Abstract
The forkhead transcription factor Fkh2p acts in a DNA-bound complex with Mcm1p and the coactivator Ndd1p to regulate cell cycle-dependent expression of the CLB2 gene cluster in Saccharomyces cerevisiae. Here, we demonstrate that Fkh2p is a target of cyclin-dependent protein kinases and that phosphorylation of Fkh2p promotes interactions between Fkh2p and the coactivator Ndd1p. These phosphorylation-dependent changes in the Fkh2p-Ndd1p complex play an important role in the cell cycle-regulated expression of the CLB2 cluster. Our data therefore identify an important regulatory target for cyclin-dependent kinases in the cell cycle and further our molecular understanding of the key cell cycle regulatory transcription factor Fkh2p.
Cell cycle progression in Saccharomyces cerevisiae is controlled in part by the sequential transcription of specific gene clusters whose expression peaks at different times during the cell cycle (5, 24; see reference 7 for a review). Cyclin-dependent kinases (Cdks) are important regulators of many cell cycle processes, including these gene expression patterns. However, despite the identification of cell cycle stage-specific Cdk complexes and transcription factors important for cell cycle-dependent gene expression there is much to learn regarding the interface between these different cell cycle components in eukaryotes.
In S. cerevisiae nine different cyclins, Cln1 to -3 and Clb1 to -6, associate with the catalytic subunit Cdc28p to phosphorylate target molecules during the cell cycle (for a review see reference 2). The Clnp cyclins act in G1, while the Clbp cyclins act in S, G2, and M phases. The expression of these cyclins changes during the cell cycle. For example, the expression of the CLB5 and CLB6 genes is induced in late G1, while the expression of the CLB1 and CLB2 genes is induced in G2 phase (2). Consistent with this timing of expression, Clb5p and Clb6p have been shown to be important for DNA replication, whereas Clb1p and Clb2p are important for the completion of later cell cycle events. Cdc28p-dependent phosphorylation preferentially occurs on serine and threonine residues located within the minimum consensus S/TP, with basic residues often present at the C terminus of the site (23). Although the in vivo targets of these cyclin-Cdc28p complexes are largely unknown, Clb2p has been linked with the regulation of gene expression. For example, the activity of SBF, a transcription factor complex consisting of Swi4p and Swi6p (see reference 13 for a review), is repressed through a mechanism dependent on Clb1p and Clb2p (1), and Clb2p coimmunoprecipitates with Swi4p from extracts of mitotically arrested cells. In addition, Cdc28p-Clb2p was found to be required for the cell cycle-dependent expression of CLB1 and CLB2 in G2 phase (1).
The CLB1 and CLB2 genes are coincidentally expressed with a group of genes in G2 and M phases, which includes SWI5 and ACE2, known as the CLB2 gene cluster (see references 7 and 8 for reviews). Previous studies have shown that this gene cluster is regulated by the SFF (Swi five factor) complex (17, 18). The Mcm1p-SFF ternary complex contains the MADS box protein Mcm1p, which forms a promoter-bound complex with the forkhead transcription factor Fkh2p (14, 15, 19, 29; reviewed in references 4 and 7). Fkh2p is closely related to the Fkh1p forkhead protein in S. cerevisiae and has particularly high similarity in the forkhead DNA-binding domain and the forkhead-associated (FHA) domain. However, Fkh1p lacks the extended C-terminal domain found in Fkh2p and, importantly, lacks a motif required for cooperative DNA binding with Mcm1p (3, 12). The Mcm1p-Fkh2p complex activates cell cycle-regulated transcription in conjunction with the coactivator Ndd1p (14). Recent results have elucidated some aspects of the mechanism(s) of activation of the Mcm1p-Fkh2p-Ndd1p complex during the cell cycle. The expression of NDD1 is cell cycle regulated, with peak expression occurring during S phase (16). Moreover, Ndd1p is a target of the Cdc28p-Clb2p kinase, and this phosphorylation is important for the recruitment of Ndd1p and the activation of the Mcm1p-Fkh2p-Ndd1p complex (6, 20). Cell cycle-dependent phosphorylation of Fkh2p, which peaks coincidentally with the onset of Mcm1p-Fkh2p-dependent gene expression, has also been detected (19). However, the role of Fkh2p phosphorylation in complex assembly and activation is unclear.
In this study, we have investigated the cell cycle-dependent regulation of Fkh2p. We demonstrate that Fkh2p is phosphorylated by Cdc28p-Clbp complexes and that this phosphorylation regulates the interaction of Fkh2p with Ndd1p. Fkh2p phosphorylation triggers important changes in intermolecular interactions within the Fkh2p-Ndd1p complex. This has important consequences for the activation of expression of genes in the CLB2 cluster. Our data therefore provide important insights into the function of the Fkh2p-Ndd1p complex and, more generally, the mechanism(s) by which Cdks regulate transcriptional programs during the cell cycle.
MATERIALS AND METHODS
Plasmid construction and mutagenesis.
The following bacterial expression vectors were created: pAS1751 [encoding glutathione S-transferase (GST)-Fkh2(1-254)], pAS1609 [encoding GST-Fkh2(292-325)], pAS1752 [encoding GST-Fkh2(254-458)], pAS1753 [encoding GST-Fkh2(458-862)], and pAS1755 [encoding GST-Fkh2(254-862)]. These vectors were constructed by inserting EcoRI/XhoI-cleaved PCR fragments (generated with primers ADS945 and ADS949, ADS950 and ADS948, ADS950 and ADS871, ADS947 and ADS862, and ADS945 and ADS862, respectively) into the same sites in pGEXKG. pAS1757 (encoding full-length Fkh2p amino acids 1 to 862 fused to Flag and His6 tags) and pAS1769 (encoding GST fused to full-length Ndd1p amino acids 1 to 554) were described previously (6). pAS1775(S683A), pAS1776(T697A), and pAS1777(S772A), encoding GST-Fkh2(458-863) m1, m2, and m3, respectively, were constructed by inserting EcoRI/XhoI-cleaved two-step PCR fragments (primers ADS1113, ADS947, and ADS862; ADS1114, ADS947, and ADS862; and ADS1115, ADS947, and ADS862) into the same sites in pGEXKG (10).
The pBluescript KS(+)-derived plasmids pAS1242 (encoding Fkh2p amino acids 1 to 862), pAS1243 (encoding Fkh2p amino acids 1 to 458), pAS1241 (encoding Fkh2p amino acids 458 to 862), and pAS1247 (encoding Fkh2p amino acids 254 to 458) were described previously (3).
The following vectors were used for yeast expression. pMW20-Clb2-6HISFlag and pAS1760 [encoding Gal-myc-Fkh2(458-862)] were described previously (6). To construct pMW20-Clb5-6HISFlag, a PCR fragment containing the CLB5 gene (generated with primers Clb5BamHI forward and Clb56HISFlagBamHI) was digested with BamHI and ligated with BamHI-digested pMW20. pUS454 (pGAL1-HA3-NDD1) (16) was kindly provided by U. Surana. pAS1771 [encoding Gal-myc-Fkh2(1-180)] and pAS1773 [encoding Gal-myc-Fkh2(108-254)] were created by inserting NcoI-EcoRI-cleaved PCR fragments (generated with primers ADS857 and ADS986 and primers ADS985 and ADS960, respectively) into the same sites in the pGBKT7 vector (Clontech). pAS1771 and pAS1773 were also used to generate in vitro-translated proteins. pAS1794 [encoding Myc-Fkh2(1-862)] and pAS1795 [encoding Myc-Fkh2(1-458)] were created by inserting SalI/EcoRI-cleaved PCR fragments, generated with primer-template combinations ADS1162-ADS962-pAS1761 and primers ADS1162-ADS961-pAS1762, respectively, into the same sites of YCplac111 and YCplac22 (9), respectively. pAS1761 and pAS1762 were constructed by inserting either NcoI- or NcoI/EcoRI-cleaved PCR fragments, generated with primers ADS857 and ADS858 and primers ADS857 and ADS961 with template pAS1242 (3), into the same sites in pAS1760 and pGBKT7, respectively. pAS1970 and pAS1971 [encoding myc-Fkh2(1-862)(S683A) and myc-Fkh2(1-862)(T697A), respectively] were created with the QuikChange site-directed mutagenesis kit (Stratagene) using the primer pair ADS1183 and ADS1184 and primer pair ADS1185 and ADS1186 on the template pAS1794. pDL58, containing the PSWI5-LacZ reporter gene cassette, was kindly provided by D. Lydall (17).
Yeast strains, growth conditions, LacZ reporter gene assays, RNA analysis, and chromatin immunoprecipitations (ChIPs).
All yeast strains were haploids and derived from the W303-1a genetic background. AP11 (MATa ade2-1 trp1-1 leu2-3,112 his3-11 ura3 fkh1::HIS3 fkh2::URA3) and AP16 (MATa ade2-1 trp1-1 leu2-3,112 his3-11 ura3 FKH2-13Myc KanR) was described previously (19). AP136 (MATa clb1Δ clb2ts clb3::TRP1 clb4::HIS3 FKH2-13Myc KanR) was from a cross between US373 (MATa clb1Δ clb2ts clb3::TRP1 clb4::HIS3 ura3 gal:CLB2; kindly provided by U. Surana) and AP17 (MATα ade2-1 trp1-1 leu2-3,112 his3-11 ura3 FKH2-13Myc KanR). AP153 (MATa ade2-1 trp1-1 leu2-3,112 his3-11 ura3 clb5::URA3 FKH2-13Myc KanR), AP166 (MATa ade2-1 trp1-1 leu2-3,112 his3-11 ura3 clb6::LEU2 FKH2-13Myc KanR), and Y583-K3353-7B (AP144) were described previously (6). AP188 (cdc28-1N FKH2-13Myc KanR) was obtained from a cross between the cdc28-1N strain (MATa ade2 leu2 trp1 ura3 cdc28-1N; kindly provided by L. Johnston) and AP17. AP142 (CDC28-HA FKH2-13Myc KanR) was from a cross between AP17 and CDC28-HA (kindly supplied by L. Johnston).
Yeast cells were grown, GAL1 promoter-driven constructs were induced, and transformations were performed as described previously (3, 6).
Yeast cultures were synchronized by treatment with α factor (Calbiochem; 5 μg/ml) for 3 h (G1), hydroxyurea (Sigma; 15.2 mg/ml) for 60 to 90 min (S), or nocodazole (Sigma; 15 μg/ml) for 120 to 150 min (M), respectively. Block-and-release experiments were carried out as described previously (6).
LacZ reporter gene assays were carried out as described previously (6).
RNA extraction and Northern blot analysis were performed and analyzed as described previously (19) with cells treated with α factor or nocodazole. Probes were PCR-generated internal fragments of the genes (details of PCR primers available on request). A probe for the RPB4 gene was used as a loading control. Details of probes are available on request.
ChIP assays were done according to the protocol described at http://www.fhcrc.org/labs/breeden/Methods/chromatinIP.html using the following primer pairs; ADS1197 and ADS1198 (CLB2), ADS1195 and ADS1196 (SWI5), and ADS1193 and ADS1194 (ACE2) (details available on request).
Protein production, pulldown assays, and Western blotting.
Wild-type Fkh2p and truncated derivatives (with wheat germ lysates; Promega) were produced by coupled in vitro transcription and translation and subsequently analyzed and quantified by phosphorimaging. His-tagged Fkh2p was expressed in Escherichia coli BL21 and purified with nickel-nitrilotriacetic acid (NTA)-agarose resin according to standard procedures. GST fusion proteins were prepared and GST pulldown assays with in vitro-translated proteins were carried out as described previously (21).
Pulldowns with purified, immobilized GST-Ndd1p or His-tagged Fkh2p fusion proteins and proteins from yeast cell extracts were carried out and analyzed as described previously (6).
To analyze phosphorylation-dependent shifts in Fkh2p mobility, protein samples were resolved on sodium dodecyl sulfate-8, 12.5, or 15% polyacrylamide gel electrophoresis gels, and transferred to nitrocellulose. To detect Myc-tagged Fkh2p derivatives, anti-Myc (Santa Cruz Biotechnology) was used. Standard Western blotting and immunoprecipitations were also performed using antihemagglutinin (anti-HA; Cancer Research UK), anti-Flag (Sigma), or a control antibody against tubulin (kindly provided by D. Lydall).
Coimmunoprecipitations were carried out as described previously (6).
Protein kinase production and kinase assays.
HA-tagged Cdc28p and Flag-tagged Clb2p- and Clb5p-kinase complexes were purified from yeast cells harboring CDC28-HA (chromosomally integrated), pMW20-Clb2-6HISFlag, or pMW20-Clb5-6HISFlag, respectively, with HA or Flag antibodies coupled to protein G-Sepharose beads. Subsequent protein kinase assays were performed as described previously (6). Two-dimensional (2D) mapping was carried out with a Hunter thin-layer peptide mapping electrophoresis system according to the manufacturer's instructions (C.B.S. Scientific Company, Inc.).
RESULTS
Fkh2p is phosphorylated by Cdc28p-Clbp kinases.
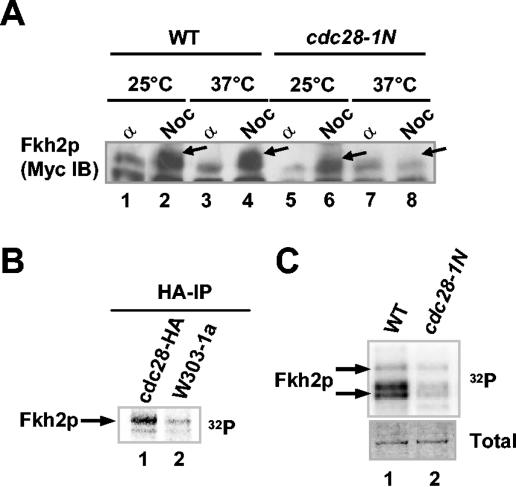
Fkh2p was previously shown to be phosphorylated in vivo in a cell cycle-dependent manner, and this phosphorylation increases during S and G2 phases, preceding the activation of Fkh2p target genes such as SWI5 (19). We therefore wished to identify the protein kinase(s) involved in the phosphorylation of Fkh2p. An obvious candidate protein kinase is Clbp cyclin-associated Cdc28p. Previous studies have shown that a temperature-sensitive cdc28-1N mutant (25) is blocked after S phase but prior to mitosis at the nonpermissive temperature. The induction of a mitotic block with nocodazole induces the appearance of slower-mobility phosphorylated Myc-tagged Fkh2p protein in wild-type cells at both 25 and 37°C (Fig. 1A, compare lanes 2 and 4). However, this phosphorylation is reduced in cdc28-1N cells grown at 25°C and blocked at the nonpermissive temperature (Fig. 1A, compare lanes 6 and 8). To further probe a possible link between Cdc28p and Fkh2p phosphorylation, HA-tagged Cdc28p-containing complexes were immunoprecipitated from mid-log-phase cells and their ability to phosphorylate Fkh2p was analyzed. Efficient phosphorylation of Fkh2p by HA-precipitated Cdc28p was observed (Fig. 1B, lane 1). Furthermore, to detect kinases that associate with Fkh2p, purified His-tagged Fkh2p was immobilized on agarose beads and incubated with total-cell extracts and, after nonspecifically bound proteins were washed away, a kinase assay was performed with the remaining proteins. This assay revealed the presence of a Fkh2p-associated kinase in extracts from wild-type cells (Fig. 1C, lane 1). However, much-reduced Fkh2p-associated kinase activity was detected in extracts from nocodazole-treated cdc28-1N cells (Fig. 1C, lane 2). These data show that Cdc28p is required for phosphorylation of Fkh2p.
FIG. 1.
Fkh2p is phosphorylated by Cdc28p kinase. (A) Western blot of Myc-tagged Fkh2p in extracts isolated from α factor (α)- and nocodazole (Noc)-treated wild-type (WT; AP16) cells (lanes 1 to 4) and cdc28-1N mutant (AP188) cells (lanes 5 to 8) grown at 25 and 37°C. Arrows, locations of bands corresponding to hyperphosphorylated Fkh2p. (B) Full-length, His-tagged Fkh2p was phosphorylated in vitro by HA-tagged Cdc28p-containing complexes immunoprecipitated (IP) from wild-type cells (W303-1a) or cells containing CDC28-HA. (C) Activity of Fkh2p-associated kinase in a cdc28-1N mutant. His-tagged Fkh2p immobilized on agarose beads was incubated with extracts isolated from W303-1a (lane 1) and cdc28-1N (lane 2) cells treated with nocodazole, and kinase reactions were then carried out with the precipitated kinases. The total amounts of Fkh2p protein in the assays are shown in the Coomassie-stained gel (bottom).
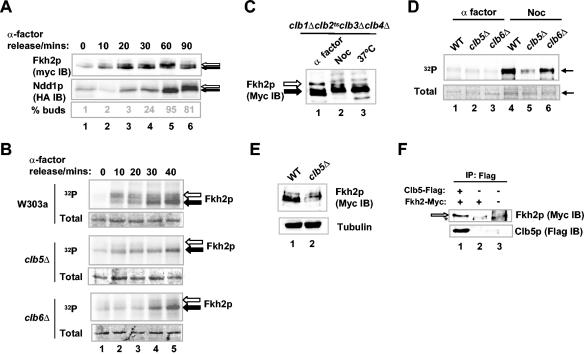
Cdc28p sequentially associates with a series of partner cyclin proteins at different points in the cell cycle. To begin to investigate which cyclin(s) may act with Cdc28p to phosphorylate Fkh2p, the timing of the onset of Fkh2p phosphorylation was compared with that of the Cdc28p-Clb2p substrate Ndd1p (6). Phosphorylation of Fkh2p is first detectable 10 to 20 min after α factor release, compared to 60 min for Ndd1p, and peaks at 60 min compared to 90 min for Ndd1p (Fig. 2A). Thus, the phosphorylation of Fkh2p is initiated earlier in the cell cycle than that of Ndd1p and also peaks at an earlier time point. Next, we investigated whether kinase activity that associates with Fkh2p and varies during the cell cycle can be identified. Extracts were prepared from wild-type W303-1a cells that had been synchronized with α factor and subsequently released into the cell cycle. His-tagged, full-length, bacterially purified Fkh2p, immobilized on nickel-NTA-agarose beads, was then added to the extracts, and interacting proteins were isolated. The resulting complexes were then subjected to a kinase assay by the addition of radioactively labeled ATP. The activity of the Fkh2p-associated kinase was detectable 10 min after release from α factor and increased for the next 30 min (Fig. 2B, top), consistent with the onset of Fkh2p phosphorylation in vivo. A previous study indicated that the mitotic cyclins Clb1p to -4p do not appear to be required for Fkh2p phosphorylation (6). Indeed, Fkh2p hyperphosphorylation was detected in clb1Δ clb2ts clb3Δ clb4Δ cells grown at the nonpermissive temperature (Fig. 2C, compare lanes 2 and 3).
FIG. 2.
Role of Clb5p in Cdc28p-mediated Fkh2p phosphorylation. (A) Western blot analysis of Myc-tagged Fkh2p and HA-tagged Ndd1p in AP16 cells harboring the vector pGAL1-HA3-NDD1 at the indicated times after release from α factor block. Solid and open arrows, hypo- and hyperphosphorylated species, respectively. A budding index is shown at the bottom. IB, immunoblot. (B) In vitro kinase assays of His-tagged, full-length Fkh2p by coprecipitated kinases using extracts isolated from wild-type (AP16), clb5Δ (AP153), or clb6Δ (AP166) cells at the indicated times following release from α factor-induced arrest. Arrows, bands corresponding to full-length Fkh2p; open arrows, phosphorylation-shifted Fkh2p; solid arrows, hypophosphorylated Fkh2p. Phosphorimages (32P) and Coomassie-stained gels (total) of the input Fkh2p proteins are shown. (C) Western blot of Myc-tagged Fkh2p in clb1Δ clb2ts clb3Δ clb4Δ cells (AP136) arrested with α factor or nocodazole (Noc) or following incubation at 37°C. Solid arrow, Fkh2p; open arrow, hyperphosphorylated Fkh2p. (D) Activity of Fkh2p kinase in cells blocked in G1 and M phases. Fkh2p immobilized on nickel-NTA-agarose beads was incubated with extracts isolated from wild-type (WT; AP16; lanes 1 and 4), AP153 (clb5Δ, lanes 2 and 5), or AP166 (clb6Δ, lanes 3 and 6) cells treated with α factor or nocodazole. Kinase reactions were then carried out with the precipitated kinases. A phosphorimage (32P) and Coomassie-stained gel (total) of the input Fkh2p proteins are shown. Arrows, bands corresponding to Fkh2p. (E) Western blot analysis of Fkh2p (anti-Myc) in extracts isolated from mid-log-phase cultures of wild-type (AP16) and clb5Δ (AP153) strains. Tubulin levels (antitubulin) are shown as a control. (F) Fkh2p forms a complex with Clb5p. Immunoprecipitations (IPs) were performed with the anti-Flag antibody using extracts isolated from cells (AP16) expressing Myc-tagged Fkh2p in the presence (lane 1) and absence (lane 2) of coexpressed Flag-tagged Clb5p. Control IPs were also done in wild-type W303-1a cells (lane 3). Precipitated Fkh2p and Clb5p were detected with anti-Myc and anti-Flag antibodies, respectively. Open arrow, coprecipitated Fkh2p; *, nonspecific band appearing in the control lysate.
The timing of Fkh2p phosphorylation and the lack of requirement for the mitotic cyclins Clb1p to -4p suggested a potential role for Clb5p and/or Clb6p. These cyclins are activated after Start and are important for the initiation of S phase. In contrast to extracts from wild-type control cells, extracts from clb5Δ mutant cells had small amounts of kinase activity that could associate with Fkh2p (Fig. 2B, middle), while intermediate levels of phosphorylation were observed in extracts from clb6Δ mutant cells (Fig. 2B, bottom). We next analyzed the activity of Fkh2p-associated kinase using extracts isolated from wild-type, clb5Δ, or clb6Δ cells that had been blocked in G1 by α factor or in M phase by nocodazole (Fig. 2D). As expected, little phosphorylation of Fkh2p occurred when G1 extracts from all three strains were used (Fig. 2D, lanes 1 to 3) and high levels of kinase activity were observed when extracts from nocodazole-treated wild-type cells were used (Fig. 2D, lane 4). However, in comparison, lower levels of kinase activity were observed when extracts isolated from clb5Δ mutant cells were used, while extracts isolated from clb6Δ mutant cells retained high levels (Fig. 2D, lanes 5 and 6). Significantly, little difference in kinase activity towards Ndd1p was observed when extracts isolated from nocodazole treated wild-type, clb5Δ, and clb6Δ cells were used (6). The mobility of Fkh2p was also examined by Western blot analysis using extracts isolated from mid-log-phase cultures of wild-type and clb5Δ strains containing Myc-tagged Fkh2p (Fig. 2E). In clb5Δ extracts, both the mobility and the amount of Fkh2p were altered (Fig. 2E, lane 2), which is most apparent when the relative intensities of the lower-mobility bands are compared to that of the fastest-migrating band. This suggests that the deletion of the CLB5 gene has important consequences for modification of Fkh2p. Thus, these data suggest that a Clb5p-dependent kinase activity is involved in the phosphorylation of Fkh2p. Indeed, immunoprecipitation of Flag-tagged Clb5p resulted in coprecipitation of Fkh2p, demonstrating that these proteins can be detected in the same complex in vivo (Fig. 2F, lane 1).
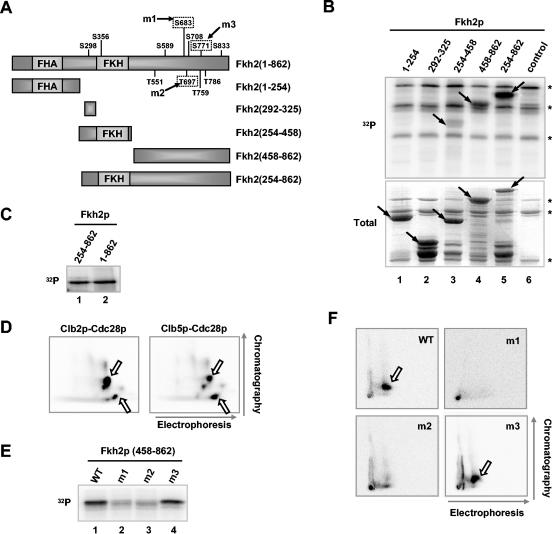
It was possible that the effects of Clb5p on Fkh2p phosphorylation were indirect. Hence, to establish that Cdc28p-Clb5p complexes can directly phosphorylate Fkh2p, the ability of Flag-tagged Clb5p-containing complexes, immunoprecipitated from W303-1a cells containing pMW20-Clb5-6HISFlag, to phosphorylate purified GST-Fkh2p was examined. Phosphorylation of the C-terminal, but not the N-terminal, region of Fkh2p could readily be observed (data not shown). Although phosphorylation of Fkh2p was reduced by loss of Clb5p, it was not abolished (Fig. 2D and E), suggesting that another Clbp(s) is responsible for phosphorylation of Fkh2p in the absence of Clb5p. Indeed, similar to Clb5p-containing complexes, Flag-tagged Clb2p-containing complexes immunoprecipitated from W303-1a cells containing pMW20-Clb2-6HISFlag could phosphorylate Fkh2p (Fig. 3B and C). To establish whether the same sites are targeted by Clb2p- and Clb5p-kinase complexes, we carried out phosphotryptic analysis of Fkh2p that had been phosphorylated by these kinases. Two major phosphopeptides and a number of similar lower-intensity spots were detected in both cases (Fig. 3D), demonstrating that similar sites were targeted.
FIG. 3.
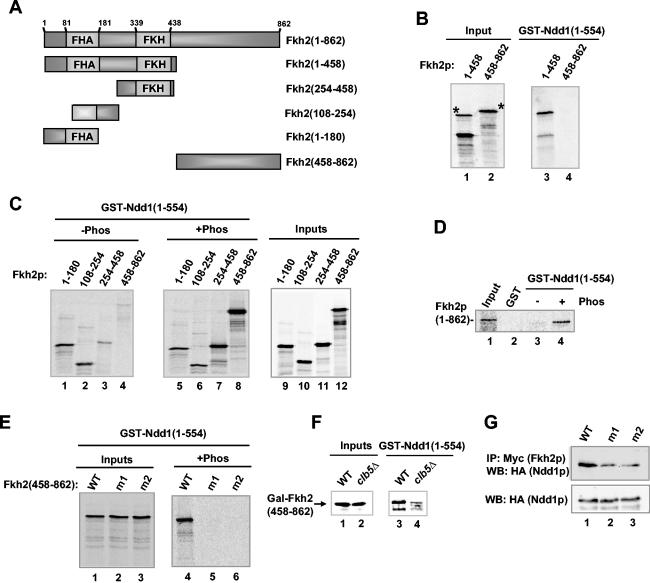
Mapping Clbp-associated kinase phosphorylation sites in Fkh2p. (A) Schematic representation of full-length Fkh2p and a series of deletion constructs. The locations of the forkhead DNA binding (FKH) and FHA domains, the potential Cdc28p phosphorylation sites (Ser/ThrPro motifs), and those sites that were mutated (m1 to m3), are indicated. (B) Kinase reaction of the indicated GST-Fkh2p constructs with Flag-tagged Clb2p-associated kinase complexes immunoprecipitated from W303-1a cells containing pMW20-Clb2-6HISFlag. Control represents GST alone. A phosphorimage (32P) and Coomassie-stained gel (total) of the input Fkh2p proteins are shown. Arrows, locations of bands corresponding to full-length GST fusion proteins; *, bands which arise from the kinase-containing extract alone. (C) Kinase reaction of GST-Fkh2(254-862) and His-tagged Fkh2(1-862) with immunoprecipitated Flag-tagged Clb2p-associated kinase complexes. (D) 2D mapping of the peptides phosphorylated in Fkh2p by immunoprecipitated Flag-tagged Clb2p- or Clb5p-associated kinase complexes. Following phosphorylation, Fkh2p(458-862) was digested with trypsin and subjected to 2D gel electrophoresis. Arrows, directions of the first dimension (horizontal) and second dimension (vertical); open arrows, spots representing the major phosphopeptides. (E) Residues located within the C-terminal region of Fkh2p are required for Clbp kinase-dependent phosphorylation. Shown are the results of a kinase assay of the indicated GST-Fkh2p fusion proteins with immunoprecipitated Flag-tagged Clb2p-associated kinase complexes. (F) 2D mapping of the peptides phosphorylated in GST-Fkh2p(458-862) by immunoprecipitated Flag-tagged Clb2p-associated kinase complexes. Phosphopeptides were generated by digestion with clostripain. Arrows, spots that disappear in the m1 and m2 mutant proteins.
Hence, these results demonstrate that Cdc28p has an important role in Fkh2p phosphorylation and that the Cdc28p-Clb5p complex likely represents the relevant cyclin complex involved. However, we cannot exclude a role for cyclins such as Clb2p. Indeed, Clb2p may have a latent ability to phosphorylate Fkh2p, as has been observed for Clb1p to -4p in regulating DNA replication in the absence of Clb5p and Clb6p (reviewed in reference 26).
Mapping the phosphorylation sites in Fkh2p.
Cdks phosphorylate serine and threonine residues within the minimum recognition sequence Ser/ThrPro, although other surrounding residues can influence the efficiency of phosphorylation (23). Fkh2p contains multiple potential Cdk phosphorylation sites corresponding to this minimum consensus sequence (Fig. 3A). To locate Cdk-mediated Fkh2p phosphorylation sites, we investigated whether Clbp-kinase complexes could phosphorylate a series of GST fusion proteins (Fig. 3B). In these experiments, we used Clb2p-kinase complexes, as higher levels of Clb2p-associated kinase activity could be immunoprecipitated in comparison to immunoprecipitations with Clb5p. No phosphorylation of amino acids 1 to 254 was detected (Fig. 3B, lane 1). In contrast, weak phosphorylation of amino acids 254 to 458 was observed (Fig. 3B, lane 3), with stronger activity towards amino acids 458 to 862 (Fig. 3B, lane 4). Two potential Cdk sites are located within amino acids 254 to 458 (Fig. 3A, Ser298 and Ser356). However, amino acids 292 to 325 are not phosphorylated (Fig. 3B, lane 2), suggesting that Ser356 is the site targeted in this region. Maximal activity towards amino acids 254 to 862 (Fig. 3B, lane 5), which was equivalent to the phosphorylation of full-length Fkh2p, was seen (Fig. 3C). Similar results were obtained with immunoprecipitated Flag-tagged Clb5p-associated kinase (data not shown). These data suggest that the majority of Clbp-kinase target sites are located in the C-terminal region (amino acids 458 to 862) of Fkh2p, although the site at position Ser356 within the N terminus is also a likely in vitro target. A number of potential Cdk sites are present in the C-terminal region of Fkh2p (Fig. 3A). However, a comparison of the sequence of Fkh2p with a translation of the genome sequence of the yeast Candida albicans (http://genolist.pasteur.fr/CandidaDB/) suggested that the closest homologue of Fkh2p in C. albicans has a very limited region of potential homology to the C-terminal region of Fkh2p, which includes Ser683 and Thr697. Indeed, this region is highly conserved among Saccharomyces species and the more distantly related Ashbya gossypii. Thus, Ser683 and Thr697 may be important for the function of Fkh2p. Hence, Ser683 and Thr697, as well as another potential phosphorylation site outside this region of homology, Ser771, were individually replaced with alanine residues (Fig. 3A). Wild-type and mutant GST-Fkh2p fusion proteins were then tested as substrates for immunoprecipitated Clb2p-associated kinase complexes. In comparison to wild-type Fkh2p, the m1 and m2 mutant proteins both exhibited much-reduced phosphorylation whereas the m3 mutant protein retained nearly wild-type levels of phosphorylation (Fig. 3E). To further study these phosphorylation events, 2D phosphopeptide mapping was performed. Wild-type and mutant versions of the GST-Fkh2p fusion proteins were phosphorylated in vitro by immunoprecipitated Clb2p-associated kinase complexes, digested with clostripain, and analyzed by 2D gel electrophoresis. One major phosphopeptide was observed in the wild-type Fkh2p, and this peptide was retained in the m3 mutated version of Fkh2p (Fig. 3F). Interestingly, this phosphopeptide was lost in either the m1 or the m2 mutant proteins (Fig. 3F). Thus, the m1 and m2 mutations define the primary sites of phosphorylation by Clb2p-associated kinase complexes in vitro and, furthermore, suggest an interdependency of each site for phosphorylation. Similar results were also obtained with alternative proteolytic enzymes and immunoprecipitated Clb5p-associated kinase complexes (data not shown).
Taken together, these data reveal that the C-terminal region of Fkh2p (amino acids 458 to 862) contains the major sites, Ser683 and Thr697, for phosphorylation by Clbp-kinase complexes.
The Cdc28p-Clbp-dependent phosphorylation sites in Fkh2p are required for the assembly and function of the Fkh2p-Ndd1p complex in vivo.
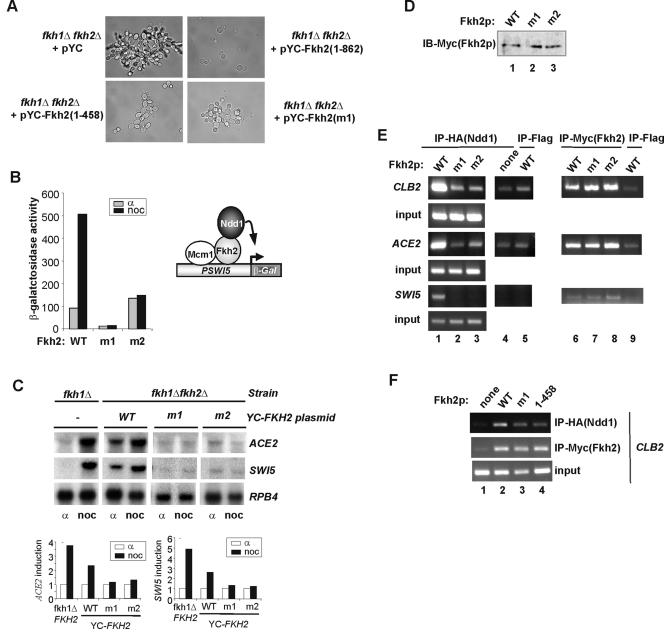
The observation that the C-terminal region of Fkh2p contains the major Cdc28p-Clbp-dependent phosphorylation sites suggests an important role for this domain in regulating Fkh2p activity. Indeed, this extended C-terminal tail is unique to Fkh2p and is not present in Fkh1p. To establish whether this unique C-terminal region of Fkh2p plays an important physiological role, we attempted to rescue the pseudohyphal phenotype of fkh1Δ fkh2Δ cells (AP11) with Fkh2(1-458). In contrast to full-length Fkh2p, Fkh2(1-458) was unable to fully rescue the pseudohyphal phenotype (Fig. 4A). Interestingly, as observed with the full-length protein, the presence of the truncated Fkh2(1-458) protein is lethal in the absence of Ndd1p, suggesting a role for this domain in Ndd1p interactions (3). To establish whether Cdc28p-Clbp phosphorylation sites within the C terminus of Fkh2p are important for the function of Fkh2p in vivo, we also examined the ability of the S683A (m1) or T697A (m2) versions of Fkh2p to rescue pseudohyphal growth. As observed with the complete C-terminal deletion, the m1 and m2 mutant proteins were unable to fully rescue pseudohyphal growth (Fig. 4A and data not shown).
FIG. 4.
Ser683 and Thr697 play critical roles in the assembly and function of the Fkh2p-Ndd1p complex in vivo. (A) Differential interference contrast images of fkh1Δ fkh2Δ cells (AP11) that were transformed with plasmids encoding HA-tagged Ndd1p and either the empty pYC vector or pYC vectors encoding Fkh2p(1-862), Fkh2p(1-458), or Fkh2p(m1). (B) Reporter gene analysis using a SWI5 promoter-driven LacZ gene was carried out with the fkh1Δ fkh2Δ strain containing the indicated plasmid-borne Fkh2p derivatives (pAS1794, pAS1970, and pAS1971). Cells were synchronized with either α factor or nocodazole (noc). WT, wild type. (C) Northern blot analysis of ACE2, SWI5, and RPB4 expression in a fkh1Δ (left) or fkh1Δ fkh2Δ strain (AP11) containing the indicated plasmid-borne Fkh2p derivatives. Cells were blocked with either α factor or nocodazole (noc). The results were normalized for the expression of RPB4 and are presented as fold induction in nocodazole-arrested cells relative to that in α factor-arrested cell (taken as 1). (D) Western analysis of the expression of mutant Fkh2p proteins in an fkh1Δ fkh2Δ strain (AP11) transformed with plasmids encoding HA-tagged Ndd1p and the indicated Myc-tagged Fkh2p derivatives. Fkh2p was detected by immunoprecipitation with anti-Myc antibodies followed by detection by immunoblotting (IB) with anti-Myc antibodies. (E and F) ChIP analysis of Ndd1p and Fkh2p occupancy at the promoters of the indicated genes. Immunoprecipitations were carried out with anti-HA (to detect Ndd1p) or anti-Myc (to detect Fkh2p) antibodies in extracts from formaldehyde-treated fkh1Δ fkh2Δ cells (AP11) containing plasmids encoding HA-tagged Ndd1p or the indicated Myc-tagged Fkh2p derivatives. A control immunoprecipitation (IP) was carried out with a nonspecific antibody (Flag; lanes 5 and 9). Ndd1p expression was induced by growth on galactose, and experiments in all panels were carried out under the same conditions for consistency.
Next, we wished to further probe the molecular basis to the phenotypic defects caused by mutation of the phosphorylation sites in the C terminus of Fkh2p. First, we examined the activities of the wild-type, S683A (m1), and T697A (m2) Fkh2p proteins on a β-galactosidase reporter driven by the SWI5 promoter, a known Fkh2p target (Fig. 4B). In the presence of wild-type Fkh2p, the expression of this reporter was induced by nocodazole treatment, consistent with the enhanced activity associated with the Fkh2p-Ndd1p complex in mitotically arrested cells. In contrast, the SWI5-LacZ reporter was not inducible in strains containing either the m1 or the m2 mutated versions of Fkh2p. We extended this analysis by using Northern blotting to investigate the expression of the endogenous SWI5 gene and another target gene, ACE2 (Fig. 4C). While both genes were inducible by nocodazole treatment in cells containing wild-type Fkh2p, inducible expression was virtually abolished in strains containing either the m1 or the m2 mutant versions of Fkh2p. Importantly, the reduced activity of the mutant Fkh2p derivatives was not due to differences in expression (Fig. 4D).
A key event in the regulation of genes in the CLB2 cluster is the recruitment of the coactivator Ndd1p by Fkh2p to their promoters (14, 20). Cell cycle-dependent regulatory events are likely to be important for controlling this interaction. One facet of this regulation is the Cdc28p-Clb2p-mediated phosphorylation of Ndd1p that promotes interactions with the FHA domain of Fkh2p (6, 20). However, a role for cell cycle-dependent phosphorylation of Fkh2p is also possible. We therefore used ChIP analysis to test whether the mutated Fkh2p proteins could recruit Ndd1p to the promoters of the Fkh2p target genes CLB2, ACE2, and SWI5. The recruitment of Ndd1p to all of these promoters was compromised in strains containing either the m1 or the m2 mutated versions of Fkh2p (Fig. 4E, compare lane 1 with lanes 2 and 3). In contrast, the recruitment of the Fkh2p proteins to these promoters was unaffected by the presence of phosphorylation site mutations (Fig. 4E, lanes 6 to 8).
Finally, we examined whether deletion of the C-terminal region of Fkh2p causes similar defects in the recruitment of Ndd1p to promoters, as observed with the phosphoacceptor site mutant proteins. ChIP analysis of occupancy at the CLB2 promoter revealed similar decreases in the recruitment of Ndd1p in strains containing either the m1 or C-terminal deletion versions of Fkh2p (Fig. 4F).
In summary, these data therefore demonstrate that the Cdc28p phosphorylation sites that we have identified in the C-terminal region of Fkh2p play a critical role in permitting Fkh2p to recruit the coactivator Ndd1p to promoters and hence regulate cell cycle-dependent gene expression.
Phosphorylation-dependent intermolecular interactions between Fkh2p and Ndd1p.
One obvious molecular mechanism for enhancing Ndd1p occupancy at promoters of genes in the CLB2 cluster would be to increase the intermolecular interactions between Fkh2p and Ndd1p. Indeed, enhanced interactions between purified nonphosphorylated Ndd1p and Myc-tagged Fkh2p isolated from extracts of nocodazole-blocked cells, in comparison with α factor-blocked cells, were detected (data not shown). Hence, we next investigated whether interactions between Ndd1p and Fkh2p are affected by Clbp kinase-dependent phosphorylation of Fkh2p.
First, the potential role of Clbp kinase phosphorylation in regulating complex formation between Ndd1p and Fkh2p was tested. In the absence of phosphorylation, interactions of Ndd1p could be detected with a region(s) in the N-terminal part of Fkh2p (amino acids 1 to 458) but not with the C-terminal region (amino acids 458 to 862) of Fkh2p (Fig. 5B, lanes 3 and 4). To further probe these interactions and examine the effect of phosphorylation, a series of truncated Fkh2p derivatives were translated in vitro and a pulldown reaction was performed. In the absence of prior phosphorylation, Ndd1p bound to fragments from the N-terminal region of Fkh2p (Fig. 5C, lanes 1 and 2). Little binding to the region encompassing the FKH domain or the C-terminal region of Fkh2p was observed (Fig. 5C, lanes 3 and 4). However, the inclusion of Flag-tagged Clb2p-associated kinase complexes in the pulldown reaction led to a large increase in binding of Ndd1p to the C-terminal region of Fkh2p (amino acids 458 to 862) and to amino acids 254 to 458 but did not affect the interactions with the fragments spanning amino acids 1 to 254 (Fig. 5C, lanes 5 to 8). This phosphorylation-dependent interaction between Fkh2p and Ndd1p could also be recapitulated with full-length Fkh2p (Fig. 5D).
FIG. 5.
Phosphorylation-dependent regulation of intermolecular interactions between Fkh2p and Ndd1p. (A) Schematic representation of full-length Fkh2p and a series of deletion constructs. The locations of the forkhead DNA binding (FKH) and FHA domains are indicated. (B to E) GST pulldowns of GST-Ndd1(1-554) with the indicated Fkh2p constructs obtained by in vitro translation. (B) Mapping the Ndd1p binding region on Fkh2p. *, positions of the full-length proteins. (C to E) Phosphorylation dependency of the interactions between Fkh2p and Ndd1p. Prior to the GST pulldown experiment, Fkh2p derivatives were phosphorylated (+phos) in vitro with immunoprecipitated Flag-tagged Clb2p-associated kinase complexes. Twenty (B, C, and E) and 50% (D) inputs are shown. (F) Clb5p-dependent Fkh2p phosphorylation in vivo is important for promoting Ndd1p interactions. GST pulldowns were carried out with GST-Ndd1(1-554) and extracts obtained from logarithmically growing W303-1a (WT, lane 1) or Y583-K3353-7B (clb5Δ; lane 2) cells containing pAS1760. Bound Gal-Fkh2p fusions were detected by Western blotting with an anti-Myc antibody. Ten percent of inputs are shown. (G) Phosphorylation site-dependent formation of Fkh2p-Ndd1p complexes. Myc-tagged Fkh2p was immunoprecipitated (IP) with an anti-Myc antibody from extracts of nocodazole-treated fkh1Δ fkh2Δ cells (AP11) expressing the indicated Myc-tagged Fkh2p derivatives and HA-tagged Ndd1p. Coprecipitating Ndd1p was detected by Western analysis with an anti-HA antibody. Total Ndd1p present in the lysates was identified by Western blot (WB) analysis with the same antibody (bottom). WT, wild type.
Next, we tested whether the key phosphorylation sites located within the C-terminal region of Fkh2p were important for the ability of Clbp kinase-dependent phosphorylation to stimulate binding to Ndd1p. As expected, in the absence of phosphorylation, neither the wild-type nor mutant Fkh2(458-862) proteins bound to Ndd1p (data not shown), while Clbp kinase-dependent phosphorylation stimulated interaction with the wild-type Fkh2p protein (Fig. 5E, lanes 1 to 4). In contrast, the binding of the m1 or the m2 versions of Fkh2(458-862) was not stimulated by Clb2p kinase-mediated phosphorylation (Fig. 5E, lanes 5 and 6).
To establish whether the activity of Clb5p-kinase complexes was required for interactions between the C terminus of Fkh2p and Ndd1p in vivo, we first tested the ability of GST-Ndd1p to bind to the Fkh2p C-terminal region using extracts prepared from cultures of wild-type and clb5Δ strains expressing Gal-Fkh2(458-862). In agreement with the observation that Clbp kinase complex-dependent phosphorylation of the C-terminal region of Fkh2p is important for interaction with Ndd1p, interactions between Ndd1p and the Gal-Fkh2(458-862) fusion protein were greatly reduced when extracts from strains lacking Clb5p were used (Fig. 5F, compare lanes 3 and 4). Finally, we compared the ability of wild-type and mutant versions of Fkh2p to interact with Ndd1p in vivo by coimmunoprecipitation analysis. Myc-tagged Fkh2p derivatives and HA-tagged Ndd1p derivatives were coexpressed in nocodazole-treated fkh1Δ fkh2Δ ndd1Δ cells, Fkh2p was immunoprecipitated, and coprecipitating Ndd1p was detected by Western blotting. In comparison to those of wild-type Fkh2p, the interactions of the m1 and m2 mutated versions of Fkh2p with Ndd1p were much reduced (Fig. 5G).
In summary, these data demonstrate that Clbp-dependent phosphorylation of sites in the C-terminal region of Fkh2p promotes intermolecular interactions with Ndd1p and hence activation of genes in the CLB2 cluster.
DISCUSSION
A complex containing the transcription factors Mcm1p and Fkh2p and the coactivator protein Ndd1p plays a pivotal role in regulating genes in the late G2 and M phases of the cell cycle in S. cerevisiae (14, 15, 19, 29; see references 4 and 7 for reviews). Here, we show that Fkh2p is a target of cell cycle-regulated Cdc28p-Clbp Cdk complexes. This is consistent with results of a recent large-scale in vitro screen that showed that Fkh2p is a target of Cdc28p (27). Functionally, phosphorylation regulates Fkh2p interactions with Ndd1p. This study, in combination with recent observations that Cdc28p-Clb2p-dependent phosphorylation of Ndd1p is important for the formation of Fkh2p-Ndd1p complexes (6, 20), reveals direct links between cell cycle-regulated kinases and cell cycle-regulated transcription factors.
Several lines of evidence strongly suggest that Fkh2p is a direct target of Cdc28p kinase in combination with the cyclin Clb5p. (i) The timing of Fkh2p phosphorylation in the cell cycle and the in vitro activity of Fkh2p-associated kinase in extracts isolated from different points in the cell cycle correspond to time frames when Cdc28p-Clb5p is active (Fig. 2A and B) (19). (ii) Phosphorylation of Fkh2p in vivo in a cdc28-1N mutant is much reduced when cells were grown at the nonpermissive temperature (Fig. 1A). (iii) The activity of the Fkh2p-associated kinase in extracts isolated from a clb5Δ mutant is greatly diminished (Fig. 2B and D). (iv) Fkh2p associates with Clb5p in cell extracts (Fig. 2F). (v) Immunoprecipitated HA-tagged Cdc28p and Clb5p-associated kinase complexes phosphorylate Fkh2p in vitro (Fig. 1B and 3D). (vi) The activity of the Fkh2p-associated kinase in extracts isolated from a cdc28-1N mutant is greatly diminished (Fig. 1C). (vii) Interactions with Ndd1p were found to be dependent on Clb5p, as judged by studies using extracts from wild-type and clb5Δ strains (Fig. 5F). Fkh2p plays a key role in regulating the expression of CLB2, and thus our data may partly explain why the early-acting cyclin Clb5p, along with Clb3p and Clb4p, regulates the accumulation of Clb1p/Clb2p (28).
It is possible that, in addition to Clb5p, other Clbp cyclins important for S, G2, and M phase progression play roles in Fkh2p phosphorylation. Indeed, although phosphorylation of Fkh2p was reduced by loss of Clb5p, it was not abolished (Fig. 2B, D, and E), suggesting that another kinase(s) is capable of phosphorylating Fkh2p. One possibility is that Clb2p has a latent ability to phosphorylate Fkh2p in the absence of Clb5p. Indeed, although Clb2p-associated kinase complexes can also phosphorylate Fkh2p in a similar manner to Clb5p complexes (Fig. 3D), unlike the loss of Clb5p function, inactivation of Clb2p in clb1Δ clb2ts clb3Δ clb4Δ mutant cells does not reduce the activity of Fkh2p-associated kinase(s) in extracts (6). Although we cannot discount the possibility that Clb2p has a role in vivo in Fkh2p regulation, the fact that Clb5p is active before Clb2p in the cell cycle suggests that this is the more likely Clbp for Fkh2p phosphorylation. In this context, it is interesting that Clb1p to -4p have a latent ability to regulate DNA replication in the absence of Clb5p and Clb6p (see reference 26 for a review).
Our mapping studies of Fkh2p localized the majority of cyclin-dependent kinase phosphorylation sites to the unique C-terminal region, absent in Fkh1p (Fig. 3A). The importance of this C-terminal region for the function of Fkh2p in the cell cycle is emphasized by the observation that Fkh2(1-458) is incapable of completely rescuing the pseudohyphal growth phenotype of an fkh1Δ fkh2Δ mutant (Fig. 4A). However, the expression of Fkh2(1-458), like that of the wild-type protein, cannot be tolerated in an fkh2Δ ndd1Δ background, suggesting that Fkh2(1-458) is biologically active (3). This residual function may be attributable to the ability of Fkh2(1-458) to still repress transcription in the absence of Ndd1p, presumably by recruiting corepressors. Indeed, the Sin3p corepressor was isolated from a global protein-protein interaction screen as a binding partner for Fkh2p (11), and this binding is mediated through the N-terminal region of Fkh2p (our unpublished data). Our results contrast with recent data that show that deletion of the C-terminal region of Fkh2p to amino acid 584 overcomes the lethality of the ndd1 deletion (20). The basis of this discrepancy is unclear. However, results similar to ours have been observed in an independent study (J. Veis, H. Klug, M. Koranda, and G. Ammerer, personal communication) where it was also demonstrated that deletion of the C terminus of Fkh2p to various positions between amino acids 443 and 776 (including 584) had no effect on the lethality caused by ndd1 deletion. However, despite these differences, both studies point to an important role for the C-terminal region in Fkh2p function.
The data presented in this paper suggest that phosphorylation of the C-terminal region of Fkh2p plays an important role in cell cycle-dependent regulation of the Mcm1p-Fkh2p-Ndd1p complex. Our data apparently contradict a recent study (20) suggesting that phosphorylation of Fkh2p is unimportant for function. However, as described above, we see different phenotypic consequences of deleting the C terminus of Fkh2p, and expression data for the CLB2 cluster were presented only for C-terminal Fkh2p proteins in the study by Reynolds et al. Moreover, the recruitment of Ndd1p to promoters is not abolished in strains containing a mutant version of Ndd1p that lacks a crucial Cdc28p phosphorylation site (20). This suggests that there is an alternative recruitment mechanism for Ndd1p that acts in an Ndd1p phosphorylation-independent manner. Indeed, here we show that cell cycle-dependent phosphorylation of the C-terminal region of Fkh2p promotes intermolecular interactions with Ndd1p in vivo and in vitro (Fig. 5). Clbp-mediated phosphorylation of Fkh2p also appears to alter intramolecular interactions in Fkh2p (data not shown), suggesting that phosphorylation of the C terminus of Fkh2p may promote a conformational change in Fkh2p. It is tempting to speculate that such regulation triggers activation of the complex. Current evidence using promoter mutations suggests that Ndd1p acts through the Mcm1p-Fkh2p complex to participate in G2 phase-specific gene regulation (14). Thus, the lack of a phosphorylatable C-terminal domain in Fkh1p may be a key difference that specifically permits Fkh2p to be activated and to recruit Ndd1p in a cell cycle-dependent manner.
ChIP studies have implicated Ndd1p as a Fkh2p binding partner (14, 22). Our study reveals that Ndd1p can interact with the N terminus of Fkh2p in the absence of phosphorylation. This binding can be further augmented by Clbp-dependent phosphorylation of Ndd1p and subsequent association with the FHA domain of Fkh2p (6, 20). However, we also show that phosphorylation of the C-terminal domain of Fkh2p promotes new interactions between Ndd1p and Fkh2p (Fig. 5C). Moreover, the analysis of the phosphorylation site mutations of the C terminus of Fkh2p demonstrated that phosphorylation leads to enhanced stability of Fkh2p-Ndd1p complexes in vivo (Fig. 5G) and is required for the efficient recruitment of Ndd1p to promoters (Fig. 4E). This is consistent with previous observations that enhanced recruitment of Ndd1p to promoters of genes in the CLB2 cluster does not correlate merely with the expression levels of Ndd1p (14).
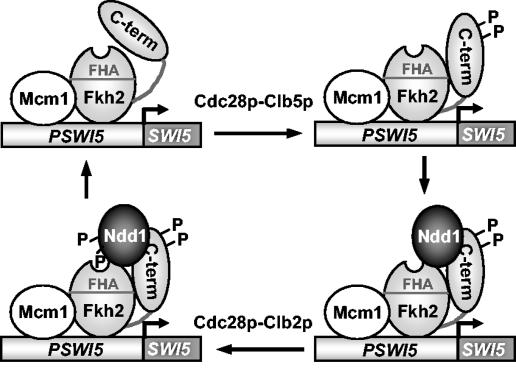
In combination with previous data (6, 20), the results presented here suggest that contacts between Fkh2p and Ndd1p change during the cell cycle (Fig. 6). In the G1 and early S phases, Ndd1p is not present; hence Fkh2p-Ndd1p complexes are not formed. The synthesis of Ndd1p and phosphorylation-induced changes in Fkh2p then permit Ndd1p recruitment and stabilization of the Fkh2p-Ndd1p complex. This complex is then further stabilized by Clb2p-dependent phosphorylation of Ndd1p (6, 20). Indeed, consistent with this model, Fkh2p and Ndd1p can be detected by ChIP analysis of occupancy at the promoters of the CLB2 gene cluster irrespective of the activity of Clb2p and the phosphorylation status of the key site in Ndd1p (T319) (20). Further enhancement of binding is then seen in nocodazole-treated cells as Ndd1p becomes phosphorylated. It is tempting to speculate that these structural changes lead to activation of the complex. Currently, it is unclear how phosphorylation of Fkh2p affects the function of the Fkh2p-Ndd1p complex in regulating transcription. It is possible that one function of the phosphorylation-driven changes in Fkh2p might also be to promote activation and/or recruitment of chromatin-remodeling machines and/or the basal machinery to the transcription factor complex.
FIG. 6.
Model for Cdc28p-Clbp regulation of the Mcm1p-Fkh2p-Ndd1p complex. Interactions between the C terminus of Fkh2p and its N-terminal region are altered by phosphorylation (P) by Cdc28p-Clb5p kinase. Ndd1p can then be recruited to the promoter. Subsequent phosphorylation of Ndd1p by Cdc28p-Clb2p enhances interactions between Ndd1p and Fkh2p through phosphopeptide-mediated interactions with the FHA domain of Fkh2p, activating gene expression (6, 20).
In summary, in combination with recent studies, we have demonstrated an important point of integration of the cell cycle control machinery and the regulation of transcription factor complexes that influence cell cycle-dependent gene expression. Future work will be directed towards understanding further how phosphorylation regulates the activities of the Fkh2p-Ndd1p complex.
Acknowledgments
We thank Anne Clancy and June Saunders for excellent technical assistance. We are grateful to Peter March for advice, Steve Sedgwick, David Lydall and members of our laboratories for comments on the manuscript and helpful discussions; Gustav Ammerer for discussion of his unpublished data; and Lee Johnston, David Lydall, Richard Reece, and Uttam Surana for reagents.
This work was supported by Cancer Research UK, the BBSRC, the Wellcome Trust, and the Lister Institute of Preventive Medicine.
REFERENCES
- 1.Amon, A., M. Tyers, B. Futcher, and K. Nasmyth. 1993. Mechanisms that help the yeast cell cycle clock tick: G2 cyclins transcriptionally activate G2 cyclins and repress G1 cyclins. Cell 74:993-1007. [DOI] [PubMed] [Google Scholar]
- 2.Andrews, B., and V. Measday. 1998. The cyclin family of budding yeast: abundant use of a good idea. Trends Genet. 14:66-72. [DOI] [PubMed] [Google Scholar]
- 3.Boros, J., F. L. Lim, Z. Darieva, A. Pic-Taylor, R. Harman, B. A. Morgan, and A. D. Sharrocks. 2003. Molecular determinants of the cell-cycle regulated Mcm1p-Fkh2p transcription factor complex. Nucleic Acids Res. 31:2279-2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Breeden, L. L. 2000. Cyclin transcription: timing is everything. Curr. Biol. 10:R586-R588. [DOI] [PubMed] [Google Scholar]
- 5.Cho, R. J., M. J. Campbell, E. A. Winzeler, L. Steinmetz, A. Conway, L. Wodicka, T. G. Wolfsberg, A. E. Gabrielian, D. Landsman, D. J. Lockhart, and R. W. Davis. 1998. A genome-wide transcriptional analysis of the mitotic cell cycle. Mol. Cell 2:65-73. [DOI] [PubMed] [Google Scholar]
- 6.Darieva, Z., A. Pic-Taylor, J. Boros, A. Spanos, M. Geymonat, R. J. Reece, S. G. Sedgwick, A. D. Sharrocks, and B. A. Morgan. 2003. Cell cycle regulated transcription through the FHA domain of Fkh2p and the coactivator Ndd1p. Curr. Biol. 13:1740-1745. [DOI] [PubMed] [Google Scholar]
- 7.Futcher, B. 2000. Microarrays and cell cycle transcription in yeast. Curr. Opin. Cell Biol. 12:710-715. [DOI] [PubMed] [Google Scholar]
- 8.Futcher, B. 2002. Transcriptional regulatory networks and the yeast cell cycle. Curr. Opin. Cell Biol. 14:676-683. [DOI] [PubMed] [Google Scholar]
- 9.Gietz, R. D., and A. Sugino. 1988. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene 74:527-534. [DOI] [PubMed] [Google Scholar]
- 10.Guan, K., and J. E. Dixon. 1991. Eukaryotic proteins expressed in Escherichia coli: an improved thrombin cleavage and purification procedure of fusion proteins with glutathione S-transferase. Anal. Biochem. 192:262-267. [DOI] [PubMed] [Google Scholar]
- 11.Ho, Y., A. Gruhler, A. Heilbut, G. D. Bader, L. Moore, S. L. Adams, A. Millar, P. Taylor, K. Bennett, K. Boutilier, L. Yang, C. Wolting, I. Donaldson, S. Schandorff, J. Shewnarane, M. Vo, J. Taggart, M. Goudreault, B. Muskat, C. Alfarano, D. Dewar, Z. Lin, K. Michalickova, A. R. Willems, H. Sassi, P. A. Nielsen, K. J. Rasmussen, J. R. Andersen, L. E. Johansen, L. H. Hansen, H. Jespersen, A. Podtelejnikov, E. Nielsen, J. Crawford, V. Poulsen, B. D. Sorensen, J. Matthiesen, R. C. Hendrickson, F. Gleeson, T. Pawson, M. F. Moran, D. Durocher, M. Mann, C. W. Hogue, D. Figeys, and M. Tyers. 2002. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415:180-183. [DOI] [PubMed] [Google Scholar]
- 12.Hollenhorst, P. C., G. Pietz, and C. A. Fox. 2001. Mechanisms controlling differential promoter-occupancy by the yeast forkhead proteins Fkh1p and Fkh2p: implications for regulating the cell cycle and differentiation. Genes Dev. 15:2445-2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koch, C., and K. Nasmyth. 1994. Cell cycle regulated transcription in yeast. Curr. Opin. Cell Biol. 6:451-459. [DOI] [PubMed] [Google Scholar]
- 14.Koranda, M., A. Schleiffer, L. Endler, and G. Ammerer. 2000. Forkhead-like transcription factors recruit Ndd1 to the chromatin of G2/M-specific promoters. Nature 406:94-98. [DOI] [PubMed] [Google Scholar]
- 15.Kumar, R., D. M. Reynolds, A. Shevchenko, A. Shevchenko, S. D. Goldstone, and S. Dalton. 2000. Forkhead transcription factors, Fkh1p and Fkh2p, collaborate with Mcm1p to control transcription required for M-phase. Curr. Biol. 10:896-906. [DOI] [PubMed] [Google Scholar]
- 16.Loy, C. J., D. Lydall, and U. Surana. 1999. NDD1, a high-dosage suppressor of cdc28-1N, is essential for expression of a subset of late-S-phase-specific genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:3312-3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lydall, D., G. Ammerer, and K. Nasmyth. 1991. A new role for MCM1 in yeast: cell cycle regulation of SWI5 transcription. Genes Dev. 5:2405-2419. [DOI] [PubMed] [Google Scholar]
- 18.Maher, M., F. Cong, D. Kindelberger, K. Nasmyth, and S. Dalton. 1995. Cell cycle-regulated transcription of the CLB2 gene is dependent on Mcm1 and a ternary complex factor. Mol. Cell. Biol. 15:3129-3137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pic, A., F.-L. Lim, S. J. Ross, A. L. Johnson, R. A. Sultan, A. G. West, L. H. Johnston, A. D. Sharrocks, and B. A. Morgan. 2000. The forkhead protein Fkh2 is a component of the yeast cell cycle transcription factor SFF. EMBO J. 19:3750-3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reynolds, D., B. J. Shi, C. McLean, F. Katsis, B. Kemp, and S. Dalton. 2003. Recruitment of Thr 319-phosphorylated Ndd1p to the FHA domain of Fkh2p requires Clb kinase activity: a mechanism for CLB cluster gene activation. Genes Dev. 17:1789-1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shore, P., and A. D. Sharrocks. 1994. The transcription factors Elk-1 and serum response factor interact by direct protein-protein contacts mediated by a short region of Elk-1. Mol. Cell. Biol. 14:3283-3291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Simon, I., J. Barnett, N. Hannett, C. T. Harbison, N. J. Rinaldi, T. L. Volkert, J. J. Wyrick, J. Zeitlinger, D. K. Gifford, T. S. Jaakkola, and R. A. Young. 2001. Serial regulation of transcriptional regulators in the yeast cell cycle. Cell 106:697-708. [DOI] [PubMed] [Google Scholar]
- 23.Songyang, Z., S. Blechner, N. Hoagland, M. F. Hoekstra, H. Piwnica-Worms, and L. C. Cantley. 1994. Use of an oriented peptide library to determine the optimal substrates of protein kinases. Curr. Biol. 4:973-982. [DOI] [PubMed] [Google Scholar]
- 24.Spellman, P. T., G. Sherlock, M. Q. Zhang, V. R. Iyer, K. Anders, M. B. Eisen, P. O. Brown, D. Botstein, and B. Futcher. 1998. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol. Biol. Cell 9:3273-3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Surana, U., H. Robitsch, C. Price, T. Schuster, I. Fitch, A. B. Futcher, and K. Nasmyth. 1991. The role of CDC28 and cyclins during mitosis in the budding yeast S. cerevisiae. Cell 65:145-161. [DOI] [PubMed] [Google Scholar]
- 26.Toone, W. M., B. Aerne, B. A. Morgan, and L. H. Johnston. 1997. Getting started: regulating the initiation of DNA replication in yeast. Annu. Rev. Microbiol. 51:125-149. [DOI] [PubMed] [Google Scholar]
- 27.Ubersax, J. A., E. L. Woodbury, P. N. Quang, M. Paraz, J. D. Blethrow, K. Shah, K. M. Shokat, and D. O. Morgan. 2003. Targets of the cyclin-dependent kinase Cdk1. Nature 425:859-864. [DOI] [PubMed] [Google Scholar]
- 28.Yeong, F. M., H. H. Lim, Y. Wang, and U. Surana. 2001. Early expressed Clb proteins allow accumulation of mitotic cyclin by inactivating proteolytic machinery during S phase. Mol. Cell. Biol. 21:5071-5081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhu, G., P. T. Spellman, T. Volpe, P. O. Brown, D. Botstein, T. N. Davis, and B. Futcher. 2000. Two yeast forkhead genes regulate the cell cycle and pseudohyphal growth. Nature 406:90-94. [DOI] [PubMed] [Google Scholar]