Abstract
Gene trapping in mouse embryonic stem cells is an important genetic approach that allows simultaneous mutation of genes and generation of corresponding mutant mice. We designed a selection scheme with drug selection markers and Cre/loxP technology which allows screening of gene trap events that responded to a signaling molecule in a 96-well format. Nine hundred twenty gene trap clones were assayed, and 258 were classified as gene traps induced by in vitro differentiation. Sixty-five of the in vitro differentiation-inducible gene traps were also responsive to retinoic acid treatment. In vivo analysis revealed that 85% of the retinoic acid-inducible gene traps trapped developmentally regulated genes, consistent with the observation that genes induced by retinoic acid treatment are likely to be developmentally regulated. Our results demonstrate that the inducible gene trapping system described here can be used to enrich in vitro for traps in genes of interest. Furthermore, we demonstrate that the cre reporter is extremely sensitive and can be used to explore chromosomal regions that are not detectable with neo as a selection cassette.
Phenotype-driven genetic screens are used to select mutants with similar phenotypes from large-scale mutagenesis experiments. Analysis of these mutants helps to functionally dissect genes involved in common pathways. Phenotype-driven, genome-wide screens are routinely carried out in Saccharomyces cerevisiae, Drosophila melanogaster, and Caenorhabditis elegans. However, large-scale phenotype-driven genetic screens in mice have been restricted to laboratories with large mouse facilities and strong support (18, 20, 31, 33). The discovery of mouse embryonic stem (ES) cells and the demonstration that they can be used to reconstruct mice have created an unprecedented opportunity for mouse geneticists (6). Because each individual ES cell has the potential to be converted into an animal, theoretically a screen of millions of individuals can be performed in a single plate if a phenotype is selectable or can be identified at the ES cell stage (23).
Since most autosomal mutations are recessive and require disruption of both alleles to show a phenotype, one of the challenges in performing genetic screens in ES cells is to overcome the diploid nature of the mouse genome to elicit the phenotypic consequences of recessive mutations. One way to overcome the problem is to increase the mitotic recombination rate in ES cells so that heterozygous mutations can be converted to a homozygous state. ES cells with an 18-fold increase in the rate of loss of homozygosity have been described (26). This cell line has been used successfully as a genetic background for in vitro screens (16). Mitotic recombination rates can also be elevated by placing loxP sites on homologous chromosomes so that Cre-catalyzed chromosome-specific mitotic recombination can be achieved efficiently (25). This system will be extremely useful when a chromosome-specific mutagenesis approach is available (M. Wentland and A. Bradley, unpublished data).
One way to bypass the problem of screening for recessive mutations in a diploid genome is to couple the generation of a recessive mutation to the activation of a dominant marker that enables screening or selection. For example, 5′ gene trap mutagenesis links the mutated gene to a marker which can report on the expression pattern of a disrupted gene, enabling gene expression-based screening or selection protocols (2, 10, 12, 29, 30, 41, 50). Thus, 5′ gene trapping in ES cells provides an opportunity to perform a high-throughput, cost-effective screening procedure on mutant ES cells in vitro (2, 10, 12, 29, 30, 41, 50). This in vitro phenotyping method makes it possible to categorize gene trapping events so that gene traps of interest can be identified prior to their cloning and functional assessment in vivo.
5′ gene trapping has been applied to identify developmentally regulated genes (2, 10, 12, 29, 30, 41, 50). Many key signaling pathways are active during embryogenesis, and thus developmental mutants provide valuable tools to dissect these pathways genetically. With the development of in vitro differentiation protocols for embryonic stem cells, genes which are responsive to developmental signaling molecules, such as retinoic acid, can be identified by 5′ gene trapping (2, 4, 10, 12, 13, 37, 44). In contrast to phenotype-driven screens, 5′ gene trapping is cost-effective (18, 20, 31, 33). Compared to other molecular methods, such as subtractive cDNA cloning, differential display, and expression arrays, 5′ gene trapping has the advantage of generating mutant ES cells which can be used to produce mutant mice at the same time that the gene is identified (14, 40, 47).
In this study, we used a cre-trapping retroviral vector and a drug resistance-switching responder construct to build an inducible gene trapping system which allowed phenotypic scoring in vitro. This system was used to trap developmentally regulated genes, based on the assumption that genes induced during in vitro differentiation and retinoic acid treatment are likely to be developmentally regulated (2, 4, 10, 12, 13, 37, 44). Three different conditions were used to treat replicas of the same set of “gene-trapped” ES cell clones. Comparison of the data generated from different treatments identified 65 gene traps that were induced by both in vitro differentiation and retinoic acid treatment. Furthermore, 11 of the 13 in vitro differentiation- and retinoic acid-inducible gene traps examined in vivo proved to be developmentally regulated. We also provide experimental evidence to show that the cre reporter is extremely sensitive. Thus, it is possible to use cre as a reporter to explore chromosomal regions in which the PGKneobpA selection cassette is inactive.
MATERIALS AND METHODS
DNA constructs.
The gene trap vector was a 1.6-kb DNA fragment containing a synthetic splice acceptor followed by cre and the phosphoglycerate kinase (PGK) polyadenylation signal released from pOG231 (a gift from S. O'Gorman) by SacII and SalI digestion. This fragment was ligated upstream of the PGKneobpA cassette (52) in the same transcriptional orientation as the PGKneobpA construct. To generate the retroviral gene trap vector pROSAcre, the gene trap construct described above was inserted into a unique XhoI site in the pGen retroviral vector backbone (43) in the orientation opposite that of retroviral transcription.
Responder locus.
The responder locus targeting vector pYTC86 was constructed by placing a responder construct (from pYTC83) containing PGK-loxP- bsd-bpA-loxP-puroΔtk-bpA into a KpnI site in the RIV6.8-oligo Hprt targeting vector. RIV6.8-oligo is a derivative of pRIV6.0 (17) containing an oligonucleotide with multiple cloning sites in the endogenous XhoI site within exon 3 of Hprt. The transcriptional orientation of the responder construct is opposite that of the endogenous Hprt locus. A bsd-bpA fragment was generated from the bsd coding region derived from pUCSVBSD (22) and modified to contain a Kozak ATG and bpA from pPGKneobpA. This bsd cassette was cloned into the position flanked by loxP sites in pYTC77. The loxP-bsd-bpA-loxP fragment was cloned between PGK and puΔtk-bpA in pYTC66, a plasmid derived from pYTC37 (7). The function of the loxP sites was tested by transforming plasmids into Escherichia coli BNN132 bacteria (BD Biosciences Clontech, Mountain View, Calif.), and the recombined products were examined by restriction enzyme digestion.
ES cell culture and retrovirus infection.
The procedures for ES cell culture, electroporation, and drug selection have been described previously (36). Briefly, AB1 ES cells were cultured in M15 medium (Dulbecco's modified Eagle's medium plus 15% fetal calf serum) and maintained on irradiated SNL76/7 feeders (28) or SNLPB-7/4 feeders, a derivative of SNL76/7 which carry the PGKbsdbpA cassette. pYTC86 was linearized by XmnI partial digestion and electroporated into AB1 ES cells, and 107 cells were plated onto 90-mm SNLPB-7/4 feeder plates in M15 medium. Blasticidin S (ICN 150477) selection (20 μg/ml) was initiated 24 h after electroporation, followed 6 days later by 6-thioguanine selection (10 μM). ES cell colonies were picked 13 days after electroporation. DNA from the targeted clones was restricted with HindIII, and Southern analysis was performed with phpt2 (52) as an external probe.
To generate retrovirus-producing cells, pROSAcre was linearized with ScaI and electroporated into the GP+E86 cell line (27). G418 selection (1 mg/ml) was initiated 24 h after electroporation. Forty-eight G418-resistant clones were screened for retrovirus production. One of the subcloned lines showed a retrovirus titer close to 103 CFU/ml when assayed on ES cells and was used for subsequent experiments.
Infection of ES cells with the ROSAcre trapping virus was performed as described previously (43). Briefly, 3.2 × 106 ES cells were seeded onto a 90-mm feeder plate on day 0. On day 1, 0.45 μM filtered ROSAcre virus-containing supernatant collected overnight (2 ml) was mixed with 8 ml of fresh M15 medium and applied to ES cells in the presence of 4 μg of Polybrene per ml for 24 h. G418 selection (180 μg of active ingredient per ml) was initiated 48 h after infection to identify proviral integration events. Colonies resistant to G418 were picked after 7 days of selection.
In vitro differentiation and drug selection.
G418-resistant ES cell clones were cultured on 96-well master plates for 6 days, and six replicas were generated. One set of plates was frozen at −80°C for chimera generation. The second set of plates was used for genomic DNA extraction to examine proviral integration events by Southern analysis. The third and fourth sets of plates were maintained under normal ES cell conditions for 1 week before either puromycin or 1-(−2-deoxy-2-fluoro-1-β-d-arabinofuranosyl)-5-iodouracil (FIAU) and blasticidin S dual selection was initiated. The ES cells seeded on the fifth and sixth plates were plated on gelatinized plates for 1 h to remove the feeders and then seeded onto fresh gelatinized plates. From 1 day after seeding, the fifth set of plates were maintained in differentiation medium, which contains only 1% fetal calf serum, for 1 week before initiating puromycin selection. The sixth set of plates were maintained in retinoic acid induction medium which comprises differentiation medium plus 10 μM all-trans-retinoic acid (Sigma, St. Louis, Mo.) for 7 days (10, 12, 13, 37). Puromycin selection was initiated after the treatment and maintained for another 7 days. The plates were then stained with 2% methylene blue in 70% ethanol for 5 min and rinsed in tap water for 10 min. The absorbance at 600 nm was measured with the Bio-Rad model 2550 enzyme immunoassay reader (Bio-Rad, Hercules, Calif.). Data analysis was performed with Microsoft Excel software.
Chimera generation and germ line transmission.
Inducible gene-trapped clones were injected into blastocysts from the C57BL/6 Tyrc-Brd mouse strain by standard procedures (36). Injected blastocysts were transferred to the uteri of pseudopregnant female B6/CBA F1 recipients. Approximately 40 blastocysts were injected for each clone. The resulting high-percentage chimeras were mated with an albino variant of R26R females (42) to allow cell lineage tracing simultaneous with identification of pups derived from the injected ES cells.
In vivo expression, cell lineage tracing by R26R, and X-Gal staining.
Timed matings were set up between albino R26R females and high-percentage chimeras. Pregnant females were sacrificed at 10 days post coitum. Embryos were collected and fixed in 2% paraformaldehyde in 1× phosphate-buffered saline (pH 7.4) with 2 mM MgCl2 and 625 nM EGTA for 1.5 to 2 h at room temperature. Fixed embryos were permeabilized for 1.5 h at room temperature with a solution containing 1× phosphate-buffered saline (pH 7.4), 2 mM MgCl2, 0.02% NP-40, and 0.01% sodium deoxycholate. Permeabilized embryos were equilibrated in X-Gal buffer, which contains 25 mM K4Fe(CN)6, 25 mM K3Fe(CN)6O · 3H2O, 2 mM MgCl2, 0.01% Nadex, and 0.02% NP-40 in 1× phosphate-buffered saline. After 1 h, X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) (Sigma) was added to the reaction buffer at a final concentration of 1 mg/ml, and the reaction was incubated overnight at room temperature. Stained embryos were washed with phosphate-buffered saline and fixed in 4% paraformaldehyde. Paraffin sections were prepared and cut on a Jung Histocut 820 microtome (Leica Microsystems, Buffalo, N.Y.) with standard histology methods. Eosin Y aqueous solution (Sigma) was used for counterstaining.
5′ RACE, RT-PCR, and inverse PCR.
5′ rapid amplification of cDNA end (RACE) reactions were carried out with Micro-fast track (Invitrogen, Carlsbad, Calif.) on purified polyadenylated RNA that was purified from a mixture of differentiated ES cells (cultured in differentiation medium for 1, 4, or 7 days). A 5′ RACE kit (5′ RACE System, version 2.0; Invitrogen) and three ROSAcre-specific primers were used to amplify fusion transcripts. The primer for the reverse transcription (RT) reaction is 5659 Cre-RT28, 5′-TTCTCC CAC CGT CAG TAC GTG AGA TAT C-3′; the primers for the first RACE reaction are 5657 Cre-GSP28, 5′-CCC TGA TCC TGG CAA TTT CGG CTA TAC G-3′ and AAP 5′-GGC CAC GCG TCG ACT AGT ACG GGI IGG GII GGG IIG-3′; the primers for the second RACE reaction are 5658 Cre-NGSP28, 5′-CGA CCG GTA ATG CAG GCA AAT TTT GGT G-3′ and AUAP 5′-GGC CAC GCG TCG ACT AGT AC-3′).
The 5′ RACE products were either sequenced directly with gsp2 (provided by the kit) as a primer or subcloned with TOPO TA cloning kits (Invitrogen) and subsequently sequenced. RT-PCR was performed with Superscript One-Step RT-PCR with Platinum Taq (Invitrogen). The primers used to amplify the fusion transcript of gene trap clone 6E6 were 5657 and 6E6-RT10, 5′-CCT TTG TGT TGC AAC CCT CAG CAT-3′. The primers used to amplify the fusion transcript of gene trap clone 9G3 were 5657 and 9G3-AK122413-f4, 5′-GAT GAA GGA CAA ACA GAA GAG GAA G-3′. Inverse PCR was carried out with BglII-digested ES cell DNA as the template. Ten microliters (400 U) of T4 DNA ligase (Invitrogen) was used to catalyze self-circularization of 0.5 to 2.5 μg of BglII-digested genomic DNA fragments in a 500-μl reaction at 16°C overnight, followed by heat inactivation, phenol-chloroform extraction, and ethanol precipitation.
Precipitated DNA was used for three runs of nested, long-template PCR with AccuTaq La DNA polymerase mix (Sigma) following the manufacturer's instructions. PCR products were gel purified and then subcloned with TOPO TA cloning kits for subsequent sequencing steps. The primers used for inverse PCR were as follows: the primers for the primary PCR were 2677, 5′-AAA ACT GCA GCC AAC GCC ACC ATG GGG ATG GGA TCG GCC ATT GAA CA-3′, and 4627 5′-CCT GAT CCT GGC AAT TTC GGC TA-3′; the primers for the secondary PCR were 2430, 5′-GAT CCC CCG GGG GAT CAG CCT CGA CTG TGC CTT CTA GT-3′ and 3056 5′-ACC TTC GAA GTC GAT GAC GGC AGA TTT AGA G-3′; and the primers for the tertiary PCR were 3030, 5′-ATC TGC CGT CAT CGA CTT CGA AGG TTC G −3′, and 3031, 5′-GAC GCG CCG CTG TAA AGT GTT ACG TTG AG-3′.
RESULTS
Construction of responder ES cells.
To establish an ES cell line that responds to transient Cre expression by permanent drug resistance switching, a responder construct which contained a phosphoglycerate kinase-1 (PGK) promoter and two selectable markers was generated (Fig. 1a). The blasticidin S deaminase gene (bsd) from Aspergillus terreus, which provides resistance to blasticidin S (21, 22), was placed between two loxP sites and cloned next to the PGK promoter. The positive-negative selectable marker puΔtk (7) was cloned downstream of bsd and is not transcribed until bsd is deleted by the action of Cre. The puΔtk cassette allows complete and clean positive selection with puromycin and also enables the use of 1-(−2-deoxy-2-fluoro-1-β-d-arabinofuranosyl)-5-iodouracil (FIAU) for negative selection to select against Cre-catalyzed recombination events. The deleted form of herpes simplex virus thymidine kinase used in the puΔtk cassette does not interfere with germ line transmission (7), and thus the ES cell clones identified in the in vitro screen can be efficiently transmitted into the mouse germ line.
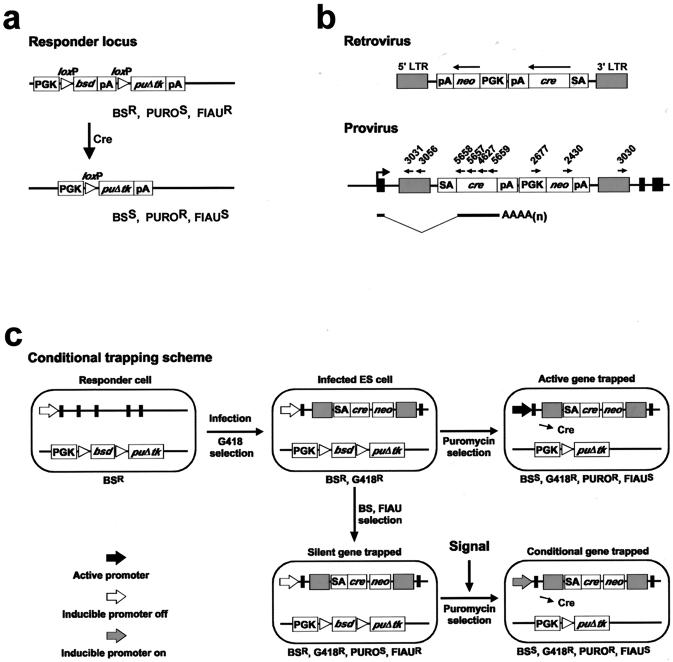
FIG. 1.
Design of the inducible gene trapping system. (a) The first component is an ES cell carrying a responder construct which allows the cell to respond to transient Cre expression by permanent drug resistance switching. In the uninduced state, the responder construct provides blasticidin S resistance. Cre-catalyzed recombination between the loxP sites switches the responder construct to provide resistance to puromycin. (b) The second component of the system is a ROSAcre retroviral vector, which carries cre as a reporter gene which is activated by 5′ gene trapping. The expression pattern of the promoterless cre reporter in the provirus will reflect the expression pattern of the trapped gene. (c) Two possible pathways activate the cre reporter and lead to permanent drug resistance switching in the responder ES cells. If the ROSAcre vector traps a gene which is active in ES cells, the cre reporter will be activated and catalyze recombination between the loxP sites, rendering the cell puromycin resistant. If the ROSAcre vector traps a silent gene and the trapped gene can respond to a signaling molecule or a biological event of interest, the cre reporter will be activated and puromycin resistance will be obtained. LTR, long terminal repeats; SA, splice acceptor; cre, cyclization recombinase coding region; pA, polyadenylation signal; PGK, phosphoglycerate kinase 1 promoter; neo, neomycin phosphotransferase; bsd, blasticidin S deaminase; puΔtk, fusion-selectable marker between puromycin N-acetyltransferase and a truncated version of herpes simplex virus 1 thymidine kinase. For clarity, the PGK promoter and polyadenylation signals are omitted from panel c.
To produce a stable locus for the responder construct, the cassette was targeted into the endogenous X-linked Hprt locus (Fig. 2). This has two advantages. First, the X chromosome is very stable in XY ES cells because it is not subject to loss of heterozygosity, and in vivo this X linkage allows the responder locus to be traced based on gender, reducing molecular genotyping efforts. The Hprt gene is also disrupted by the targeting event, allowing the use of the Hprt gene as another positive selectable marker with the system in the future.
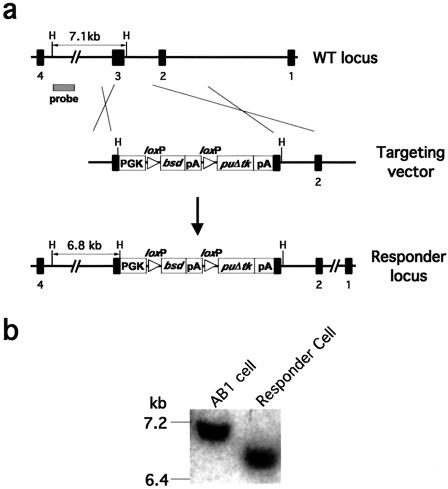
FIG. 2.
Construction of the responder ES cell line. (a) Targeting strategy. (b) Southern blot of wild-type and responder ES cell DNAs digested with HindIII and probed with a 5′ external probe, showing the 7.1-kb wild-type and 6.8-kb responder alleles.
The responder locus was tested by transient Cre expression. Five targeted clones were electroporated with the plasmid pOG231 and seeded at low density (2 × 103 cells/10-cm plate) so that each individual recombination event could be detected as a puromycin-resistant colony. One-third of the electroporated cells switched to puromycin resistance when selection was initiated 24 h after electroporation. Thus, the responder ES cells exhibited efficient drug resistance switching in response to Cre expression.
Leaky expression of downstream markers before loxP recombination has been described (42, 45). To examine this possibility, puromycin selection was applied to responder ES cells without introducing Cre. One hundred percent of the cells were killed in these tests (>107 cells tested), and thus leaky expression of the downstream marker did not occur in this responder allele. Two responder cell lines were tested for their germ line potential. One line, BC86-G2, which consistently generated germ line chimeras was used in all subsequent experiments.
ROSAcre gene trap virus.
To build a retrovirus-based 5′ gene trap vector, the cre-trapping cassette was cloned in pGen opposite to the direction of transcription of the virus (Fig. 1b). The virus was named ROSAcre (reverse orientation splice acceptor cre) (11). The cre-trapping cassette consists of a synthetic splice acceptor sequence derived from an IgG variable region (5, 19) cloned 5′ to cre. Translation of functional Cre requires splicing, but because Cre has its own ATG, an in-frame fusion between the Cre and the endogenous gene coding region is not required (38). A retroviral producer line was established in the GP+E86 packaging cell line (27), and a viral titer of 103 G418-resistant CFU/ml was achieved when assayed on ES cells. This titer was comparable to that of other gene trap retroviral vectors described previously (11).
Retrovirus infection, in vitro differentiation, and drug selection scheme.
The experimental scheme is summarized in Fig. 3. The responder cells were infected with ROSAcre virus, and 48 h later G418 selection was initiated to select for integration events. A total of 960 G418-resistant colonies were arrayed in 10 × 96 well master plates. Southern analysis of 10% of the clones revealed that the multiplicity of infection was 1.1 in this experiment.
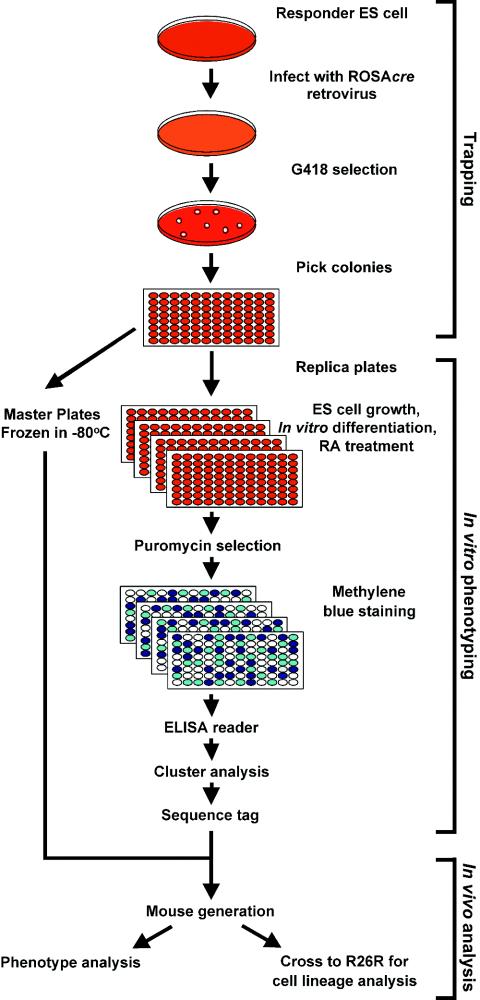
FIG. 3.
Flowchart illustrating the inducible gene trapping system to screen for gene traps of interest in vitro. Responder ES cells were infected with the ROSAcre trapping virus, followed by selection of trapping vector integration events in G418. G418-resistant colonies were picked and replicated, and a set of master plates were frozen at −80°C for future in vivo experiments. Replica plates were treated with different inducing conditions or exposed to various signaling molecules. After the treatment, puromycin selection was applied to each clone. The number of cells surviving puromycin selection represents the accumulated Cre-catalyzed drug resistance events induced by the conditions or signaling molecule. The absorbance readings at 600 nm of methylene blue-stained plates reflect the number of cells which survived drug selection, which is extrapolated to be an indicator of the expression level of the cre reporter. Multiple readings from different clones were used for cluster analysis. Gene trap clones from the subgroup of interest were used to obtain sequence tags with either 5′ RACE or inverse PCR. Clones of interest were used to generate mutant mice, allowing in vivo functional characterization of the trapped gene. ROSAcre-trapped mice can be crossed to R26R mice to trace the cell lineages of the trapped gene.
Replica plates were prepared for the different treatments. In vitro differentiation and retinoic acid treatment experiments were conducted according to protocols described previously (2, 4, 10, 12, 13, 37, 44). Enlarged, differentiated cells were observed in wells cultured with differentiation medium. At the end of retinoic acid treatment, a very low percentage of neuron-like cells were observed in retinoic acid-treated wells. Cells were maintained under in vitro differentiation or retinoic acid treatment conditions for 7 days, and puromycin selection was then initiated. This time period was used because we found that 4 to 7 days are required for at least one cell in a clone to undergo Cre-mediated drug resistance switching in a majority of the individual active gene trap clones (Y.-T. Chen and A. Bradley, unpublished data). This relatively long period reflects the dynamic range of the expression levels of trapped genes resulting from random insertional mutagenesis.
After the selection was complete, the 96-well plates were stained with methylene blue and scored with an enzyme-linked immunosorbent assay reader at a wavelength of 600 nm. The absorbance reading from a well is a measure of the number of cells surviving puromycin selection. This reflects the product of the accumulated Cre-catalyzed recombination events and the proliferation rate. The protocols used for in vitro differentiation and retinoic acid treatment were not elaborate, and thus we believe that similar cell types with similar proliferation rates were generated in different wells. To a first approximation, the absorbance reflects the accumulated Cre-mediated recombination events, which indirectly reflects cre reporter expression levels. However, the absolute readings from wells under different treatments are not directly comparable. Different treatments will drive the ES cells through different differentiation pathways, resulting in differentiated cell types that will have different proliferation rates.
Quantification of induction after in vitro differentiation and retinoic acid treatment.
A total of 3,840 absorbance readings were used for analysis. The results are summarized in Table 1 and Fig. 4. One set of readings were from plates that went through blasticidin S and FIAU dual selection, and these are responder cells with an unrecombined responder construct (data not shown). The second set of readings were from plates cultured under ES cell conditions followed by puromycin selection, which represent the accumulated Cre-catalyzed recombination events in ES cells. The third set of readings were from plates cultured with differentiation medium for 1 week followed by puromycin selection, which represent the accumulated Cre-catalyzed recombination events in differentiating ES cells. Finally, the fourth set of readings were taken from plates treated with retinoic acid followed by puromycin selection, which represent the accumulated Cre-catalyzed recombination events in retinoic acid-treated ES cells.
TABLE 1.
Summary of inducible gene trapping results
| Type of gene trap | Criteriaa | No. of clones examined | % of clones |
|---|---|---|---|
| G418 resistance | (ABSBS + ABSES + ABSDiff + ABSRA) > 0.5 | 920 | 100.0 |
| Active | ABSES > 0.75 | 392 | 42.6 |
| Silent | ABSES ≤ 0.75 | 528 | 57.4 |
| Differentiation inducible | ABSES ≤ 0.75, ABSDiff > 0.5 | 258 | 28.0 |
| Retinoic acid inducible | ABSES ≤ 0.75, ABSRA > 0.15 | 67 | 7.3 |
| Differentiation and retinoic acid inducible | ABSES ≤ 0.75, ABSDiff > 0.5, ABSRA > 0.15 | 65 | 7.1 |
ABSBS, ABSES, ABSDiff, and ABSRA represent absorbance readings at a wavelength of 600 nm from wells cultured under ES cell conditions followed by either blasticidin S selection (ABSBS) or puromycin selection (ABSES) and wells treated with differentiating medium (ABSDiff) or retinoic acid (ABSRA) followed by puromycin selection.
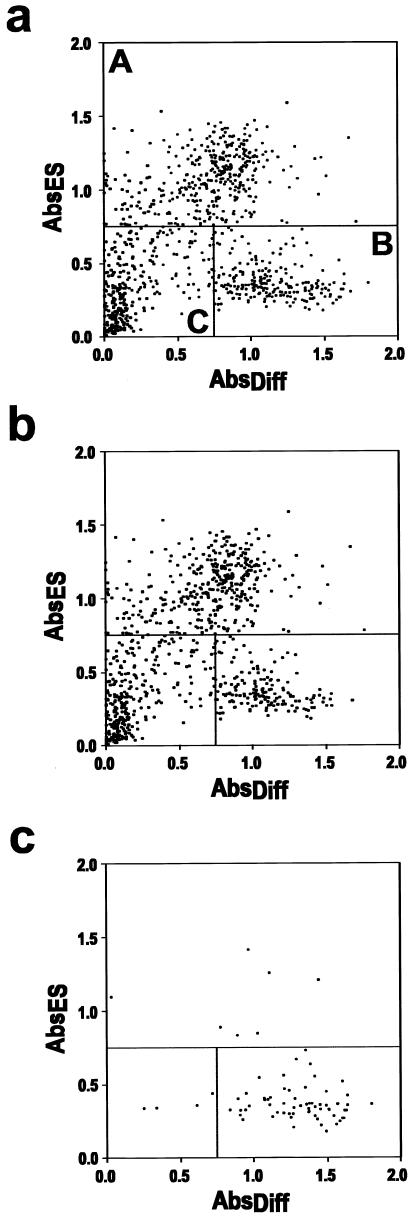
FIG. 4.
Cluster analysis identifies distinguishable subgroups of gene traps. (a) Absorbance readings of 920 G418-resistant clones cultured under different conditions, including normal ES cell conditions, in vitro differentiation, and retinoic acid (RA) treatment. Each data point is from a different gene trap clone. Its position on the plot is determined by readings obtained from undifferentiated ES cell culture and in vitro differentiating conditions. The gene trap clones can be divided into three different subgroups according to their positions on the plot. The first group, A, comprise gene trap clones that have the highest absorbance reading after growth in normal ES cell conditions (ABSES > 0.75). This group represents clones in which the ROSAcre vector trapped active genes. The second group, B (ABSES ≤ 0.75 and ABSDiff. > 0.5), represents gene traps that are inactive in ES cells but are induced during the in vitro differentiation process. We refer to these as in vitro differentiation-inducible gene traps. The third group of clones, C, comprise integrations which do not trap genes or integrations in genes which are not activated by the conditions used in this study. (b) Gene traps with absorbance reading from the retinoic acid treatment set of less than 0.15. (c) Gene traps with absorbance readings greater than 0.15 following retinoic acid treatment. The majority of the gene trap clones that give higher readings in the retinoic acid-treated plates (ABSRA > 0.15) fall into the subgroup that are in vitro differentiation-inducible (subgroup B in plot a). The details of the gene trap results are summarized in Table 1.
The absorbance results from the ES cells cultured in ES cell conditions (ABSES or ABSBS), in vitro differentiation medium (ABSDiff), or retinoic acid treatment (ABSRA) are presented in a two-dimensional plot in Fig. 4. The spatial distribution of the dots on the plot separates the gene trap clones into three distinguishable subgroups (Fig. 4a). The first group, A, comprises gene trap clones that have the highest absorbance reading after growth under standard ES cell conditions (ABSES > 0.75). This group represents clones in which the ROSAcre vector trapped active genes. The second group, B (ABSES ≤ 0.75 and ABSDiff. > 0.5), are clones that have gene traps which are inactive in ES cells but are inducible during the in vitro differentiation process. We refer to these as in vitro differentiation-inducible gene traps. The third group, C, comprises clones in which the virus did not trap a gene (non-gene traps) or the trapped gene was not induced by the conditions used in this experiment.
Sensitivity of the detection scheme.
The two-element gene trap system with Cre and loxP appears to be extremely sensitive. The proportion of active gene traps detected in these experiments was 42.6%, which is much higher than that reported with the ROSAβ-gal gene trap vector (11.6%). This phenomenon can be explained by the sensitivity of the reporter gene (cre or β-gal) relative to the selectability of the cassette used to detect retroviral vector integration events (PGKneobpA). The Cre/loxP system allows the identification of transiently expressed genes or those which are expressed at low levels (45), and therefore it is expected that cre would be more sensitive as a reporter.
The sum of the active gene traps and the in vitro differentiation-inducible gene traps constituted approximately 70% of the G418-resistant clones. This proportion is much higher than expected because not all insertions will be in genes, only a portion of those that are will be in the correct orientation, and only a portion of these are expected to produce a functional chimeric transcript. One possible explanation for the high trapping efficiency is that the gene trap events are a nonrandom sample from the genome, selected because of the detection limit of the PGKneobpA and/or biases associated with retroviral integration sites.
To test the detection limit of the selectable marker, an experiment was performed in which G418 selection for viral integration was omitted. Clones which had undergone Cre-mediated drug resistance switching were selected in puromycin, and these were subsequently assessed for resistance to G418. Thirty-one of 80 puromycin-resistant colonies tested were G418 resistant, implying that more than 60% of the gene trap events were excluded in the previous experiments because they were killed by G418 selection. All 80 clones were confirmed by Southern analysis to have an integrated provirus. The clones that did not survive G418 selection are likely to be those in which ROSAcre integrates into chromosomal regions that would not allow PGKneobpA to transcribe neo at a level sufficient to provide G418 resistance. Therefore, the gene trap events analyzed in this work are biased toward chromosomal regions that are compatible with PGKneobpA selection. Proviral integration site preference cannot be assessed until a large sample of integration loci are cloned.
Retinoic acid differentiation.
A third set of readings from the retinoic acid treatment plates were analyzed. Readings from the retinoic acid treatment plates are represented in Fig. 4b and c. The majority of these clones fell into the category of in vitro differentiation-inducible gene traps (62 of 71); however, not all of the in vitro differentiation-inducible gene traps were induced by retinoic acid treatment.
One obvious difference between the retinoic acid-treated plates and the others is that the average absorbance reading was significantly lower (ABSBS = 0.77 ± 0.32, ABSES = 0.63 ± 0.4, ABSDiff. = 0.62 ± 0.44, and ABSRA = 0.05 ± 0.11). This phenomenon may occur if the retinoic acid treatment strongly induces differentiation and stops ES cells from proliferating. Thus, the Cre-mediated drug resistance switching signal is not amplified.
Cell lineage tracing.
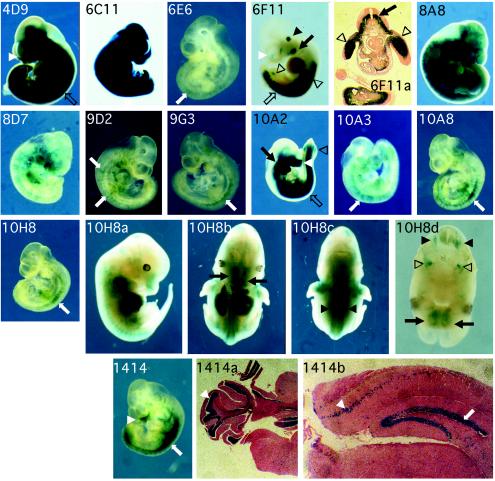
To demonstrate that the in vitro phenotyping method can enrich for gene traps that are developmentally regulated, one active gene trap line, 16 retinoic acid- and in vitro differentiation-inducible gene trap clones, and a pool of enriched silent gene trap clones were used to generate chimeras for in vivo studies. Fourteen of 17 clones were established in the germ line. The germ line chimeras or F1 males carrying the gene trap were crossed to R26R females (42). The R26R mouse carries a cre-inducible β-gal transgene, which allows cells to permanently turn on β-gal in response to Cre expression during development. Thus, the Cre-expressing cells and all subsequent derived lineages will express β-galactosidase. We examined embryos at embryonic day 10.5 by X-Gal staining (Fig. 5). Eleven of 13 (85%) retinoic acid- and in vitro differentiation-inducible lines examined showed X-Gal staining in restricted cell lineages, and ubiquitous X-Gal staining was observed in the active gene trap line 6C11.
FIG. 5.
Whole-mount X-Gal histochemical stains of 10.5 day post coitum compound heterozygous embryos derived from retinoic acid- and in vitro differentiation-inducible gene trap alleles from crosses with R26R females. Ubiquitous X-Gal staining was observed in the active gene trap line 6C11. Inducible gene trap lines 6E6, 9D2, 9G3, 10A3, 10A8, and 10H8 and a silent gene trap line, 1414, showed X-Gal staining in spatially defined regions along the anterior-posterior axis associated with the spinal cord (white arrows). 8A8 and 8D7 showed a scattered staining pattern. 4D9, 6F11, 10A2, and 1414 showed X-Gal staining in mesenchymal tissues of the facial nasal region (white arrowheads), branchial arches (black arrow), limb buds (open arrowheads), and posterior trunk (open arrows). In 6F11, staining was also observed in the otic vesicles (black arrowhead). A cross section of a 10.5 day post coitum 6F11-R26R compound heterozygous embryo is shown in picture 6F11a. X-Gal staining was distributed in the motor neuron region of the anterior spinal cord (black arrow). In the posterior spinal cord, X-Gal staining is found across the ventral half of the spinal cord (data not shown). Lateral, ventral, and dorsal views of a 12.5 day post coitum 10H8-R26R compound heterozygous embryo and a cross section from the head region of the same embryo. Inducible gene trap line 10H8 showed X-Gal staining associated with the peripheral nervous system, including the trigeminal ganglia (open arrowheads), major olfactory epithelium (black arrows), and dorsal root ganglia (black arrowheads) in 12.5 day post coitum embryos. Coronal sections of X-Gal-stained 1414-R26R compound heterozygous adult cerebellum and cerebrum. X-Gal staining of the 1414 adult brain was restricted to the granular layer of the cerebral cortex (white arrowhead), dentate gyrus of the hippocampus (white arrow), and the pleats in the cortex of the cerebellum (white arrowhead).
Several in vitro differentiation- and retinoic acid-inducible lines, including 6E6, 9D2, 9G3, 10A3, 10A8, and 10H8, showed X-Gal staining in spatially defined regions along the anterior-posterior axis associated with the spinal cord. Two other lines, 8A8 and 8D7, showed a scattered staining pattern which might have resulted from transient Cre expression in the neural crest. The remaining three lines, 4D9, 6F11, and 10A2, showed X-Gal staining in mesenchymal tissues of the facial nasal region, branchial arches, limb buds, and posterior trunk. These X-Gal staining patterns reflect β-gal expression that is initiated after a Cre-catalyzed recombination event, and therefore the staining pattern reflects both the previous and current expression patterns of the trapped gene. The X-Gal staining patterns revealed in this study will be different from the real-time expression patterns obtained when β-gal or β-geo is used as a gene trap reporter.
One interesting X-Gal staining pattern was observed in line 6F11. Staining was observed in the posterior spinal cord, mesenchyme of the posterior trunk, brachial arches, limb buds, otic vesicles, and facial nasal mesenchyme (Fig. 5, 6F11). Histological analysis revealed that the cell lineages derived from the cells in which the trapped gene was expressed were distributed in the motor neuron region of the spinal cord. Transverse sections of the posterior spinal cord revealed X-Gal staining across the ventral half of the spinal cord (Fig. 5, 6F11a). Another inducible gene trap line, 10H8, showed X-Gal staining associated with the peripheral nervous system, including the retina, trigeminal ganglia, major olfactory epithelium, and dorsal root ganglia in 12.5 day post coitum embryos (Fig. 5, 10H8, a, b, c, and d).
To enrich for silent gene traps, a pool of traps were generated and selected in blasticidin S and FIAU. Line 1414 (Fig. 5, 1414 and 1414a and -b) was generated from one of the trapped alleles in the silent gene trap-enriched pool. Examination of the 1414 adult brain revealed that X-Gal staining was restricted to the granular layer of the cerebral cortex, dentate gyrus of the hippocampus, and the pleats in the cortex of the cerebellum. All these are neuron-dense regions, suggesting that in this line, the trapped gene is expressed in neuronal lineages.
Identification of trapped genes.
The cloning results are summarized in Table 2. Sequence tags from seven retinoic acid- and in vitro differentiation-inducible gene traps were obtained. With the help of the mouse genome sequence (15), all seven gene traps were mapped to unique chromosomal regions. They were confirmed by Southern analysis with cloned genomic fragments as probes. The mapping results from two of the gene traps were also confirmed by linkage analyses. The trapping vector insertion site in transgenic line 9C10 was mapped to the distal end of mouse chromosome X, and, as expected, genotyping data confirmed X linkage. Another locus, 9G3, was mapped to chromosome 2, 2 centimorgans from the Agouti locus. In this case, the vector insertion was tightly linked to the 129 agouti allele and was not separated in backcrosses to C57BL/6 nonagouti mice.
TABLE 2.
Inducible gene trap cloning results
| Clone no. | Gene trap sequence taga | Mouse genome assembly NCBI m30b | Gene trap orientationb | Transcript hit description |
|---|---|---|---|---|
| 6E6 | AY685649 | 3.135953836-7 (I) | + | ENSMUSG00000037994; Na+ H+ exchangerb |
| AY685650 | 3.135952700-835 (R) | |||
| AY685651 | ||||
| 6F11 | AY68552 | 7.86044621-2 (I) | + | ENSMUSG00000035623; HBV pX-associated |
| AY68553 | 7.86021589-739 (R) | protein (Hbxap)b | ||
| AY68554 | ||||
| 8A8 | AY68555 | 4.114651507-8 (I) | + | Novel transcript |
| 4.114651426-507 (R) | ||||
| 8D7 | AY68556 | 3.35576857-8 (I) | − | EST clone AA867886a |
| 3.35577029-76858 (R) | ||||
| 9C10 | AY68557 | X.10757488-9 (I) | − | Not available |
| AY68558 | ||||
| 9G3 | AY68559 | 2.155175924-5 (I) | + | ENSMUSG00000042548; additional sex |
| AY68560 | 2.155174799-856 (R) | combs-like protein-1 (Asx1)b | ||
| 10C6 | AY68561 | 5.116663090-1 (I) | + | Novel transcript |
| 5.116662993-3090 (R) |
http://www.ncbi.nlm.nih.gov/.
http://mouse30.ensembl.org/Mus_musculus/. I, insertion site (determined by inverse PCR); R, 5′ RACE or RT-PCR product hit.
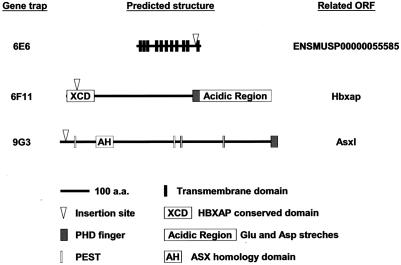
The cloning of the integration site of 9G3 revealed that the gene trap virus had inserted in the first intron of an Ensembl annotated gene (ENSMUSG00000042548; Asxl) which encodes a 1,513-amino-acid protein, a homolog of putative human polycomb group protein ASXL1 (additional sex combs-like 1) (9). The gene trap event is an in-frame fusion between the cre reporter sequence and the coding region, resulting in a chimeric reporter protein with the first 19 amino acids derived from the trapped gene (Fig. 6).
FIG. 6.
Predicted structures and gene trap positions in three Ensembl-annotated proteins. Schematics show the predicted protein structure based on Ensembl prediction and published literature (9, 24, 49).
The molecular cloning result of the inducible gene trap clone 6E6 revealed that the ROSAcre provirus had landed within intron 7 of a novel gene annotated by Ensembl (ENSMUSG00000037994 [37994]). The gene prediction is supportedby spliced expressed sequence tag evidence from different species, and its gene structure is conserved among fly, mouse, rat, macaca, and human. A predicted transcript encodes a peptide of 447 amino acids, which contains 11 transmembrane domains and belongs to an Na+/H+ exchanger subfamily (InterPRO database; accession number IPR006153) (8, 34, 35). The proviral insertion point predicts that Cre is fused in-frame with the predicted protein 392 amino acids from the N terminus of the protein, which is predicted to result in truncation of the protein after the tenth transmembrane domain (Fig. 6).
In the inducible gene trap line 6F11, the trapping vector was inserted in the first intron of an Ensembl annotated gene (ENSMUSG00000035623) which encodes the mouse homolog of the human plant homology domain (PHD) finger-containing gene HBXAP (ENSMUSP00000042399; hepatitis B virus protein X-associated protein) (39). Recently it was shown that HBXAP is p325, the largest subunit of human initiation switch 1 (ISWI)-containing factor RSF (remodeling and spacing factor), which mediates nucleosome deposition and generates regularly spaced nucleosome arrays (24). The gene trap insertion is predicted to truncate Hbxap after 53 amino acids (Fig. 6), so most likely this is a null allele. The X-Gal staining pattern of this insertion argues for a role during development.
In the inducible gene trap line 8A8, the vector was integrated 235 bp upstream of the transcription start of a known gene, Akr1a4. This insertion has trapped a novel transcript transcribed in the opposite orientation. Mouse Akr1a4 is the homologue of human AKR1A1, which encodes an aldo-ketoreductase which has been implicated in neural transmitter metabolism (46). The human AKR1A1 promoter region has been characterized as bidirectional in a hepatocyte cell line (3). Whether our gene trap vector insertion affects endogenous Akr1a4 expression is not clear at this moment.
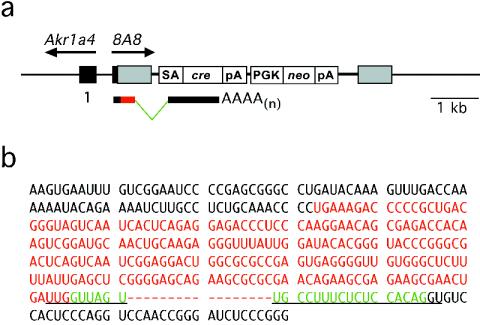
Sequence tags from 5′ RACE and RT-PCR experiments provided transcript information for six of seven retinoic acid- and in vitro differentiation-inducible gene traps cloned. One of the sequence tags, 8D7, can be identified in the mouse expressed sequence tag database (GenBank accession number AA867886) and appears to be spliced appropriately. The transcripts obtained from 8A8 and 10C6 revealed a cryptic splice donor site in the U5 region of the provirus. These sequences reveal that the transcripts have read through the genomic DNA-provirus junction. The splicing events occurred between the cryptic splice donor in the U5 region and the synthetic splice acceptor cloned 5′ of the cre reporter (Fig. 7). The cryptic splice donor site (UUG|GUUAGU) is very similar to the consensus splice donor sequence ([C/A]AG|GU[A/G]AGU). Six of nine nucleotides of the cryptic splice donor site are identical to the splice donor consensus sequence (1). Similar cryptic splicing events have been reported previously (29).
FIG. 7.
Cryptic splice donor sequence identified in the long terminal repeat of the ROSAcre gene trap virus. (a) ROSAcre has integrated 235 bp upstream of the transcription start of a known gene, Akr1a4. This insertion has trapped a novel transcript transcribed in the opposite orientation. (b) The transcript sequence obtained from 8A8 reveals a cryptic splice donor site in the U5 region of the provirus. The transcript is a readthrough of the genomic DNA-provirus junction. Splicing occurs between the cryptic splice donor (underlined) in the U5 region and the synthetic splice acceptor (underlined) cloned 5′ of the cre reporter. Green letters indicate the sequence spliced out of the transcript.
DISCUSSION
The inducible gene trapping system described here was designed to replace laborious screening steps with simple drug selection. Our original plan was to eliminate colony picking and directly apply drug selection to a pool of gene traps so that large-scale high-throughput screening could be achieved with minimum effort. However due to the complexity of the mouse genome, the dynamics of Cre activity, and ES cell proliferation, it was not possible to achieve this goal.
Three important variables prevent screening without colony isolation. First, the expression level of the trapped genes varies. In most of the gene trap clones, the level of Cre available to catalyze recombination is less than that produced by transient expression of CMVcre in our control experiments. This is not unexpected, given that transient transfection introduces multiple copies of the cassette, resulting in high levels of cre expression. Most of the active gene trap clones produce their first drug resistance switching event 4 days after retrovirus infection. Second, the gene trap clone is expanding throughout the period of drug selection. This will result in drug resistance heterogeneity within a gene trap clone if Cre-catalyzed drug resistance switching does not occur before clonal expansion. For example, if an active gene trap clone produced its first Cre-catalyzed recombination event 4 days after retrovirus infection, only 1 of 16 cells in the clone will be drug resistant. This heterogeneity is the main reason why we were not able to achieve clean clonal selection by drug selection on a mixture of gene trap clones. Third, Cre-catalyzed drug resistance switching is an irreversible reaction; each responder ES cell only allows detection of reporter expression once. Therefore, a gene trap that is active in ES cells can no longer be examined for its inducibility, which becomes a limitation of this system. However, irreversible switching does allow us to calculate the accumulated Cre-catalyzed drug resistance events at different times after induction.
Because of the difficulty in achieving clean selection without colony isolation, individual gene trap events were cloned so that Cre levels could be determined by examining the accumulated recombination events within populations. A high-throughput, standardized screening protocol was established. We took advantage of drug selection and used the number of cells which had undergone drug resistance switching as an indicator of the level of reporter expression, which could be evaluated with a 96-well plate reader. This quantitative experimental data are easy to process and analyze, making this trapping system more amenable to large-scale screening than other 5′ gene trapping protocols.
In the results reported here, close to 85% of the differentiation- and retinoic acid-inducible gene traps identified by the inducible gene trapping system were developmentally regulated in vivo, whereas only 15 to 30% of the gene trap clones described in other random gene trap experiments were observed to show restricted expression during development (11, 51). The inducible gene trapping system can be applied to trap genes involved in the differentiation of a specific cell lineage, if a differentiation protocol is available, as well as genes induced by a signaling molecule or a transcription factor. Since not all signaling molecules or transcription factors have their receptors or cofactors expressed in ES cells, in some cases it may not be possible to use this system directly without first modifying the ES cells to express the missing components. Alternatively, the inducible gene trapping experiments can be performed in different types of cells isolated from a mouse carrying the responder construct.
cre has been used previously as a reporter in conjunction with a loxP-containing construct as a responder (38, 45, 48, 50). One important feature of the two-component Cre/loxP system is that it allows the detection of transient gene expression by permanently switching the responder drug resistance. Another feature of a two-component gene trapping system is the uncoupling of the trapped gene's expression level from the drug resistance provided by the responder locus. This uncoupling strongly enhances the potential of this system to identify genes expressed transiently and at low levels, which are hard to detect by other means.
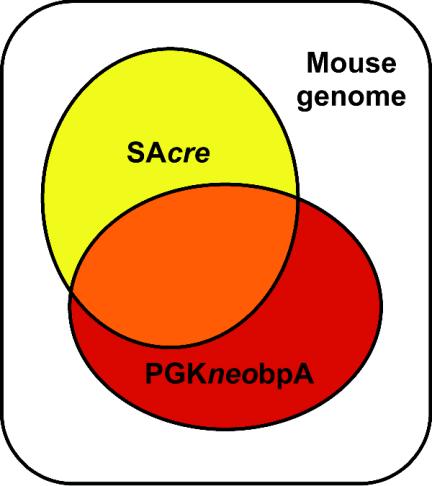
One important observation in these experiments is the sensitivity of Cre compared with conventional selection cassettes. In Fig. 8, we illustrate the detection limits of the 5′ gene trap reporter SAcre and the drug-selectable maker used to detect retrovirus integration events, PGKneobpA. The SAβ-gal reporter has been reported to work as an active trap in 11.6% of retrovirus integration events selected for integration with a PGKneobpA cassette (10). In our study, 42.6% of integration events selected by PGKneobpA were classified as active gene trap events, presumably because the Cre/loxP system allows detection of traps in genes that are expressed at low levels or only transiently (45). We also demonstrated that cre can report integration events that are not selected by the PGKneobpA cassette. In our experiments, just 40% of the cre-detectable active gene trap events were within the PGKneobpA detection limit. Therefore, the use of cre allows us to explore chromosomal regions that are not detectable with other reporters.
FIG. 8.
Schematic representation of the detection limits of the 5′ gene trap reporter SAcre and the PGKneobpA cassette.
The inducible gene trapping system described here has the potential to help to construct a comprehensive and informative gene trap library. First, the use of cre as the 5′ gene trapping reporter extends the trappable chromosomal regions to those not detectable with the PGKneobpA cassette as the selectable marker. This sensitivity reflects the nature of the Cre/loxP system, which allows detection of genes that are only transiently expressed or expressed at low levels. Such genes are usually underrepresented in conventional cDNA libraries. This observation is consistent with the fact that less than two-thirds of the transcripts trapped with the inducible gene trapping system have been identified previously. Therefore, the use of the Cre/loxP system allows us to build a comprehensive gene trap repository which includes as yet unannotated genes. Second, in vitro phenotyping provides the expression profiles for trapped genes. In vitro phenotyping information should be helpful in selecting genes of interest. Third, the draft mouse genome sequence (49) enables gene trap sequence tags to be rapidly assigned to their corresponding genes.
The cluster analysis of our experimental results indicates that there is a distinct set of genes which are not expressed in ES cells but are activated during in vitro differentiation. This provides a starting point to examine the embryonic stem cell differentiation process. Finally, the gene trap lines generated here carry a cre gene under the control of endogenous regulatory elements, and the mice generated from these clones are conditional cre lines, each of which is a valuable tool for conditional genetics experiments (32).
Acknowledgments
We thank Hugo J. Bellen, Xiao-Zhong Wang, and Meredith Wentland for critical reading of the manuscript. We also thank Sukeshi Vaishnav, Sandra Rivera, Gabriele C. Schuster, and Janis Wesley for technical assistance in histological analysis, feeder cell preparation, chimera generation, and mouse colony maintenance and Sarah Quakkelaar-Howard for help preparing the manuscript.
This work was supported by grants to A.B. from the NIH National Cancer Institute.
REFERENCES
- 1.Alberts, B., D. Bray, J. Lewis, M. Raff, K. Roberts, and J. D. Watson. 1994. Molecular biology of the cell, 3rd ed. Garland Publishing, Inc., New York, N.Y.
- 2.Baker, R. K., M. A. Haendel, B. J. Swanson, J. C. Shambaugh, B. K. Micales, and G. E. Lyons. 1997. In vitro preselection of gene-trapped embryonic stem cell clones for characterizing novel developmentally regulated genes in the mouse. Dev. Biol. 185:201-214. [DOI] [PubMed] [Google Scholar]
- 3.Barski, O. A., K. H. Gabbay, and K. M. Bohren. 1999. Characterization of the human aldehyde reductase gene and promoter. Genomics 60:188-198. [DOI] [PubMed] [Google Scholar]
- 4.Bonaldo, P., K. Chowdhury, A. Stoykova, M. Torres, and P. Gruss. 1998. Efficient gene trap screening for novel developmental genes using IRES beta geo vector and in vitro preselection. Exp. Cell Res. 244:125-136. [DOI] [PubMed] [Google Scholar]
- 5.Bothwell, A. L., M. Paskind, M. Reth, T. Imanishi-Kari, K. Rajewsky, and D. Baltimore. 1981. Heavy chain variable region contribution to the NPb family of antibodies: somatic mutation evident in a gamma 2a variable region. Cell 24:625-637. [DOI] [PubMed] [Google Scholar]
- 6.Bradley, A., M. Evans, M. H. Kaufman, and E. Robertson. 1984. Formation of germ-line chimaeras from embryo-derived teratocarcinoma cell lines. Nature 309:255-256. [DOI] [PubMed] [Google Scholar]
- 7.Chen, Y. T., and A. Bradley. 2000. A new positive/negative selectable marker, puDeltatk, for use in embryonic stem cells. Genesis 28:31-35. [DOI] [PubMed] [Google Scholar]
- 8.Dibrov, P., and L. Fliegel. 1998. Comparative molecular analysis of Na+/H+ exchangers: a unified model for Na+/H+ antiport? FEBS Lett. 424:1-5. [DOI] [PubMed] [Google Scholar]
- 9.Fisher, C. L., J. Berger, F. Randazzo, and H. W. Brock. 2003. A human homolog of Additional sex combs, Additional Sex Combs-like 1, maps to chromosome 20q11. Gene 306:115-126. [DOI] [PubMed] [Google Scholar]
- 10.Forrester, L. M., A. Nagy, M. Sam, A. Watt, L. Stevenson, A. Bernstein, A. L. Joyner, and W. Wurst. 1996. An induction gene trap screen in embryonic stem cells: Identification of genes that respond to retinoic acid in vitro. Proc. Natl. Acad. Sci. USA 93:1677-1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Friedrich, G., and P. Soriano. 1991. Promoter traps in embryonic stem cells: a genetic screen to identify and mutate developmental genes in mice. Genes Dev. 5:1513-1523. [DOI] [PubMed] [Google Scholar]
- 12.Gajovic, S., K. Chowdhury, and P. Gruss. 1998. Genes expressed after retinoic acid-mediated differentiation of embryoid bodies are likely to be expressed during embryo development. Exp. Cell Res. 242:138-143. [DOI] [PubMed] [Google Scholar]
- 13.Gajovic, S., L. St-Onge, Y. Yokota, and P. Gruss. 1997. Retinoic acid mediates Pax6 expression during in vitro differentiation of embryonic stem cells. Differentiation 62:187-192. [DOI] [PubMed] [Google Scholar]
- 14.Gesemann, M., E. D. Litwack, K. T. Yee, U. Christen, and D. D. O'Leary. 2001. Identification of candidate genes for controlling development of the basilar pons by differential display PCR. Mol. Cell Neurosci. 18:1-12. [DOI] [PubMed] [Google Scholar]
- 15.Gregory, S. G., M. Sekhon, J. Schein, S. Zhao, K. Osoegawa, C. E. Scott, R. S. Evans, P. W. Burridge, T. V. Cox, C. A. Fox, R. D. Hutton, I. R. Mullenger, K. J. Phillips, J. Smith, J. Stalker, G. J. Threadgold, E. Birney, K. Wylie, A. Chinwalla, J. Wallis, L. Hillier, J. Carter, T. Gaige, S. Jaeger, C. Kremitzki, D. Layman, J. Maas, R. McGrane, K. Mead, R. Walker, S. Jones, M. Smith, J. Asano, I. Bosdet, S. Chan, S. Chittaranjan, R. Chiu, C. Fjell, D. Fuhrmann, N. Girn, C. Gray, R. Guin, L. Hsiao, M. Krzywinski, R. Kutsche, S. S. Lee, C. Mathewson, C. McLeavy, S. Messervier, S. Ness, P. Pandoh, A. L. Prabhu, P. Saeedi, D. Smailus, L. Spence, J. Stott, S. Taylor, W. Terpstra, M. Tsai, J. Vardy, N. Wye, G. Yang, S. Shatsman, B. Ayodeji, K. Geer, G. Tsegaye, A. Shvartsbeyn, E. Gebregeorgis, M. Krol, D. Russell, L. Overton, J. A. Malek, M. Holmes, M. Heaney, J. Shetty, T. Feldblyum, W. C. Nierman, J. J. Catanese, T. Hubbard, R. H. Waterston, J. Rogers, P. J. de Jong, C. M. Fraser, M. Marra, J. D. McPherson, and D. R. Bentley. 2002. A physical map of the mouse genome. Nature 418:743-750. [DOI] [PubMed] [Google Scholar]
- 16.Guo, G., W. Wang, and A. Bradley. 2004. Mismatch repair genes identified using genetic screens in Blm-deficient embryonic stem cells. Nature 429:891-895. [DOI] [PubMed] [Google Scholar]
- 17.Hasty, P., J. Rivera-Perez, and A. Bradley. 1991. The length of homology required for gene targeting in embryonic stem cells. Mol. Cell. Biol. 11:5586-5591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hrabe de Angelis, M. H., H. Flaswinkel, H. Fuchs, B. Rathkolb, D. Soewarto, S. Marschall, S. Heffner, W. Pargent, K. Wuensch, M. Jung, A. Reis, T. Richter, F. Alessandrini, T. Jakob, E. Fuchs, H. Kolb, E. Kremmer, K. Schaeble, B. Rollinski, A. Roscher, C. Peters, T. Meitinger, T. Strom, T. Steckler, F. Holsboer, T. Klopstock, F. Gekeler, C. Schindewolf, T. Jung, K. Avraham, H. Behrendt, J. Ring, A. Zimmer, K. Schughart, K. Pfeffer, E. Wolf, and R. Balling. 2000. Genome-wide, large-scale production of mutant mice by ENU mutagenesis. Nat. Genet. 25:444-447. [DOI] [PubMed] [Google Scholar]
- 19.Huang, M. T., and C. M. Gorman. 1990. Intervening sequences increase efficiency of RNA 3′ processing and accumulation of cytoplasmic RNA. Nucleic Acids Res. 18:937-947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Justice, M. J., J. K. Noveroske, J. S. Weber, B. Zheng, and A. Bradley. 1999. Mouse ENU mutagenesis. Hum. Mol. Genet. 8:1955-1963. [DOI] [PubMed] [Google Scholar]
- 21.Kimura, M., T. Kamakura, Q. Z. Tao, I. Kaneko, and I. Yamaguchi. 1994. Cloning of the blasticidin S deaminase gene (BSD) from Aspergillus terreus and its use as a selectable marker for Schizosaccharomyces pombe and Pyricularia oryzae. Mol. Gen. Genet. 242:121-129. [DOI] [PubMed] [Google Scholar]
- 22.Kimura, M., A. Takatsuki, and I. Yamaguchi. 1994. Blasticidin S deaminase gene from Aspergillus terreus (BSD): a new drug resistance gene for transfection of mammalian cells. Biochim. Biophys. Acta 1219:653-659. [DOI] [PubMed] [Google Scholar]
- 23.Kuehn, M. R., A. Bradley, E. J. Robertson, and M. J. Evans. 1987. A potential animal model for Lesch-Nyhan syndrome through introduction of HPRT mutations into mice. Nature 326:295-298. [DOI] [PubMed] [Google Scholar]
- 24.LeRoy, G., A. Loyola, W. S. Lane, and D. Reinberg. 2000. Purification and characterization of a human factor that assembles and remodels chromatin. J. Biol. Chem. 275:14787-14790. [DOI] [PubMed] [Google Scholar]
- 25.Liu, P., N. A. Jenkins, and N. G. Copeland. 2002. Efficient Cre-loxP-induced mitotic recombination in mouse embryonic stem cells. Nat. Genet. 30:66-72. [DOI] [PubMed] [Google Scholar]
- 26.Luo, G., I. M. Santoro, L. D. McDaniel, I. Nishijima, M. Mills, H. Youssoufian, H. Vogel, R. A. Schultz, and A. Bradley. 2000. Cancer predisposition caused by elevated mitotic recombination in Bloom mice. Nat. Genet. 26:424-429. [DOI] [PubMed] [Google Scholar]
- 27.Markowitz, D., S. Goff, and A. Bank. 1988. A safe packaging line for gene transfer: separating viral genes on two different plasmids. J. Virol. 62:1120-1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McMahon, A. P., and A. Bradley. 1990. The Wnt-1 (int-1) proto-oncogene is required for development of a large region of the mouse brain. Cell 62:1073-1085. [DOI] [PubMed] [Google Scholar]
- 29.Medico, E., G. Gambarotta, A. Gentile, P. M. Comoglio, and P. Soriano. 2001. A gene trap vector system for identifying transcriptionally responsive genes. Nat. Biotechnol. 19:579-582. [DOI] [PubMed] [Google Scholar]
- 30.Muth, K., R. Bruyns, I. S. Thorey, and H. von Melchner. 1998. Disruption of genes regulated during hematopoietic differentiation of mouse embryonic stem cells. Dev. Dyn. 212:277-283. [DOI] [PubMed] [Google Scholar]
- 31.Nadeau, J. H., R. Balling, G. Barsh, D. Beier, S. D. Brown, M. Bucan, S. Camper, G. Carlson, N. Copeland, J. Eppig, C. Fletcher, W. N. Frankel, D. Ganten, D. Goldowitz, C. Goodnow, J. L. Guenet, G. Hicks, M. H. de Angelis, I. Jackson, H. J. Jacob, N. Jenkins, D. Johnson, M. Justice, S. Kay, D. Kingsley, H. Lehrach, T. Magnuson, M. Meisler, A. Poustka, E. M. Rinchik, J. Rossant, L. B. Russell, J. Schimenti, T. Shiroishi, W. C. Skarnes, P. Soriano, W. Stanford, J. S. Takahashi, W. Wurst, and A. Zimmer. 2001. Sequence interpretation. Functional annotation of mouse genome sequences. Science 291:1251-1255. [DOI] [PubMed] [Google Scholar]
- 32.Nagy, A. 2000. Cre recombinase: the universal reagent for genome tailoring. Genesis 26:99-109. [PubMed] [Google Scholar]
- 33.Nolan, P. M., J. Peters, M. Strivens, D. Rogers, J. Hagan, N. Spurr, I. C. Gray, L. Vizor, D. Brooker, E. Whitehill, R. Washbourne, T. Hough, S. Greenaway, M. Hewitt, X. Liu, S. McCormack, K. Pickford, R. Selley, C. Wells, Z. Tymowska-Lalanne, P. Roby, P. Glenister, C. Thornton, C. Thaung, J. A. Stevenson, R. Arkell, P. Mburu, R. Hardisty, A. Kiernan, A. Erven, K. P. Steel, S. Voegeling, J. L. Guenet, C. Nickols, R. Sadri, M. Nasse, A. Isaacs, K. Davies, M. Browne, E. M. Fisher, J. Martin, S. Rastan, S. D. Brown, and J. Hunter. 2000. A systematic, genome-wide, phenotype-driven mutagenesis programme for gene function studies in the mouse. Nat. Genet. 25:440-443. [DOI] [PubMed] [Google Scholar]
- 34.Numata, M., K. Petrecca, N. Lake, and J. Orlowski. 1998. Identification of a mitochondrial Na+/H+ exchanger. J. Biol. Chem. 273:6951-6959. [DOI] [PubMed] [Google Scholar]
- 35.Orlowski, J., and S. Grinstein. 1997. Na+/H+ exchangers of mammalian cells. J. Biol. Chem. 272:22373-22376. [DOI] [PubMed] [Google Scholar]
- 36.Ramirez-Solis, R., A. C. Davis, and A. Bradley. 1993. Gene targeting in embryonic stem cells. Methods Enzymol. 225:855-878. [DOI] [PubMed] [Google Scholar]
- 37.Rohwedel, J., K. Guan, and A. M. Wobus. 1999. Induction of cellular differentiation by retinoic acid in vitro. Cells Tissues Organs 165:190-202. [DOI] [PubMed] [Google Scholar]
- 38.Russ, A. P., C. Friedel, K. Ballas, U. Kalina, D. Zahn, K. Strebhardt, and H. von Melchner. 1996. Identification of genes induced by factor deprivation in hematopoietic cells undergoing apoptosis using gene-trap mutagenesis and site-specific recombination. Proc. Natl. Acad. Sci. USA 93:15279-15284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shamay, M., O. Barak, and Y. Shaul. 2002. HBXAP, a novel PHD-finger protein, possesses transcription repression activity. Genomics 79:523-529. [DOI] [PubMed] [Google Scholar]
- 40.Shimono, A., and R. R. Behringer. 2000. Differential screens with subtracted PCR-generated cDNA libraries from subregions of single mouse embryos. Methods Mol. Biol. 136:333-344. [DOI] [PubMed] [Google Scholar]
- 41.Skarnes, W. C., J. E. Moss, S. M. Hurtley, and R. S. Beddington. 1995. Capturing genes encoding membrane and secreted proteins important for mouse development. Proc. Natl. Acad. Sci. USA 92:6592-6596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Soriano, P. 1999. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat. Genet. 21:70-71. [DOI] [PubMed] [Google Scholar]
- 43.Soriano, P., G. Friedrich, and P. Lawinger. 1991. Promoter interactions in retrovirus vectors introduced into fibroblasts and embryonic stem cells. J. Virol. 65:2314-2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stoykova, A., K. Chowdhury, P. Bonaldo, M. Torres, and P. Gruss. 1998. Gene trap expression and mutational analysis for genes involved in the development of the mammalian nervous system. Dev. Dyn. 212:198-213. [DOI] [PubMed] [Google Scholar]
- 45.Thorey, I. S., K. Muth, A. P. Russ, J. Otte, A. Reffelmann, and H. von Melchner. 1998. Selective disruption of genes transiently induced in differentiating mouse embryonic stem cells by using gene trap mutagenesis and site-specific recombination. Mol. Cell. Biol. 18:3081-3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Turner, A. J., J. A. Illingworth, and K. F. Tipton. 1974. Simulation of biogenic amine metabolism in the brain. Biochem. J. 144:353-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Valerius, M. T., L. T. Patterson, D. P. Witte, and S. S. Potter. 2002. Microarray analysis of novel cell lines representing two stages of metanephric mesenchyme differentiation. Mech. Dev. 112:219-232. [DOI] [PubMed] [Google Scholar]
- 48.Wan, Y., and S. K. Nordeen. 2002. Identification of genes differentially regulated by glucocorticoids and progestins using a Cre/loxP-mediated retroviral promoter-trapping strategy. J. Mol. Endocrinol. 28:177-192. [DOI] [PubMed] [Google Scholar]
- 49.Waterston, R. H., K. Lindblad-Toh, E. Birney, J. Rogers, J. F. Abril, P. Agarwal, R. Agarwala, R. Ainscough, M. Alexandersson, P. An, S. E. Antonarakis, J. Attwood, R. Baertsch, J. Bailey, K. Barlow, S. Beck, E. Berry, B. Birren, T. Bloom, P. Bork, M. Botcherby, N. Bray, M. R. Brent, D. G. Brown, S. D. Brown, C. Bult, J. Burton, J. Butler, R. D. Campbell, P. Carninci, S. Cawley, F. Chiaromonte, A. T. Chinwalla, D. M. Church, M. Clamp, C. Clee, F. S. Collins, L. L. Cook, R. R. Copley, A. Coulson, O. Couronne, J. Cuff, V. Curwen, T. Cutts, M. Daly, R. David, J. Davies, K. D. Delehaunty, J. Deri, E. T. Dermitzakis, C. Dewey, N. J. Dickens, M. Diekhans, S. Dodge, I. Dubchak, D. M. Dunn, S. R. Eddy, L. Elnitski, R. D. Emes, P. Eswara, E. Eyras, A. Felsenfeld, G. A. Fewell, P. Flicek, K. Foley, W. N. Frankel, L. A. Fulton, R. S. Fulton, T. S. Furey, D. Gage, R. A. Gibbs, G. Glusman, S. Gnerre, N. Goldman, L. Goodstadt, D. Grafham, T. A. Graves, E. D. Green, S. Gregory, R. Guigo, M. Guyer, R. C. Hardison, D. Haussler, Y. Hayashizaki, L. W. Hillier, A. Hinrichs, W. Hlavina, T. Holzer, F. Hsu, A. Hua, T. Hubbard, A. Hunt, I. Jackson, D. B. Jaffe, L. S. Johnson, M. Jones, T. A. Jones, A. Joy, M. Kamal, E. K. Karlsson, et al. 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420:520-562. [DOI] [PubMed] [Google Scholar]
- 50.Wempe, F., J. Y. Yang, J. Hammann, and H. von Melchner. 27June2001, posting date. Gene trapping identifies transiently induced survival genes during programmed cell death. Genome Biol. 2:research0023.1-0023.10/http://genomebiology.com/2001/2/7/research/0023. [DOI] [PMC free article] [PubMed]
- 51.Wurst, W., J. Rossant, V. Prideaux, M. Kownacka, A. Joyner, D. P. Hill, F. Guillemot, S. Gasca, D. Cado, A. Auerbach, and et al. 1995. A large-scale gene-trap screen for insertional mutations in developmentally regulated genes in mice. Genetics 139:889-899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang, H., P. Hasty, and A. Bradley. 1994. Targeting frequency for deletion vectors in embryonic stem cells. Mol. Cell. Biol. 14:2404-2410. [DOI] [PMC free article] [PubMed] [Google Scholar]