Abstract
Retinoid X receptor (RXR) plays a central role in the regulation of intracellular receptor signaling pathways by acting as a ubiquitous heterodimerization partner of many nuclear receptors, including the orphan receptor Nur77 (also known as thyroid hormone receptor 3 or NGFI-B), which translocates from the nucleus to mitochondria, where it interacts with Bcl-2 to induce apoptosis. Here, we report that RXRα is required for nuclear export and mitochondrial targeting of Nur77 through their unique heterodimerization that is mediated by dimerization interfaces located in their DNA-binding domain. The effects of RXRα are attributed to a putative nuclear export sequence (NES) present in its carboxyl-terminal region. RXRα ligands suppress NES activity by inducing RXRα homodimerization or altering RXRα/Nur77 heterodimerization. The RXRα NES is also silenced by RXRα heterodimerization with retinoic acid receptor or vitamin D receptor. Consistently, we were able to show that the mitochondrial targeting of the RXRα/Nur77 heterodimer and its induction of apoptosis are potently inhibited by RXR ligands. Together, our results reveal a novel nongenotropic function of RXRα and its involvement in the regulation of the Nur77-dependent apoptotic pathway.
Retinoid X receptors (RXRs) belong to the nuclear receptor superfamily, consisting of a large number of ligand-regulated transcription factors that mediate the diverse physiological functions of their ligands, such as steroid hormones, retinoids, thyroid hormone, and vitamin D3, in embryonic development, growth, differentiation, apoptosis, and homeostasis (29, 46). The superfamily also includes many orphan receptors whose ligands remain to be identified. All nuclear receptors consist of three major domains: the variable length N-terminal domain, the well-conserved DNA-binding domain (DBD), and the ligand-binding domain (LBD) (29, 46). The C-terminal LBD is multifunctional and, in addition to harboring a ligand-binding site, contains regions for receptor dimerization and the ligand-dependent transactivation function (AF-2). The DBD also contains a dimerization interface that determines target gene specificity (38, 55, 60, 85). RXRs mediate retinoid signaling through the RXR/retinoic acid receptor (RAR) heterodimer and the RXR/RXR homodimer (29, 46, 89). In addition, RXRs form heterodimers with many members of the subfamily 1 nuclear receptors, including vitamin D receptor (VDR), peroxisome proliferator-activated receptor (PPAR), and thyroid hormone receptor (TR), as well as several orphan receptors, such as liver X receptor, pregnane X receptor, constitutively activated receptor, and Nur77 (TR3 or NGFI-B) (29, 46). RXRs, therefore, play an essential role in the regulation of multiple nuclear hormone-signaling pathways through their unique and potent dimerization capacity. The vitamin A metabolite, 9-cis-retinoic acid (9-cis-RA), is a high-affinity ligand for RXRs (22, 39). It induces transactivation of the RXR homodimer (26, 29, 46, 88, 90) and certain RXR heterodimers, such as the RXR/Nur77 heterodimer (16, 54, 76).
Heterodimerization of RXR with its partners dramatically enhances their DNA binding and subsequently transcriptional regulation (29, 46, 89). On binding DNA, some nuclear receptors repress transcription of target genes through their interaction with transcriptional corepressors in the absence of ligands. Ligand binding by a transcriptional agonist causes a conformational change in the receptors, allowing dissociation of transcriptional corepressors and association of transcriptional coactivators (80). In addition to DNA binding and interaction with receptor cofactors, recent studies suggest that subcellular distribution of RXR and its dimerization partners represents another mechanism that regulates their biological activity. Despite being localized in the nucleus in many cell types, RXR (12, 27) and its partners, including RARs (12). TRs (2, 6, 92), VDR (57, 58), and Nur77 (30, 40), were found in the cytoplasm in certain cell types and stages during development or under different cellular environments. Interestingly, RXR heterodimerization promotes nuclear localization of TR (2) and VDR (57).
Orphan receptor Nur77 (7, 20, 51) is an immediate-early response gene whose expression is rapidly induced by a variety of extracellular stimuli, including growth factors, the phorbol ester 12-O-tetradecanoyl-13-phorbol acetate (TPA) and cyclic-AMP-dependent pathways. Nur77 and its closely related family members, Not-1 (also called Nurr1 and RNR-1) (36, 45) and NOR-1 (also called MINOR and TEC) (21, 47, 52), constitute a distinct subfamily within the nuclear receptor superfamily (29, 46, 48). Nur77 was originally recognized for its role in cell proliferation and differentiation. Paradoxically, Nur77 was later found to be a potent proapoptotic molecule (48, 73, 84, 86). The expression of Nur77 was rapidly induced during the apoptosis of immature thymocytes and T-cell hybridomas, as well as various types of cancer cells (24, 25, 28, 40, 41, 44, 62, 66, 74, 78). Overexpression of a dominant-negative Nur77 protein or inhibition of Nur77 expression by antisense Nur77 inhibited apoptosis, whereas constitutive expression of Nur77 resulted in massive apoptosis (40, 41, 44, 66, 69, 74, 75).
Nur77 can function in the nucleus as a transcription factor to regulate the gene expression necessary to alter the cellular phenotype in response to various stimuli. Consistently, Nur77 response elements (NBRE or NurRE) have been identified (56, 72). In addition to its heterodimerization with RXR (16, 54), Nur77 can interact with the orphan receptor COUP-TF (77) that binds to the RARβ promoter and is required for efficient RARβ expression (42). Through its interaction with RXR and COUP-TF, Nur77 can modulate RARβ expression and the growth response of cells to retinoids (8, 77). Interestingly, the EWS/TEC (Nor-1) fusion protein generated by the t(9;22) chromosomal translocation in extraskeletal myxoid chondrosarcoma is ∼270-fold more active than the native receptor in activating a reporter containing the NBRE (34, 35). This result suggests that the cancer-associated TEC (Nor-1) fusion receptor exerts its oncogenic activity by inducing the expression of target genes involved in cell proliferation. Thus, Nur77 may confer its growth-promoting activities through its action in the nucleus. This is supported by our recent observation that DNA binding and transactivation of Nur77 are required for its induction of cell proliferation (31).
Recent studies have demonstrated that Nur77 can also act outside the nucleus to mediate several important biological functions, including apoptosis and differentiation. Nur77, in response to apoptotic stimuli, translocates from the nucleus to the cytoplasm where it targets mitochondria to induce cytochrome c release and apoptosis (40). Our investigation of this phenomenon demonstrated that the proapoptotic effect of Nur77 does not require its transcriptional activity and DNA binding. Furthermore, Nur77 targets mitochondria through its interaction with Bcl-2, resulting in conversion of Bcl-2 from an antiapoptotic to a proapoptotic molecule (43). Nur77 targets mitochondria in prostate cancer (40, 71), lung cancer (10, 31), colon cancer (71), ovarian cancer (24), and gastric cancer (28, 79) cells, and its mitochondrial localization is involved in Sindbis virus-induced apoptosis (37). The cytoplasmic action of Nur77 was also demonstrated by the translocation of NGFI-B from the nucleus to the cytoplasm in response to nerve growth factor (NGF) treatment in PC12 pheochromocytoma cells, suggesting that the cytoplasmic action of NGFI-B is involved in NGF-induced PC12 cell differentiation (30). Thus, the diverse biological activities of Nur77 depend on its subcellular localization. The importance of TR3 pathways in regulating cancer cell growth is supported by the positive correlation between Nor-1 expression and survival in diffuse large B-cell lymphoma patients on chemotherapy (63) and a recent observation that TR3 is 1 of the 17 signature genes associated with metastasis of primary solid tumors (59).
Because Nur77 heterodimerizes with RXRα, we investigated the role of RXRα and its ligands in the regulation of the Nur77-dependent apoptotic pathway. Here, we report that RXRα migrates from the nucleus to mitochondria as an RXRα/Nur77 heterodimer in response to apoptotic stimuli. The migration is mediated by a putative nuclear export sequence (NES) present in the LBD of RXRα. The RXRα NES is active in the RXRα/Nur77 heterodimer formed through dimerization interfaces in their DBDs. Interestingly, in the presence of RXRα ligands, the RXRα/Nur77 heterodimer is formed through dimerization interfaces in their LBDs. Such an RXRα ligand-induced switch of RXRα/Nur77 heterodimerization interfaces silences the RXRα NES. Consistently, RXRα ligands effectively inhibit the mitochondrial targeting of the RXRα/Nur77 heterodimer and apoptosis. Together, our results demonstrate that RXRα and its ligands plays a critical role in the regulation of the Nur77-dependent apoptotic pathway through their heterodimerization.
MATERIALS AND METHODS
Plasmid constructs.
The construction of green fluorescent protein (GFP)-RXRα and Nur77 fusion, as well as GFP-Nur77/ΔDBD and GFP-Nur77/Δ1, has been described (40). GFP-RXRα and GFP-Nur77 mutants were generated by cloning RXRα or Nur77 mutants into pGFP-N2 or pGFP-C2 vector (Clontech). RXRα and Nur77 mutants were generated by standard restriction enzyme digestion or amplified by PCR. The following oligonucleotides were used as primers: GAA GAT CTT CGA CAC CAA ACA TTT CCT GCC GCT C (upstream) and CGG AAT TCC GGC AGG CCT AAG TCA TTT GGT GCG G (downstream) for RXRα, CGG AAT TCC GTC GAG AGC CCC TTG GAG TCA GGG (downstream) for RXRα/385, CGG AAT TCC GCA AAG ATG GCG CCC ACC CCT GCG C (downstream) for RXRα/347, CCG GAA TTC CGG GAT GTG CTT GGT GAA GGA AGC (downstream) for RXRα/135, GAA GAT CTT CAC CAG CAG CGC CAA CGA GGA CAT G (upstream) and CGG AAT TCC GGC AGG CCT AAG TCA TTT GGT GCG G (downstream) for RXRα/C1, 5′-CGG GAT CCC GGT GCG CCA TCT GCG GGG ACC GC-3′ (upstream) and 5′-CGG AAT TCG ACC CGC TTG TCA ATC AGG CAG TC-3′ (downstream) for RXRα/182, 5′-CGG AAT TCG ACC ATG CCC ATG GCC ACG CAC TTC-3′ (downstream) for RXRα/200, 5′-CGG AAT TCG ACG CCA CGC TGC CGC TCC TCC TG-3′ (downstram) for RXRα/212, 5′-GAA GAT CTT CAT GCC CTG TAT CCA AGC CCA ATA TG-3′ (upstream) and 5′-CGG AAT TCC GGC ACC AAG TCC TCC AGC TTG AGG TAG-3′ (downstream) for Nur77, 5′-CGG AAT TCC GGT AGC ACC AGG CCT GAG CAG AAG ATG-3′ (downstream) for Nur77/467, 5′-CGG AAT TCC GCG GAG AGC AGG TCG TAG AAC TGC TG (downstream) for Nur77/410, 5′-CGG AAT TCG CCC ACC GCC AGG CAC TTC TGG-3′ (downstream) for Nur77/332, 5′-CGG AAT TCC GCT CCA CTG TGC GCT TGA AGA AGC CCT G-3′ (downstream) for Nur77/296, 5′-CGG AAT TCC GGC AGG CCT TCG TAA GTC TGG CTG GGC (downstream) for Nur77/170, 5′-GAA GAT CTT CAT GGC CCT GTC CTC CAG TGG CTC TGA C-3′ (upstream) for Nur77/Δ2, 5′-GAA GAT CTT CTC CGG TTC TCT GGA GGT CAT CCG C-3′ (upstream) and 5′-CGG AAT TCC AGC ACC AGG CCT GAG CAG AAG ATG-3′ (downstream) for Nur77/ΔC2, and 5′-GAA GAT CTT CAT GGG CCT GGT GCT ACA CCG GCT G-3′ (upstream) and 5′-CGG AAT TCC GGC ACC AAG TCC TCC AGC TTG AGG TAG-3′ (downstream) for Nur77/ΔC3. The BamHI-EcoRI fragment (324 to 462), BamHI and StuI fragment (324 to 402), and BglII and BamHI fragment (1 to 235) of RXRα were cloned into pGFP-C2 to generate GFP-RXRα/C2, GFP-RXRα/C3, and GFP-RXRα/235, respectively. Nur77 or RXRα mutants were also cloned into pcDNA3, pECE-Flag, or pCMV-myc vector. For NES-RXRα, 5′-GAT CTT CAG GGT GCT GAC GGA GCT TGT GTC CAA GAT GCG GGA CAT GCA GAT GGA CAA GAC GGA GCT GGG CG-3′ and AAT TCG CCC AGC TCC GTC TTG TCC ATC TGC ATG TCC CGC ATC TTG GAC ACA AGC TCC GTC AGC ACC CTG AA-3′ were annealed and ligated into pGFP-C2. NESm-RXRα was generated with QuikChange site-directed mutagenesis kit (Stratagene) with primers 5′-AAG GCA CGG GAC GCA CAG ATG GAC AAG-3′ and CTG TGC GTC CCG TGC CTT GGA CAC AAG-3′. RXRα/LLL was described previously (90). Bcl-2 expression vector and antibody were kindly provided by John C. Reed at the Burnham Institute Cancer Center.
RXRα siRNA.
Small interfering RNAs (siRNAs) used in the experiments were obtained from Dharmacon Research, Inc. The following siRNA sequences were used: RXRα siRNA, 5′-AAG CAC UAU GGA GUG UAC AGC-3′, 5′-GCU GUA CAC UCC AUA GUG CUU-3′. A 2.5-μl aliquot of 20 μmol of siRNA/liter/per well was transfected into cells grown in 12-well plates by using oligofectamine reagent (Invitrogen) according to the manufacturer's recommendations. Two days after transfection, the cells were harvested for Western blotting or prepared for immunostaining.
Cell culture.
LNCaP prostate cancer cells and H460 lung cancer cells were maintained in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS). CV-1 cells and HEK293T embryonic kidney cells were grown in Dulbecco modified Eagle’s medium supplemented with 10% FBS.
Protein-protein interaction assays.
Glutathione S-transferase (GST)-pull-down and gel shift assays have been described previously (77, 87). For GST pull-down assays, GST and GST fusion proteins were expressed in Escherichia coli BL21 and purified by using glutathione-agarose affinity chromatography. The GST fusion proteins were analyzed by sodium dodecyl sulfate-10% polyacrylamide gel electrophoresis (SDS-10% PAGE) for integrity. [35S]methionine-labeled RXRα or Nur77 was produced by TNT-coupled transcription-translation system (Promega) and visualized by SDS-PAGE. In vitro binding assays were performed with glutathione-agarose beads (20 μl) coated with 10 μg of GST fusion protein and 2 to 5 μl of [35S]methionine-labeled protein in 200 μl of binding buffer (20 mM HEPES [pH 7.9], 100 mM KCI, 1 mM MgC12, 10 μM ZnC12 2 mM dithiothreitol, 10% glycerol, 0.05% Triton X-100, 40 μg of leupeptin/ml). The reaction was allowed to proceed overnight at 4°C with rocking. Beads were then collected by centrifugation and washed five times with 1 ml of NETN buffer (100 mM NaC1, 1 mM EDTA, 20 mM Tris-HCl [pH 8.0], 0.5% NP-40, 40 μg of leupeptin/ml). The beads were resuspended in 20 μl of SDS-PAGE sample buffer and boiled for 5 min. The eluted proteins were loaded on an SDS-PAGE gel, dried, and autoradiographed at −70°C. For coimmunoprecipitation assays, HEK293T cells grown in 10-cm dishes were transfected with 7.5 μg of Flag-tagged Nur77 and 7.5 μg of GFP-tagged RXRα/385. Transfected HEK293T cells were lysed in 300 μl of TAE buffer (10 mM Tris-HCI [pH 8.0], 10 mM NaC1, 10 mM EDTA, and 1% NP-40 containing protease inhibitors [Sigma]). Lysate (300 μl) was incubated with 2 μg of anti-Flag monoclonal antibody (M2; Sigma) at 4°C for 2 h. Immunocomplexes were then precipitated with 40 μl of protein A/G-Sepharose (Santa Cruz Biotechnology, Inc.). After an extensive washing with TAE buffer, the beads were boiled in 40 μl of loading buffer and analyzed by Western blotting. Mouse immunoglobulin G (IgG; Santa Cruz Biotechnology) was used as a negative control.
Nondenaturing gel electrophoresis.
Nuclear and cytoplasmic fractions of HEK293T cells expressing GFP-RXRα/C1 treated with or without 9-cis-RA were prepared and separated on a 8% nondenaturing Tris-glycine gel (Invitrogen), followed by immunoblotting.
Confocal microscopy.
Cells were seeded on the chamber slides overnight and treated with apoptotic agents in medium containing 0.5% FBS. After treatment, the cells were fixed in 4% paraformaldehyde in phosphate-buffered saline (PBS) for 10 min and washed twice with PBS. Cells were then permeabilized with 1% triton X-100 in PBS for 5 min. Fixed cells were preincubated for 30 min in PBS containing 5% bovine serum albumin at room temperature. Cells were stained with polyclonal anti-RXRα antibody (D20 [Santa Cruz Biotechnology] at 1:500) or monoclonal anti-Nur77 antibody (1:500; Abgent) before detection of fluorescein isothiocyanate (FITC)-labeled anti-rabbit IgG (1:500; Sigma). For mitochondrial staining, cells were incubated with anti-Hsp60 goat IgG (1:500; Santa Cruz Biotechnology), followed by anti-goat IgG conjugated with Cy3 (1:1,000; Sigma). For cytochrome c staining, cells were incubated with monoclonal anti-cytochrome c IgG (1:300; Pharmingen), followed by anti-mouse IgG conjugated with Cy5 (1:500; Amersham). For staining transfected protein, 1 μg of Myc-tagged TR3, Flag-tagged RXRα, or Bcl-2 was transfected into HEK293T cells grown in chamber slides in six-well plates overnight and treated with apoptotic agents. After treatment, cells were fixed and incubated with monoclonal anti-Myc IgG antibody (1:300; 9E10; Santa Cruz Biotechnology), followed by FITC-conjugated anti-mouse IgG (1:500; Sigma) or Cy3-conjugated anti-mouse IgG (1:1,000; Sigma). For Bcl-2 staining, cells were incubated with polyclonal anti-Bcl-2 antibody (67), followed by Cy5-conjugated goat anti-rabbit antibody (1:500; Sigma).
Cell fractionation.
Subcellular fractionation was performed as described with minor modifications (40). Briefly, cells (107 cells) suspended in 0.5 ml of hypotonic buffer (250 mM sucrose, 20 mM HEPES-KOH [pH 7.4], 10 mM KCl, 10 mM MgCl2, 0.5 mM EGTA, 1.5 mM EDTA [pH 8.0], 1 mM dithiothreitol) with proteinase inhibitors were homogenized, and cell extracts were centrifuged at 800 × g for 10 min. The pellet containing nuclei was resuspended in 200 μl of 1.6 M sucrose in hypotonic buffer plus protease inhibitors and laid over 1 ml of 2.0 M sucrose in the same buffer and then centrifuged at 150,000 × g for 90 min at 4°C to obtain the nuclear fraction. The supernatant was centrifuged at 10,000 × g for 30 min at 4°C to obtain the heavy membrane (HM) and cytosolic fractions. Nuclear and HM fractions were resuspended in 100 μl of lysis buffer (10 mM Tris [pH 7.4], 150 mM NaCl, 1% Triton X-100, 5 mM EDTA [pH 8.0]) with a cocktail of proteinase inhibitors for Western blotting analysis as described previously (40).
Transient-transfection assays.
Cells (105 cells/well) seeded in 24-well plates were transiently transfected by using a modified calcium phosphate precipitation procedure as described previously (42). For GFP studies, 1 μg of GFP or GFP constructs was transfected into cells seeded in six-well plates by calcium phosphate precipitation.
Apoptosis assays.
For terminal deoxynucleotidyltransferase (TdT) assays, cells were treated with or without indicated agents overnight after pretreatment with RXRα ligand 9-cis-RA or SR11237 for 12 h, treated with trypsin, washed with PBS, fixed in 1% formaldehyde, and then resuspended in 70% ice-cold ethyl alcohol. Cells were then labeled with biotin-16-deocyuridine 5′ triphosphate by TdT and stained with avidin-fluorescein isothiocyanate (Boehringer Mannheim). For nuclear morphological change analysis, LNCaP cells were treated with trypsin, washed with PBS, fixed with 3.7% paraformaldehyde, and stained with DAPI (4′,6′-diamidino-2-phenylindole; 50 μg/ml) to visualize nuclei by fluorescence microscopy.
Western blotting.
Cell lysates were boiled in SDS sample buffer, resolved by SDS-12.5% PAGE, and transferred to nitrocellulose. After transfer, the membranes were blocked in 5% milk in TBST (10 mM Tris-HCl [pH 8.0], 150 mM NaCl, 0.05% Tween 20) containing antibody. The membranes were then washed three times with TBST and then incubated for 1 h at room temperature in TBST containing horseradish peroxide-linked anti-immunoglobulin. After three washes in TBST, immunoreactive products were detected by using enhanced chemiluminescence (Amersham). Anti-GFP antibody was purchased from Santa Cruz Biotechnology.
RESULTS
RXRα targets mitochondria in response to apoptotic stimuli.
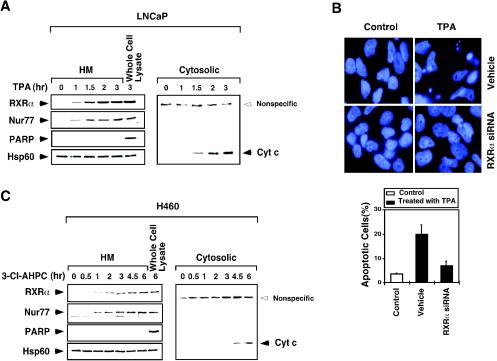
Nur77 migrates from the nucleus to mitochondria to induce apoptosis in response to certain apoptotic stimuli (10, 24, 31, 37, 40, 43, 71, 78). Since RXRα heterodimerizes with Nur77 (16, 54), we studied whether RXRα also targeted mitochondria. Subcellular localization of RXRα in LNCaP prostate cancer cells in the absence or presence of TPA, which potently induces LNCaP cell apoptosis (40), was examined by confocal microscopy analysis. Immunostaining showed that RXRα predominantly localized in the nucleus in the absence of TPA treatment (Fig. 1A). However, when cells were treated with TPA, RXRα was found in the cytoplasm. To study whether RXRα was associated with mitochondria, cells were stained for heat shock protein 60 (Hsp60), a mitochondrion-specific protein (Fig. 1A). The extensive overlap in the distribution patterns of Hsp60 and RXRα suggested the association of RXRα with mitochondria. Similarly, treatment with TPA also resulted in mitochondrial localization of Nur77 (Fig. 1B), as previously reported (40). The enhanced staining of Nur77 in TPA-treated cells was due to induction of endogenous Nur77 expression by TPA. Thus, RXRα and Nur77 associate with mitochondria in LNCaP cells undergoing apoptosis.
FIG. 1.
Nur77 and RXR comigrate from the nucleus to the cytoplasm. (A/B) RXRα (A) or Nur77 (B) targets mitochondria in response to apoptotic stimulus. LNCaP prostate cancer cells were treated with TPA (100 ng/ml) for 1 h and then immunostained with either anti-RXRα (Santa Cruz Biotechnology) (A) or anti-Nur77 (Active Motif) (B) antibody, followed by Cy3-conjugated secondary antibody (Sigma) to detect RXRα or Nur77 or with anti-Hsp60 (Santa Cruz Biotechnology), followed by FITC-conjugated secondary antibody (Sigma) to detect mitochondria. RXRα, Nur77, and mitochondria (Hsp60) were visualized by using confocal microscopy, and the images of RXRα or Nur77 with those of mitochondria were overlaid (overlay). About 80% of cells displayed mitochondrial targeting of RXRα and Nur77 when cells were treated with TPA. One of three similar experiments is shown. (C) Nur77 and RXRα comigrate from the nucleus to the cytoplasm. LNCaP cells were treated with or without TPA for 1 h and then immunostained with anti-RXRα antibody, followed by FITC-conjugated secondary antibody, or with anti-Nur77 antibody (Abgent, San Diego, Calif.), followed by Cy3-conjugated secondary antibody. RXRα and Nur77 were visualized by using confocal microscopy and the images were overlaid (overlay). Approximately 80% of TPA-treated cells showed RXRα colocalization with Nur77. One of three similar experiments is shown. (D) 3-Cl-AHPC induces mitochondrial localization of transfected RXRα and Nur77. Expression vectors for myc-Nur77 and RXRα weretransfected into LNCaP cells. Cells were then treated with 3-Cl-AHPC for 3 h and then immunostained with anti-myc antibody (9E10; Santa Cruz Biotechnology), followed by FITC-conjugated secondary antibody, anti-RXRα antibody, followed by Cy3-conjugated secondary antibody or anti-Hsp60 antibody, followed by Cy5-conjugated secondary antibody (Jackson Immunoresearch). Myc-Nur77, RXRα, and Hsp60 were visualized and the images were overlaid. Overlay 1 is the merger of myc-Nur77 and RXRα images, and overlay 2 is the merger of myc-Nur77, RXRα, and Hsp60 images. About 30% of transfected cells exhibited the colocalization presented. One of three similar experiments is shown. (E) Mitochondrial localization of RXRα is Nur77 dependent. LNCaP cells or LNCaP cells stably expressing Nur77 antisense RNA (Nur77/antisense) (40) were treated with or without TPA for 1 h and then immunostained with anti-RXRα antibody, followed by Cy3-conjugated secondary antibody. Approximately 70% of cells displayed the effect presented. One of two similar experiments is shown. (F) Effect of RXR siRNA on RXRα levels and Nur77 localization. LNCaP cells were transfected with or without RXRα siRNA or control siRNA for 72 h. Cell extracts were prepared and analyzed for RXRα expression by Western blotting. Cells were also analyzed for subcellular localization of RXRα and Nur77 by confocal microscopy. One of two similar experiments is shown. (G) Nur77/Δ1 prevents RXRα mitochondrial targeting. LNCaP cells were transfected with the GFP-Nur77/Δ1 expression vector and then treated with TPA for 1 h and immunostained with anti-RXRα antibody. RXRα and GFP-Nur77/Δ1 distributions were analyzed by confocal microscopy. About 80% of nontransfected cells showed cytoplasmic localization of RXRα after treatment with TPA, whereas >70% of cells transfected with GFP-Nur77/Δ1 showed RXRα nuclear localization with the same treatment. One of three similar experiments is shown. (H) 3-Cl-AHPC induces mitochondrial localization of RXRα and Nur77 in H460 lung cancer cells. H460 cells were treated with 3-Cl-AHPC (10−6 M) for 3 h and then immunostained with anti-RXRα antibody, followed by FITC-conjugated secondary antibody, anti-Nur77 followed by Cy3-conjugated secondary antibody, or anti-Hsp60 antibody, followed by Cy5-conjugated secondary antibody (Jackson Immunoresearch). RXRα, Nur77, and Hsp60 were visualized by confocal microscopy, and the images were overlaid. Overlay 1 represents the merger of RXRα and Nur77 images. Overlay 2 indicates merged RXRα, Nur77, and Hsp60 images. Approximately 80% of 3-Cl-AHPC-treated cells showed the patterns presented. One of three similar experiments is shown.
RXRα and Nur77 mitochondrial targeting are mutually dependent.
To study whether Nur77 and RXRα targeted mitochondria as a heterodimer, subcellular localization of Nur77 and RXRα was examined in LNCaP cells treated with or without TPA. In the absence of TPA, both Nur77 and RXRα resided mainly in the nucleus (Fig. 1C). However, when cells were treated with TPA, Nur77 and RXRα were colocalized in the cytoplasm and their distribution patterns overlaid extensively (Fig. 1C), suggesting their association in the cytoplasm. We also examined whether transfected RXRα and Nur77 targeted mitochondria in response to apoptosis induction. When expression vectors for RXRα and Nur77 were transfected into LNCaP cells, the expressed RXRα and Nur77 resided in the nucleus. However, when cells were treated with 3-Cl-AHPC, a apoptosis-inducing retinoid (91), both RXRα and Nur77 were found in the cytoplasm, and their distributions were overlaid (Fig. 1D). It is noteworthy that we have consistently observed that endogenous RXRα and Nur77 targeted mitochondria with much higher efficiency than the transfected receptors, suggesting the possible involvement of other protein factors in their targeting. We next determined whether RXRα cytoplasmic localization depended on Nur77 expression by examining its subcellular localization in LNCaP cells stably expressing Nur77 antisense RNA (40). Expression of Nur77 antisense RNA strongly inhibited TPA-induced Nur77 expression in LNCaP cells (40). In contrast to that observed in wild-type LNCaP cells (Fig. 1A), RXRα was found only in the nucleus in the Nur77 antisense stable clone, even though the cells were treated with TPA (Fig. 1E). To study whether Nur77 mitochondrial targeting required RXRα, we used the siRNA approach (14) to inhibit RXRα expression in LNCaP cells and then examined the subcellular localization of Nur77. Transfection of LNCaP cells with RXRα siRNA strongly reduced RXRα protein levels (Fig. 1F). In cells transfected with RXRα siRNA, Nur77 was mainly confined in the nucleus despite TPA treatment (Fig. 1F). Thus, cytoplasmic localization of Nur77 and RXRα is mutually dependent. We previously reported that Nur77 with its N terminus deleted (Nur77/Δ1) acted as a dominant-negative mutant, which inhibited Nur77 mitochondrial targeting and apoptosis (40). Therefore, we examined whether Nur77/Δ1 also interfered with RXRα mitochondrial targeting. GFP-Nur77/Δ1 was transfected into LNCaP cells, which were then treated with TPA. RXRα immunostaining showed that RXRα was confined to the nucleus in cells transfected with GFP-Nur77/Δ1 in the absence or presence of TPA treatment (Fig. 1G). Thus, Nur77/Δ1 retained RXRα in the nucleus probably through their heterodimerization, and the Nur77 N-terminal sequences are important for the TPA effect. It is noteworthy that the N terminus of Nur77 is enriched with serine, threonine, and tyrosine residues, suggesting the potential regulation of Nur77 activity by phosphorylation. To determine whether mitochondrial targeting of RXRα and Nur77 was specific to LNCaP cells and to the TPA treatment, we treated H460 lung cancer cells with 3-Cl-AHPC, which potently induced the apoptosis of lung cancer cells (31). Similar to what we observed with the LNCaP cells, the treatment of H460 cells with 3-Cl-AHPC resulted in extensive targeting of RXRα and Nur77 to mitochondria (Fig. 1H).
To further characterize the mitochondrial targeting of RXRα and Nur77, we conducted a time course analysis of their targeting in LNCaP cells treated with TPA (Fig. 2A). Immunoblotting of mitochondrion-enriched HM fractions showed that both RXRα and Nur77 began to be associated with mitochondria as early as 1 h after cells were treated with the apoptotic stimulus. This result is consistent with our previous observation showing that Nur77 targeted mitochondria after LNCaP cells were treated with TPA for 1 h (40). The simultaneous targeting of RXRα and Nur77 to mitochondria again suggested that they targeted mitochondria as a heterodimer. We previously reported that mitochondrial targeting of Nur77 initiated the release of cytochrome c from mitochondria (40). Consistently, we observed significant amounts of cytochrome c in the cytosolic fractions after LNCaP cells were treated with TPA for 1.5 h. These results indicate that mitochondrial targeting of RXRα and Nur77 precedes the release of cytochrome c, further demonstrating the role of Nur77/RXRα mitochondrial targeting in triggering cytochrome c release. The requirement of RXRα in Nur77-dependent apoptosis is also illustrated by our observation that inhibition of RXRα expression by the expression of RXRα siRNA diminished the apoptotic effect of TPA in LNCaP cells (Fig. 2B). Similar results were also obtained in H460 lung cancer cells treated with 3-Cl-AHPC (Fig. 2C).
FIG. 2.
Time course analysis of mitochondrial targeting of RXRα and Nur77 and the release of cytochrome c from mitochondria. (A) Time course analysis of LNCaP cells in response to TPA. Cells were treated with TPA (100 ng/ml) for the indicated times. Mitochondrion-enriched HM and cytosolic fractions were prepared and analyzed for the presence of RXRα, Nur77, and cytochrome c as indicated. As a control, the whole-cell extract was also analyzed. Expression of mitochondrion-specific Hsp60 protein and nucleus-specific PARP protein was determined to control the purity of HM fractions. One of two similar experiments is shown. (B) Inhibition of RXRα expression suppresses the apoptotic effect of TPA. LNCaP cells were transfected with RXRα siRNA, followed by treatment with TPA (100 ng/ml) for 3 h. Cells were stained with DAPI and analyzed for nuclear morphological changes. Apoptotic cells were scored by examining 300 cells for apoptotic morphology from three different experiments. (C) Time course analysis of H460 cells in response to 3-Cl-AHPC. Cells were treated with 3-Cl-AHPC (10−6 M) and analyzed as described for panel A. One of two similar experiments is shown.
Subcellular localization of RXRα and Nur77 mutants.
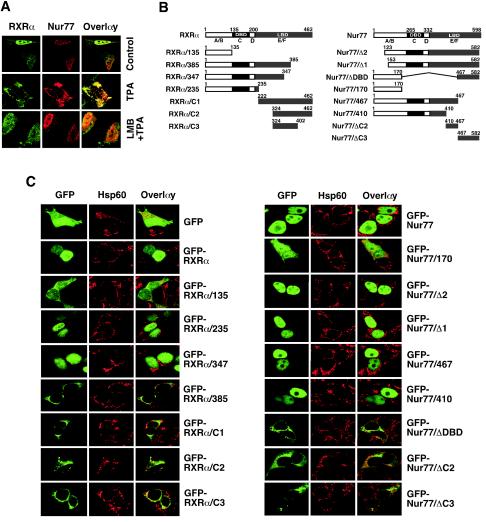
Previous studies showed that the cytoplasmic localization of NGFI-B and RXR was mediated through a CRM1-dependent nuclear export process (30). To study whether the translocation of the RXRα/Nur77 heterodimer from the nucleus to the cytoplasm is mediated through this process, we examined the effect of leptomycin B (LMB), an inhibitor of CRM1-dependent nuclear export (33). Treatment of LNCaP cells with LMB completely blocked TPA-induced cytoplasmic localization of both Nur77 and RXRα (Fig. 3A). This result suggests that the nuclear export of the RXRα/Nur77 heterodimer during apoptosis is mediated by a CRM1-dependent mechanism, a finding similar to that observed for the RXR/NGFI-B heterodimer (30). To identify the putative NES responsible for the nuclear export of RXRα/Nur77 heterodimer, various Nur77 and RXRα mutants were constructed (Fig. 3B) and fused to GFP. The fusions were then transfected into HEK293T cells, and their cellular localization was examined by confocal microscopy (Fig. 3C). HEK293T cells were used for these studies because of their high transfection efficiency. GFP was equally distributed in both the nucleus and the cytoplasm of HEK293T cells, indicating the absence of a nuclear localization signal (NLS) and NES. GFP-Nur77 and GFP-RXRα, however, were found predominantly in the nucleus. The RXRα and Nur77 mutants with only the A/B domain, RXRα/135 and Nur77/170 were diffusely distributed in both the nucleus and cytoplasm. However, mutants that contain the A/B, C, and D domains and a portion of the E domain—RXRα/235, RXRα/347, Nur77/467, and Nur77/410—were confined to the nucleus, with the exception of RXRα/385 (see below). The nuclear localization of these mutants may reflect the presence of an NLS in the DBD of the receptors (11). Indeed, an NLS was identified in the DBD of RXRα (57). Consistently, deletion of the DBD from Nur77 resulted in a mutant (Nur77/ΔDBD) that exclusively localized in the cytoplasm. Interestingly, C-terminus mutants of RXRα or Nur77—RXRα/C1, RXRα/C2, RXRα/C3, Nur77/ΔC2, and Nur77/ΔC3—were found predominantly in the cytoplasm.
FIG. 3.
Identification of domains in RXRα and Nur77 required for their nuclear export. (A) Cytoplasmic localization of RXRα/Nur77 is mediated by CRM1-dependent nuclear export. LNCaP cells were treated with TPA (100 ng/ml) in the absence (control) or presence of LMB (2.5 ng/ml; Sigma) and analyzed by confocal microscopy as described for Fig. 1C. About 80% cells showed the cytoplasmic localization of RXRα and Nur77 after treatment with TPA, whereas >50% of cells showed nuclear localization of RXRα and Nur77 when pretreated with LMB. One of two similar experiments is shown. (B) Schematic representations of RXRα and Nur77 mutants. The DBD, LBD, and A to F domains are indicated. (C) Analysis of subcellular localization of Nur77 and RXRα mutants. The indicated plasmids were transfected into HEK293T cells and analyzed by confocal microscopy as described in Fig. 1. More than 90% of transfected cells showed diffused distribution of GFP and GFP-RXRα/135. Nuclear localization of GFP-RXRα, RXRα/235, and RXRα/347 was found in 80, 85, and 60% of transfected cells, respectively. More than 90% of transfected cells showed exclusive cytoplasmic localization of RXRα/385, RXRα/C2, and RXRα/C3, whereas cytoplasmic localization of RXRα/C1 was found in 70% of transfected cells. Nuclear localization of GFP-Nur77 and its mutants, GFP-Nur77/Δ2, GFP-Nur77/Δ1, GFP-Nur77/467, and GFP-Nur77/410, was found in >90% of transfected cells, whereas cytoplasmic localization of GFP-Nur77/ΔDBD, cytoplasmic localization of GFP-Nur77/ΔDBD, GFP-Nur77/ΔC2, and GFP-Nur77/ΔC3 was observed in 80, 60 and 70% of transfected cells, respectively. One of four similar experiments is shown.
Identification of a putative NES in RXRα.
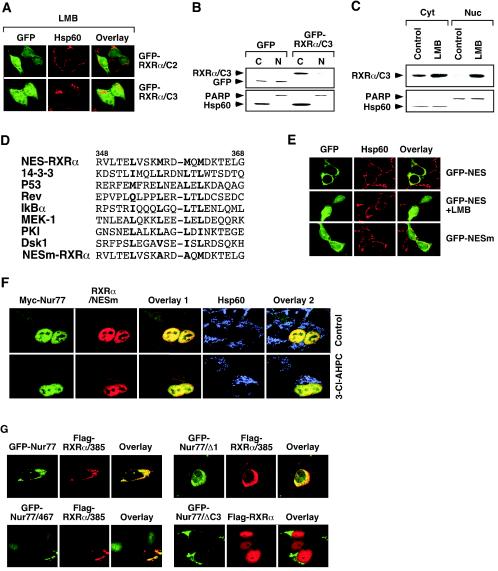
A previous study (30) demonstrated that the C-terminal half of Nur77 contains several NES sequences. To determine whether the cytoplasmic localization of the RXRα C-terminal-domain mutants was mediated by a CRM1-dependent mechanism, the effect of LMB on their cellular localization was examined (Fig. 4A). RXRα/C3 and RXRα/C2 accumulated exclusively in the cytoplasm of HEK293T cells. However, treatment with LMB resulted in their diffuse distribution (compare Fig. 3C and Fig. 4A). The inhibitory effect of LMB on the cytoplasmic localization of RXRα/C3 was further confirmed by immunoblotting of nuclear and cytoplasmic fractions prepared from HEK293T cells transfected with GFP-RXRα/C3. Although GFP was equally distributed in both nuclear and cytoplasmic fractions (Fig. 4B), which is consistent with our confocal microscopy results (Fig. 3C), GFP-RXRα/C3 was only found in the cytoplasm (Fig. 4B). The exclusive cytoplasmic presence of GFP-RXRα/C3 was prevented when cells were treated with LMB (Fig. 4C). Our results are consistent with a previous observation that mutation of the RXRα NLS resulted in its predominant cytoplasmic localization (57) and suggest that RXRα/C3 contains an active NES.
FIG. 4.
Identification of a nuclear export sequence in RXRα. (A) Effect of LMB on subcellular localization of RXRα mutants. The indicated expression vector for GFP fusion proteins was transfected into HEK293T cells and analyzed by confocal microscopy. Approximately 70% of transfected cells displayed diffused distribution of RXRα/C2 and RXRα/C3 after treatment with LMB. One of four similar experiments is shown. (B) RXRα/C3 exclusively resides in the cytoplasm. GFP-RXRα/C3 was transfected into HEK293T cells, and its localization was analyzed by cellular fractionation, followed by Western blotting with anti-GFP antibody (Santa Cruz Biotechnology). One of three similar experiments is shown. (C) The effect of LMB on subcellular localization of RXRα/C3. RXRα/C3 was transfected into HEK293T cells, which were then treated with or without LMB (2.5 ng/ml) for 6 h. Localization of RXRα/C3 was analyzed as described in panel B. One of three similar experiments is shown. (D) Schematic representation of the RXRα NES. The identified RXRα NES is compared to known NESs identified in the indicated genes. The boldface letters indicate conserved amino acid residues. (E) The RXRα NES is capable of directing GFP to the cytoplasm. The putative RXRαNES (RVLTELVSKMRDMQMDKTELG) or its mutant (RVLTELVSKARDAQMDKTELG) (NESm) was fused to GFP, and the expression vectors were transfected into HEK293T cells. Cells were treated with or without LMB for 6 h and then stained for Hsp60 and analyzed by confocal microscopy. Approximately 90% of GFP-NES-transfected cells exhibited cytoplasmic localization, whereas <20% of GFP-NESm-transfected cells displayed the same cytoplasmic localization. One of three similar experiments is shown. (F) Mutation of the RXRα NES impairs the 3-Cl-AHPC-induced mitochondrial localization of RXRα/Nur77 heterodimer. Myc-Nur77 was cotransfected with RXRα/NESm into LNCaP cells, which were then treated with 3-Cl-AHPC (10−6 M) for 3 h. Cells were stained for Nur77 by anti-myc antibody and for RXRα/NESm by anti-RXRα antibody and analyzed by the confocal microscopy. About 90% of transfected cells showed nuclear localization of Myc-Nur77 and RXRα/NESm, even after treatment with 3-Cl-AHPC. (G) RXRα NES is required for cytoplasmic localization of Nur77. The indicated expression vectors were cotransfected into HEK293T cells. Their subcellular localization was analyzed by confocal microscopy as described in Fig. 1A. Less than 10% of transfected cells showed cytoplasmic localization of GFP-Nur77, Nur77/467, and Nur77/Δ1, which was increased to 30, 40, and 40% upon cotransfection with Flag-RXRα/385, respectively. About 80% of transfected cells showed nuclear localization of Flag-RXRα, with or without Nur77/ΔC3 cotransfection. One of three similar experiments is shown.
The CRM1-dependent nuclear export is mediated by a Leu-rich NES (70). Inspection of RXR/C3 revealed the presence of a methionine-rich sequence, which had significant homology to previously identified NESs (Fig. 4D). To determine whether the RXRα sequence represented a putative NES, the DNA sequence representing the RXRα amino acids 348 to 368 was fused to GFP. The resulting GFP fusion, GFP-NES, was transfected into HEK293T cells. As shown in Fig. 4E, the GFP-NES protein was found exclusively in the cytoplasm, whereas the fusion was diffusely distributed in cells in the presence of LMB. Furthermore, replacing Met357 and Met360 in the NES with Ala (Fig. 4D) largely abolished its nuclear export activity (Fig. 4E). Thus, the RXRα amino acid 348 to 368 sequence represents a putative NES. This conclusion is also supported by our observation that GFP-RXRα/385 was exclusively localized in the cytoplasm, whereas the removal of amino acid residues 348 to 385 from the mutant abolished its cytoplasmic localization, as indicated by the nuclear localization of GFP-RXRα/347 (Fig. 3C). To determine the role of the RXRα NES in mediating the nuclear export of Nur77 and RXRα, both Met357 and Met360 in full-length RXRα were replaced with Ala. The resulting mutant, RXRα/NESm, was cotransfected with Nur77 into LNCaP cells. Unlike RXRα and Nur77, which targeted mitochondria in response to 3-Cl-AHPC treatment (Fig. 1D), both RXRα/NESm and Nur77 remained in the nucleus despite 3-Cl-AHPC treatment (Fig. 4F). Thus, the RXRα NES is required for apoptosis-induced nuclear export of the RXRα/Nur77 heterodimer.
Our observation that RXRα/385 was exclusively confined to the cytoplasm even in the absence of an apoptotic stimulus (Fig. 3C) was intriguing since this mutant contains both the NLS and NES. This result suggests that removal of the C-terminal sequences (amino acid residues 386 to 462) strongly activates the RXRα NES and/or silences its NLS. To determine whether the superactivation of RXRα NES in RXRα/385 could confer cytoplasmic localization to Nur77, RXRα/385 was cotransfected into HEK293T cells with Nur77 or its mutant Nur77/Δ1 or Nur77/467, all of which alone resided in the nucleus (Fig. 3C). Our data showed that coexpression of RXRα/385 with Nur77 or either mutant resulted in their colocalization in the cytoplasm (Fig. 4G). The extensive colocalization of RXRα/385 with Nur77, Nur77/467, or Nur77/Δ1 suggests that they comigrated to the cytoplasm as a heterodimer. Thus, RXRα/385 was able to shuttle Nur77 to the cytoplasm. In contrast, Nur77/ΔC3 could not confer its cytoplasmic localization to RXRα when they were coexpressed (Fig. 4G). Thus, the RXRα NES plays a critical role in the nuclear export of the RXRα/Nur77 heterodimer.
Role of Bcl-2 in RXRα mitochondrial targeting.
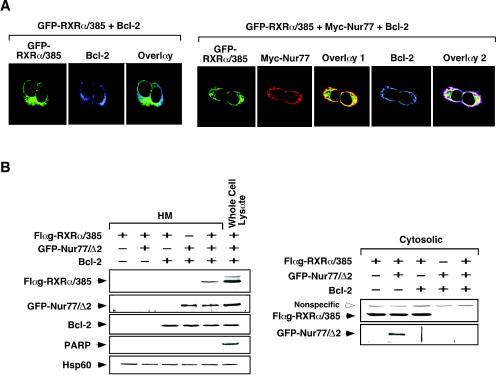
We recently reported that TR3 mitochondrial targeting requires its interaction with Bcl-2 (43). Therefore, we examined whether Bcl-2 expression modulated RXRα mitochondrial targeting. GFP-RXRα/385 was cotransfected with Bcl-2, and their distribution patterns were visualized by confocal microscopy. Figure 5A shows that expression of Bcl-2 did not alter the diffuse distribution of GFP-RXRα/385. However, when Nur77 was cotransfected, GFP-RXRα/385 colocalized with both Nur77 and Bcl-2, as evidenced by punctate staining. This result indicated that Bcl-2 interacted with Nur77 but not RXRα. The requirement of Nur77 and Bcl-2 for RXR mitochondrial targeting was also illustrated by our cellular fractionation assay (Fig. 5B). GFP-RXRα/385 expressed in HEK293T cells did not show any accumulation in the mitochondrion-enriched HM fraction in the absence or presence of Bcl-2 coexpression. However, when Nur77/Δ2, a Nur77 mutant lacking the N-terminal 122 amino acid residues, was coexpressed, a significant amount of GFP-RXRα/385 was found in the HM fraction, whereas its cytosolic level decreased (Fig. 5B). Together, these results demonstrate that mitochondrial targeting of RXRα requires both Nur77 and Bcl-2 and that the RXRα/Nur77 heterodimer interacts with Bcl-2.
FIG. 5.
Role of Bcl-2 in mitochondrial targeting of RXRα and Nur77 mutants. (A) Mitochondrial targeting of RXRα/385 in HEK293T cells requires Nur77 and Bcl-2. Expression vectors of GFP-RXRα/385 and Bcl-2 were transfected, together with or without myc-Nur77 vector into HEK293T cells. Cells were then immunostained with anti-myc antibody (9E10; Santa Cruz Biotechnology), followed by Cy3-conjugated secondary antibody, or with anti-Bcl-2 antibody, followed by Cy5-conjugated secondary antibody. GFP-RXRα/385, myc-Nur77, and Bcl-2 were visualized by confocal microscopy. Overlay 1 represents the merger of the GFP-RXRα/385 and myc-Nur77 images, and overlay 2 is a merge of the GFP-RXRα/385, myc-Nur77, and Bcl-2 images. Approximately 70% of cells transfected with GFP-RXRα/385 and myc-Nur77 exhibited colocalization with Bcl-2, whereas <5% of cells transfected with GFP-RXRα/385 alone showed colocalization with Bcl-2. (B) Accumulation of RXRα/385 at mitochondria in HEK293T cells requires both Nur77 and Bcl-2. Flag-RXRα/385, GFP-Nur77/Δ2, and Bcl-2 were transfected into HEK293T cells as indicated. HM and cytosolic fractions were prepared and levels of Flag-RXRα/385, GFP-Nur77/Δ2, and Bcl-2 were determined by immunoblotting with anti-Flag (Sigma), anti-GFP, and anti-Bcl-2 antibody, respectively. For control, levels of Hsp60 and PARP were also determined. One of two similar experiments is shown.
Mitochondrial targeting and apoptotic effect of RXRα and Nur77 mutants.
To study the role of cytoplasmic mutants of Nur77 and RXRα in apoptosis, GFP-RXRα/C1, GFP-RXRα/385, GFP-Nur77/ΔC3, and GFP-Nur77/ΔDBD were transfected into LNCaP cells. Cells were then analyzed for nuclear morphological changes by DAPI staining (Fig. 6A). Interestingly, cells transfected with the Nur77 mutants exhibited extensive nuclear fragmentation and condensation, which are characteristics of apoptotic cells. In contrast, cells transfected with the RXRα mutants displayed normal nuclear morphology. Consistently, expression of Nur77/ΔDBD in LNCaP cells targeted mitochondria and triggered extensive cytochrome c release, while expression of RXR/C1 did not (Fig. 6B). The mitochondrial targeting of Nur77/ΔDBD in LNCaP cells but not in HEK293T cells likely reflects the expression of Bcl-2 in LNCaP cells but not in HEK293T cells (43). Thus, the cytoplasmic Nur77 mutants are capable of targeting mitochondria and inducing cytochrome c release and apoptosis in LNCaP cells, whereas the RXRα mutants are not. These results suggest that RXRα may mainly act as a helper factor to facilitate the translocation of Nur77 from the nucleus to the cytoplasm.
FIG. 6.
Mitochondrial targeting and apoptosis effects of cytoplasmic RXRα and Nur77 mutants. (A) Cytoplasmic Nur77 mutants but not RXRα mutants induce apoptosis. The indicated GFP-RXRα or GFP-Nur77 mutant was transfected into LNCaP cells. After 48 h, cells were stained by DAPI and analyzed for nuclear morphological change by microscopy. Arrows indicate cells with extensive nuclear condensation or fragmentation. Percentages of apoptotic cells were determined by examining 200 GFP-positive cells for nuclear fragmentation and/or chromatin condensation. Bars represent averages ± the means from three experiments. (B) Cytoplasmic Nur77 mutant but not RXRα mutant induces cytochrome c (Cyt) release. GFP-Nur77/ΔDBD or GFP-RXRα/C1 expression vector was transfected into LNCaP cells. Cells were then stained for mitochondria (Hsp60) and cytochrome c and analyzed by confocal microscopy. Cytochrome c release was observed in 80% of cells showing Nur77/ΔDBD mitochondrial targeting, whereas it was not found in cells transfected with RXRα/C1. One of two similar experiments is shown.
Regulation of the RXRα NES activity by ligand binding and RXRα homodimerization.
To determine whether RXRα ligands inhibited RXRα NES activity through their induction of RXRα dimerization (88, 90), we first examined their effects on the cytoplasmic accumulation of RXRα/C1 in HEK293T cells. Treatment of GFP-RXRα/C1-transfected cells with the RXRα ligand 9-cis-RA or SR11237 resulted in the diffuse distribution of RXRα/C1 throughout the cells, whereas treatment with the RAR ligand trans-RA and RARα subtype-selective Am80 produced no such effect (Fig. 7A). The inhibitory effect of the RXR ligands on the cytoplasmic accumulation of RXRα/C1 was confirmed by immunoblotting analysis (Fig. 7B), which showed an equal distribution of GFP-RXRα/C1 in both cytoplasmic and nuclear fractions after cells were treated with 9-cis-RA or SR11237 but not with RARα ligand Am80. In contrast, GFP-RXRα/C1 was only found in the cytoplasm in nontreated cells.
FIG. 7.
Regulation of RXRα nuclear export by its ligands and homodimerization. (A and B) Analysis of subcellular localization of RXRα/C1 in the absence or presence of retinoids. (A) GFP-RXRα/C1 was transfected into HEK293T cells, which were then treated with the indicated retinoid, stained with Hsp60, and analyzed by confocal microscopy. The inhibitory effect of RXR ligands on the cytoplasmic localization of RXRα/C1 was observed in 80% of transfected cells, whereas >90% of transfected cells failed to respond to RAR ligands. (B) Nuclear and cytoplasmic extracts were also prepared and analyzed for expression of GFP-RXRα/C1 by Western blotting with anti-GFP antibody. One of three similar experiments is shown. (C) RXRα/C1 dimerization status determines its subcellular localization. GFP-RXRα/C1 was transfected into HEK293T cells, which were not treated or treated with 9-cis-RA (10−7 M). Nuclear and cytoplasmic extracts were prepared and analyzed by using nondenaturing PAGE and anti-GFP antibody. The same extracts were analyzed by denaturing PAGE for the expression of PARP and Hsp60 to ensure fraction purity. One of two similar experiments is shown. (D) Confocal microscopy analysis of RXR homodimerization-defective mutants. GFP-RXRα/385, GFP-RXRα/LLL, or GFP-RXRα/C1/C432R was transfected into HEK293T cells. Cells were treated with or without 9-cis-RA (10−7 M) and analyzed by confocal microscopy. Approximately 80% of transfected cells showed nuclear localization of GFP-RXRα, which was slightly increased to 85% after treatment with 9-cis-RA. Cytoplasmic localization of GFP-RXRα/385 (90%), GFP-RXRα/LLL (65%), and GFP-RXRα/C1/C432R (85%) was not affected by 9-cis-RA treatment. One of three similar experiments is shown.
We next examined the cellular distribution of GFP-RXRα/C1 expressed in HEK293T cells in the absence or presence of 9-cis-RA by nondenaturing PAGE. As shown in Fig. 7C, GFP-RXRα/C1 was found only as a monomer in the cytoplasmic fraction. After cells were treated with 9-cis-RA, the monomeric form of GFP-RXRα/C1 in the cytoplasmic fraction disappeared, and only the homodimeric form was detected in the nuclear fraction. These studies demonstrate that monomeric RXRα exists in the cytoplasm, whereas liganded-homodimeric RXRα is in the nucleus. Thus, the RXR ligands 9-cis-RA and SR11237 inhibit RXRα nuclear export probably by inducing RXRα homodimerization. We also analyzed the subcellular localization of two RXRα mutants: RXRα/385, which lacks the major homodimerization domain, and RXRα/LLL, which has Leu418, Leu420, and Leu430 in helix 10 replaced with Ala and fails to homodimerize in response to 9-cis-RA (90). Interestingly, both homodimerization-defective RXRα mutants were exclusively localized in the cytoplasm regardless of the presence of 9-cis-RA (Fig. 7D). We also used another RXRα mutant, RXRα/C1/C432R, to study the role of ligand in the regulation of RXRα subcellular localization. Cys432 is involved in the formation of the RXRα ligand-binding pocket and primarily responsible for the shorter length of this L-shaped pocket compared to the linear RAR I-shaped pocket (13). Replacing Cys432 with Arg (RXRα/C1/C432R) impaired homodimerization in response to 9-cis-RA (data not shown). RXR/C1/C432R expressed in HEK293T cells was stained exclusively in the cytoplasm despite 9-cis-RA treatment (Fig. 7D). Together, these data demonstrate that subcellular localization of RXRα depends on its dimerization status and that homodimerization suppresses RXRα nuclear export.
Regulation of RXRα NES activity by heterodimerization.
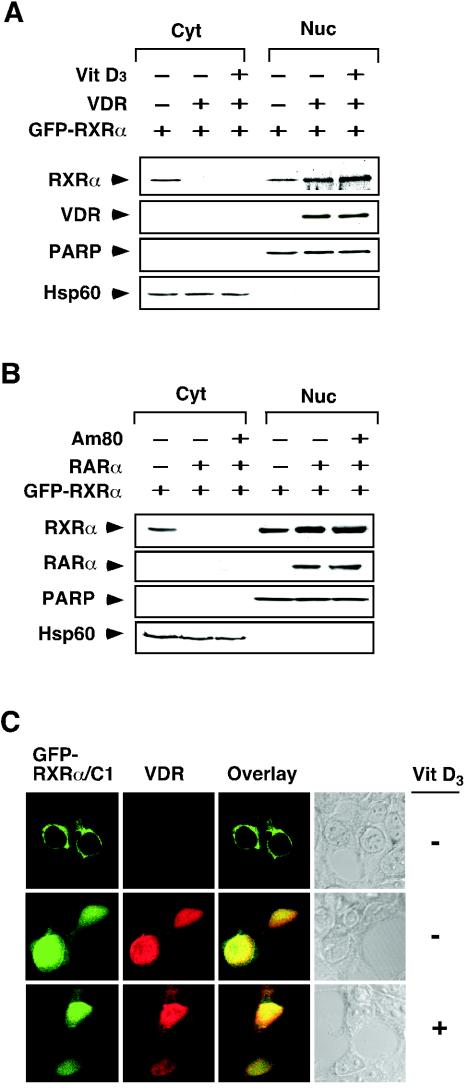
Heterodimerization of RXRα with other receptors, such as RAR and VDR, is required for their efficient DNA binding and transactivation (29, 46, 89). Therefore, we analyzed the subcellular localization of RXRα/VDR and RXRα/RAR heterodimers. GFP-RXRα was transfected alone or with either VDR (Fig. 8A) or RARα (Fig. 8B) into HEK293T cells. Without cotransfection, GFP-RXRα was distributed in both the cytoplasm and the nucleus. However, on cotransfection of VDR or RARα, the cytoplasmic localization of GFP-RXRα was completely abolished and the levels of nuclear GFP-RXRα increased. Treatment of cells with the VDR ligand 1α,25-dihydroxyvitamin D3 or the RAR ligand trans-RA did not further modulate the cellular distribution of cotransfected GFP-RXRα. The inhibitory effect of VDR was also examined by studying the subcellular localization of RXRα/C1 in the absence or presence of VDR coexpression. In the absence of VDR, RXRα/C1 exclusively localized in the cytoplasm of HEK293T cells. However, when VDR was cotransfected, RXRα/C1 was confined to the nucleus and its distribution pattern overlaid with that of VDR. Again, addition of 1α,25-dihydroxyvitamin D3 had no effect on localization of both VDR and RXRα/C1 (Fig. 8C). Together, these results demonstrate that heterodimerization of RXRα with RAR and VDR suppresses RXRα NES activity in a ligand-independent manner.
FIG. 8.
Regulation of RXRα nuclear export by heterodimerization. (A and B) Regulation of RXR nuclear export by VDR (A) and RARα (B). Expression vector for VDR or RARα was transfected into HEK293T cells, together with GFP-RXRα expression vector. Cells were then treated with the indicated ligand, Vit D3 (10−7 M) or Am80 (10−6 M). Nuclear and cytoplasmic extracts were prepared and analyzed for expression of transfected GFP-RXRα by anti-GFP antibody. Expression of transfected VDR or RARα was also determined. One of two similar experiments is shown. (C) Confocal analysis of the effect of VDR expression on RXR localization. The GFP-RXRα/C1 expression vector was transfected into HEK293T cells together with or without the VDR expression vector. Cells were then treated with VD3 (10−7 M),stained with anti-VDR antibody (Santa Cruz Biotechnology), followed by Cy3-conjugated secondary antibody (Sigma) and analyzed by confocal microscopy. About 70% of transfected cells showed cytoplasmic localization of RXRα/C1, whereas >80% of cotransfected cells showed nuclear localization of RXRα/C1 and VDR, which was not enhanced by VD3 treatment. One of three similar experiments is shown.
Unique RXRα heterodimerization with Nur77.
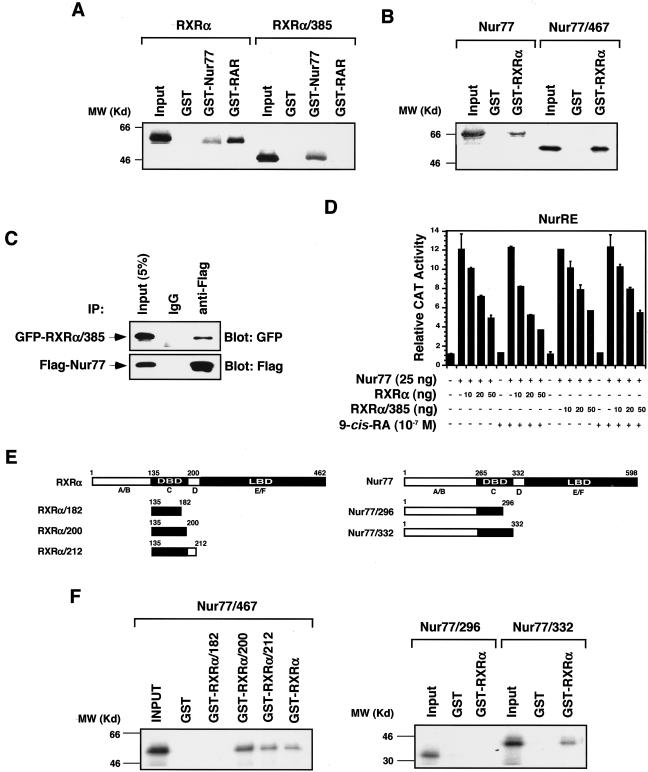
The data presented above demonstrate that RXRα NES activity was suppressed upon RXRα homodimerization and RXRα heterodimerization with VDR and RAR. Ironically, RXRα was required for the cytoplasmic localization of Nur77 through their heterodimerization. One possible explanation is that the RXRα/Nur77 heterodimer may be different from other RXRα heterodimers. Three types of RXR heterodimers have been described (29, 46). In some heterodimers, such as the RXR/VDR, RXR is a completely silent partner. In others, such as the RXR/RAR, RXR is a conditionally silent partner. In contrast, RXR acts as a fully active and competent partner of heterodimers with certain orphan receptors, such as Nur77 (16, 54). Heterodimerization of RXRα with RARα in solution largely depends on their dimerization interfaces localized in their LBDs and has been mapped to a region in the carboxyl terminus, corresponding to helices 9 and 10 in the canonical nuclear receptor LBD structure (5, 13, 17, 18). The results of studies on RXR/Nur77 heterodimerization that indicated different interaction modalities depending on systems and approaches (1, 61, 68) suggest that RXR and Nur77 may interact differently under different conditions. Our finding that RXRα/385 was able to shuttle Nur77/467 from the nucleus to the cytoplasm (Fig. 4G) suggests that RXRα may utilize regions other than the LBD C terminus for binding Nur77. This possibility was studied by using GST pull-down assays. Full-length RXRα was pulled down by either GST-Nur77 or GST-RARα (Fig. 9A), whereas RXRα/385 was only pulled down by GST-Nur77 but not by GST-RARα. Thus, in solution the RXRα C terminus is required for its interaction with RARα but not with Nur77. Similarly, as Nur77/467 was effectively pulled down by GST-RXRα (Fig. 9B), the Nur77 C terminus was also dispensable for interaction with RXRα in solution. The interaction between RXRα/385 and Nur77 was also revealed by in vivo coimmunoprecipitation, which showed the efficient precipitation of GFP-RXRα/385 by anti-Flag antibody when Flag-Nur77 was coexpressed in HEK293T cells (Fig. 9C). In the reporter gene assays, both RXRα/385 and RXRα similarly inhibited the transactivation of Nur77 homodimer activity in the absence of 9-cis-RA (Fig. 9D). The addition of 9-cis-RA slightly enhanced the inhibitory effect of RXRα but not RXRα/385. The RXRα DBD also contains a dimerization interface (38, 55, 60, 85). To determine whether the dimerization interfaces in the DBDs of RXRα and Nur77 are involved in their heterodimerization in solution, several RXRα and Nur77 mutants were constructed (Fig. 9E) and their interaction was analyzed. The RXRα DBD alone (RXRα/135-200) was as efficient as the whole-length RXRα in pulling down Nur77/467. Removal of the C-terminal portion from the RXRα DBD completely abolished its interaction with Nur77/467 (Fig. 9F). Similarly, Nur77 deleted with the D-E/F domains (Nur77/332) retained the ability to bind RXRα, whereas further deletion of the C-terminal portion from the Nur77 DBD completely abolished its ability to bind RXRα. Together, these results demonstrate that the dimerization interfaces in the DBDs of RXRα and Nur77 are mainly responsible for their interaction in solution.
FIG. 9.
C termini of RXRα and Nur77 are not required for RXRα/Nur77 interaction in solution. (A and B) GST pull-down assays for determination of RXRα/Nur77 heterodimerization. GST-Nur77, GST-RXRα, or GST control protein immobilized on glutathione-Sepharose (20 μl) was incubated with in vitro synthesized 35S-labeled Nur77, RXRα or their mutants (5 μl) as indicated. Bound proteins were analyzed by SDS-PAGE and autoradiography. One of three similar experiments is shown. (C) Coimmunoprecipitation assay for Nur77 and RXRα/385 interaction. Expression vectors for Flag epitope-tagged-Nur77 (Flag-Nur77) and GFP-RXRα/385 were cotransfected into HEK293T cells. The expressed Flag-Nur77 and GFP-RXRα/385 were then immunoprecipitated by using either anti-Flag antibody or control IgG, and immunoprecipitates were examined by Western blotting with anti-GFP antibody. The same membranes were also blotted with anti-Flag antibody to determine precipitation specificity and efficiency. Input represents 5% of total cell extract used in the precipitation assays. One of two similar experiments is shown. (D) Reportergene assay. (NurRE)2-tk-chloramphenicol acetyltransferase (CAT) (100 ng), β-galactosidase (β-Gal; 100 ng), and Nur77 (25 ng) expression vectors were transiently transfected into HEK293T cells together with or without RXRα or RXRα/385. Cells were treated with or without 9-cis-RA (10−7 M) as indicated. CAT activity was determined and normalized relative to β-Gal activity. Bars represent averages ± deviations from three different experiments. (E) Schematic representations of RXRα and Nur77 mutants. The DBD, LBD, and A to F domains are indicated. (F) GST pull-down assays for determination of RXRα/Nur77 heterodimerization. The indicated GST-RXRα, its mutants, or GST control protein was immobilized on glutathione-Sepharose (20 μl) and incubated with in vitro-synthesized 35S-labeled Nur77 or its mutants (5 μl) as indicated. Bound proteins were analyzed by SDS-PAGE and autoradiography. One of three similar experiments is shown.
Modulation of RXRα/Nur77 heterodimerization by RXR ligands.
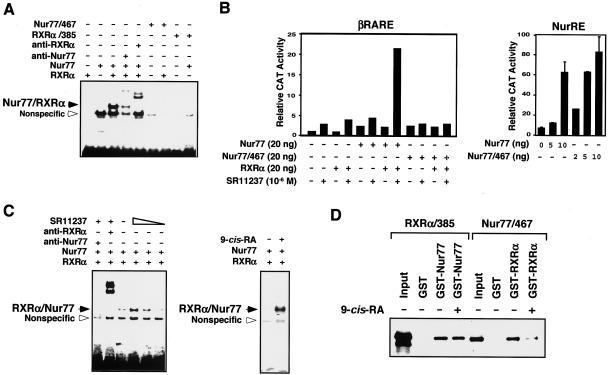
The RXR/Nur77 heterodimer binds and activates DNA sequences consisting of AGGTCA or like motifs arranged as a direct repeat with a 5-bp spacing (the DR-5 response element) (16, 54, 76). To characterize RXRα/Nur77 heterodimerization on DNA, gel shift assays were conducted by using the βRARE, a DR-5 element, as a probe (Fig. 10A). In vitro-synthesized Nur77 and RXRα bound to the βRARE as a heterodimer (Fig. 10A), as reported previously (76, 77). However, when Nur77/467, which heterodimerizes with RXRα in solution (Fig. 9B), was incubated with RXRα, we did not detect any heterodimeric complex binding to the βRARE (Fig. 10A). Similarly, coincubation of RXRα/385 and Nur77 did not result in formation of a stable heterodimer on the βRARE (Fig. 10A). Thus, the C termini of both Nur77 and RXRα are required for the formation of a stable RXRα/Nur77 heterodimer complex on the βRARE. This finding is also supported by our analysis of Nur77 mutants for their transactivation activity on the NurRE and βRARE (Fig. 10B). Removal of the Nur77 C terminus did not affect its transactivation on NurRE as Nur77 and Nur77/467 similarly activated the NurRE (Fig. 10B). These results are consistent with recent reports that the major transactivation function of Nur77 is located in its N terminus (68). In contrast, Nur77/467 failed to activate the βRARE on cotransfection with RXRα in either the absence or presence of RXR ligand SR11237, whereas cotransfection of full-length Nur77 and RXRα strongly activated the βRARE in response to SR11237 (Fig. 10B). The lack of transactivation activity displayed by Nur77/467 is likely a reflection of its inability to form βRARE- bound heterodimers with RXRα (Fig. 10A).
FIG. 10.
9-cis-RA modulates RXRα/Nur77 heterodimerization and promotes DNA binding. (A) The C-terminal domains of Nur77 and RXRα are required for the formation of RXRα/Nur77 heterodimer on DNA. The indicated Nur77, RXRα, or their mutants were synthesized in vitro and analyzed for binding to the βRARE by gel shift assays. One of three similar experiments is shown. (B) Differential requirement of the Nur77 C terminus for transactivation of the Nur77 response element NurRE and the βRARE. (NurRE)2-tk-CAT (100 ng) or βRARE-tk-CAT (100 ng), β-Gal expression vector (100 ng), and the expression vector for Nur77 or a Nur77 mutant (20 ng) were transiently transfected into HEK293T cells with or without the RXRα expression vector. The CAT activity was determined and normalized relative to the β-Gal activity. One of three similar experiments is shown. (C) Effect of RXR ligands on binding of RXRα-Nur77 heterodimers to the βRARE. Equivalent amounts of in vitro synthesized Nur77 and RXRα were incubated alone or together with or without RXR ligand 9-cis-RA (10−7 M) or SR11237 (10−6 M) and analyzed by gel retardation assays with the βRARE as a probe. One of two similar experiments is shown. (D) RXR ligand modulates RXRα/Nur77 heterodimerization in solution. GST pull-down assays for determination of RXRα/Nur77 heterodimerization. The indicated GST fusions immobilized on glutathione-Sepharose (20 μl) were incubated with in vitro synthesized 35S-labeled receptor protein (5 μl) as indicated. Bound proteins were analyzed by SDS-PAGE and/or autoradiography. One of two similar experiments is shown.
To determine how RXR ligands influenced RXRα/Nur77 heterodimer binding to the βRARE, RXRα protein was exposed to its ligand SR11237 or 9-cis-RA prior to incubation with Nur77. Although the RXRα/Nur77 heterodimer bound to the βRARE in the absence of an RXR ligand, the addition of SR11237 or 9-cis-RA strongly enhanced binding by the heterodimer (Fig. 10C). These data suggest that RXRα/Nur77 heterodimers in solution may be incompetent for DNA binding and that RXR ligand binding may induce a C-terminal-stabilized heterodimer conformation that favors DNA binding. To address the possibility that an RXR ligand can modulate the RXRα/Nur77 heterodimerization interface, we used GST pull-down assays to study the effect of 9-cis-RA on the interaction between Nur77/467 and RXRα (Fig. 10D). Incubation of GST-RXRα with 9-cis-RA strongly reduced the ability of GST-RXRα to interact with Nur77/467, suggesting that 9-cis-RA binding may mask or alter a putative dimerization interface required for RXRα binding to Nur77 in solution. In comparison, the addition of 9-cis-RA did not affect the binding of RXRα/385 to GST-Nur77. Thus, the dimerization interface for the formation of the RXRα/Nur77 heterodimer in solution is different from that required for the formation of the DNA-bound heterodimer and is regulated by RXR ligand binding.
RXRα mitochondrial localization is inhibited by RXRα ligands.
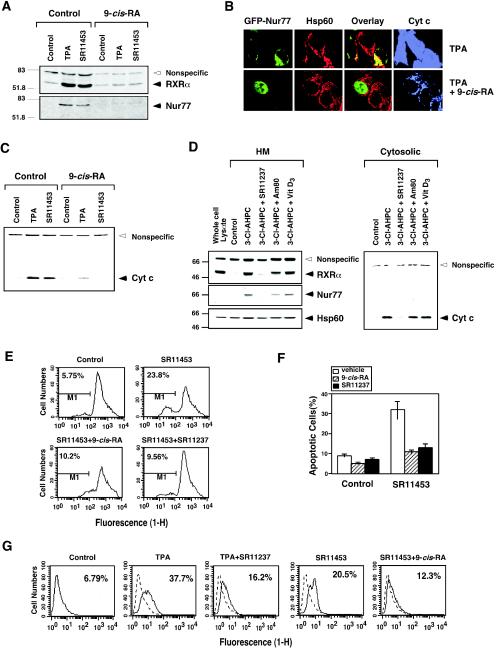
Inhibition of RXRα mitochondrial localization by its ligands was demonstrated by immunoblotting HM fractions from LNCaP cells. The accumulation of RXRα in the HM fraction after cells were treated with TPA or an analog of AHPN/CD437 (SR11453) (40) was inhibited by pretreatment with 9-cis-RA (Fig. 11A). Similar to its effect on RXRα, mitochondrial accumulation of Nur77 was also abolished by 9-cis-RA pretreatment (Fig. 11A). The observation that 9-cis-RA inhibited Nur77 mitochondrial targeting further suggests the role of RXRα in the regulation of Nur77 mitochondrial targeting.
FIG. 11.
Effect of RXR ligands on RXR mitochondrial targeting and apoptosis. (A) Effect of 9-cis-RA on mitochondrial localization of RXRα and Nur77. HM fractions were prepared from LNCaP cells treated with or without TPA or SR11453 (10−6 M) for 3 h with or without a 9-cis-RA (10−7 M) pretreatment for 12 h and then analyzed for expression of Nur77 or RXRα by immunoblotting. One of two similar experiments is shown. (B) 9-cis-RA inhibits Nur77-dependent release of cytochrome c from mitochondria. GFP-Nur77 expression vector was transfected into LNCaP cells, treated with or without 9-cis-RA for 12 h before TPA treatment (1 h). Cells were stained for mitochondria (Hsp60) and cytochrome c, and analyzed by confocal microscopy. 100% of cells displayed diffused cytochrome c staining when treated with TPA, whereas 60% of cells showed punctate cytochrome c staining when cells were cotreated with TPA and 9-cis-RA. (C) Inhibition of cytochrome c release by 9-cis-RA. LNCaP cells were treated with or without 9-cis-RA (10−7 M) for 12 h before treatment with TPA (100 ng/ml) or SR11453 (10−6 M) for 1 h. Cytosolic fractions were analyzed for cytochrome c by immunoblotting. A nonspecific band at ∼70 kDa served as a control for equal loading of proteins. One of three similar experiments is shown. (D) RAR and VDR ligands fail to inhibit mitochondrial localization of RXRα and Nur77 and cytochrome c release. LNCaP cells were treated with 3-Cl-AHPC (10−6 M) for 3 h after pretreatment with SR11237 (10−6 M), RAR ligand Am80 (10−6 M), or VDR ligand vitamin D3 (10−7 M). HM and cytosolic fractions were prepared and analyzed for mitochondrial localization of RXRα and Nur77, as well as the cytochrome c release described above. (E) RXR ligands prevent SR11453 induced mitochondrial membrane potential change (Δψm). LNCaP cells were pretreated with 9-cis-RA or SR11237 for 12 h before treatment with SR11453 (10−6 M) for 18 h. Cells were then incubated with Rh123 for 30 min and analyzed by FACScalibur cytometry. One of three similar experiments is shown. (F) Inhibition of apoptosis by RXR ligands. LNCaP cells were pretreated with 9-cis-RA or SR11237 for 12 h before treatment with SR11453 for 48 h. Apoptosis was determined by nuclear staining with DAPI. The percentages of cells fluorescing within the range of Rh123 were considered as depolarized (i.e., Δψm disrupted). Bars represent averages ± the standard deviations from two experiments. (G) LNCaP cells were pretreated 9-cis-RA or SR11237 for 12 h before treatment with SR11453 or TPA for 24 h. Apoptosis was determined by the TdT assay. One of two similar experiments is shown. Cyt c, cytochrome c.
Since RXRα comigrated with Nur77 from the nucleus to mitochondria in cells undergoing apoptosis, we investigated the effect of 9-cis-RA on cytochrome c release and apoptosis. Treatment of LNCaP cells with TPA induced the massive release of cytochrome c from mitochondria, as revealed by cytochrome c staining, which was diffusely distributed (Fig. 11B). However, on pretreatment with 9-cis-RA, cytochrome c staining was only detected in the mitochondria, since its distribution colocalized with that of Hsp60. LNCaP cells were also transfected with GFP-Nur77 to monitor the effect of 9-cis-RA on Nur77 mitochondrial targeting (Fig. 11B). TPA-induced cytochrome c release was accompanied with the mitochondrial localization of transfected GFP-Nur77. However, when cells were pretreated with 9-cis-RA, GFP-Nur77 was confined in the nucleus and cytochrome c release was inhibited (Fig. 11B). These data further demonstrate the role of Nur77 mitochondrial targeting in the induction of cytochrome c release and the inhibitory effect of RXR ligands on Nur77-dependent apoptosis. The inhibition of cytochrome c release by 9-cis-RA was further revealed by immunoblotting of cytosolic fractions prepared from LNCaP cells treated with TPA or the AHPN analog SR11453 (Fig. 11C). In the absence of 9-cis-RA, significant cytoplasmic cytochrome c was detected in treated cells, whereas preexposure of cells to 9-cis-RA prevented cytochrome c release (Fig. 11C). We also examined the effects of RAR and VDR ligands on mitochondrial localization of RXRα and Nur77 and cytochrome c release in LNCaP cells treated with 3-Cl-AHPC. Unlike the inhibitory effect of RXR ligand, treatment of LNCaP cells, which express RARs and VDR, with either RAR ligand Am80 or VDR ligand had no effect on 3-Cl-AHPC-induced mitochondrial localization of RXRα and Nur77 and cytochrome c release (Fig. 11D), a finding in agreement with our cotransfection results (Fig. 8). Consistent with their effects on cytochrome c release, 9-cis-RA and SR11237 strongly antagonized the effect of SR11453 on mitochondrial membrane potential change (Fig. 11E) and apoptosis of LNCaP cells revealed by DAPI staining (Fig. 11F) and the TdT assay (Fig. 11G). Thus, 9-cis-RA inhibits cytochrome c release and apoptosis in LNCaP cells by preventing nuclear export of the RXRα/Nur77 heterodimer.
DISCUSSION
We previously reported that Nur77 translocated from the nucleus to the cytoplasm, where it targeted mitochondria to induce apoptosis (40). Our results presented here demonstrate that the translocation of Nur77 requires its heterodimerization with RXRα. This is illustrated by our observations that Nur77 and RXRα colocalized in the cytoplasm (Fig. 1C) and that they targeted mitochondria simultaneously in LNCaP prostate cancer cells (Fig. 2). In addition, their mitochondrial targeting depended on their coexpression (Fig. 1). Moreover, expression of cytoplasmic RXRα mutant RXRα/385 conferred cytoplasmic localization to Nur77 (Fig. 4G), whereas overexpression of Nur77/Δ1 retained RXRα in the nucleus (Fig. 1G). These results are consistent with our observation that Nur77 mitochondrial targeting and its induction of cytochrome c release and apoptosis were suppressed by RXR ligands (Fig. 11). The translocation of the RXRα/Nur77 heterodimer from the nucleus to mitochondria is a rapid process, occurring 1 h after cells are exposed to an apoptotic stimulus, and precedes the release of cytochrome c from mitochondria (Fig. 2). These results agree with our previous observation (40) and are consistent with the notion that the mitochondrial targeting of RXRα/Nur77 triggers apoptosis. Apoptosis induction by Nur77 mitochondrial targeting occurs in different types of cancer cells (10, 24, 28, 31, 37, 40, 43, 71, 79). Our results demonstrated that RXRα was also required for Nur77 translocation in H460 lung cancer cells (Fig. 1H and 2C). Thus, it is likely that RXRα and its ligands play a critical role in regulating Nur77-dependent apoptosis in various cancer cells.
We recently reported that Nur77 mitochondrial targeting is mediated through its interaction with Bcl-2, which mainly resides on the mitochondrial outer membrane (43). Consistently, Nur77 mutants, such as Nur77/ΔDBD, targeted mitochondria in LNCaP cells (Fig. 6B), which express Bcl-2, but not in HEK293T cells (Fig. 3C), which do not. Interestingly, mitochondrial targeting by RXRα occurred in HEK293T cells only when both Nur77 and Bcl-2 were coexpressed (Fig. 5), while expression of Bcl-2 alone did not result in RXRα mitochondrial targeting. Transfection of Nur77 mutants, but not RXRα mutants, into LNCaP cells targeted mitochondria and induced apoptosis (Fig. 6). These results suggest that RXRα does not interact with Bcl-2 but rather functions as a shuttling protein in the Nur77-dependent apoptotic pathway.
The migration of RXRα/Nur77 heterodimers from the nucleus to the cytoplasm is CRM1 dependent. By extensive mutational analysis, we have identified an NES in the RXRα LBD that is required for the efficient nuclear export of RXRα and RXRα/Nur77 heterodimers (Fig. 4). A recent study suggested that NGFI-B has several NESs that are required for RXR/NGFI-B nuclear export in PC12 cells (30). The fact that RXRα also possesses an NES, which alone is capable of translocating GFP from the nucleus to the cytoplasm, offers an explanation for the efficient nuclear export of the RXRα/Nur77 heterodimer.
One unique property of the RXRα NES is that it lies in helix 7 of the RXR LBD, which undergoes an unusual conformational changes upon homodimerization, homotetramerization, and heterodimerization with RAR and PPAR (5, 17, 18). The RXRα helix 7 is an α-helix structure in the RXRα monomer (4). However, in the context of RXRα dimer, tetramer, or heterodimer with PPARγ or RAR, a π-helix is formed due to the presence of a glutamic acid residue (E352) in the middle of helix 7 (5, 17, 18). The transformation of helical geometry in the region, wherein the RXRα NES lies, suggests that RXRα NES activity is subject to regulation by RXRα homodimerization and heterodimerization. Indeed, subcellular localization of RXRα is highly regulated by its dimerization. This finding is clearly illustrated by our analysis of RXRα distribution patterns by using nondenaturing PAGE (Fig. 7C) that shows that RXRα/C1 existed as a monomer in the cytoplasm but in response to 9-cis-RA resided as a homodimer in the nucleus. RXRα mutants unable to homodimerize predominantly resided in the cytoplasm (Fig. 7D). Heterodimerization with certain nuclear receptors, including RARα and VDR, also suppressed RXR NES activity (Fig. 8). Thus, the RXRα NES is active in the RXRα monomer but is silenced by RXRα homodimerization and certain heterodimerizations. Such a conformational-change-mediated regulation of RXRα NES activity represents a unique regulatory mechanism that dictates subcellular localization of various RXRα homodimers and heterodimers, ultimately allowing efficient transcriptional regulation by RXRα homodimers and certain RXRα heterodimers. Our results are consistent with previous observations that RXRs play a critical role in determining nuclear localization of TR (2) and VDR (57, 58) through their heterodimerization.
Very intriguingly, heterodimers formed by RXRα and VDR or RAR reside in the nucleus, whereas RXRα/Nur77 is found in the cytoplasm when cells were treated with apoptotic stimuli (Fig. 1). Our results demonstrate that nuclear export of the RXRα/Nur77 heterodimer is due to its unique heterodimerization in solution (Fig. 9 and 10). RXR possesses two dimerization interfaces, which are located in the DBD and the LBD (55, 86). The strong dimerization interface in the LBD enables RXR homodimerization and heterodimerization with certain receptors, such as RAR and VDR in solution. In contrast, the weak dimerization interface in the DBD does not allow the formation of the RXR homodimer or heterodimer with RAR in the absence of a specific response element (60). Our data indicate that deletion of the major dimerization interface from the RXRα LBD (RXRα/385) did not impair the interaction of RXRα with Nur77 in solution, as revealed by an in vitro GST pull-down assay (Fig. 9A) and an in vivo coimmunoprecipitation assay (Fig. 9C), although the deletion completely abolished RXRα/385 interaction with RAR (Fig. 9A). Similarly, deletion of the C-terminal sequence from Nur77 had no effect on its interaction with RXRα (Fig. 9B). Our observation that the RXRα DBD alone was sufficient to interact with Nur77 (Fig. 9F) demonstrated that the formation of the RXRα/Nur77 heterodimer in solution is mediated by dimerization interfaces in their DBDs. Given the fact that the RXRα NES is active in the RXRα monomer conformation, we envision that the unique RXRα/Nur77 heterodimer formed in solution through their DBD dimerization interfaces will ensure that the RXRα NES is situated in its active conformation, resulting in their nuclear export. This notion is consistent with our observation that RXRα/385 is able to shuttle Nur77 to the cytoplasm (Fig. 4G). Thus, the unique property of RXR dimerization interfaces allows cross talk among RXR heterodimerization partners with respect to their subcellular localization and function (Fig. 8).
In contrast to their heterodimerization in solution, the C-terminal sequences of Nur77 and RXRα are required for the efficient binding of the RXRα/Nur77 heterodimer to their specific response element βRARE (Fig. 10A) and transactivation (Fig. 10B), as is found for the RXR/RAR and RXR/TR heterodimers (87). Our observation that an RXR ligand can promote RXRα/Nur77 heterodimer binding to the βRARE (Fig. 10C) is interesting since RXR ligands have not been previously shown to modulate DNA binding of other RXR heterodimers at least in vitro (87). This finding suggests that ligand binding can modulate RXRα/Nur77 interaction to favor DNA binding and transactivation. Such a modulation is reflected by the inhibition by 9-cis-RA of the heterodimerization of RXRα with Nur77/467 in solution (Fig. 10D). Thus, the induction of RXRα/Nur77 heterodimer DNA binding and transactivation by 9-cis-RA is associated with its inhibition of their DBD-mediated dimerization. It is tempting to speculate that ligand binding allows RXRα to interact with Nur77 through their LBD dimerization interfaces, which may silence the RXRα NES, as does RXR/VDR or RXR/RAR heterodimerization (Fig. 8). Thus, the nuclear export of the RXRα/Nur77 heterodimer may be suppressed by 9-cis-RA through its induction RXRα homodimerization or modulation of RXRα/Nur77 heterodimerization interfaces.
We found that RXR ligands 9-cis-RA and SR11237 effectively inhibited the release of cytochrome c induced by TPA or SR11453 in LNCaP cells (Fig. 11). Since the inhibition was accompanied by the prevention of Nur77 and RXRα mitochondrial targeting (Fig. 11B), we suggest that RXR ligands suppress apoptosis by inhibiting mitochondrial targeting of the RXRα/Nur77 heterodimer. The inhibitory effect of RXR ligands on apoptosis has been reported previously (3, 30, 65, 81-83). 9-cis-RA is known to potently inhibit the activation-induced apoptosis of T cells and thymocytes (3, 30, 65, 81-83). The inhibitory effect of 9-cis-RA was enhanced in T-cell hybridomas overexpressing RXRβ but attenuated in cells overexpressing dominant-negative RXRβ (82). The present study and the fact that Nur77 expression is necessary for activation-induced T-cell apoptosis suggest that inhibition of activation-induced apoptosis by RXR ligands may be mediated by their modulation of RXRα/Nur77 heterodimer activity.
RXRα/Nur77 nuclear export is also regulated by apoptotic stimuli. Endogenously expressed RXRα and Nur77 were found mainly in the nucleus (Fig. 1A) but resided in the mitochondria after cells were treated with apoptotic stimuli (Fig. 1). How an apoptotic stimulus activates RXRα NES activity remains unknown. Our observations that RXRα and Nur77 nuclear export is highly regulated by their heterodimerization point to the possibility that the apoptotic stimulus may regulate an RXRα/Nur77 heterodimerization interface switch. This is consistent with observations that Nur77 activities are highly regulated by phosphorylation status (9, 15, 19, 23, 49, 53, 69) and many apoptotic stimuli act through various kinase pathways. Our premise is also consistent with the recent observation that Akt, a potent antiapoptotic kinase, phosphorylates Nur77 (49, 53). Interestingly, the location of the Akt phosphorylation site in the Nur77 C-terminal extension points to the possibility that Akt phosphorylation may modulate the interaction of Nur77 with other proteins.
Different RXRα/Nur77 heterodimers may exist in a dynamic equilibrium depending on their cellular environment. Under normal conditions, both the DBD and the LBD dimerization interfaces may participate in RXRα/Nur77 heterodimer formation, so that the dimers exist in both the nucleus and cytoplasm. In response to apoptotic stimuli, RXRα and Nur77 may preferentially heterodimerize through their DBD dimerization interfaces to activate the RXRα NES, resulting in their cytoplasmic localization. In contrast, binding by RXR ligands may induce a dimerization interface switch that silences the RXRα NES to ensure RXRα/Nur77 nuclear localization and efficient transcriptional regulation.
Evidence has been accumulating to demonstrate that many nuclear receptors act nongenotropically to regulate important biological processes. The estrogen receptor and androgen receptor modulate the Src/Shc/Erk signaling pathway in a ligand-dependent manner to regulate cell proliferation. These effects can be dissociated from their transcriptional activity (32, 50). The estrogen receptor can also act outside the nucleus to activate phosphatidylinositol-3-OH kinase activity in a ligand-dependent and transcriptional regulation-independent manner (64). The results presented here demonstrate that RXRα also acts nongenotropically by migrating from the nucleus to mitochondria to trigger cytochrome c release and apoptosis in response to apoptotic stimuli. Thus, nongenotropic action appears to be an important mechanism by which nuclear receptors exert their biological effects. Our identification of a putative RXRα NES in helix 7 reveals an interesting regulatory mechanism that dictates subcellular localization of RXRα and its heterodimerization partners through ligands and dimerization. Our observation that RXR ligands regulate apoptosis by modulating RXRα/Nur77 heterodimer nuclear export provides a novel approach for developing RXR-based apoptosis regulators.
Acknowledgments
We thank L. Frazer for preparation of the manuscript and members of the Zhang laboratory for helpful discussions.
This study was supported in part by grants to X.-K. Zhang and M. I. Dawson from the National Institute of Health, the U.S. Army Medical Research and Material Command, the California Tobacco-Related Diseases Research Program, and the California Breast Cancer Research Program.
REFERENCES
- 1.Aarnisalo, P., C. H. Kim, J. W. Lee, and T. Perlmann. 2002. Defining requirements for heterodimerization between the retinoid X receptor and the orphan nuclear receptor Nurr1. J. Biol. Chem. 277:35118-35123. [DOI] [PubMed] [Google Scholar]
- 2.Baumann, C. T., P. Maruvada, G. L. Hager, and P. M. Yen. 2001. Nuclear cytoplasmic shuttling by thyroid hormone receptors. multiple protein interactions are required for nuclear retention. J. Biol. Chem. 276:11237-11245. [DOI] [PubMed] [Google Scholar]
- 3.Bissonnette, R. P., T. Brunner, S. B. Lazarchik, N. J. Yoo, M. F. Boehm, D. R. Green, and R. A. Heyman. 1995. 9-cis retinoic acid inhibition of activation-induced apoptosis is mediated via regulation of fas ligand and requires retinoic acid receptor and retinoid X receptor activation. Mol. Cell. Biol. 15:5576-5585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bourguet, W., M. Ruff, P. Chambon, H. Gronemeyer, and D. Moras. 1995. Crystal structure of the ligand-binding domain of the human nuclear receptor RXR-α. Nature 375:377-382. [DOI] [PubMed] [Google Scholar]
- 5.Bourguet, W., V. Vivat, J. M. Wurtz, P. Chambon, H. Gronemeyer, and D. Moras. 2000. Crystal structure of a heterodimeric complex of RAR and RXR ligand-binding domains. Mol. Cell 5:289-298. [DOI] [PubMed] [Google Scholar]
- 6.Bunn, C. F., J. A. Neidig, K. E. Freidinger, T. A. Stankiewicz, B. S. Weaver, J. McGrew, and L. A. Allison. 2001. Nucleocytoplasmic shuttling of the thyroid hormone receptor alpha. Mol. Endocrinol. 15:512-533. [DOI] [PubMed] [Google Scholar]
- 7.Chang, C., J. Kokontis, S. S. Liao, and Y. Chang. 1989. Isolation and characterization of human TR3 receptor: a member of steroid receptor superfamily. J. Steroid Biochem. 34:391-395. [DOI] [PubMed] [Google Scholar]
- 8.Chen, G. Q., B. Lin, M. I. Dawson, and X. K. Zhang. 2002. Nicotine modulates the effects of retinoids on growth inhibition and RAR beta expression in lung cancer cells. Int. J. Cancer 99:171-178. [DOI] [PubMed] [Google Scholar]
- 9.Davis, I. J., T. G. Hazel, R. H. Chen, J. Blenis, and L. F. Lau. 1993. Functional domains and phosphorylation of the orphan receptor Nur77. Mol. Endocrinol. 7:953-964. [DOI] [PubMed] [Google Scholar]
- 10.Dawson, M. I., P. D. Hobbs, V. J. Peterson, M. Leid, C. W. Lange, K. C. Feng, G. Chen, J. Gu, H. Li, S. K. Kolluri, X. Zhang, Y. Zhang, and J. A. Fontana. 2001. Apoptosis induction in cancer cells by a novel analogue of 6-[3-(1-adamantyl)-4-hydroxyphenyl]-2-naphthalenecarboxylic acid lacking retinoid receptor transcriptional activation activity. Cancer Res. 61:4723-4730. [PubMed] [Google Scholar]
- 11.DeFranco, D. B. 1999. Regulation of steroid receptor subcellular trafficking. Cell Biochem. Biophys. 30:1-24. [DOI] [PubMed] [Google Scholar]
- 12.Dufour, J. M., and K. H. Kim. 1999. Cellular and subcellular localization of six retinoid receptors in rat testis during postnatal development: identification of potential heterodimeric receptors. Biol. Reprod. 61:1300-1308. [DOI] [PubMed] [Google Scholar]
- 13.Egea, P. F., A. Mitschler, N. Rochel, M. Ruff, P. Chambon, and D. Moras. 2000. Crystal structure of the human RXRα ligand-binding domain bound to its natural ligand: 9-cis retinoic acid. EMBO J. 19:2592-2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elbashir, S. M., J. Harborth, W. Lendeckel, A. Yalcin, K. Weber, and T. Tuschl. 2001. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411:494-498. [DOI] [PubMed] [Google Scholar]
- 15.Fahrner, T. J., S. L. Carroll, and J. Milbrandt. 1990. The NGFI-B protein, an inducible member of the thyroid/steroid receptor family, is rapidly modified posttranslationally. Mol. Cell. Biol. 10:6454-6459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Forman, B. M., K. Umesono, J. Chen, and R. M. Evans. 1995. Unique response pathways are established by allosteric interactions among nuclear hormone receptors. Cell 81:541-550. [DOI] [PubMed] [Google Scholar]
- 17.Gampe, R. T., Jr., V. G. Montana, M. H. Lambert, A. B. Miller, R. K. Bledsoe, M. V. Milburn, S. A. Kliewer, T. M. Willson, and H. E. Xu. 2000. Asymmetry in the PPARγ/RXRα crystal structure reveals the molecular basis of heterodimerization among nuclear receptors. Mol. Cell 5:545-555. [DOI] [PubMed] [Google Scholar]
- 18.Gampe, R. T., Jr., V. G. Montana, M. H. Lambert, G. B. Wisely, M. V. Milburn, and H. E. Xu. 2000. Structural basis for autorepression of retinoid X receptor by tetramer formation and the AF-2 helix. Genes Dev. 14:2229-2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hazel, T. G., R. Misra, I. J. Davis, M. E. Greenberg, and L. F. Lau. 1991. Nur77 is differentially modified in PC12 cells upon membrane depolarization and growth factor treatment. Mol. Cell. Biol. 11:3239-3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hazel, T. G., D. Nathans, and L. F. Lau. 1988. A gene inducible by serum growth factors encodes a member of the steroid and thyroid hormone receptor superfamily. Proc. Natl. Acad. Sci. USA 85:8444-8448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hedvat, C. V., and S. G. Irving. 1995. The isolation and characterization of MINOR, a novel mitogen-inducible nuclear orphan receptor. Mol. Endocrinol. 9:1692-1700. [DOI] [PubMed] [Google Scholar]
- 22.Heyman, R. A., D. J. Mangelsdorf, J. A. Dyck, R. B. Stein, G. Eichele, R. M. Evans, and C. Thaller. 1992. 9-cis retinoic acid is a high-affinity ligand for the retinoid X receptor. Cell 68:397-406. [DOI] [PubMed] [Google Scholar]
- 23.Hirata, Y., K. Kiuchi, H. C. Chen, J. Milbrandt, and G. Guroff. 1993. The phosphorylation and DNA binding of the DNA-binding domain of the orphan nuclear receptor NGFI-B. J. Biol. Chem. 268:24808-24812. [PubMed] [Google Scholar]
- 24.Holmes, W. F., D. R. Soprano, and K. J. Soprano. 2003. Comparison of the mechanism of induction of apoptosis in ovarian carcinoma cells by the conformationally restricted synthetic retinoids CD437 and 4-HPR. J. Cell Biochem. 89:262-278. [DOI] [PubMed] [Google Scholar]
- 25.Holmes, W. F., D. R. Soprano, and K. J. Soprano. 2002. Elucidation of molecular events mediating induction of apoptosis by synthetic retinoids using a CD437-resistant ovarian carcinoma cell line. J. Biol. Chem. 277:45408-45419. [DOI] [PubMed] [Google Scholar]
- 26.Ijpenberg, A., N. S. Tan, L. Gelman, S. Kersten, J. Seydoux, J. Xu, D. Metzger, L. Canaple, P. Chambon, W. Wahli, and B. Desvergne. 2004. In vivo activation of PPAR target genes by RXR homodimers. EMBO J. 23:2083-2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Janssen, J. J., E. D. Kuhlmann, A. H. van Vugt, H. J. Winkens, B. P. Janssen, A. F. Deutman, and C. A. Driessen. 1999. Retinoic acid receptors and retinoid X receptors in the mature retina: subtype determination and cellular distribution. Curr. Eye Res. 19:338-347. [DOI] [PubMed] [Google Scholar]
- 28.Jeong, J. H., J. S. Park, B. Moon, M. C. Kim, J. K. Kim, S. Lee, H. Suh, N. D. Kim, J. M. Kim, Y. C. Park, and Y. H. Yoo. 2003. Orphan nuclear receptor Nur77 translocates to mitochondria in the early phase of apoptosis induced by synthetic chenodeoxycholic acid derivatives in human stomach cancer cell line SNU-1. Ann. N. Y. Acad. Sci. 1010:171-177. [DOI] [PubMed] [Google Scholar]
- 29.Kastner, P., M. Mark, and P. Chambon. 1995. Nonsteroid nuclear receptors: what are genetic studies telling us about their role in real life? Cell 83:859-869. [DOI] [PubMed] [Google Scholar]
- 30.Katagiri, Y., K. Takeda, Z. X. Yu, V. J. Ferrans, K. Ozato, and G. Guroff. 2000. Modulation of retinoid signalling through NGF-induced nuclear export of NGFI-B. Nat. Cell Biol. 2:435-440. [DOI] [PubMed] [Google Scholar]
- 31.Kolluri, S. K., X. Cao, N. Bruey-Sedano, B. Lin, F. Lin, Y. Han, M. I. Dawson, and X. Zhang. 2003. Mitogenic effect of orphan receptor TR3 and its regulation by MEKK1 in lung cancer cells. Mol. Cell. Biol. 23:8651-8667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kousteni, S., T. Bellido, L. I. Plotkin, C. A. O'Brien, D. L. Bodenner, L. Han, K. Han, G. B. DiGregorio, J. A. Katzenellenbogen, B. S. Katzenellenbogen, P. K. Roberson, R. S. Weinstein, R. L. Jilka, and S. C. Manolagas. 2001. Nongenotropic, sex-nonspecific signaling through the estrogen or androgen receptors: dissociation from transcriptional activity. Cell 104:719-730. [PubMed] [Google Scholar]
- 33.Kudo, N., N. Matsumori, H. Taoka, D. Fujiwara, E. P. Schreiner, B. Wolff, M. Yoshida, and S. Horinouchi. 1999. Leptomycin B inactivates CRM1/exportin 1 by covalent modification at a cysteine residue in the central conserved region. Proc. Natl. Acad. Sci. USA 96:9112-9117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Labelle, Y., J. Bussieres, F. Courjal, and M. B. Goldring. 1999. The EWS/TEC fusion protein encoded by the t(9;22) chromosomal translocation in human chondrosarcomas is a highly potent transcriptional activator. Oncogene 18:3303-3308. [DOI] [PubMed] [Google Scholar]
- 35.Labelle, Y., J. Zucman, G. Stenman, L. G. Kindblom, J. Knight, C. Turc-Carel, B. Dockhorn-Dworniczak, N. Mandahl, C. Desmaze, M. Peter, et al. 1995. Oncogenic conversion of a novel orphan nuclear receptor by chromosome translocation. Hum. Mol. Genet. 4:2219-2226. [DOI] [PubMed] [Google Scholar]
- 36.Law, S. W., O. M. Conneely, F. J. DeMayo, and B. W. O'Malley. 1992. Identification of a new brain-specific transcription factor, NURR1. Mol. Endocrinol. 6:2129-2135. [DOI] [PubMed] [Google Scholar]
- 37.Lee, J. M., K. H. Lee, M. Weidner, B. A. Osborne, and S. D. Hayward. 2002. Epstein-Barr virus EBNA2 blocks Nur77-mediated apoptosis. Proc. Natl. Acad. Sci. USA 99:11878-11883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee, M. S., S. A. Kliewer, J. Provencal, P. E. Wright, and R. M. Evans. 1993. Structure of the retinoid X receptor alpha DNA binding domain: a helix required for homodimeric DNA binding. Science 260:1117-1121. [DOI] [PubMed] [Google Scholar]
- 39.Levin, A. A., L. J. Sturzenbecker, S. Kazmer, T. Bosakowski, C. Huselton, G. Allenby, J. Speck, C. Kratzeisen, M. Rosenberger, A. Lovey, et al. 1992. 9-cis retinoic acid stereoisomer binds and activates the nuclear receptor RXR alpha. Nature 355:359-361. [DOI] [PubMed] [Google Scholar]
- 40.Li, H., S. K. Kolluri, J. Gu, M. I. Dawson, X. Cao, P. D. Hobbs, B. Lin, G. Chen, J. Lu, F. Lin, Z. Xie, J. A. Fontana, J. C. Reed, and X. Zhang. 2000. Cytochrome c release and apoptosis induced by mitochondrial targeting of nuclear orphan receptor TR3. Science 289:1159-1164. [DOI] [PubMed] [Google Scholar]
- 41.Li, Y., B. Lin, A. Agadir, R. Liu, M. I. Dawson, J. C. Reed, J. A. Fontana, F. Bost, P. D. Hobbs, Y. Zheng, G. Q. Chen, B. Shroot, D. Mercola, and X. K. Zhang. 1998. Molecular determinants of AHPN (CD437)-induced growth arrest and apoptosis in human lung cancer cell lines. Mol. Cell. Biol. 18:4719-4731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lin, B., G. Q. Chen, D. Xiao, S. K. Kolluri, X. Cao, H. Su, and X. K. Zhang. 2000. Orphan receptor COUP-TF is required for induction of retinoic acid receptor beta, growth inhibition, and apoptosis by retinoic acid in cancer cells. Mol. Cell. Biol. 20:957-970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lin, B., S. Kolluri, F. Lin, W. Liu, Y. H. Han, X. Cao, M. I. Dawson, J. C. Reed, and X. K. Zhang. 2004. Conversion of Bcl-2 from protector to killer by interaction with nuclear orphan receptor Nur77/TR3. Cell 116:527-540. [DOI] [PubMed] [Google Scholar]
- 44.Liu, Z. G., S. W. Smith, K. A. McLaughlin, L. M. Schwartz, and B. A. Osborne. 1994. Apoptotic signals delivered through the T-cell receptor of a T-cell hybrid require the immediate-early gene nur77. Nature 367:281-284. [DOI] [PubMed] [Google Scholar]
- 45.Mages, H. W., O. Rilke, R. Bravo, G. Senger, and R. A. Kroczek. 1994. NOT, a human immediate-early response gene closely related to the steroid/thyroid hormone receptor NAK1/TR3. Mol. Endocrinol. 8:1583-1591. [DOI] [PubMed] [Google Scholar]
- 46.Mangelsdorf, D. J., and R. M. Evans. 1995. The RXR heterodimers and orphan receptors. Cell 83:841-850. [DOI] [PubMed] [Google Scholar]
- 47.Maruyama, K., T. Tsukada, S. Bandoh, K. Sasaki, N. Ohkura, and K. Yamaguchi. 1995. Expression of NOR-1 and its closely related members of the steroid/thyroid hormone receptor superfamily in human neuroblastoma cell lines. Cancer Lett. 96:117-122. [DOI] [PubMed] [Google Scholar]
- 48.Maruyama, K., T. Tsukada, N. Ohkura, S. Bandoh, T. Hosono, and K. Yamaguchi. 1998. The NGFI-B subfamily of the nuclear receptor superfamily. Int. J. Oncol. 12:1237-1243. [DOI] [PubMed] [Google Scholar]
- 49.Masuyama, N., K. Oishi, Y. Mori, T. Ueno, Y. Takahama, and Y. Gotoh. 2001. Akt inhibits the orphan nuclear receptor Nur77 and T-cell apoptosis. J. Biol. Chem. 276:32799-32805. [DOI] [PubMed] [Google Scholar]
- 50.Migliaccio, A., G. Castoria, M. Di Domenico, A. de Falco, A. Bilancio, M. Lombardi, M. V. Barone, D. Ametrano, M. S. Zannini, C. Abbondanza, and F. Auricchio. 2000. Steroid-induced androgen receptor-oestradiol receptor beta-Src complex triggers prostate cancer cell proliferation. EMBO J. 19:5406-5417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Milbrandt, J. 1988. Nerve growth factor induces a gene homologous to the glucocorticoid receptor gene. Neuron 1:183-188. [DOI] [PubMed] [Google Scholar]
- 52.Ohkura, N., M. Hijikuro, A. Yamamoto, and K. Miki. 1994. Molecular cloning of a novel thyroid/steroid receptor superfamily gene from cultured rat neuronal cells. Biochem. Biophys. Res. Commun. 205:1959-1965. [DOI] [PubMed] [Google Scholar]
- 53.Pekarsky, Y., C. Hallas, A. Palamarchuk, A. Koval, F. Bullrich, Y. Hirata, R. Bichi, J. Letofsky, and C. M. Croce. 2001. Akt phosphorylates and regulates the orphan nuclear receptor Nur77. Proc. Natl. Acad. Sci. USA 98:3690-3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Perlmann, T., and L. Jansson. 1995. A novel pathway for vitamin A signaling mediated by RXR heterodimerization with NGFI-B and NURR1. Genes Dev. 9:769-782. [DOI] [PubMed] [Google Scholar]
- 55.Perlmann, T., K. Umesono, P. N. Rangarajan, B. M. Forman, and R. M. Evans. 1996. Two distinct dimerization interfaces differentially modulate target gene specificity of nuclear hormone receptors. Mol. Endocrinol. 10:958-966. [DOI] [PubMed] [Google Scholar]
- 56.Philips, A., S. Lesage, R. Gingras, M. H. Maira, Y. Gauthier, P. Hugo, and J. Drouin. 1997. Novel dimeric Nur77 signaling mechanism in endocrine and lymphoid cells. Mol. Cell. Biol. 17:5946-5951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Prufer, K., and J. Barsony. 2002. Retinoid X receptor dominates the nuclear import and export of the unliganded vitamin D receptor. Mol. Endocrinol. 16:1738-1751. [DOI] [PubMed] [Google Scholar]
- 58.Prufer, K., A. Racz, G. C. Lin, and J. Barsony. 2000. Dimerization with retinoid X receptors promotes nuclear localization and subnuclear targeting of vitamin D receptors. J. Biol. Chem. 275:41114-41123. [DOI] [PubMed] [Google Scholar]
- 59.Ramaswamy, S., K. N. Ross, E. S. Lander, and T. R. Golub. 2003. A molecular signature of metastasis in primary solid tumors. Nat. Genet. 33:49-54. [DOI] [PubMed] [Google Scholar]
- 60.Rastinejad, F., T. Wagner, Q. Zhao, and S. Khorasanizadeh. 2000. Structure of the RXR-RAR DNA-binding complex on the retinoic acid response element DR1. EMBO J. 19:1045-1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sacchetti, P., H. Dwornik, P. Formstecher, C. Rachez, and P. Lefebvre. 2002. Requirements for heterodimerization between the orphan nuclear receptor Nurr1 and retinoid X receptors. J. Biol. Chem. 277:35088-35096. [DOI] [PubMed] [Google Scholar]
- 62.Sakaue, M., H. Adachi, M. Dawson, and A. M. Jetten. 2001. Induction of Egr-1 expression by the retinoid AHPN in human lung carcinoma cells is dependent on activated ERK1/2. Cell Death Differ. 8:411-424. [DOI] [PubMed] [Google Scholar]
- 63.Shipp, M. A., K. N. Ross, P. Tamayo, A. P. Weng, J. L. Kutok, R. C. Aguiar, M. Gaasenbeek, M. Angelo, M. Reich, G. S. Pinkus, T. S. Ray, M. A. Koval, K. W. Last, A. Norton, T. A. Lister, J. Mesirov, D. S. Neuberg, E. S. Lander, J. C. Aster, and T. R. Golub. 2002. Diffuse large B-cell lymphoma outcome prediction by gene-expression profiling and supervised machine learning. Nat. Med. 8:68-74. [DOI] [PubMed] [Google Scholar]
- 64.Simoncini, T., A. Hafezi-Moghadam, D. P. Brazil, K. Ley, W. W. Chin, and J. K. Liao. 2000. Interaction of estrogen receptor with the regulatory subunit of phosphatidylinositol-3-OH kinase. Nature 407:538-541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Szondy, Z., U. Reichert, and L. Fesus. 1998. Retinoic acids regulate apoptosis of T lymphocytes through an interplay between RAR and RXR receptors. Cell Death Differ. 5:4-10. [DOI] [PubMed] [Google Scholar]
- 66.Uemura, H., and C. Chang. 1998. Antisense TR3 orphan receptor can increase prostate cancer cell viability with etoposide treatment. Endocrinology 139:2329-2334. [DOI] [PubMed] [Google Scholar]
- 67.Wang, H. G., U. R. Rapp, and J. C. Reed. 1996. Bcl-2 targets the protein kinase Raf-1 to mitochondria. Cell 87:629-638. [DOI] [PubMed] [Google Scholar]
- 68.Wansa, K. D., J. M. Harris, and G. E. Muscat. 2002. The activation function-1 domain of Nur77/NR4A1 mediates transactivation, cell specificity, and coactivator recruitment. J. Biol. Chem. 277:33001-33011. [DOI] [PubMed] [Google Scholar]
- 69.Weih, F., R. P. Ryseck, L. Chen, and R. Bravo. 1996. Apoptosis of nur77/N10-transgenic thymocytes involves the Fas/Fas ligand pathway. Proc. Natl. Acad. Sci. USA 93:5533-5538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wen, W., J. L. Meinkoth, R. Y. Tsien, and S. S. Taylor. 1995. Identification of a signal for rapid export of proteins from the nucleus. Cell 82:463-473. [DOI] [PubMed] [Google Scholar]
- 71.Wilson, A. J., D. Arango, J. M. Mariadason, B. G. Heerdt, and L. H. Augenlicht. 2003. TR3/Nur77 in colon cancer cell apoptosis. Cancer Res. 63:5401-5407. [PubMed] [Google Scholar]
- 72.Wilson, T. E., T. J. Fahrner, M. Johnston, and J. Milbrandt. 1991. Identification of the DNA binding site for NGFI-B by genetic selection in yeast. Science 252:1296-1300. [DOI] [PubMed] [Google Scholar]
- 73.Winoto, A., and D. R. Littman. 2002. Nuclear hormone receptors in T lymphocytes. Cell 109(Suppl.):S57-S66. [DOI] [PubMed] [Google Scholar]
- 74.Woronicz, J. D., B. Calnan, V. Ngo, and A. Winoto. 1994. Requirement for the orphan steroid receptor Nur77 in apoptosis of T-cell hybridomas. Nature 367:277-281. [DOI] [PubMed] [Google Scholar]
- 75.Woronicz, J. D., A. Lina, B. J. Calnan, S. Szychowski, L. Cheng, and A. Winoto. 1995. Regulation of the Nur77 orphan steroid receptor in activation-induced apoptosis. Mol. Cell. Biol. 15:6364-6376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wu, Q., M. I. Dawson, Y. Zheng, P. D. Hobbs, A. Agadir, L. Jong, Y. Li, R. Liu, B. Lin, and X. K. Zhang. 1997. Inhibition of trans-retinoic acid-resistant human breast cancer cell growth by retinoid X receptor-selective retinoids. Mol. Cell. Biol. 17:6598-6608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wu, Q., Y. Li, R. Liu, A. Agadir, M. O. Lee, Y. Liu, and X. Zhang. 1997. Modulation of retinoic acid sensitivity in lung cancer cells through dynamic balance of orphan receptors nur77 and COUP-TF and their heterodimerization. EMBO J. 16:1656-1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wu, Q., S. Liu, X. F. Ye, Z. W. Huang, and W. J. Su. 2002. Dual roles of Nur77 in selective regulation of apoptosis and cell cycle by TPA and ATRA in gastric cancer cells. Carcinogenesis 23:1583-1592. [DOI] [PubMed] [Google Scholar]
- 79.Wu, W. S., Z. X. Xu, R. Ran, F. Meng, and K. S. Chang. 2002. Promyelocytic leukemia protein PML inhibits Nur77-mediated transcription through specific functional interactions. Oncogene 21:3925-3933. [DOI] [PubMed] [Google Scholar]
- 80.Xu, L., C. K. Glass, and M. G. Rosenfeld. 1999. Coactivator and corepressor complexes in nuclear receptor function. Curr. Opin. Genet. Dev. 9:140-147. [DOI] [PubMed] [Google Scholar]
- 81.Yang, Y., J. Bailey, M. S. Vacchio, R. Yarchoan, and J. D. Ashwell. 1995. Retinoic acid inhibition of ex vivo human immunodeficiency virus-associated apoptosis of peripheral blood cells. Proc. Natl. Acad. Sci. USA 92:3051-3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yang, Y., S. Minucci, K. Ozato, R. A. Heyman, and J. D. Ashwell. 1995. Efficient inhibition of activation-induced Fas ligand up-regulation and T-cell apoptosis by retinoids requires occupancy of both retinoid X receptors and retinoic acid receptors. J. Biol. Chem. 270:18672-18677. [DOI] [PubMed] [Google Scholar]
- 83.Yang, Y., M. S. Vacchio, and J. D. Ashwell. 1993. 9-cis-Retinoic acid inhibits activation-driven T-cell apoptosis: implications for retinoid X receptor involvement in thymocyte development. Proc. Natl. Acad. Sci. USA 90:6170-6174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zamzami, N., and G. Kroemer. 2001. The mitochondrion in apoptosis: how Pandora's box opens. Nat. Rev. Mol. Cell. Biol. 2:67-71. [DOI] [PubMed] [Google Scholar]
- 85.Zechel, C., X. Q. Shen, J. Y. Chen, Z. P. Chen, P. Chambon, and H. Gronemeyer. 1994. The dimerization interfaces formed between the DNA binding domains of RXR, RAR, and TR determine the binding specificity and polarity of the full-length receptors to direct repeats. EMBO J. 13:1425-1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhang, X. K. 2002. Vitamin A and apoptosis in prostate cancer. Endocrinol. Relat. Cancer 9:87-102. [DOI] [PubMed] [Google Scholar]
- 87.Zhang, X. K., B. Hoffmann, P. B. Tran, G. Graupner, and M. Pfahl. 1992. Retinoid X receptor is an auxiliary protein for thyroid hormone and retinoic acid receptors. Nature 355:441-446. [DOI] [PubMed] [Google Scholar]
- 88.Zhang, X. K., J. Lehmann, B. Hoffmann, M. I. Dawson, J. Cameron, G. Graupner, T. Hermann, P. Tran, and M. Pfahl. 1992. Homodimer formation of retinoid X receptor induced by 9-cis retinoic acid. Nature 358:587-591. [DOI] [PubMed] [Google Scholar]
- 89.Zhang, X. K., and M. Pfahl. 1993. Hetero- and homodimeric receptors in thyroid hormone and vitamin A action. Receptor 3:183-191. [PubMed] [Google Scholar]
- 90.Zhang, X. K., G. Salbert, M. O. Lee, and M. Pfahl. 1994. Mutations that alter ligand-induced switches and dimerization activities in the retinoid X receptor. Mol. Cell. Biol. 14:4311-4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhang, Y., M. I. Dawson, R. Mohammad, A. K. Rishi, L. Farhana, K. C. Feng, M. Leid, V. Peterson, X. K. Zhang, M. Edelstein, D. Eilander, S. Biggar, N. Wall, U. Reichert, and J. A. Fontana. 2002. Induction of apoptosis of human B-CLL and ALL cells by a novel retinoid and its nonretinoidal analog. Blood 100:2917-2925. [DOI] [PubMed] [Google Scholar]
- 92.Zhu, X. G., J. A. Hanover, G. L. Hager, and S. Y. Cheng. 1998. Hormone-induced translocation of thyroid hormone receptors in living cells visualized using a receptor green fluorescent protein chimera. J. Biol. Chem. 273:27058-27063. [DOI] [PubMed] [Google Scholar]