growth (49). Recently, it has been demonstrated that the members of the FoxO subfamily of forkhead transcription factors AFX, FKHR, and FKHR-L1 (which have recently been renamed FoxO4, FoxO1a, and FoxO3a, respectively) are directly phosphorylated by PKB (also called Akt) (6, 25, 47). When cells are stimulated with serum or growth factors, FoxO transcription factors are phosphorylated by activated PKB and exported from the nucleus to the cytoplasm, resulting in the inhibition of target gene transcription (6, 25, 47). In contrast, when cells are deprived of serum or growth factors, FoxO factors become dephosphorylated, translocate into the nucleus, and activate transcription of target genes.
Whereas it is clear that the molecular programs regulated by the forkhead family of transcription factors are critical for cell cycle progression, the genes that are regulated by these proteins are largely unknown. It has been reported previously that FoxO factor-induced withdrawal from the cell cycle occurs in G1 phase and is the result of increased transcription of the cdk inhibitor p27kip1 (32). More recently, cell cycle inhibition by FoxO factors has also been shown to involve down-regulation of cyclin D1 and cyclin D2 (42). This effect was demonstrated to be mediated through transcriptional repression, although the exact mechanism is unclear. The promoter region of the D-type cyclins does not contain any obvious FoxO binding sites, suggesting that transcriptional regulation either takes place through the interaction with other transcription factors or is indirectly mediated through FoxO-dependent induction of a transcriptional repressor protein.
The BCR-ABL chimeric oncogenes encode the constitutively active p230, p210, and p185 BCR-ABL tyrosine kinases, which play essential roles in the pathogenesis of chronic myeloid leukemia (CML) and Philadelphia (Ph1) acute lymphoblastic leukemia. BCR-ABL exerts diverse actions on hematopoietic cells regarding cell transformation, protection of apoptosis, cell cycle progression, altered cell migration, and adhesion to extracellular matrix (reviewed in reference 9). The expression of BCR-ABL replaces the requirement for growth factors and activates multiple signaling cascades, including the signal transducers and activators of transcription (STATs; STAT1 and STAT5), Ras, and PI3-K pathways (36, 41). Among these, it has been demonstrated that PI3-K activity is required for growth, transformation, and survival of Ph1 chromosome-positive cells (45, 46).
Interestingly, a direct relationship between BCR-ABL activity and cyclin D2 expression in BCR-ABL-positive cells has been demonstrated (10, 36); these reports suggest the importance of cyclin D2 in mediating the proliferative signals from BCR-ABL and show that BCR-ABL regulates cyclin D2 expression at the transcriptional level. A recent study showed that the FoxO3a transcription factor lies downstream of the BCR-ABL signaling pathway and has a negative role in cell growth mediated by the BCR-ABL fusion protein (24). Here, we have used the lymphoid CML cell line BV173 and BCR-ABL-expressing BaF3 cells as model systems to study the molecular mechanisms whereby the transcription factor FoxO3a regulates cyclin D2 expression.
We show that the mechanism of regulation of cyclin D2 by FoxO3a involves induction of the transcriptional repressor BCL6. In BCR-ABL-positive cells, BCR-ABL inhibition will lead to activation of FoxO3a, and this will induce expression of BCL6, ultimately causing transcriptional repression of cyclin D2 by interaction of BCL6 with the STAT5/BCL6 consensus site of the cyclin D2 promoter. Importantly, the present study defines a potential mechanism by which FoxO proteins regulate cyclin D2 expression and confirms that cyclin D2 is a bona fide downstream target of FoxO transcription factors.
MATERIALS AND METHODS
Cell culture.
The human lymphoid cell line BV173 was maintained in RPMI 1640 medium supplemented with 10% fetal calf serum (FCS), 2 mM l-glutamine, 50 μM 2-mercaptoethanol, and 100 U of penicillin-streptomycin/ml. All experiments were performed on exponentially growing cells kept at low concentrations (<1 × 106 cells/ml). STI571 was added to the medium at a final concentration of 1 μM from a 100 mM stock solution in dimethyl sulfoxide. BaF3 and BaF3 · FoxO3a(A3):ER cells were cultured in RPMI 1640 medium supplemented with 10% FCS, 2 mM l-glutamine, 100 U of penicillin-streptomycin/ml, and 10% (vol/vol) WEHI-3B-conditioned medium as a source of interleukin-3. 4-Hydroxytamoxifen (4-OHT; Sigma) was added to the cells at a final concentration of 200 nM. The BaF3 · FoxO3a(A3):ER and the BaF3/BCR-ABL cell lines have been described previously (13, 36).
Plasmid constructs.
The human full-length BCL6 cDNA (2.2 kb) was generated by PCR from cDNA derived from primary human B cells using the primers 5′-CTCGCCGGCTGACAGCT-3′ and 5′-TATCCTTTGGGTAGATTCTGAGAAGG-3′ and was cloned into the pGEM-T Easy vector (Promega). The identity of the BCL6 cDNA was confirmed by sequencing. A NotI fragment consisting of the full-length 2.2-kb cDNA was subcloned into the expression vector pcDNA3 (Invitrogen) and used for subsequent Northern blotting. The following pGL3 plasmids have been previously described (31): −1624D2Luc, −1303D2Luc, −1204D2Luc, −1303mtSp1D2Luc, −1303mtStat5D2Luc, and −1227/−1168D2Luc. The pGL3 plasmids −892D2Luc, −637D2Luc, and −444D2Luc were generated by excising the corresponding XcmI/HindIII, BamHI/HindIII, and PvuII/HindIII fragments from the pGL2-1624D2Luc human cyclin D2 promoter plasmid (obtained from Dov Shiffman, CV Therapeutics, Palo Alto, Calif.) and subcloning them into the pGL3 basic vector (Promega). The −1296BCL6pGL3 and −1296mtBCL6pGL3 constructs are kind gifts from Tracy Tang and Laurence Lasky (Genentech Inc., South San Francisco, Calif.) (48). The FoxO3a expression vectors pLPC-FoxO3a(wt) and pLPC-FoxO3a(A3) were derived from pECE-FKHR-L1(wt) and pECE-FKHR-L1(A3), respectively (6). The cDNAs were excised from pECE-FKHR-L1(wt) and pECE-FKHR-L1(A3) by HindIII and XbaI restriction digestion and subcloned into the retroviral pLPC vector (12).
Western blot analysis and antibodies.
Whole-cell extracts for Western blotting were prepared by lysing cells with 4× the packed cell volume of lysis buffer (1% Nonidet P-40, 100 mM NaCl, 20 mM Tris-HCl [pH 7.4], 10 mM NaF, 1 mM Na3VO4, and Complete protease inhibitors purchased from Roche) on ice for 15 min. Protein yield was quantified by a Dc protein assay kit (Bio-Rad Laboratories, Richmond, Calif.). Fifty micrograms of lysate was separated by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis and transferred to nitrocellulose membranes (Protan, Schleicher & Schuell), and specific proteins were recognized by specific antibodies. The antibodies against BCL6 (C-19) and cdk4 (C-22) were from Santa Cruz Biotechnology, anti-STAT5 was from Zymed, anti-phospho-STAT5 (Tyr694) was from Cell Signaling, and anti-FoxO3a and anti-phospho-FoxO3a (Thr32) were purchased from Upstate Biotechnology. The cyclin D2 monoclonal antibody (DCS 3.1+DCS 5.2) was acquired from NeoMarkers. The c-Abl antibody (Ab-3) was from Calbiochem. The beta-tubulin antibody was from PharMingen. The antibodies were detected by using horseradish peroxidase-linked goat anti-mouse or anti-rabbit immunoglobulin G (Dako) and visualized with the enhanced chemiluminescent detection system (Amersham Biosciences).
RNA isolation and analysis.
Total RNA was isolated by using the RNeasy Kit (QIAGEN), and the concentration and purity of each sample were assessed by absorbance at 260 nm and by the 260 nm/280 nm ratio, respectively. The integrity of the RNA was verified by observing the rRNA bands in ethidium bromide-stained gel under UV irradiation. Twenty micrograms of RNA, prepared as described above, was resolved on 1.5% formaldehyde-agarose gels. Following electrophoresis, RNAs were transferred to Hybond-N+ membrane (Amersham Biosciences) and subjected to Northern blotting as previously described (28). Cyclin D2 and BCL6 were detected by hybridization with their respective full-length 32P-labeled human (BV173) or mouse (BaF3) cDNA probe generated by reverse transcription (RT)-PCR, as previously described. The probes were labeled with Ready-To-Go DNA labeling beads (Amersham Biosciences) and purified with ProbeQuant G-50 Micro columns (Amersham Biosciences).
Real-time quantitative RT-PCR.
Total RNA was isolated as described earlier and DNase I treated. Equal amounts of total RNA (2 μg) were reverse transcribed by using the Superscript First-Strand synthesis system for RT-PCR (Invitrogen), and the resulting first-strand cDNA was diluted and used as template in the real-time quantitative PCR analysis. All measurements were performed in triplicate. The following gene-specific primer pairs were designed by using the ABI Primer Express software: human L19-sense, 5′-GCGGAAGGGTACAGCCAAT-3′; human L19-antisense, 5′-GCAGCCGGCGCAAA-3′; human cyclin D2-sense, 5′-CTGTGTGCCACCGACTTTAAGTT-3′; human cyclin D2-antisense, 5′-GATGGCTGCTCCCACACTTC-3′; mouse BCL6-sense, 5′-AGTGCCAGGCAAGTCCCTAAT-3′; mouse BCL6-antisense, 5′-CGAGTAGATGTTGCTGTGACACAA-3′; mouse cyclin D2-sense, 5′-GCGTGCAGAAGGACATCCA-3′; mouse cyclin D2-antisense, 5′-CACTTTTGTTCCTCACAGACCTCTAG-3′; mouse L19-sense, 5′-GGAAAAAGAAGGTCTGGTTGGA-3′; and mouse L19-antisense, 5′-TGATCTGCTGACGGGAGTTG-3′. The specificity of each primer was determined with the National Center for Biotechnology Information BLAST module. The levels of cyclin D2, BCL6, and L19 mRNAs were analyzed. L19 represents a nonregulated gene, and its expression served as internal control and was used to normalize for variances in input cDNA. Detection of cyclin D2, BCL6, and L19 expression was performed with SYBR Green (Applied Biosystems) and an ABI PRISM 7700 sequence detection system (Applied Biosystems) using the relative standard curve method. All measurements were performed in triplicate.
Transfection.
A total of 15 × 106 BV173 cells were resuspended in 500 μl of RPMI 1640 medium containing 10% FCS, 2 mM l-glutamine, antibiotics, and 10 μg of reporter plasmid DNA, plus 1 μg of pRLTK Renilla plasmid (Promega) as a transfection control. After a 5-min incubation, cells were electroporated by using a Gene Pulser (Bio-Rad) at 350 V and 950 μF. Cells were then transferred to 10 ml of RPMI 1640 medium and left to recover for 24 h before treatment. Then, cells were harvested, washed with phosphate-buffered saline (PBS), and lysed in 100 μl of Passive lysis buffer (Promega) for 5 min at room temperature; after one freeze-thaw cycle, insoluble cell debris was spun down in a microcentrifuge for 1 min, and the supernatant fraction was assayed for luciferase activity with the Dual-Luciferase Reporter assay system (Promega).
Extraction of DNA-binding proteins.
DNA-binding protein extracts from BV173 cells were prepared essentially as described previously (1). Briefly, 25 × 106 cells were centrifuged at 4,000 × g for 10 min at 4°C, washed with ice-cold PBS, and resuspended in 400 μl of cold buffer A (10 mM HEPES-KOH [pH 7.9], 1.5 mM MgCl2, 10 mM KCl, 0.5 mM dithiothreitol, 0.2 mM 4-2-aminoethylbenzenesulfonyl hydrochloride [ABSF]) by gently flicking the tube. The cells were allowed to swell on ice for 10 min. After a brief vortex, samples were centrifuged for 2 min at 4°C, and the supernatant fraction was discarded. The pellet was resuspended in 30 to 40 μl of cold buffer C (20 mM HEPES-KOH [pH 7.9], 25% [vol/vol] glycerol, 420 mM NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 0.5 mM dithiothreitol, 0.2 mM ABSF) and incubated on ice for 20 min for high-salt extraction. Cellular debris was removed by centrifugation (13,000 × g, 2 min, 4°C). The supernatant fraction contains the DNA-binding proteins. Protein yield was quantified by a Dc protein assay kit (Bio-Rad).
Electrophoretic mobility shift and supershift analyses.
Gel retardation assays were performed essentially as described previously (28), using a double-stranded oligonucleotide probe (5′-CGGGCCATTTCCTAGAAAGCTGCATC-3′ and 3′-GCCCGGTAAAGGATCTTTCGACGTA-5′) encompassing the human cyclin D2 promoter STAT5 binding site. The oligonucleotides were annealed, and the resulting double-stranded oligonucleotide was radiolabeled with a fill-in reaction using [α-32P]dGTP (Amersham Biosciences) and DNA polymerase (Klenow fragment). Fifteen micrograms of DNA-binding protein extract was incubated with 1 to 2 ng of 32P-labeled oligonucleotide probe in a total volume of 20 μl at 30°C for 10 min. The reactions were electrophoresed on 4% polyacrylamide gels in 0.33× Tris-buffered EDTA at 4°C. Gels were then dried and exposed to X-ray films. Supershift assays were performed by adding 1 μl of concentrated antibodies before the addition of the probe. Mouse anti-STAT5 was from Zymed, and anti-BCL6 (C-19) and anti-BCL6 (N-3) were from Santa Cruz Biotechnology.
Chromatin immunoprecipitation (ChIP) assay.
BV173 cells cultured at 1 × 106 cells per ml of medium were either untreated or treated with 5 μM STI571 for 4 h and then collected by centrifugation and resuspended in 10 ml of cold PBS. After adjusting to a final formaldehyde concentration of 1% (wt/vol), the cells were incubated for 10 min at 37°C, followed by the addition of glycine to 0.136 M and incubation for 10 min. After being washed with cold PBS, the cells were sonicated in six 10-s pulses at maximum power in 300 μl of sonication buffer (1% SDS, 10 mM EDTA, 50 mM Tris-HCl [pH 8.1], 1 mM phenylmethyl sulfonyl fluoride, 1× protease inhibitor cocktail [Roche]). The supernatant was cleared by centrifugation at 13,000 × g and diluted five times with sonication buffer, of which 1/10 was retained as input control. DNA shearing was controlled by running part of the sonicated chromatin on an agarose gel to ensure that fragmented DNA is 200 to 500 bp. To preclear the chromatin solution, 2 μg of sonicated single-strand herring sperm DNA (Sigma) and 45 μl of a 50% (vol/vol) slurry of protein G-Sepharose (Pharmacia) beads were added to the rest of the sample. After incubation with agitation for 30 min at room temperature, the beads were removed by brief centrifugation, and the supernatant was collected. The retrieved supernatant was incubated with either anti-BCL6 (C-19; Santa Cruz), anti-STAT5α (Zymed), or an isotype control antibody (Babco) for 2 h at room temperature with rotation in the presence of 2 μg of sonicated single-strand herring sperm DNA (Sigma) and 45 μl of a 50% (vol/vol) slurry of protein G-Sepharose (Pharmacia) beads. The beads were then washed sequentially with PBS, sonication buffer, Tris-SDS-EDTA buffer (TSE) I, TSE II, TSE III, and PBS, as described previously (3). The immunoprecipitated DNA (or the input control) was extracted with 300 μl of extraction solution (1% SDS and 1.1 M NaHCO3), incubated at 65°C for 6 h to reverse the cross-links, and purified with the QIAGEN PCR purification kit according to the manufacturer's protocol. PCRs were then performed on the purified DNA according to the QIAGEN Taq PCR handbook in the presence of 2.5 mM MgCl2 at 55°C for 28 cycles and with the following primers: primer A sense, 5′-GACTCAAGGATGCGTTAGAGCAC-3′; primer A antisense, 5′-CCCGAGGTGTCTGCGC-3′; primer B sense, 5′-TCTGAGGTCACCCCATCTTCA-3′; primer B antisense, 5′-CATCCAGCCTGCATGCC-3′; primer C sense, 5′-CTCCGTTCGGGCCTCAAT-3′; primer C antisense, 5′-CAGGAACACGAACAGCAACATT-3′; primer D sense, 5′-GGCCTTTAGAATTCCCTCCGGC-3′; primer D antisense, 5′-GCTGAAGTGTGTCTCTCCTGCAC-3′; primer E sense, 5′-AGCTCGATCTGCTGAGTTTATGGG-3′; and primer E antisense, 5′-TTTCACTGGCCGGCATATTTCGAG-3′. Analysis of the PCR products was performed on a standard 2% (wt/vol) agarose gel by electrophoresis in Tris-acetate-EDTA buffer.
Southern blot analysis of ChIP samples.
The agarose gel with the resolved PCR samples was soaked in 0.4 M NaOH for 30 min and Southern blotted onto a Hybond-N+ membrane (Amersham) overnight in the presence of 0.4 M NaOH according to the manufacturer's protocol. The promoter region of human cyclin D2 gene (−1624/+6) was excised from the pGL2 human wild-type cyclin D2 promoter (4) and used as a template for generating a 32P-labeled hybridization probe according to the manufacturer's directions (Ready-To-Go DNA labeling beads; Amersham Biosciences). The probe was added to the Southern blot membrane and hybridized in Church and Gilbert buffer at 45°C for 16 h as described above for Northern blotting. The Southern blot signals were detected by the Typhoon 8600 Phosphoimager and quantified with the ImageQuant program.
siRNA.
The mammalian expression vector pSUPER (5) was used for gene silencing of FoxO3a in BaF3/BCR-ABL cells. The gene-specific insert specifies a 19-nucleotide sequence corresponding to nucleotides GACCTGCTTGCTTCAGACT, which is separated by a 9-nucleotide noncomplementary spacer (TCTCTTGAA) from the reverse complement of the same 19-nucleotide sequence. This construct (10 μg) was transiently transfected in BOSC cells by calcium phosphate coprecipitation. The viral supernatant was harvested 48 h after transfection, filtered, and added to BaF3/BCR-ABL cells. Infected BaF3/BCR-ABL cells were selected by addition of puromycin (1 μg/ml) to the growth medium. After 14 days of selection, cells were expanded and analyzed for FoxO3a expression. Small interfering RNA (siRNA) oligonucleotides were transfected into BV173 cells by nucleofection with the Nucleofector system from Amaxa GmbH (Cologne, Germany). After centrifugation, 10 × 106 cells were resuspended in 100 μl of prewarmed Nucleofector solution kit R containing either a combination of two different siRNA oligonucleotides (2 μg each) against human BCL6 (oligo 1 sense, 5′-GUCGAGACAUCUUGACUGA(dTdT)-3′; oligo 1 antisense, 5′-UCAGUCAAGAUGUCUCGAC(dTdT)-3′; oligo 2 sense, 5′-UGUACACAUCUCGGCUCAA(dTdT)-3′; and oligo 2 antisense, 5′-UUGAGCCGAGAUGUGUACA(dTdT)-3′) or a control oligonucleotide (control oligo sense, 5′-UCAAGCCUCCUCGUGAAGA(dTdT)-3′; control oligo antisense, 5′-UCUUCACGAGGAGGCUUGA(dTdT)-3′) and transferred to the provided electroporation cuvettes. The cells were transfected with the electrical setting T-16. After nucleofection, the cells were transferred immediately into prewarmed complete RPMI 1640 medium, left overnight, and analyzed the day after for BCL6 expression. The siRNA oligonucleotides were purchased from Dharmacon Research Inc. (Lafayette, Colo.).
Cell cycle analysis.
Cell cycle analysis was performed by using propidium iodide staining as described previously (50). Briefly, cells were washed in PBS and then fixed in 90% ethanol. Fixed cells were then washed twice in PBS and stained in 50 μM propidium iodide containing 5 μg of DNase-free RNase/ml for 1 h, analyzed by flow cytometry with a FACScan (BD Biosciences), and analyzed with Cell Quest software (BD Biosciences).
RESULTS
STI571 inhibits cyclin D2 expression and affects FoxO3a phosphorylation in BV173 cells.
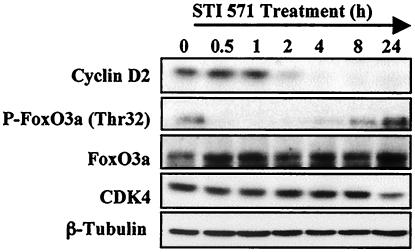
In order to investigate the molecular mechanisms responsible for regulation of cyclin D2 expression, we used the BV173 cell line, a B-lymphoid cell line derived from a patient with Philadelphia chromosome (Ph1)-positive acute leukemia (38). In this and a number of BCR-ABL-expressing cell lines, the oncogenic BCR-ABL tyrosine kinase has been shown to target cyclin D2 expression via the PI3-K/PKB signaling pathway in order to promote cell cycle progression (10, 36). In BV173 cells, expression of cyclin D2 is inhibited by treatment with STI571, a 2-phenylaminopyrimidine derivative that selectively inhibits the tyrosine kinase activity of c-ABL and BCR-ABL (7, 14). STI571 has been shown to block proliferation and induce apoptosis in BCR-ABL-positive cell lines and tumors (7, 8, 14, 16). We treated BV173 cells with STI571 for various times and examined the expression of FoxO3a and cyclin D2. Consistent with previous findings, Western blot analysis showed that STI571 down-regulated the expression of cyclin D2 as early as 2 h after treatment (Fig. 1). The FoxO3a transcription factor was constitutively phosphorylated in cycling BV173 cells, as previously reported for other BCR-ABL-expressing cell lines (24). Expression of other FoxO family members was not detectable. Down-regulation of cyclin D2 expression by STI571 was accompanied by dephosphorylation of FoxO3a 30 min after STI571 addition. The FoxO3a level was also up-regulated after 30 min of exposure to the drug. The expressions of CDK4 and tubulin were used as a loading control and shown to be unchanged. These preliminary results raise the possibility that the transcription factor FoxO3a could be responsible for the down-regulation of cyclin D2 by STI571 treatment.
FIG. 1.
Expression of signaling proteins and cell cycle regulators in BV173 cells after STI571 treatment. BV173 cells were treated for the indicated times with STI571. Cell lysates were prepared at the times indicated, separated on polyacrylamide gels, and immunoblotted with specific antibodies. The expressions of cyclin D2, FoxO3a, cdk4, and tubulin were analyzed by Western blotting.
STI571 treatment of BV173 cells causes a decrease in cyclin D2 mRNA levels.
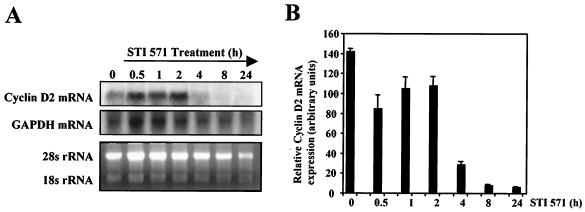
On the basis of these results, we decided to examine whether the effect of STI571 on cyclin D2 protein levels was a consequence of changes in the expression of cyclin D2 at mRNA levels. To this end, BV173 cells were treated with STI571 for different times, and the levels of cyclin D2 mRNA were analyzed by Northern blotting (Fig. 2A) and RT-PCR (Fig. 2B). The levels of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA and 28S rRNA were used as a control for loading. These two experimental approaches showed similar results, a marked decrease in the cyclin D2 mRNA levels 4 h after STI571 addition. This decrease in cyclin D2 mRNA expression was maintained 24 h after treatment, correlating with the pattern of expression of cyclin D2 obtained in the Western blot analysis. These results were in agreement with previous data showing a strong down-regulation of cyclin D2 mRNA upon exposure of BCR-ABL-positive cells to STI571 (10, 36), confirming that BCR-ABL modulates the expression of cyclin D2 predominantly at the transcriptional level.
FIG. 2.
Effect of STI571 treatment on the mRNA levels of cyclin D2 in BV173 cells. (A) BV173 cells were treated with STI571 for the indicated times, and total RNA was extracted and analyzed for expression of cyclin D2. Also shown are the expressions of GAPDH mRNA and 28S rRNA as a control for RNA loading. (B) Expression of cyclin D2 RNA was also analyzed in parallel by real-time PCR and normalized to the level of L19. The results shown are the average of triplicate results from two independent sets of samples.
Inhibition of BCR-ABL affects the transcriptional activity of the human cyclin D2 promoter.
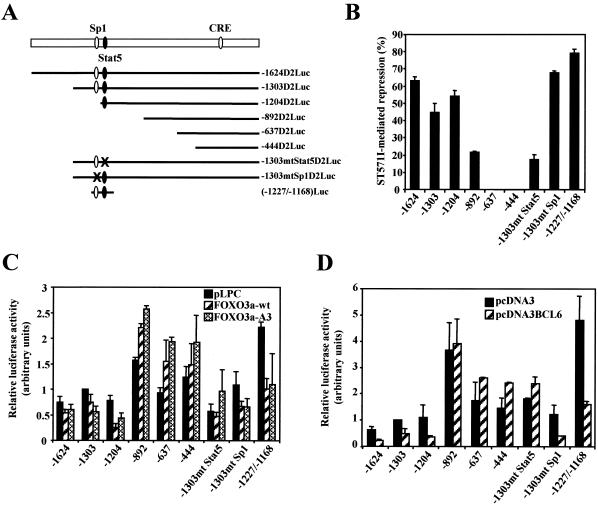
In order to investigate whether the down-regulation of cyclin D2 expression occurs at the gene promoter level, we examined the effect of STI571 on a set of deletion constructs of the human cyclin D2 promoter (referred to as D2Luc) (Fig. 3A) (4, 31). As shown in Fig. 3B, the activities of the −1624D2Luc, −1303D2Luc, and −1204D2Luc deletion mutants were inhibited by 50 to 60% in STI571-treated cells; the −1624D2Luc and −1303D2Luc constructs contain both the Sp1 and the STAT5 binding sites, while the −1204D2Luc construct lacks the Sp1 site. Interestingly, promoter inhibition by STI571 was reduced to 20% in the −892D2Luc construct, which lacks the STAT5 binding site. No effect of STI571 was observed on the activity of the −637D2Luc and −444D2Luc constructs. Mutation of the STAT5 site (AA to CC at nucleotides −1192 and −1191; −1303mtStat5D2Luc) significantly inhibited the inhibitory effect of STI571 on the promoter activity. In contrast, mutation of the Sp1 site (CTCC to AGAA at nucleotides −1217 to −1214; −1303mtSp1D2Luc) slightly increased this inhibition. The activity of the −1227/−1168D2Luc construct containing only the STAT5 and Sp1 binding regions was inhibited by 80% after STI571 treatment. These results highlight the importance of the STAT5 binding site in the STI571-mediated repression of the cyclin D2 promoter.
FIG. 3.
Effect of treatment with STI571 and expression of FoxO3a and BCL6 on cyclin D2 promoter activity in BV173 cells. (A) Schematic representation of the 5′ deletion mutants of the human cyclin D2 promoter used in this study. CRE, cAMP-responsive element. (B) BV173 cells were transiently transfected with pGL3cyclinD2 promoter constructs (10 μg) and incubated the day after with STI571 for 24 h. Cells were harvested and assayed for luciferase activity. Values are corrected for cotransfected Renilla activity and expressed as the percentage of inhibition by STI571 compared with the nontreated conditions. (C and D) BV173 cells were transiently transfected with pGL3cyclinD2 promoter constructs (10 μg) together with 5 μg of pLPC-FoxO3a(wt) or pLPC-FoxO3a(A3) (C), 5 μg of pcDNA3BCL6 (D), or empty vector. Cells were harvested 24 h after transfection and assayed for luciferase activity. Values are corrected for cotransfected Renilla activity, and the relative luciferase activity is standardized to that observed with the empty vector and the full-length promoter. All data shown represent the averages of results from three independent experiments, and the error bars show the standard deviation.
Inhibition of the cyclin D2 promoter by FoxO3a requires the STAT5 binding site.
To examine the role of the FoxO3a transcription factor in the regulation of cyclin D2 expression in our system, we transiently transfected BV173 cells with expression vectors encoding the wild-type FoxO3a (FoxO3a-wt) or a nonphosphorylatable, constitutively active mutant (FoxO3a-A3), together with the different 5′ deletion mutants of the human cyclin D2 promoter. Interestingly, the activities of −1624D2Luc, −1303D2Luc, and −1204D2Luc were partially inhibited by expression of the wild-type and active forms of FoxO3a (Fig. 3C). Deletion or mutation of the promoter region containing the STAT5 consensus site (−892D2Luc and −1303mtStat5D2Luc) blocked the promoter inhibition by FoxO3a, while mutation of the Sp1 site was dispensable for the inhibitory effect. To test whether the Sp1/STAT5 enhancer region is sufficient for gene repression by FoxO3a, we used the −1227/−1168D2Luc construct containing both consensus sites. The results showed that activity of this construct can be inhibited by coexpression of FoxO3a. Together these results suggest that the STAT5 site is required for repression of the cyclin D2 promoter by FoxO3a.
Inhibition of the cyclin D2 promoter by BCL6 requires the STAT5 binding site.
Interestingly, the DNA-binding sequence of the STAT family conforms to the BCL6 consensus binding sequence (11), and the BCL6 transcriptional repressor has been shown to bind with various affinities to different STAT sites (21). In order to investigate the putative role of BCL6 in the regulation of the cyclin D2 promoter, we next performed transient transfection studies of BV173 cells using the various 5′ deletion mutants of the promoter in combination with a BCL6 expression vector. Of all the constructs tested, the activities of −1624D2Luc, −1303D2Luc, and −1204D2Luc were markedly inhibited by coexpression of BCL6, while −892D2Luc and subsequent deletions of the promoter were not affected by the transcriptional repressor (Fig. 3D). Interestingly, mutation of the STAT5 site in the −1303mtStat5D2Luc construct reversed inhibition by BCL6, while mutation of the Sp1 site had no effect. Activity of the −1227/−1168D2Luc construct containing both STAT5 and Sp1 sites was down-regulated by BCL6. These results suggest the involvement of BCL6 in the transcriptional repression of cyclin D2 and raise the possibility that BCL6 might exert its action through the STAT5 consensus binding site.
STI571 treatment and expression of FoxO3a activates expression of BCL6 in BV173 cells.
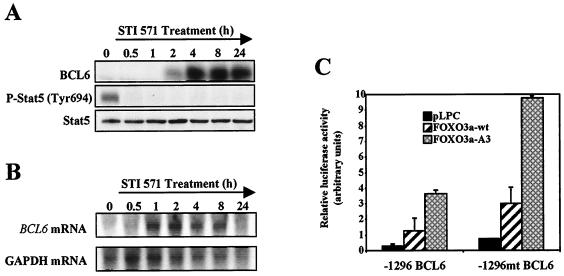
The deletion and cotransfection studies of the cyclin D2 promoter suggest that the STAT5/BCL6 site is important for both STI571- and FoxO3a-mediated repression of cyclin D2 expression. To further investigate this line of evidence, we next studied the effects of STI571 treatment on BCL6 and STAT5 expression in BV173 cells. To this end, BV173 cells were treated with STI571 for various times, and the expression of these molecules was examined (Fig. 4A). Western blot studies revealed a very significant up-regulation in the levels of BCL6 that was maintained over the time course. The kinetics of BCL6 induction were very similar to those previously observed for cyclin D2 inhibition. This result was accompanied by dephosphorylation of STAT5 30 min after STI571 treatment, while STAT5 levels were not affected. These results suggest a role for STAT5 and BCL6 transcription factors in the regulation of cyclin D2 by BCR-ABL.
FIG. 4.
Expression of BCL6 and STAT5 in BV173 cells after STI571 treatment and effect of FoxO3a expression on BCL6 promoter activity. (A) BV173 cells were treated for the indicated times with STI571. Cell lysates were prepared at the indicated times, separated on polyacrylamide gels, and subjected to immunoblotting with specific antibodies for BCL6 and STAT5. (B) BV173 cells were treated with STI571 for the times indicated, and total RNA was extracted and analyzed for expression of BCL6. Also shown is the expression of GAPDH as a control for RNA loading and blotting. (C) BV173 cells were transiently transfected with pGL3.1296 or pGL3.1296.mutBCL6 promoter reporter constructs (10 μg) along with pLPC-FoxO3a(wt), pLPC-FoxO3a(A3), or the empty vector (5 μg). Cells were harvested 24 h after transfection and assayed for luciferase activity. Values are corrected for cotransfected Renilla activity and expressed as arbitrary units. The data shown represent the averages of results from two independent experiments, and the error bars show the standard deviation.
We then examined whether the STI571-mediated induction of BCL6 protein levels was a consequence of an increase in the levels of BCL6 mRNA. BV173 cells were treated for different times with STI571, total RNA was extracted, and the levels of BCL6 mRNA were analyzed by Northern blotting (Fig. 4B). The result obtained showed an increase in the level of BCL6 mRNA after STI571 treatment. This increase was apparent 1 h after treatment and was maintained for 8 h. By 24 h the mRNA levels declined, which could be due to the existence of a consensus BCL6 binding site in the BCL6 gene promoter that may be acting as a negative autoregulatory loop.
Since the human cyclin D2 promoter does not contain any obvious FoxO binding sites, transcriptional regulation of cyclin D2 by this transcription factor can be indirectly mediated through FoxO3a-mediated induction of a transcriptional repressor protein. One plausible candidate is BCL6, as this potent transcriptional repressor has been shown recently to be induced by FoxO4 (48). In order to determine whether in our system BCL6 expression can be up-regulated by FoxO3a activity, we transiently transfected BV173 cells with −1296BCL6pGL3, a reporter construct containing 1,296 nucleotides upstream of the TATA box of the BCL6 promoter (48), in combination with either the empty vector, the FoxO3a-wt, or the constitutively active mutant FoxO3a-A3. As shown in Fig. 4C, the activity of the BCL6 promoter was up-regulated by coexpression of FoxO3a, especially when the active form, FoxO3a-A3, was used. This result supports the idea that FoxO3a may be affecting cyclin D2 expression through an indirect mechanism that involves induction of the BCL6 trancriptional repressor. The BCL6 binding site in the BCL6 promoter might negatively regulate FoxO-induced BCL6 expression, as reported by Tang et al. (48). This was confirmed by cotransfection experiments using the −1296mtBCL6pGL3 construct that contains a mutation in the BCL6 binding site of the BCL6 promoter. Mutation of this site significantly enhances the response of the promoter to transfected FoxO3a (Fig. 4C). This negative feedback mechanism may account for the down-regulation of BCL6 mRNA after the initial rapid accumulation following STI571 treatment.
STI571 treatment affects nuclear protein binding to the STAT5 site of the cyclin D2 promoter.
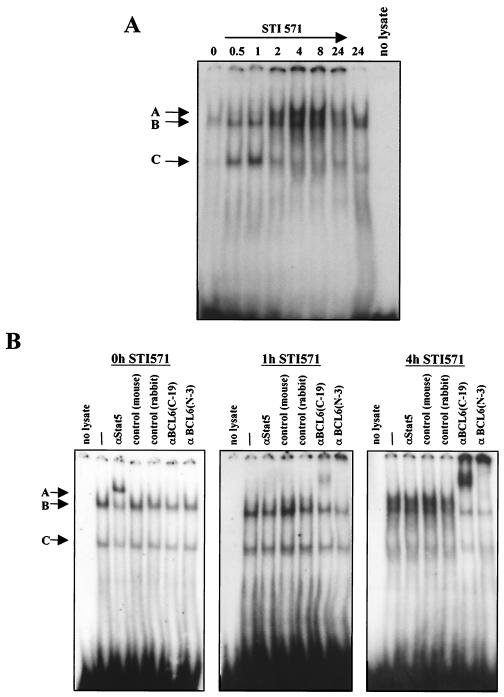
All of the results obtained from transfection studies support the interpretation that the STI571-mediated inhibition of the cyclin D2 promoter can take place through the combined action of FoxO3a, STAT5, and BCL6 on the STAT5 consensus binding site of the promoter. It was important, then, to characterize the functional complexes that bind to this site in CML cells before and after STI571 treatment. For this purpose, we performed mobility shift analyses on extracts derived from BV173 cells treated with STI571. The oligonucleotide probe used in these experiments contained the consensus STAT5 binding region of the cyclin D2 promoter. The mobility shift experiment (Fig. 5A) identified at least three species of DNA-binding complexes (complexes A to C) by virtue of the differences in their mobilities. Of these, two (B and C) were present in nontreated cells; upon STI571 treatment, the levels of both complexes increased, and a third low-migrating complex (complex A) that peaked 4 h after addition of the drug appeared. In order to identify the components of these STAT5 site-binding complexes, we performed antibody supershift experiments on extracts corresponding to BV173 cells treated with STI571 for 0, 1, and 4 h, when apparent changes in mobility shift complexes occur. As shown in Fig. 5B, in untreated cells (0 h) addition of STAT5 antibody partially supershifted both complex B and complex C, while the BCL6 antibodies failed to shift these complexes at all. As the cells were treated with STI571 for 1 h, STAT5 antibody had no effect on any of the three complexes observed (A to C), while two different BCL6 antibodies raised against different regions of the protein shifted (C-19) or disrupted (N-3) the slower-migrating complex (complex A). When cells were treated with STI571 for 4 h, complex A was almost completely shifted by the BCL6 antibodies, and no effect was observed for the STAT5 antibody. These results led us to conclude that in BV173 cells STAT5 is binding to the STAT5 consensus site in the cyclin D2 promoter and that it is replaced by the BCL6 transcriptional repressor when BCR-ABL activity is inhibited by STI571.
FIG. 5.
Electrophoretic mobility shift analysis of STAT5 DNA-binding complexes after STI571 treatment of BV173 cells. (A) DNA-binding protein extracts were prepared from BV173 cells at the indicated times after STI571 treatment. The extracts were then used for gel mobility shift experiments with a 32P-labeled oligonucleotide probe containing the STAT5 binding site in the human cyclin D2 promoter. (B) Antibody supershift analysis of components of STAT5 complexes at different times after STI571 treatment. Supershifts were performed using specific antibodies against STAT5 and BCL6, as indicated, on whole-cell extracts prepared from cells at 0, 1, and 4 h after STI571 treatment.
STI571 treatment affects STAT5/BCL6 binding to the cyclin D2 promoter and promotes FoxO binding to the BCL6 promoter.
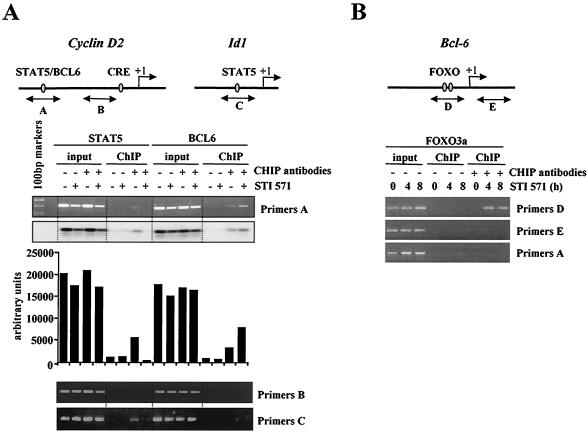
In order to confirm that STI571 treatment affects STAT5, BCL6, and FoxO binding in vivo, we adopted a different approach, to study whether STI571 treatment influences occupation of the STAT5/BCL6 site of the cyclin D2 promoter by ChIP assay. BV173 cells were treated with STI571, protein-DNA complexes were formaldehyde cross-linked, and sites bound by STAT5 or BCL6 were immunoprecipitated with the appropriate antibodies. PCR primer pairs were designed to selectively detect the STAT5/BCL6 site in the human cyclin D2 gene (A primers). We found that the cyclin D2 promoter bound predominantly to STAT5 complexes and to a lesser extent to BCL6 in nontreated cells (Fig. 6A). Interestingly, STI571 treatment completely abolished STAT5 association with the cyclin D2 promoter, while binding of BCL6 increased. The relative levels of STAT5 and BCL6 binding to the cyclin D2 promoter detected in the ChIP assays therefore correlated with the levels associated with STAT5 complexes detected by electrophoretic mobility shift assays (Fig. 5B). To ensure proper DNA shearing, we used primers detecting an unrelated region of the cyclin D2 promoter (B primers, about 0.7 kbp downstream of the STAT5/BCL6 site). As shown in Fig. 6, the PCR results showed no specific chromatin precipitation with either the anti-STAT5 or the anti-BCL6 antibodies. We also performed a positive control using the promoter region of the Id1 gene, which consists of a STAT5 but not a BCL6 consensus site (C primers) (Fig. 6A). As expected, we detected STI571-sensitive binding of STAT5 but failed to observe BCL6 binding to the STAT5 site of the Id1 promoter. These experiments therefore demonstrate that BCR-ABL activity has an important role in promoting binding of active STAT5 complexes to the STAT5 site of the cyclin D2 promoter and that upon STI571 treatment the STAT5 transcriptional activator is replaced by the BCL6 transcriptional repressor. It has been suggested that members of the FoxO family can interact with promoters through direct or indirect binding to promoter elements others than their consensus sequences (39). We have shown the STAT5/BCL6 site to be responsible for STI571- and FoxO3a-induced repression of the cyclin D2 promoter. In order to analyze for the presence of FoxO3a binding to the STAT5/BCL6 site of the cyclin D2 promoter, we performed new ChIP assays using a FoxO3a-specific antibody. As shown in Fig. 6B, no binding of FoxO3a was observed either in the absence or in the presence of STI571 (A primers), reinforcing the hypothesis that FoxO3a-mediated regulation of cyclin D2 expression is via an indirect mechanism.
FIG. 6.
ChIP assay of BV173 cells after STI571 treatment. (A) BV173 cells were treated with STI571 for 4 h. Protein-DNA complexes were formaldehyde cross-linked in vivo. Chromatin fragments from these cells were subjected to immunoprecipitation with antibodies to STAT5 or BCL6 as indicated. After cross-link reversal, the coimmunoprecipitated DNA was amplified by PCR using the A (−1446/−1149), B (−713/−499), and C (−548/−323) primers and resolved in 2% agarose gel. The PCR products obtained with A primers were also detected by Southern blotting, and shown is the quantitation by phosphorimaging of the results obtained. (B) BV173 cells were treated with STI571 for 4 or 8 h. Protein-DNA complexes were formaldehyde cross-linked in vivo. Chromatin fragments from these cells were subjected to immunoprecipitation with a specific antibody to FoxO3a. After cross-link reversal, the coimmunoprecipitated DNA was amplified by PCR using the A (−1446/−1149), D (−839/−501), and E (+38/+349) primers and resolved in 2% agarose gel.
As a plausible mechanism of regulation of cyclin D2 by FoxO3a is through induction of BCL6, we sought to investigate the in vivo occupancy of the FoxO3a consensus binding sites of the BCL6 promoter in STI571-treated cells. The human BCL6 promoter contains a number of potential FoxO binding sites, of which a few have been shown to bind avidly to FoxO (48). We designed PCR primer pairs to selectively detect two of the major FoxO target sites (D primers). The ChIP analysis results revealed association of FoxO3a with the BCL6 promoter upon STI571 treatment, while no binding was detected in the untreated condition (Fig. 6B). As a control for DNA shearing we used primers detecting an unrelated region of the BCL6 promoter (E primers, 1 kbp downstream of the FoxO sites) that showed no specific chromatin precipitation with the FoxO3a antibody. These results confirm that BCL6 is a target of FoxO3a in STI571-treated cells and provide further evidence for the involvement of BCL6 in the indirect mechanism of regulation of cyclin D2 by FoxO3a. This finding also provides a positive control for the earlier FoxO3a ChIP experiment (Fig. 6A).
Induction of FoxO3a affects cyclin D2 and BCL6 expression.
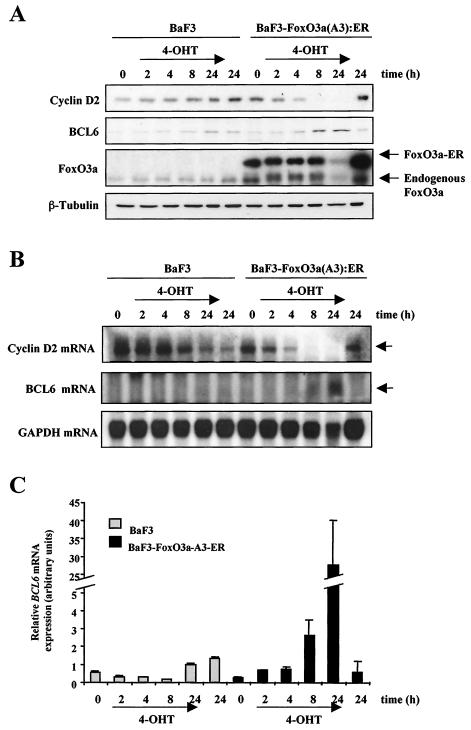
To further confirm the results obtained for BV173 cells, we used a murine BaF3 cell line expressing a 4-OHT-inducible FoxO3a-A3 construct, BaF3 · FoxO3a(A3):ER (13), to specifically analyze the consequence of FoxO3a activation in more detail. BaF3 · FoxO3a(A3):ER cells were grown in parallel with the parental cell line, BaF3, cells were treated for the indicated times with 4-OHT, and the expression of cyclin D2 and BCL6 was examined. As shown in Fig. 7A, Western blot analysis revealed down-regulation of cyclin D2 expression when FoxO3a activity was induced by treatment of BaF3 · FoxO3a(A3):ER cells with 4-OHT. This effect was obvious 4 h after addition of the drug, and by 8 h expression of cyclin D2 was completely abolished. Also, cells treated with 4-OHT for 24 h did not express cyclin D2, while the nontreated cells showed high levels of expression after 24 h of culture with growth medium alone. In the parental BaF3 cell line, addition of 4-OHT had no effect on the expression of cyclin D2 in the examined conditions. When BCL6 expression was analyzed, the results obtained showed up-regulation of protein levels in BaF3 · FoxO3a(A3):ER cells 8 h after addition of 4-OHT. BCL6 was undetectable in BaF3 cells for all conditions examined. Expression of FoxO3a-ER in the BaF3 · FoxO3a(A3):ER cell line was confirmed by Western blotting using a FoxO3a antibody. The level of expression of FoxO3a was constant in the BaF3 cell line, while in BaF3 · FoxO3a(A3):ER cells FoxO3a expression was low after treatment with 4-OHT for 24 h. This result can be due to apoptosis caused by prolonged FoxO3a activation. It is notable that down-regulation of cyclin D2 occurs as soon as 2 to 4 h following 4-OHT treatment, before any detectable signs of apoptosis (fluorescence-activated cell sorting data not shown). This result indicates that the down-regulation of cyclin D2 is independent of the apoptotic effects induced by FoxO3a expression. Tubulin levels were analyzed to confirm equal loading.
FIG. 7.
Effect of 4-OHT treatment on BaF3 and BaF3 · FoxO3a(A3):ER cells. BaF3 and BaF3 · FoxO3a(A3):ER cells were treated with 4-OHT for the indicated times. (A) Cell lysates were prepared at the times indicated, separated on polyacrylamide gels, and subjected to immunoblotting with specific antibodies. The expressions of cyclin D2, BCL6, FoxO3a, and tubulin were analyzed by Western blotting. (B) Total RNA was extracted and analyzed for expression of cyclin D2 and BCL6. Also shown is the expression of GAPDH mRNA as a control for RNA loading and blotting. (C) The expression of BCL6 mRNA was also analyzed in parallel by real-time PCR and normalized to the level of L19 RNA. The results shown are averages of triplicate results from two independent sets of samples.
Furthermore, we wanted to investigate whether induction of FoxO3a in BaF3 · FoxO3a(A3):ER cells was affecting the mRNA levels of cyclin D2 and BCL6. In accordance with results from Western blotting, the results obtained in the Northern blot analysis showed that treatment of BaF3 · FoxO3a(A3):ER cells with 4-OHT resulted in a dramatic down-regulation of cyclin D2 mRNA within 2 to 4 h (Fig. 7B) and an up-regulation of BCL6 mRNA after 4-OHT addition compared to the control parental BaF3 cell line. This finding provides compelling evidence for regulation of cyclin D2 and BCL6 by FoxO3a in vivo.
To further confirm regulation of BCL6 mRNA by FoxO3a in this cell system, we performed RT-PCR analysis on BaF3 and BaF3 · FoxO3a(A3):ER cells treated with 4-OHT (Fig. 7C). These results showed that BCL6 mRNA levels are induced by 4-OHT addition in BaF3 · FoxO3a(A3):ER cells.
Silencing of FoxO3a alters the effects of STI571 on cyclin D2 and BCL6 in BCR-ABL-expressing cells.
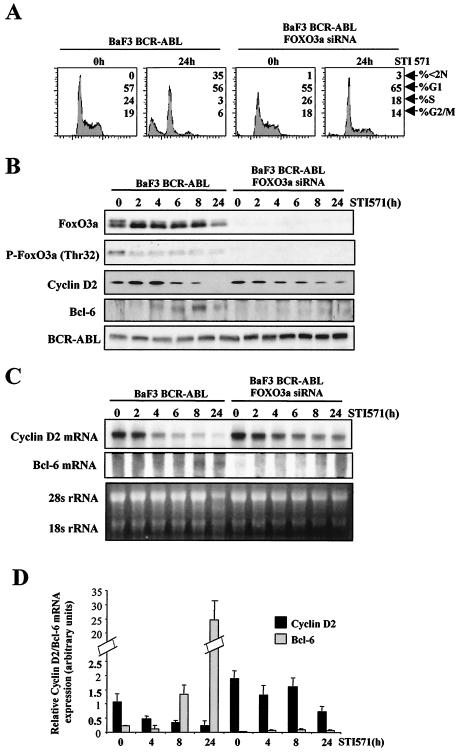
To confirm the role of FoxO3a in BCR-ABL signaling we used the pSUPER system to stably suppress the expression of FoxO3a. The BV173 cell line proved to be resistant to efficient transfection for production of stable permanent transfectants. To circumvent this problem, we used BaF3 cells expressing BCR-ABL (BaF3/BCR-ABL), a cell model system commonly used to study BCR-ABL signaling. We have already shown that BaF3/BCR-ABL cells are sensitive to STI571 and that STI571 represses the expression of cyclin D2 (36). BaF3/BCR-ABL cells were transduced with the pSUPER-FoxO3a construct, and puromycin-resistant cells were expanded and analyzed for FoxO3a expression. As shown in Fig. 8B, the resistant cells showed a complete silencing of FoxO3a expression. Flow cytometry analysis of cells revealed that STI571 treatment of BaF3/BCR-ABL cells for 24 h induced cell cycle arrest and apoptosis (Fig. 8A). In sharp contrast, the cell cycle arrest and apoptosis were inhibited in cells expressing the FOXO3a RNA interference (RNAi) vector, highlighting the biological relevance of FoxO3a in BCR-ABL signaling. In order to assess the molecular effects of FoxO3a silencing, cells were treated with STI571 for various times and analyzed for the expression of cyclin D2 and BCL6. As seen in Fig. 8B, BaF3/BCR-ABL cells showed inhibition of expression of cyclin D2 after STI571 treatment. Interestingly, the effect of STI571 on cyclin D2 was abolished by depletion of FoxO3a, particularly after 24 h, which confirms FoxO3a as an important mediator of BCR-ABL signaling to cyclin D2. Expression of BCL6 was induced in BaF3/BCR-ABL cells after 4 to 6 h of exposure to STI571, while in the FoxO3a-negative cells, BCL6 expression was undetectable for the times studied. Remarkably, in BaF3/BCR-ABL cells STI571 treatment was accompanied by dephosphorylation of FoxO3a together with up-regulation of the total FoxO3a level, which correlates with our observations of BV173 cells (Fig. 1). Expression of BCR-ABL in both cell lines was confirmed by Western blotting. To confirm that the observed effects on cyclin D2 and BCL6 were due to changes in their mRNA levels, we next performed Northern blotting and RT-PCR analyses of the two BaF3/BCR-ABL cell lines. The results obtained with both experimental approaches (Fig. 8C and D) showed that knockdown of FoxO3a expression in BaF3/BCR-ABL cells abolished the inhibitory effects of STI571 on cyclin D2 mRNA levels. Analysis of BCL6 mRNA levels showed that BCL6 was induced in BaF3/BCR-ABL cells upon STI571 treatment, while in the absence of FoxO3a the levels of BCL6 were undetectable. These results confirm cyclin D2 and BCL6 as transcriptional targets of FoxO3a in vivo.
FIG. 8.
Effect of FoxO3a gene silencing on the effects of STI571 treatment of BaF3/BCR-ABL cells. The pSUPER vector coding for FoxO3a RNAi sequence was introduced into BaF3/BCR-ABL cells in order to silence FoxO3a expression. BaF3/BCR-ABL and BaF3/BCR-ABL FoxO3a-negative cells were treated in parallel with STI571 for the indicated times. (A) Cell cultures were fixed in ethanol, and DNA content was analyzed by flow cytometry following propidium iodide staining. The percentage of cells in each phase of the cell cycle (<2N, G1, S, and G2/M) is indicated. (B) Cell lysates were prepared at the times indicated, separated on polyacrylamide gels, and subjected to immunoblotting with specific antibodies. The expressions of FoxO3a, cyclin D2, BCL6, and BCR-ABL were analyzed by Western blotting. (C) Total RNA was extracted and analyzed for expression of cyclin D2 and BCL6. Also shown is the expression of 28S rRNA as a control for RNA loading and blotting. (D) Expressions of cyclin D2 and BCL6 mRNA were analyzed in parallel by real-time PCR and normalized to the level of L19 RNA. The results shown are the averages of triplicate results from two independent sets of samples.
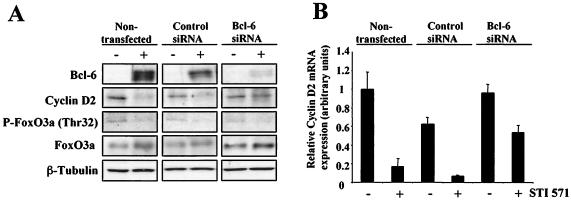
Silencing of BCL6 suppresses repression of cyclin D2 by STI571 in BCR-ABL-expressing cells.
To assess further the role of BCL6 in the observed down-regulation of cyclin D2 upon STI571 treatment, we performed gene silencing experiments on BCL6 with BV173 cells. In these experiments, cells were transfected with siRNAs specific for BCL6 and then treated with STI571. A control group consisting of cells transfected with an irrelevant siRNA was also included. Figure 9A shows the result of Western blotting experiments that clearly show an induction of BCL6 expression after nontransfected and control cells were treated with STI571. It is also clear that transfection with the BCL6-specific siRNAs reduced the expression of BCL6 in STI571-treated cells to almost undetectable levels. Importantly, knockdown of BCL6 induction by siRNA transfection resulted in the efficient inhibition of the STI571-mediated repression of cyclin D2. We also analyzed the phosphorylation status and total levels of FoxO3a in these cells, and the results obtained correlate with those shown in Fig. 1. The results also indicate that despite activation of FoxO3a by STI571, silencing of BCL6 gene expression can inhibit the effects of FoxO3a on cyclin D2. Tubulin levels were analyzed to assess equal loading.
FIG. 9.
Effect of BCL6 gene silencing on STI571 treatment of BV173 cells. BV173 cells were transfected by nucleofection with either a control oligonucleotide or a combination of two different BCL6 siRNAs and then treated with STI571 for 4 h. (A) Cell lysates were prepared, separated on polyacrylamide gels, and subjected to immunoblotting with specific antibodies. The expressions of BCL6, cyclin D2, FoxO3a, and tubulin were analyzed by Western blotting. (B) The expression of cyclin D2 RNA was analyzed in parallel by real-time PCR and normalized to the level of L19.
To confirm the Western blotting results, the mRNA levels of cyclin D2 were analyzed by RT-PCR from parallel samples. The data from Fig. 9B clearly show that transfection of BV173 cells with siRNAs specific for BCL6 blocked the reduction in cyclin D2 mRNA levels caused by STI571 treatment. Together, these results define BCL6 as a main mediator in BCR-ABL signaling to cyclin D2 and provide a better understanding of the network of regulation between FoxO3a, BCL6, and cyclin D2.
DISCUSSION
Previous studies have demonstrated that proliferative signals emanating from antigen receptor, growth factor and/or cytokine receptors, and oncogenic tyrosine kinases activate cyclin D2 transcription to mediate cell cycle progression in a number of hemopoietic cells, in particular those of the B-lymphocyte lineage. In the majority of these cell systems, the PI3-K/PKB signaling pathway has been implicated in the induction of cyclin D2 transcription. Studies of cyclin D2-deficient mice have demonstrated that cyclin D2 has a crucial role in B-cell proliferation and development (2, 18, 33). It has also been shown that B cells deficient in p85α, the regulatory subunit of classical PI3-K, fail to express cyclin D2 in response to B-cell receptor (BCR) stimulation (19), lending further support to the notion that cyclin D2 is an important downstream target of the PI3-K/PKB signaling pathway. Despite this, the molecular mechanism by which the PI3-K/PKB signaling cascade regulates cyclin D2 transcription remains undefined.
The FoxO subfamily of transcription factors is the most recently characterized and least studied component of the PI3-K/PKB signaling cascade. FoxO transcription factors are direct targets of PKB/Akt, a serine/threonine kinase located downstream of the PI3-K survival pathway. Recently, the FoxO proteins have been identified as important regulators of cell cycle regulators, including cyclin D (42), p27KIP1 (13), and p130 (26). We have discovered that the negative cell cycle regulators p27KIP1 and p130 are direct transcriptional targets of FoxO proteins. However, unlike p27KIP1 and p130, cyclin D1 and cyclin D2 promoters do not have putative FoxO binding sites and are not the direct transcriptional targets of FoxO proteins. This observation therefore implies the existence of an intermediate transcription factor that links FoxO with cyclin D expression.
In a BCR-ABL-transformed mouse pro-B BaF3 cell line, as well as a number of CML cell lines, it has been demonstrated that the anti-BCR-ABL tyrosine kinase drug STI571 represses cyclin D2 transcription through down-regulating PI3-K and PKB activities (10, 36). Here we demonstrate that STI571 induces the accumulation of the unphosphorylated forms of the FoxO3a transcription factor. This result is in agreement with recent reports showing that STI571 targets FoxO3a to induce cell cycle arrest and apoptosis in BCR-ABL-transformed cell lines (17, 24). Western and Northern blot analyses indicate that the induction of FoxO3a activity by STI571 is associated with down-regulation of cyclin D2 expression at both translational and transcriptional levels. It is also notable that in this case, the increase in FoxO3a expression as well as protein dephosphorylation contributes to the induction of FoxO3a activity in response to STI571 in BV173 cells. This induction of FOXO3a expression is not specific to the BV173 cell line only, as a similar up-regulation of FOXO3a expression is also detectable in the BaF3/BCR-ABL cells following STI571 treatment. However, the mechanism for FoxO3a induction has yet to be defined.
The fact that there is no consensus FoxO binding site in the cyclin D2 promoter suggests that FoxO3a does not regulate cyclin D2 transcription directly and an intermediate transcription factor(s) may mediate its effect. Our deletion studies of the cyclin D2 promoter have located the STI571 and FoxO3a responsive region to a putative binding site for the transcription factors STAT5 and BCL6, indicating that STAT5 and/or BCL6 could be mediating the transcription repression by FoxO3a. The Jak-STAT signaling pathway is employed by a wide range of cytokines and growth factors to mediate their signals. We confirm here that cyclin D2 is a target of STAT5, a notion in line with a previous study showing that STAT5 mediates the expression of cyclin D2 in response to interleukin-2 in the murine CD8+-T-cell line CTLL-2 (31). In cycling BV173 cells, we detected STAT5 phosphorylation by using a phospho-specific antibody, indicating that STAT5 is in its active gene-transactivating form. This result is consistent with that from previous studies demonstrating that the BCR-ABL tyrosine kinase maintains STAT5 in a constitutively active phosphorylated state (22). This finding is supported by mobility and antibody supershift experiments with nuclear extracts showing that this form of STAT5 was binding to the putative STAT5/BCL6 site of the cyclin D2 promoter in cycling BV173 cells. Upon STI571 treatment, the BV173 cells underwent cell cycle arrest at the G1 phase of the cell cycle (reference 10 and data not shown), and this was accompanied by dephosphorylation of STAT5 and its dissociation from the STAT5/BCL6 site. BCL6 expression is undetectable in cycling BV173, but its expression is induced by STI571 treatment. This increase in BCL6 expression is associated with the replacement of STAT5 by BCL6 at the STAT5/BCL6 site.
BCL6 is a transcription repressor normally expressed by germinal center B cells but not by naïve B cells or plasma cells. Gene deletion studies have shown that BCL6 has a role in maintaining lymphocytes in an undifferentiated state by preventing the expression of genes required for B-cell activation and terminal differentiation. A recent study showed that FoxO4 (AFX) can activate transcription of BCL6 to induce apoptosis (48). This observation therefore implies the possibility that FoxO3a can repress cyclin D2 transcription indirectly through inducing the expression of the transcriptional repressor BCL6. This hypothesis is validated by our finding that FoxO3a can transactivate the BCL6 promoter in BV173 cells and the observation that the STI571-induced accumulation of active FoxO3a is associated with the up-regulation of BCL6 and down-regulation of cyclin D2 expression in BV173 cells. We further show that FoxO3a protein is recruited to the endogenous BCL6 promoter in BV173 cells in response to STI571 treatment. Moreover, using transfection studies, we have identified cyclin D2 as a gene whose transcription is repressed by BCL6, a notion supported by a previous DNA microarray study identifying cyclin D2, among other genes, as a direct target of the transcriptional repressor BCL6 (43). We further demonstrated that the BCL6 protein binds directly to the promoter region of the cyclin D2 gene in vitro by mobility shift assays as well as in vivo by ChIP analysis.
Significantly, our hypothesis is further upheld by the finding that induction of FoxO3a activity by tamoxifen in the murine BaF3 · FoxO3a(A3):ER cell line causes the up-regulation of BCL6 and down-regulation of cyclin D2 at the transcriptional level in vivo. Furthermore, in BCR-ABL-expressing BaF3 cells, knockdown of FoxO3a expression by the pSUPER system abolishes the STI571-mediated repression of cyclin D2 and induction of BCL6 expression at both protein and mRNA levels. Also, the cell cycle arrest and apoptosis induced by STI571 in BaF3/BCR-ABL cells are inhibited by depletion of FoxO3a, underscoring the importance of FoxO3a in BCR-ABL signaling. In addition, gene silencing of BCL6 in BV173 cells blocks cyclin D2 repression by active FoxO3a following STI571 treatment, further demonstrating the role of BCL6 as a mediator of BCR-ABL and FoxO3a signaling to cyclin D2. These results clearly confirm FoxO3a and BCL6 as key regulators of cyclin D2 and define BCL6 as the molecule responsible for cyclin D2 repression by FoxO3a. This is, to our knowledge, the first report to establish a causal relationship between BCR-ABL activity, FoxO3a, and BCL6 and cyclin D2 expression.
In conclusion, the present report establishes the molecular mechanism by which FoxO protein regulates cyclin D2 expression. We describe a mechanism by which FoxO3a induces the expression of the transcriptional repressor BCL6, culminating in the down-regulation of cyclin D2 transcription. Upon STI571 treatment, a change that can explain the down-regulation of cyclin D2 transcription occurs in occupancy of the STAT5/BCL6 site within the cyclin D2 gene. This site is crucial for cyclin D2 regulation, as mutation of this site renders it refractory to the repression by STI571, FoxO3a, and BCL6. This STAT5/BCL6 site is conserved in the human, mouse, and rat cyclin D2 genes (4, 23, 51), suggesting that the mechanism uncovered in the present study is not unique to the human system but a common means by which cyclin D2 transcription is regulated. This is consistent with a previous report showing that cyclin D2 is a direct transcriptional target of BCL6 in B-lineage cells (43). At the same time, we also elucidate the signaling events whereby the BCR-ABL signals are relayed to the cell cycle regulator cyclin D2 to mediate cell cycle progression. BCR-ABL-induced loss of FoxO3a function may be involved in oncogenic transformation of CML, partially via the up-regulation of cyclin D2 protein.
Acknowledgments
This research was supported by grants from the Leukemia Research Fund-UK and Cancer Research-UK. I.S. is supported by a Ph.D. studentship from Fundação para a Ciência e a Tecnologia, Lisbon, Portugal, and A.E. is supported by a grant from the Algerian government.
REFERENCES
- 1.Andrews, N. C., and D. V. Faller. 1991. A rapid micropreparation technique for extraction of DNA-binding proteins from limiting numbers of mammalian cells. Nucleic Acids Res. 19:2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Banerji, L., J. Glassford, N. C. Lea, N. S. Thomas, G. G. Klaus, and E. W.-F. Lam. 2001. BCR signals target p27(Kip1) and cyclin D2 via the PI3-K signalling pathway to mediate cell cycle arrest and apoptosis of WEHI 231 B cells. Oncogene 20:7352-7367. [DOI] [PubMed] [Google Scholar]
- 3.Braunstein, M., A. B. Rose, S. G. Holmes, C. D. Allis, and J. R. Broach. 1993. Transcriptional silencing in yeast is associated with reduced nucleosome acetylation. Genes Dev. 7:592-604. [DOI] [PubMed] [Google Scholar]
- 4.Brooks, A. R., D. Shiffman, C. S. Chan, E. E. Brooks, and P. G. Milner. 1996. Functional analysis of the human cyclin D2 and cyclin D3 promoters. J. Biol. Chem. 271:9090-9099. [DOI] [PubMed] [Google Scholar]
- 5.Brummelkamp, T. R., R. Bernards, and R. Agami. 2002. A system for stable expression of short interfering RNAs in mammalian cells. Science 296:550-553. [DOI] [PubMed] [Google Scholar]
- 6.Brunet, A., A. Bonni, M. J. Zigmond, M. Z. Lin, P. Juo, L. S. Hu, M. J. Anderson, K. C. Arden, J. Blenis, and M. E. Greenberg. 1999. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell 96:857-868. [DOI] [PubMed] [Google Scholar]
- 7.Buchdunger, E., J. Zimmermann, H. Mett, T. Meyer, M. Muller, B. J. Druker, and N. B. Lydon. 1996. Inhibition of the Abl protein-tyrosine kinase in vitro and in vivo by a 2-phenylaminopyrimidine derivative. Cancer Res. 56:100-104. [PubMed] [Google Scholar]
- 8.Deininger, M. W., J. M. Goldman, N. Lydon, and J. V. Melo. 1997. The tyrosine kinase inhibitor CGP57148B selectively inhibits the growth of BCR-ABL-positive cells. Blood 90:3691-3698. [PubMed] [Google Scholar]
- 9.Deininger, M. W., J. M. Goldman, and J. V. Melo. 2000. The molecular biology of chronic myeloid leukemia. Blood 96:3343-3356. [PubMed] [Google Scholar]
- 10.Deininger, M. W., S. A. Vieira, Y. Parada, L. Banerji, E. W.-F. Lam, G. Peters, F. X. Mahon, T. Kohler, J. M. Goldman, and J. V. Melo. 2001. Direct relation between BCR-ABL tyrosine kinase activity and cyclin D2 expression in lymphoblasts. Cancer Res. 61:8005-8013. [PubMed] [Google Scholar]
- 11.Dent, A. L., A. L. Shaffer, X. Yu, D. Allman, and L. M. Staudt. 1997. Control of inflammation, cytokine expression, and germinal center formation by BCL-6. Science 276:589-592. [DOI] [PubMed] [Google Scholar]
- 12.de Stanchina, E., M. E. McCurrach, F. Zindy, S. Y. Shieh, G. Ferbeyre, A. V. Samuelson, C. Prives, M. F. Roussel, C. J. Sherr, and S. W. Lowe. 1998. E1A signaling to p53 involves the p19(ARF) tumor suppressor. Genes Dev. 12:2434-2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dijkers, P. F., R. H. Medema, C. Pals, L. Banerji, N. S. Thomas, E. W. Lam, B. M. Burgering, J. A. Raaijmakers, J. W. Lammers, L. Koenderman, and P. J. Coffer. 2000. Forkhead transcription factor FKHR-L1 modulates cytokine-dependent transcriptional regulation of p27(KIP1). Mol. Cell. Biol. 20:9138-9148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Druker, B. J., S. Tamura, E. Buchdunger, S. Ohno, G. M. Segal, S. Fanning, J. Zimmermann, and N. B. Lydon. 1996. Effects of a selective inhibitor of the Abl tyrosine kinase on the growth of Bcr-Abl positive cells. Nat. Med. 2:561-566. [DOI] [PubMed] [Google Scholar]
- 15.Dyson, N. 1998. The regulation of E2F by pRB-family proteins. Genes Dev. 12:2245-2262. [DOI] [PubMed] [Google Scholar]
- 16.Gambacorti-Passerini, C., P. le Coutre, L. Mologni, M. Fanelli, C. Bertazzoli, E. Marchesi, M. Di Nicola, A. Biondi, G. M. Corneo, D. Belotti, E. Pogliani, and N. B. Lydon. 1997. Inhibition of the ABL kinase activity blocks the proliferation of BCR/ABL+ leukemic cells and induces apoptosis. Blood Cells Mol. Dis. 23:380-394. [DOI] [PubMed] [Google Scholar]
- 17.Ghaffari, S., Z. Jagani, C. Kitidis, H. F. Lodish, and R. Khosravi-Far. 2003. Cytokines and BCR-ABL mediate suppression of TRAIL-induced apoptosis through inhibition of forkhead FOXO3a transcription factor. Proc. Natl. Acad. Sci. USA 100:6523-6528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Glassford, J., M. Holman, L. Banerji, E. Clayton, G. G. Klaus, M. Turner, and E. W.-F. Lam. 2001. Vav is required for cyclin D2 induction and proliferation of mouse B lymphocytes activated via the antigen receptor. J. Biol. Chem. 276:41040-41048. [DOI] [PubMed] [Google Scholar]
- 19.Glassford, J., I. Soeiro, S. M. Skarell, L. Banerji, M. Holman, G. G. Klaus, T. Kadowaki, S. Koyasu, and E. W. Lam. 2003. BCR targets cyclin D2 via Btk and the p85alpha subunit of PI3-K to induce cell cycle progression in primary mouse B cells. Oncogene 22:2248-2259. [DOI] [PubMed] [Google Scholar]
- 20.Grana, X., J. Garriga, and X. Mayol. 1998. Role of the retinoblastoma protein family, pRB, p107 and p130 in the negative control of cell growth. Oncogene 17:3365-3383. [DOI] [PubMed] [Google Scholar]
- 21.Harris, M. B., C.-C. Chang, M. T. Berton, N. N. Danial, J. Zhang, D. Kuehner, B. H. Ye, M. Kvatyuk, P. P. Pandolfi, G. Cattoretti, R. Dalla-Favera, and P. B. Rothman. 1999. Transcriptional repression of Stat6-dependent interleukin-4-induced genes by BCL-6: specific regulation of Iɛ transcription and immunoglobulin E switching. Mol. Cell. Biol. 19:7264-7275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ilaria, R. L., Jr., and R. A. Van Etten. 1996. P210 and P190(BCR/ABL) induce the tyrosine phosphorylation and DNA binding activity of multiple specific STAT family members. J. Biol. Chem. 271:31704-31710. [DOI] [PubMed] [Google Scholar]
- 23.Jun, D. Y., M. K. Kim, I. G. Kim, and Y. H. Kim. 1997. Characterization of the murine cyclin D2 gene: exon/intron organization and promoter activity. Mol. Cells 7:537-543. [PubMed] [Google Scholar]
- 24.Komatsu, N., T. Watanabe, M. Uchida, M. Mori, K. Kirito, S. Kikuchi, Q. Liu, T. Tauchi, K. Miyazawa, H. Endo, T. Nagai, and K. Ozawa. 2003. A member of Forkhead transcription factor FKHRL1 is a downstream effector of STI571-induced cell cycle arrest in BCR-ABL-expressing cells. J. Biol. Chem. 278:6411-6419. [DOI] [PubMed] [Google Scholar]
- 25.Kops, G. J., N. D. de Ruiter, A. M. De Vries-Smits, D. R. Powell, J. L. Bos, and B. M. Burgering. 1999. Direct control of the Forkhead transcription factor AFX by protein kinase B. Nature 398:630-634. [DOI] [PubMed] [Google Scholar]
- 26.Kops, G. J. P. L., R. H. Medema, J. Glassford, M. A. G. Essers, P. F. Dijkers, P. J. Coffer, E. W.-F. Lam, and B. M. T. Burgering. 2002. Control of cell cycle exit and entry by protein kinase B-regulated forkhead transcription factors. Mol. Cell. Biol. 22:2025-2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lam, E. W., and N. B. La Thangue. 1994. DP and E2F proteins: coordinating transcription with cell cycle progression. Curr. Opin. Cell Biol. 6:859-866. [DOI] [PubMed] [Google Scholar]
- 28.Lam, E. W.-F., and R. J. Watson. 1993. An E2F-binding site mediates cell-cycle regulated repression of mouse B-myb transcription. EMBO J. 12:2705-2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lauper, N., A. R. Beck, S. Cariou, L. Richman, K. Hofmann, W. Reith, J. M. Slingerland, and B. Amati. 1998. Cyclin E2: a novel CDK2 partner in the late G1 and S phases of the mammalian cell cycle. Oncogene 17:2637-2643. [DOI] [PubMed] [Google Scholar]
- 30.Lawlor, M. A., and D. R. Alessi. 2001. PKB/Akt: a key mediator of cell proliferation, survival and insulin responses? J. Cell Sci. 114:2903-2910. [DOI] [PubMed] [Google Scholar]
- 31.Martino, A., J. H. T. Holmes, J. D. Lord, J. J. Moon, and B. H. Nelson. 2001. Stat5 and Sp1 regulate transcription of the cyclin D2 gene in response to IL-2. J. Immunol. 166:1723-1729. [DOI] [PubMed] [Google Scholar]
- 32.Medema, R. H., G. J. P. L. Kops, J. L. Bos, and B. M. T. Burgering. 2000. AFX-like Forkhead transcription factors mediate cell cycle regulation by Ras and PKB via p27. Nature 404:782-787. [DOI] [PubMed] [Google Scholar]
- 33.Mohamedali, A., I. Soeiro, N. C. Lea, J. Glassford, L. Banerji, G. J. Mufti, E. W. Lam, and N. S. Thomas. 2003. Cyclin D2 controls B cell progenitor numbers. J. Leukoc. Biol. 74:1139-1143. [DOI] [PubMed] [Google Scholar]
- 34.Mulligan, G., and T. Jacks. 1998. The retinoblastoma gene family: cousins with overlapping interests. Trends Genet. 14:223-229. [DOI] [PubMed] [Google Scholar]
- 35.Nevins, J. R. 1998. Toward an understanding of the functional complexity of the E2F and retinoblastoma families. Cell Growth Differ. 9:585-593. [PubMed] [Google Scholar]
- 36.Parada, Y., L. Banerji, J. Glassford, N. C. Lea, M. Collado, C. Rivas, J. L. Lewis, M. Y. Gordon, N. S. Thomas, and E. W.-F. Lam. 2001. BCR-ABL and interleukin 3 promote haematopoietic cell proliferation and survival through modulation of cyclin D2 and p27Kip1 expression. J. Biol. Chem. 276:23572-23580. [DOI] [PubMed] [Google Scholar]
- 37.Pardee, A. B. 1989. G1 events and regulation of cell proliferation. Science 246:603-608. [DOI] [PubMed] [Google Scholar]
- 38.Pegoraro, L., L. Matera, J. Ritz, A. Levis, A. Palumbo, and G. Biagini. 1983. Establishment of a Ph1-positive human cell line (BV173). JNCI 70:447-453. [PubMed] [Google Scholar]
- 39.Ramaswamy, S., N. Nakamura, I. Sansal, L. Bergeron, and W. R. Sellers. 2002. A novel mechanism of gene regulation and tumor suppression by the transcription factor FKHR. Cancer Cell 2:81-91. [DOI] [PubMed] [Google Scholar]
- 40.Sardet, C., L. E. LeCam, E. Fabbrizio, and M. Vidal. 1997. E2Fs and the retinoblastoma protein family, p. 1-63. In J. Ghysdael and M. Yaniv (ed.), Oncogenes as transcriptional regulators, vol. 2. Birkhauser Verlag, Berlin, Germany. [Google Scholar]
- 41.Sattler, M., and R. Salgia. 1997. Activation of hematopoietic growth factor signal transduction pathways by the human oncogene BCR/ABL. Cytokine Growth Factor Rev. 8:63-79. [DOI] [PubMed] [Google Scholar]
- 42.Schmidt, M., S. Fernández de Mattos, A. van der Horst, R. Klompmaker, G. J. Kops, E. W. Lam, B. M. Burgering, and R. H. Medema. 2002. Cell cycle inhibition by FoxO forkhead transcription factors involves downregulation of cyclin D. Mol. Cell. Biol. 22:7842-7852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shaffer, A. L., X. Yu, Y. He, J. Boldrick, E. P. Chan, and L. M. Staudt. 2000. BCL-6 represses genes that function in lymphocyte differentiation, inflammation, and cell cycle control. Immunity 13:199-212. [DOI] [PubMed] [Google Scholar]
- 44.Sherr, C. J. 1995. D-type cyclins. Trends Biochem. Sci. 20:187-190. [DOI] [PubMed] [Google Scholar]
- 45.Skorski, T., A. Bellacosa, M. Nieborowska-Skorska, M. Majewski, R. Martinez, J. K. Choi, R. Trotta, P. Wlodarski, D. Perrotti, T. O. Chan, M. A. Wasik, P. N. Tsichlis, and B. Calabretta. 1997. Transformation of hematopoietic cells by BCR/ABL requires activation of a PI-3k/Akt-dependent pathway. EMBO J. 16:6151-6161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Skorski, T., P. Kanakaraj, M. Nieborowska-Skorska, M. Z. Ratajczak, S. C. Wen, G. Zon, A. M. Gewirtz, B. Perussia, and B. Calabretta. 1995. Phosphatidylinositol-3 kinase activity is regulated by BCR/ABL and is required for the growth of Philadelphia chromosome-positive cells. Blood 86:726-736. [PubMed] [Google Scholar]
- 47.Tang, E. D., G. Nunez, F. G. Barr, and K. L. Guan. 1999. Negative regulation of the forkhead transcription factor FKHR by Akt. J. Biol. Chem. 274:16741-16746. [DOI] [PubMed] [Google Scholar]
- 48.Tang, T. T., D. Dowbenko, A. Jackson, L. Toney, D. A. Lewin, A. L. Dent, and L. A. Lasky. 2002. The forkhead transcription factor AFX activates apoptosis by induction of the BCL-6 transcriptional repressor. J. Biol. Chem. 277:14255-14265. [DOI] [PubMed] [Google Scholar]
- 49.Testa, J. R., and A. Bellacosa. 2001. AKT plays a central role in tumorigenesis. Proc. Natl. Acad. Sci. USA 98:10983-10985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Williams, C. D., D. C. Linch, M. J. Watts, and N. S. Thomas. 1997. Characterization of cell cycle status and E2F complexes in mobilized CD34+ cells before and after cytokine stimulation. Blood 90:194-203. [PubMed] [Google Scholar]
- 51.Yang, M., Y. Hosokawa, Y. Hu, S. Kaneko, H. Kaneko, M. Tanaka, and K. Nakashima. 1997. Cloning and functional analysis of rat cyclin D2 promoter: multiple prolactin-responsive elements. Biochem. Mol. Biol. Int. 43:749-754. [DOI] [PubMed] [Google Scholar]