Abstract
PS-341, also known as Velcade or Bortezomib, represents a new class of anticancer drugs which has been shown to potently inhibit the growth and/or progression of human cancers, including head and neck squamous cell carcinoma (HNSCC). Although it has been logically hypothesized that NF-κB is a major target of PS-341, the underlying mechanism by which PS-341 inhibits tumor cell growth is unclear. Here we found that PS-341 potently activated the caspase cascade and induced apoptosis in human HNSCC cell lines. Although PS-341 could inhibit NF-κB activation, the inhibition of NF-κB was not sufficient to initiate apoptosis in HNSCC cells. Using biochemical and microarray approaches, we found that proteasome inhibition by PS-341 induced endoplasmic reticulum (ER) stress and reactive oxygen species (ROS) in HNSCC cells. The inhibition of ROS significantly suppressed caspase activation and apoptosis induced by PS-341. Consistently, PS-341 could not induce the ER stress-ROS in PS-341-resistant HNSCC cells. Taken together, our results suggest that in addition to the abolishment of the prosurvival NF-κB, PS-341 might directly induce apoptosis by activating proapoptotic ER stress-ROS signaling cascades in HNSCC cells, providing novel insights into the PS-341-mediated antitumor activity.
PS-341, also known as Velcade or Bortezomib, represents a new class of anticancer drugs which mainly inhibit 26S proteasome activity (1, 2). The 26S proteasome is a large multisubunit protein complex found in the cytoplasms and nuclei of all eukaryotic cells; its principle function is to degrade proteins via the ubiquitin pathway (26). It is composed of a 20S core cylinder, with tryptic and chymotryptic activity, and is capped at each end by a 19S regulatory particle. In most cases, proteins destined for proteasomal degradation are flagged on lysine residues with polymeric chains of a small protein known as ubiquitin (26). The ubiquitin chains are recognized by the 19S cap. The 19S cap also contains enzymatic activities that remove ubiquitin from the targeted proteins and chaperone-like activities that unfold the deubiquitinated substrates and feed them into the internal chamber of the 20S core particle. The peptidase activities (chymotryptic, tryptic, and postglutamyl hydrolyzing) then generate small peptides (1, 26). PS-341 is a small dipeptidyl boronic acid derivative that binds reversibly to the catalytic threonine and selectively blocks the chymotryptic activity of the proteasome (1, 5). The ability of PS-341 to specifically inhibit the 26S proteasome distinguishes it from other proteasome inhibitors, such as the peptide aldehydes, like MG-132, which also inhibit thiol proteases like cathepsin B and calpains. Preclinical and clinical trials have found that PS-341 is a promising chemotherapeutic adjuvant which has antitumor effects on several human cancers (7, 12, 13, 24, 33).
Most cytosolic and short-lived proteins, including many transcription factors, meet their demise at the 26S proteasome (1, 26). Therefore, it is not surprising that a myriad of cell responses and pathways are perturbed as a result of proteasome inhibition. In vitro and in vivo studies have demonstrated that the proteasome inhibitor inhibits tumor growth by inducing apoptosis in several human cancers, including myeloma, prostate cancer, and head and neck squamous cell carcinoma (HNSCC) (12, 13, 22, 37, 38, 43). PS-341-induced apoptosis was preceded by the mitochondrial release of cytochrome c, SMAC, and apoptosis-inducing factor, and cleavage of caspases has been demonstrated in cells undergoing PS-341-induced apoptosis (13, 45).
Activation of the transcription factor nuclear factor kappa B (NF-κB), a key survival factor, has been shown to be dependent on the 26S proteasome (3, 8, 23, 26, 27). Because PS-341 specifically inhibits the 26S proteasome, NF-κB has been logically considered to be a key target for PS-341-induced apoptosis (1, 27). NF-κB is a transcription activator that controls expression of genes associated with inflammation and cell proliferation and survival (3, 8). Classical NF-κB is a heterodimer composed of p50 and p65/RelA, which is retained in the cytoplasm by the IκB group of inhibitory proteins. In response to stressful stimuli, such as cancer chemotherapy, IκB is phosphorylated, ubiquitinated, and subsequently degraded by the 26S proteasome. IκB degradation results in the nuclear translocation of NF-κB and the activation of NF-κB-dependent gene transcription (3, 8). Many studies have found that PS-341 potently sensitized myeloma cells and colorectal cancer cells to DNA-damaging chemotherapeutic agents, such as doxorubicin and melphalan, by inhibiting NF-κB activation. These studies have provided a framework for clinical use of PS-341 in combination with conventional chemotherapy (12, 22, 37, 38, 43). Recently, Sunwoo et al. (37) reported that PS-341 inhibited activation of NF-κB DNA binding and functional reporter activity at physiological concentrations in HNSCC cells. Importantly, they also found that PS-341 induced apoptosis in these cells, suggesting that PS-341 may be used as a monotherapy for HNSCC.
HNSCC is the most common cancer of the head and neck region (6, 31, 36). Approximately 30,000 Americans are diagnosed with cancers affecting the head and neck and the oral cavity each year. While significant advances have been made in surgery and radiotherapy for patients with HNSCC, the 5-year survival rate is the lowest for any major cancer (6, 31, 36). Therefore, there is an urgent need to develop innovative therapies for HNSCC. To further understand the molecular mechanisms by which PS-341 induces apoptosis in HNSCC cells, we systemically dissected the signaling pathways involved in PS-341-mediated apoptosis by using biochemical and genetic approaches. We found that inhibition of the NF-κB antiapoptotic pathway was not sufficient to induce apoptosis in HNSCC cells, suggesting that PS-341 might directly activate proapoptotic signaling pathways. Interestingly, using microarray analysis, we found that a large number of genes associated with endoplasmic reticulum (ER) stress were rapidly induced following PS-341 stimulation in HNSCC cells. Consistently, caspase 12, an essential caspase in ER stress-mediated apoptosis (30), was found to be activated by PS-341 stimulation in mouse embryonic fibroblasts (MEFs). Moreover, we found that reactive oxygen species (ROS), which are frequently induced by ER stress, played a critical role in PS-341-mediated apoptosis. The inhibition of ROS significantly suppressed PS-341-induced apoptosis in HNSCC as well as expression of ER stress-associated proapoptotic genes. In support of the role of ER stress-ROS in PS-341-induced apoptosis, we found that ER stress-ROS were weakly induced in PS-341-resistant cells. These findings provide novel insight into the molecular mechanisms of PS-341-mediated apoptosis in HNSCC cells.
MATERIALS AND METHODS
Reagents and cell cultures.
PS-341 was kindly provided by Millennium Pharmaceuticals, Inc. (Cambridge, Mass.). Tiron was purchased from Sigma-Aldrich (St. Louis, Mo.). H2DCFDA, Indo-1, and ionomycin were purchased from Molecular Probes (Eugene, Ore.). HNSCC cell lines were obtained from Thomas Carey at the University of Michigan and were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum and penicillin-streptomycin from Invitrogen (Grand Island, N.Y.).
Western blot analysis.
Cells (2 × 106) were plated in 10-cm tissue culture dishes the day before PS-341 or vehicle treatment. Cells were treated with PS-341 (0.05 to 0.5 μM) for various times. Whole-cell extracts were prepared in modified radioimmunoprecipitation assay (RIPA) buffer containing phenylmethylsulfonyl fluoride and protease inhibitors (Sigma-Aldrich). Fifty micrograms of lysates were resolved on sodium dodecyl sulfate (SDS)-8 or 12% polyacrylamide gel electrophoresis (PAGE) and transferred to a polyvinylidene difluoride membrane with a Bio-Rad (Hercules, Calif.) semidry transfer apparatus. The membranes were blotted with 5% milk overnight and probed with primary antibodies. Antibodies were acquired from the following sources: phospho-PERK, caspase 9, and caspase 12 from Cell Signaling (Beverly, Mass.); ATF-4, GADD-34, GADD-153/CHOP, GRP-78, and caspase 3 from Santa Cruz (Santa Cruz, Calif.); and caspase 2 from BD PharMingen (San Diego, Calif.). After incubation with horseradish peroxidase-conjugated secondary antibody, membranes were visualized using an enhanced chemiluminescence reagent from Pierce (Rockford, Ill.).
Northern blot analysis.
Total RNAs were extracted from cells with the Trizol reagent (Invitrogen) according to the manufacturer's protocol. Five to ten micrograms of total RNAs were resolved on 1.5% agarose formaldehyde gels and transferred to nitrocellulose membranes overnight. The membranes were hybridized with 32P-labeled cDNA probes and exposed to autoradiographic film as described previously (42). cDNA probes for human GRP-78 and ATF-4 were prepared by using reverse transcription PCR with the specific primers (GRP-78 [forward, 5′-ATGTACGCCCTGCTGGGAGT-′3; reverse, 5′-CCATGTAGTTGCGAAGCTCC-′3] and ATF-4 [forward, 5′-AGATGACCTTCTGACCACG-′3; reverse, 5′-CACCCTTTACTTTTGCTGC-′3]). cDNA products were purified from agarose gels, subcloned into the pT-Easy vector from Promega according to the manufacturer's protocol, and confirmed by DNA sequencing.
Microarray.
UMSCC-23 cells were treated with PS-341 for 0, 1, 4, and 8 h. After treatment, cells were harvested, and total RNAs were extracted with Trizol reagents according to the manufacturer's protocol. To remove contaminated genomic DNA, total RNAs were passed through an RNeasy column from QIAGEN (Valencia, Calif.). Ten micrograms of total RNAs were quantitatively amplified and biotin labeled according to the manufacturer's instructions. Briefly, RNAs were converted to double-stranded cDNA, using SuperScript II reverse transcriptase (Invitrogen) with an oligo(dT) primer that has a T7 RNA polymerase site on the 5′ end. Then, the cDNAs were used in an in vitro transcription reaction in the presence of biotin-modified ribonucleotides to produce large amounts of single-stranded RNA. The biotin-labeled RNAs were fragmented. Hybridization to an Affymetrix (Santa Clara, Calif.) human U133 gene chip was performed at 45°C for 16 h in a mix that included 10 μg of fragmented RNA, 6× SSPE (1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA [pH 7.7]), 0.005% Triton X-100, and 100 μg of herring sperm DNA/ml in a total volume of 200 μl. Labeled bacterial RNAs were spiked into the hybridization mix to generate an internal standard and to allow normalization between chips. Chips were washed and stained with streptavidin R-phycoerythrin (Molecular Probes). The arrays were scanned with the GeneArray scanner (Affymetrix). Signal intensity was calculated by using the one-step Tukey biweight estimate. When computing change indices (n-fold), normalized intensities less than 20 were replaced by 20 before forming ratios in order to avoid spuriously large n-fold change values. We performed multivariate analyses to compare gene expression using analysis of variance. Genes with q values less than 5% were considered to be significantly expressed (where the q value is the multiple-comparisons equivalent of the normal P value). Because there were only two samples per group, our estimate of the error of the mean might be biased, resulting in a higher false-positive rate than expected. To account for this possibility, we imposed an additional n-fold change criterion (at least twofold) for significance.
Flow cytometry.
Cells (2 × 106) were plated in 10-cm tissue culture dishes the day before treatment with PS-341 or vehicle. After treatment, plates were washed once with phosphate-buffered saline (PBS) and cells were harvested with 1× trypsin-EDTA. Cells were subsequently washed again with PBS and then resuspended in 1 ml of PBS. Cells were incubated with 2′,7′-dichlorofluorescein diacetate (H2DCFDA; 0.5 to 1.0 μM) in the dark at 37°C for 30 min. Cells were then washed twice with PBS and analyzed at the University of Michigan Cancer Center Flow Cytometry Core Facility.
DNA laddering.
PS-341-treated cells were lysed and treated with 100 mg of proteinase K/ml for 2 h at 50°C. DNA was extracted twice with phenol-chloroform mixed 1:1 and twice with chloroform alone. Precipitated DNA was washed with 70% ethanol, and 20 to 30 μg of each sample was resolved on a 1.5% agarose gel.
Pulse-chase 35S protein labeling.
UMSCC-23 cells (105) were plated the day before treatment in 6-cm dishes. Cells were treated with PS-341, vehicle or thapsigargin (TG) as a positive control. The cells were rinsed with PBS and incubated with 35S-labeled cysteine and methionine in cysteine- and methionine-free Dulbecco's modified Eagle's medium for 15 min at 37°C. Medium was then removed, and the cells were rinsed with cold PBS. Cells were lysed with 200 μl of RIPA. Ten-microgram aliquots of each sample were resolved on an SDS-10% PAGE gel, dried, and exposed to radiographic film.
RESULTS
PS-341 activates caspase cascades and induces apoptosis in HNSCC cells.
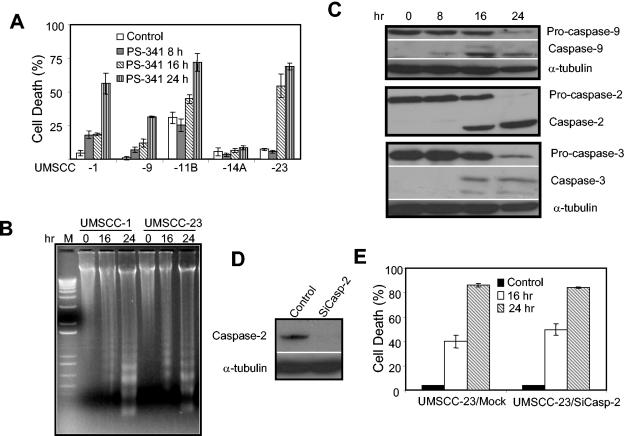
To elucidate the molecular mechanism by which PS-341 induced apoptosis, a panel of HNSCC cell lines, including UMSCC-1, -9, -11B, -14A, and -23, was treated with PS-341 for 0, 8, 16, or 24 h. Trypan blue exclusion assay found that cell death was induced in these cells, with the exception of UMSCC-14A, between 8 and 16 h following PS-341 stimulation (Fig. 1A). A DNA fragmentation assay found that PS-341 induced DNA laddering in HNSCC cells, signifying an apoptotic mechanism (Fig. 1B). Western blot analysis revealed that PS-341 sequentially induced the processing of caspases 9, 2, and 3, suggesting that caspase 9 might be an initiating caspase in HNSCC cells (Fig. 1C). While caspase 9 is considered an initiating caspase for chemotherapeutic drug-mediated apoptosis, recent studies have suggested that caspase 2 may be the apical caspase in chemotherapy-mediated apoptosis (18). To explore the role of caspase 2 in PS-341-mediated apoptosis, we utilized a small interfering RNA (siRNA) strategy to knock down caspase 2 expression as described previously (18). We found that the deletion of caspase 2 did not provide protection against cell death induced by PS-341 (Fig. 1D and E), indicating that caspase 2 was dispensable for PS-341-mediated apoptosis.
FIG. 1.
PS-341 induces caspase activation and apoptosis in HNSCC cells. (A) HNSCC cell lines UMSCC-1, -9, -11B, -14A, and -23 were treated with PS-341 (0.5 μM) for 0, 8, 16, or 24 h. Cell viability was determined by trypan blue exclusion assay. The assays were performed with triplicate samples, and the results are representative of three independent experiments. Error bars depict standard deviations. (B) UMSCC-1 and UMSCC-23 cells were treated with PS-341 for 0, 16 or 24 h. The detached and attached cells were collected, and genomic DNA was extracted with phenol-chloroform. Genomic DNA was separated on a 1.2% agarose gel. (C) UMSCC-23 cells were treated with PS-341 for 0, 8, 16 or 24 h. The whole-cell extracts were prepared with RIPA buffer, and 50-μg aliquots of protein extracts were resolved on a SDS-12% PAGE. The membrane was probed with antibodies against caspases 9, 2, and 3 (1:500). For loading control, the membrane was stripped and reprobed with anti-α-tubulin monoclonal antibodies (1:5,000). (D) UMSCC-23 cells were transfected with caspase 2 siRNA (SiCasp-2), using oligofectamine. Twenty-four hours after transfection, cells were harvested and probed with monoclonal antibodies against caspase 2. For loading control, the membrane was stripped and reprobed with α-tubulin. (E) UMSCC-23/SiCasp-2 cells and control cells were treated with PS-341 for the indicated times, and cell death was determined.
The inhibition of NF-κB by PS-341 is not sufficient to induce apoptosis in HNSCC cells.
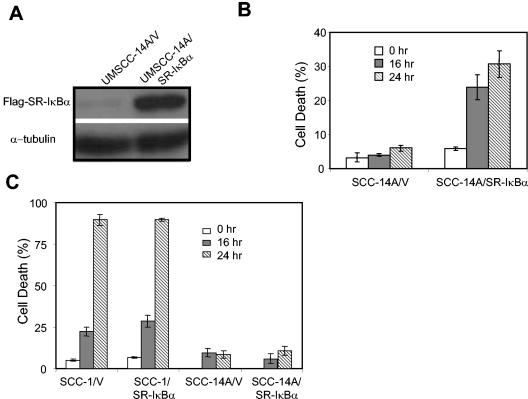
NF-κB has been found to be constitutively activated in several human cancer cell lines; the inhibition of NF-κB by biochemical and chemical inhibitors induced apoptosis in these cells. Due to its potent inhibition of NF-κB, PS-341 has been proposed to induce apoptosis in HNSCC cells by inhibiting NF-κB. Since UMSCC-14A cells were very resistant to PS-341-induced apoptosis (Fig. 1A), it was possible that PS-341 failed to inhibit NF-κB in these cells. To rule out this possibility, we utilized a biological NF-κB inhibitor, SR-IκBα. SR-IκBα is a dominant-negative mutant of IκBα in which serines 32 and 36 are mutated to alanines so that it cannot be phosphorylated and degraded by a variety of stimuli (39, 40, 43). Overexpression of SR-IκBα has been shown to specifically inhibit NF-κB activation induced by a variety of stimuli. As shown in Fig. 2A, we were able to obtain UMSCC-14A cells stably expressing SR-IκBα. UMSCC-14A cells expressing SR-IκBα were sensitive to tumor necrosis factor (TNF) killing compared with control cells, validating that SR-IκBα functioned in the inhibition of NF-κB in these cells (Fig. 2B). These results suggested that the inability of PS-341 to induce apoptosis in UMSCC-14A cells was not due to a failure of NF-κB inhibition. Moreover, like their parental cells, both UMSCC-14A cells expressing SR-IκBα and control cells were also resistant to PS-341-induced apoptosis (Fig. 2C). In contrast to UMSCC-14A cells, UMSCC-1 cells were sensitive to PS-341-induced apoptosis. We were previously able to establish SR-IκBα-expressing UMSCC-1 cells in which the inhibition of NF-κB also did not induce apoptosis in these cells (47). Since PS-341 strongly induced apoptosis in UMSCC-1 cells, it was likely that PS-341 predominantly modulated other apoptotic signaling pathways to induce apoptosis in these cells. Moreover, unlike the case with UMSCC-14A cells, PS-341 potently induced apoptosis in both UMSCC-1 cells expressing SR-IκBα (UMSCC-1/SR-IκBα) and control cells (UMSCC1/V) (Fig. 2C). Additionally, we found that the inhibition of NF-κB by the adenovirus-mediated delivery of SR-IκBα alone did not induce apoptosis in several HNSCC cell lines (3, 47). Taken together, these results suggest that the inhibition of NF-κB alone could not provide an explanation for PS-341-mediated apoptosis in HNSCC cells. PS-341 might trigger apoptosis in HNSCC cells by directly modulating other proapoptotic or antiapoptotic pathways.
FIG. 2.
The inhibition of NF-κB is not sufficient to induce apoptosis in HNSCC cells. (A) UMSCC-14A cells were transduced with retroviruses expressing SR-IκBα (UMSCC-14A/SR-IκBα) or control vector (UMSCC-14A/V) and selected with neomycin (600 μg/ml) for 1 week. Cells expressing SR-IκBα were confirmed with monoclonal antibodies against the Flag epitope (1:1,000) by Western blot analysis. For loading control, the membrane was stripped and reprobed with monoclonal antibodies against α-tubulin. (B) Both UMSCC-14A/SR-IκBα cells (SCC-14A/SR-IκBα) and control cells (SCC-14A/V) were treated with TNF (20 ng/ml) for 0, 16 or 24 h. Cell viability was determined with a trypan blue exclusion assay. (C) Cells were treated with PS-341 for 0, 16, or 24 h. Cell viability was determined with a trypan blue exclusion assay. The assay was performed in triplicate, and the results represent average values from three independent experiments.
PS-341-induced apoptosis is involved in ER stress.
To further understand how PS-341 induced apoptosis in HNSCC cells, UMSCC-23 cells were treated with PS-341 or vehicle control for short time periods and microarray analysis was performed with Affymetrix U133A gene chips to compare expression profiles. According to our statistical analysis, as shown in Table 1, 24 genes were significantly induced 4 h after PS-341 stimulation. The majority of PS-341-induced genes in UMSCC-23 cells encoded heat shock proteins, ribosomal proteins, and growth arrest and DNA-damage-inducible genes, which likely reflected a stress response (5, 29). Some of these genes were also previously identified in human multiple myeloma cells following PS-341 stimulation (29). However, unlike the case with myeloma cells, we did not find that PS-341 induced genes associated with the death receptor signaling pathways, suggesting that death receptor signaling might not be involved in PS-341-induced apoptosis in HNSCC cells. Interestingly, microarray analysis also found that heme oxygenase 1, a regulator of reactive oxygen species, and HERP, a membrane protein induced by ER stress (17, 32, 34), were also significantly induced by PS-341.
TABLE 1.
Identification of PS-341-induced genes by microarray analysis
| Gene identification | Product | Fold change | q value (%) |
|---|---|---|---|
| NM_002155 | Heat shock 70-kDa protein 6 (HSP70B′) | 43.4 | 1.1 |
| X51757 | Heat shock 70-kDa protein 6 (HSP70B′) | 16 | 1.1 |
| NM_005346 | Heat shock 70-kDa protein 1B | 8.7 | 1.1 |
| AF043337.1 | Interleukin 8 | 7 | 1.1 |
| NM_002133 | Heme oxygenase (decycling) 1 | 6.7 | 1.1 |
| BG537355 | DnaJ (Hsp40) homolog, subfamily B, member 1 | 6.2 | 1.1 |
| NM_004281 | BCL2-associated athanogene 3 (BAG3) | 6.2 | 1.1 |
| NM_006145 | DnaJ (Hsp40) homolog, subfamily B, member 1 | 5.4 | 3.9 |
| NM_014856 | KIAA0476 gene product | 4.4 | 1.1 |
| BF680255 | Ribosomal protein S11 | 3.7 | 1.1 |
| AI435828 | Stanniocalcin 2 | 3.0 | 3.8 |
| NM_024111 | Hypothetical protein MGC4504 | 2.8 | 1.1 |
| BC000023.1 | Ribosomal protein S19 | 2.6 | 1.1 |
| NM_001511 | Chemokine (C-X-C motif) ligand 1 | 2.3 | 3.8 |
| NM_014330 | Protein phosphatase 1, regulatory subunit 15A | 2.3 | 3.8 |
| U83981 | Protein phosphatase 1, regulatory subunit 15A | 2.2 | 1.1 |
| AF217990.1 | Homocysteine-inducible, ER stress-inducible, Ubiquitin-like domain member 1 (HERP) | 2.2 | 3.8 |
| BE737027 | Ribosomal protein L27a | 2.1 | 1.1 |
| NM_015675 | Growth arrest and DNA-damage-inducible, beta | 2.1 | 1.1 |
| AA320764 | ESTs, highly similar to S55918 ribosomal protein S10 | 2.1 | 1.1 |
| AF087853.1 | Growth arrest and DNA-damage-inducible, beta | 2.0 | 1.1 |
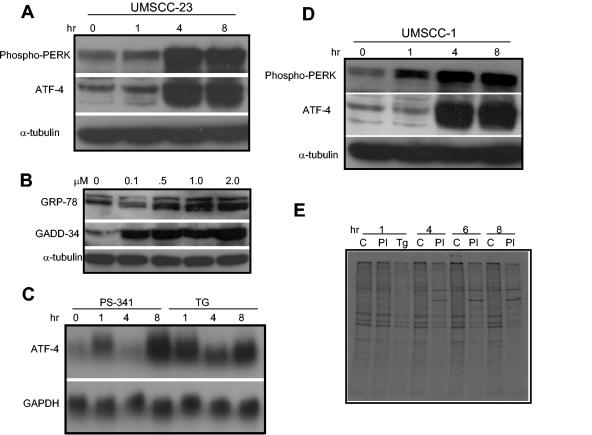
Proteins destined for the lysosome or plasma membrane and those to be secreted from the cell, approximately one-third of all cellular proteins, are posttranslationally processed in the lumen of the ER-Golgi complex (15, 17, 32, 46, 48). In the ER-Golgi complex, native proteins are glycosylated, disulfide bond formation is carefully directed, and proteins are folded into their proper three-dimensional tertiary structures. Proteins that are unable to fold properly in the ER are ubiquitinated and degraded by the 26S proteasome (17, 32). Therefore, the inhibition of the 26S proteasome by PS-341 may increase the accumulation of misfolded proteins, resulting in the ER stress. A series of poorly understood transcription and translation events lead to a small fraction, perhaps 5%, of the genome being expressed to assist in the recovery of the cell (17, 32). Comparison of our data to the gene expression profiles of ER stress (11) led us to the hypothesis that ER stress might play a significant role in PS-341-induced apoptosis in HNSCC cells. As shown in Fig. 3A, treatment of HNSCC cells with PS-341 strongly induced the phosphorylation of the ER membrane-resident stress kinase PERK, a hallmark of ER stress (17, 32, 35). To further confirm our microarray results and ER stress responses, both Northern blot and Western blot analyses were performed to examine expression of the ER stress-associated genes. As shown in Fig. 3A and C, both Western blot and Northern blot analyses demonstrated that the ER stress-dependent ATF-4 expression was induced by PS-341 in UMSCC-23 cells. Western blot analysis demonstrated that the ER stress-induced proteins GRP-78 (Bip) and GADD-34 were up-regulated by PS-341 in a dose-dependent manner in UMSCC-23 cells. Moreover, as shown in Fig. 3D, PS-341 also induced the phosphorylation of PERK in UMSCC-1 cells. The kinetics of ATF-4 expression in UMSCC-1 was nearly indistinguishable from that in UMSCC-23 cells following PS-341 stimulation.
FIG. 3.
PS-341 induces ER stress in HNSCC cells. (A and B) UMSCC-23 cells were treated with PS-341 for the indicated time periods and concentrations. Whole-cell extracts were prepared, and 50-μg aliquots of proteins were probed with polyclonal antibodies against phospho-PERK, ATF-4, GRP-78, and GADD-34. For loading control, the membranes were stripped and reprobed with monoclonal antibodies against α-tubulin. (C) UMSCC-23 cells were treated with PS-341 and thapsigargin (TG) for the indicated time periods. The total RNAs were extracted, and 10-μg aliquots of RNAs were resolved on 1.5% agarose formaldehyde gels. The membrane was probed with 32P-labeled ATF cDNA probes. For loading control, the membrane was stripped and reprobed with 32P-labeled glyceraldehyde-3-phosphate dehydrogenase (GAPDH) probe. (D) UMSCC-1 cells were treated with PS-341 for the indicated time periods, and Western blot analysis was performed as described in (A). (E) UMSCC-23 cells were treated with PS-341 or TG as a positive control for the indicated time periods. Cells were labeled with 35S for 15 min. Ten micrograms of 35S-labeled proteins were resolved by SDS-10% PAGE and exposed to autoradiographic film. C, vehicle control; PI, PS-341; Tg, TG.
In general, when a cell experiences ER stress, the response is characterized by a rapid inhibition of general protein synthesis to allow the cell to redistribute resources to facilitate recovery. Prolonged stress and the failure of the cell machinery to resume normal translation and protein folding at the ER result in apoptosis (16, 17, 19, 28, 32, 34). Thus, we were interested in whether PS-341 inhibited protein synthesis, a hallmark of ER stress. As shown in Fig. 3E, pulse-chase experiments with 35S labeling indicated that PS-341 potently inhibited general protein synthesis in UMSCC-23 cells. Within 1 h of PS-341 treatment, we observed general protein synthesis to be reduced. This effect persisted through 6 h, at which point the level of inhibition was similar to that observed with thapsigargin, a well-characterized and potent inducer of ER stress (45).
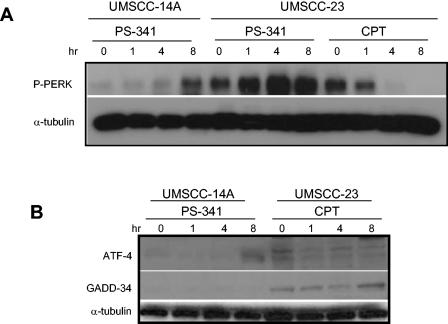
Since it is known that DNA-damaging agents can induce the expression of ATF-4 and GADD-34, we further performed experiments to compare the activation of PERK and the induction of ATF-4 and GADD-34 in UMSCC-23 cells mediated by PS-341 with camptothecin (CPT), a DNA-damaging agent. As shown in Fig. 4A, CPT stimulation did not induce the phosphorylation of PERK and ATF-4 expression and weakly induced GADD-34 expression, suggesting that the induction of ATF-4 and GADD-34 by PS-341 was mediated by ER stress in UMSCC-23 cells. Moreover, since UMSCC-14A cells were resistant to PS-341, we also examined whether PS-341 induced ER stress in these cells. As shown in Fig. 4A and B, PS-341 very weakly induced the phosphorylation of PERK in UMSCC-14A cells and the expression of ATF-4 and GADD-34. Taken together, these results strongly suggested that ER stress was involved in PS-341-induced apoptosis in HNSCC cells.
FIG. 4.
The DNA-damaging agent CPT does not induce ER stress in HNSCC cells. (A) UMSCC-23 or UMSCC-14A cells were treated with CPT (10 μM) or PS-341 for the indicated time periods. The phosphorylation of PERK was detected by Western blot analysis as described in the legend to Fig. 3A. (B) Cells were treated with PS-341 or CPT for the indicated time periods. The expression of ATF-4 and GADD-34 was examined by Western blot analysis as described in the legend to Fig. 3A.
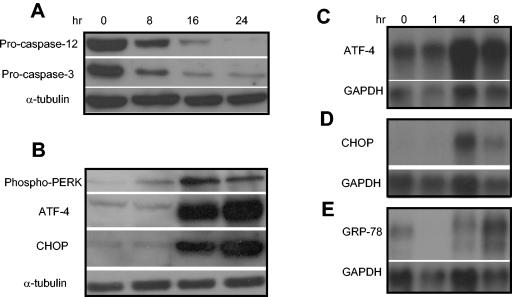
Excellent studies by Nakagawa et al. (30) have found that caspase 12 is specifically activated during ER stress-mediated apoptosis and that deletion of caspase 12 abolished ER stress-mediated apoptosis in vitro and in vivo. Since caspase 12 could not be detected in HNSCC cells, to confirm that PS-341-induced apoptosis was mediated by the ER stress, we utilized MEFs which expressed caspase 12 to determine whether PS-341 induced caspase 12 activation. As shown in Fig. 5A, cleavage of procaspase 12 in MEFs was observed within 8 h of exposure to PS-341. Procaspase 12 was fully processed by 24 h, at a time point when cells were approximately 85% dead (data not shown). Consistently, procaspase 3 also was processed after PS-341 treatment. Similarly, as shown in Fig. 5B, PS-341 induced the phosphorylation of PERK. Western blot analysis demonstrated that PS-341 induced the expression of the ER stress-dependent ATF-4 and CHOP/GADD-153 proteins. The accumulation of ATF-4, CHOP/GADD-153, and GRP-78 mRNA was also observed after PS-341 stimulation, as detected by Northern blot analysis (Fig. 5C, D, and E).
FIG. 5.
PS-341 induces ER stress and activates caspase 12 in MEFs. (A) MEFs were treated with PS-341 for the indicated time periods, and the whole-cell extracts were prepared. Fifty-microgram aliquots of proteins were probed with polyclonal antibodies against caspase 12 and caspase 3. For loading control, the membrane was stripped and reprobed with monoclonal antibodies against α-tubulin. (B) MEFs were treated with PS-341 for the indicated time periods, and the whole-cell extracts were prepared. Fifty-microgram aliquots of proteins were probed with polyclonal antibodies against ATF-4 and CHOP. For loading control, the membrane was stripped and reprobed with monoclonal antibodies against α-tubulin. (C, D, and E) MEFs were treated with PS-341 for the indicated time periods, and the total RNAs were extracted with Trizol. Ten-microgram aliquots of total RNAs were probed with 32P-labeled ATF, CHOP, and GRP-78 cDNA probes. For loading control, the membranes were stripped and reprobed with 32P-labeled glyceraldehyde-3-phosphate dehydrogenase (GAPDH) cDNA probes.
PS-341-induced apoptosis in HNSCC cells is associated with the induction of ROS.
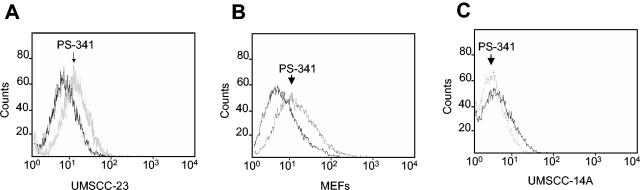
As described above, we have found that PS-341 induced heme oxygenase 1 by microarray analysis. Although heme oxygenase 1 has an antioxidant function, many studies have reported that the induction of heme oxygenase 1 is mediated by ROS, which may serve as a feedback mechanism to balance the intracellular level of ROS (9, 34). Additionally, it is well known that ER stress induces ROS (17, 32). Therefore, we were interested to know whether ROS played a critical role in PS-341-mediated apoptosis in HNSCC cells. First, we sought to determine whether PS-341 induced ROS in HNSCC cells, using fluorescence-activated cell sorting (FACS) analysis with a cell-permeable dye, H2DCFDA. In the presence of ROS, H2DCFDA is specifically cleaved to emit light at a fluorescent wavelength (44). As shown in Fig. 6A, ROS were significantly induced in HNSCC cells following PS-341 treatment. Also, PS-341 strongly induced ROS in MEFs (Fig. 6B). Interestingly, PS-341 could not induce ROS in UMSCC-14A cells, which were resistant to killing, suggesting that the induction of ROS might be associated with cell sensitivity (Fig. 6C).
FIG. 6.
PS-341 induces the production of reactive oxygen species in HNSCC cells and MEFs. (A, B, and C) UMSCC-23 cells, MEFs, and UMSCC-14A cells were treated with PS-341 or vehicle control for 4 h, respectively. After treatment, cells were harvested and incubated with the cell-permeable dye H2DCFDA. The reaction was analyzed by FACS.
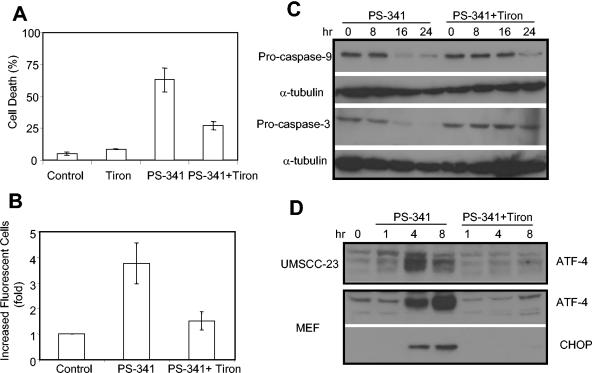
To determine whether ROS played a role in PS-341-induced apoptosis, the cell-permeable superoxide scavenger Tiron was utilized. As shown in Fig. 7A, Tiron significantly suppressed PS-341-mediated cell killing in HNSCC cells. FACS analysis also confirmed that Tiron significantly inhibited PS-341-induced ROS in HNSCC cells (Fig. 7B). In contrast, the nitric oxide synthase mRNA inhibitor pyrrolidine dithiocarbamate, also an antioxidant, did not protect cells from death (data not shown), suggesting an important role for superoxide radicals in PS-341-induced cell death. Consistent with the attenuation of cell death, Tiron greatly delayed the cleavage of procaspases 9 and 3 in HNSCC cells (Fig. 7C). Moreover, we examined the ability of the antioxidant to modulate the accumulation of the proapoptotic factors ATF-4 and CHOP/GADD-153. Unexpectedly, the addition of Tiron also abrogated the accumulation of ATF-4 and/or CHOP in HNSCC cells and MEFs following PS-341 stimulation (Fig. 7D), suggesting that ROS might play a role in ER stress-mediated gene expression. Taken together, these data suggested that ROS, probably induced by ER stress, played an important role in PS-341-induced apoptosis in HNSCC cells.
FIG. 7.
The ROS inhibitor Tiron inhibits PS-341-mediated apoptosis and gene expression. (A) UMSCC-23 cells were pretreated with or without Tiron for 30 min and then treated with PS-341 for 24 h. Cell viability was determined with a trypan blue exclusion assay. The assay was performed in triplicate, and results represent the average values from three independent experiments. (B) UMSCC-23 cells were pretreated with or without Tiron for 30 min and then treated with PS-341 for 24 h. After treatment, cells were harvested and incubated with the cell-permeable dye H2DCFDA. The reaction was analyzed by FACS. (C) UMSCC-23 cells were pretreated with or without Tiron for 30 min and then treated with PS-341 for the indicated time periods. The whole-cell proteins were probed with polyclonal antibodies against caspases 9 and 3. For loading control, the membrane was stripped and reprobed with monoclonal antibodies against α-tubulin. (D) UMSCC-23 cells or MEFs were treated with PS-341 with or without Tiron for the indicated time periods, and whole-cell extracts were prepared. Fifty-microgram aliquots were resolved on SDS-8% PAGE and probed with polyclonal antibodies for ATF-4 and CHOP.
DISCUSSION
In this study we have shown that a new chemotherapeutic drug, the proteasome inhibitor PS-341, induced the caspase cascade and apoptosis in human HNSCC cells. Although PS-341 could potently inhibit NF-κB activity, it was unlikely that PS-341-induced apoptosis was fully mediated by inactivation of NF-κB in HNSCC cells. Our results demonstrate for the first time that ER stress and the generation of ROS played a critical role in PS-341-induced apoptosis in HNSCC cells. In preclinical and clinical trials, PS-341, in combination with several chemotherapeutic drugs, has displayed strong antitumor activities against multiple cancers (1). Based on our results, it is likely that the antitumor activity of PS-341 is mediated not only by inhibition of the prosurvival NF-κB pathway but also by activation of the proapoptotic ER stress-ROS cascade. These novel findings have important implications for improving the efficacy of chemotherapy of head and neck cancer and for developing new chemotherapeutic drugs.
Our gene array analyses indicated that a group of stress-related genes was rapidly induced by PS-341 stimulation in HNSCC cells that was similar to the gene expression profile induced by ER stress. Consistently, we found that PS-341 activated the ER membrane resident stress kinase PERK. Moreover, coupled with the phosphorylation of PERK, increases in steady-state levels of the ER protein folding chaperone GRP-78 and the proapoptotic proteins ATF-4 and CHOP and a simultaneous decrease in general protein synthesis strongly indicate that the induction of ER stress might be involved in PS-341-mediated apoptosis. However, currently, it is not clear which caspase is directly activated by ER stress in HNSCC cells. Caspase 12 has been identified in ER and is considered to be specifically activated in response to ER stress. Using knockout mice, Nakagawa et al. (30) found that caspase 12 played an essential role in ER stress-mediated apoptosis. HNSCC cells, like most other human cancer cells, did not express caspase 12 (4), so we could not use the HNSCC model system to demonstrate that PS-341-mediated ER stress directly activated apoptosis. As a complementary experiment, we demonstrated that caspase 12 was activated in MEFs following PS-341 stimulation, supporting the notion that ER stress played a critical role in PS-341-mediated caspase activation and apoptosis. In the future, it will be interesting to identify the key caspase associated with the ER stress in HNSCC cells.
Upon PS-341 stimulation, caspases 9, 2, and 3 were activated in HNSCC cells. Although caspase 9 is considered an initiating caspase in the intrinsic apoptotic pathway (20), recent studies by Lassus et al. (18) have demonstrated that caspase 2 may be involved in activation of Bax/Bak and caspase 9. However, using the siRNA strategy to deplete caspase 2, we found that caspase 2 was dispensable from PS-341-mediated apoptosis. The activation of caspase 9 is dependent on mitochondrial damage and cytochrome c release (9, 10). According to our biochemical and functional analyses, PS-341-induced ROS were likely to play a critical role in caspase 9 activation through mitochondrial damage in PS-341-mediated apoptosis. Consistently, we found that the inhibition of ROS abolished PS-341-mediated caspase 9 activation. Caspase 9 was not activated in PS-341-resistant cells in which ROS were not generated upon stimulation. Interestingly, we also found that the inhibition of ROS also abolished the expression of ATF-4 and CHOP/GADD-153, suggesting that ROS may play a role in ER stress-dependent gene expression.
The transcription factor NF-κB has been found to be involved in cancer therapy resistance (14, 21, 25, 39, 40). Several antiapoptotic genes, including those encoding Bcl-2 family proteins, inhibitors of apoptosis (c-IAPs), and A-20, have been found to be transcriptionally regulated by NF-κB (27, 41, 42). Previously, members of our group and others demonstrated that several chemotherapeutic agents activated NF-κB and that PS-341 synergized with chemotherapeutic drugs to inhibit tumor cell growth in vitro and in vivo (24, 33, 43). Given the fact that PS-341 potently inhibited NF-κB activation induced by multiple stimuli, including chemotherapeutic drugs, it was logical to propose that PS-341 enhanced chemotherapeutic drug-mediated apoptosis by inhibiting NF-κB. Because PS-341 treatment alone potently induced apoptosis in HNSCC cells (37), it raised the possibility that PS-341 may be used as a monotherapy for HNSCC. In order to provide a molecular basis for this hypothesis, we systemically explored how PS-341 induced apoptosis in HNSCC cells. Although NF-κB is abnormally activated in HNSCC cells, we found that the inhibition of NF-κB by the biological inhibitor SR-IκBα did not induce apoptosis in HNSCC cells. It suggests that NF-κB was not essential for survival of HNSCC cells. In other words, the inhibition of NF-κB by PS-341 was not sufficient to induce apoptosis in HNSCC cells. Moreover, due to the functional redundancy between SR-IκBα and PS-341 in NF-κB inhibition, we found that the inhibition of NF-κB by SR-IκBα also did not significantly potentiate PS-341-mediated apoptosis. However, it should be mentioned that NF-κB has been found to be essential for cell survival for several other human cancer cell lines. Our results did not implicate that the induction of apoptosis in those cells by PS-341 was independent of the inhibition of NF-κB. Nevertheless, we found that in addition to the inhibition of prosurvival NF-κB, PS-341 activated the proapoptotic ER stress-ROS pathway to induce apoptosis in HNSCC cells. In regard to the synergistic antitumor effects between PS-341 and chemotherapeutic drugs, our findings presented here suggest that in addition to targeting NF-κB, PS-341 may enhance the efficacy of chemotherapeutic drugs through the activation of the proapoptotic ER stress-ROS pathway.
Acknowledgments
We gratefully acknowledge the technical expertise of David Adams, Tim Hale, and Anne Marie DesLauriers at the Flow Cytometry Core Facility at the University of Michigan Medical School Cancer Center for their professional data collection and analysis and Mark Rolfe at Millennium Pharmaceuticals for comments. Microarray data collection was performed by Taocong Jin at the University of Michigan School of Dentistry DNA Microarray Core Facility, and statistical analysis of gene chip data was performed by Jim McDonald at the University of Michigan Medical School Cancer Center.
This work was supported by NICDR grants DE015964, DE13848, and DE13788 to C.-Y.W. and T32-DE0757 to A.F.
REFERENCES
- 1.Adams, J. 2003. The proteasome: structure, function, and role in the cell. Cancer Treat. Rev. 29(Suppl. 1):3-9. [DOI] [PubMed] [Google Scholar]
- 2.Adams, J., V. J. Palombella, E. A. Sausville, J. Johnson, A. Destree, D. D. Lazarus, J. Mass, C. S. Pien, S. Prakash, and P. J. Elliot. 1999. Proteasome inhibitors: a novel class of potent and effective antitumor agents. Cancer Res. 59:2615-2622. [PubMed] [Google Scholar]
- 3.Chen, S., A. Fribley, and C.-Y. Wang. 2002. Potentiation of tumor necrosis factor-mediated apoptosis of oral squamous cell carcinoma cells by adenovirus-mediated gene transfer of NF-κB inhibitor. J. Dent. Res. 81:98-102. [PubMed] [Google Scholar]
- 4.Fischer, H., U. Koenig, L. Eckhart, and E. Tschachler. 2002. Human caspase 12 has acquired deleterious mutations. Biochem. Biophys. Res. Commun. 293:722-726. [DOI] [PubMed] [Google Scholar]
- 5.Fleming, J. A., E. S. Lightcap, S. Sadis, V. Thoroddsen, C. E. Bulawa, and R. K. Blackman. 2002. Complementary whole-genome technologies reveal the cellular response to proteasome inhibition by PS-341. Proc. Natl. Acad. Sci. USA 99:1461-1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Forastiere, A., W. Koch, A. Trotti, and D. Sidransky. 2001. Head and neck cancer. N. Engl. J. Med. 345:1890-1900. [DOI] [PubMed] [Google Scholar]
- 7.Frankel, A., S. Man, P. Elliott, J. Adams, and R. S. Kerbel. 2000. Lack of multicellular drug resistance observed in human ovarian and prostate carcinoma treated with the proteasome inhibitor PS-341. Clin. Cancer Res. 6:3719-3728. [PubMed] [Google Scholar]
- 8.Ghosh, S., and M. Karin. 2002. Missing pieces in the NF-kappaB puzzle. Cell 109:S81-S96. [DOI] [PubMed] [Google Scholar]
- 9.Gottlieb, E., M. G. Vander Heiden, and C. B. Thompson. 2000. Bcl-xL prevents the initial decrease in mitochondrial membrane potential and subsequent reactive oxygen species production during tumor necrosis factor alpha-induced apoptosis. Mol. Cell. Biol. 20:5680-5689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Green, D., and J. Reed. 1998. Mitochondria and apoptosis. Science 281:1309-1312. [DOI] [PubMed] [Google Scholar]
- 11.Harding, H. P., Y. Zhang, H. Zeng, I. Novoa, P. D. Lu, M. Calfon, N. Sadri, C. Yun, P. Popko, R. Paules, D. F. Stojdl, J. C. Bell, T. Hettmann, J. M. Leiden, and D. Ron. 2003. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol. Cell 11:619-633. [DOI] [PubMed] [Google Scholar]
- 12.Hideshima, T., P. Richardson, D. Chauhan, V. J. Palombella, P. J. Elliott, J. Adams, and K. C., Anderson. 2001. The proteasome inhibitor PS-341 inhibits growth, induces apoptosis, and overcomes drug resistance in human multiple myeloma cells. Cancer Res. 61:3071-3076. [PubMed] [Google Scholar]
- 13.Hideshima, T., C. Mitsiades, M. Akiyama, T. Hayashi, D. Chauhan, P. Richardson, R. Schlossman, K. Podar, N. C. Munshi, N. Mitsiades, and K. C. Anderson. 2003. Molecular mechanisms mediating antimyeloma activity of proteasome inhibitor PS-341. Blood 101:1530-1534. [DOI] [PubMed] [Google Scholar]
- 14.Huang, T. T., S. M. Wuerzberger-Davis, Z. Wu, and S. Miyamoto. 2003. Sequential modification of NEMO/IKKγ by SUMO-1 and ubiquitin mediates NF-κB activation by genotoxic stress. Cell 115:565-576. [DOI] [PubMed] [Google Scholar]
- 15.Jiang, H., S. A. Wek, B. C. McGrath, D. Lu, T. Hai, H. P. Harding, X. Wang, D. Ron, D. R. Cavener, and R. C. Wek. 2004. Activating transcription factor 3 is integral to the eukaryotic initiation factor 2 kinase stress response. Mol. Cell. Biol. 24:1365-1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jimbo, A., E. Fujita, Y. Kouroku, J. Ohnishi, N. Inohara, K. Kuida, K. Sakamaki, S. Yonehara, and T. Momoi. 2003. ER stress induces caspase-8 activation, stimulating cytochrome c release and caspase-9 activation. Exp. Cell Res. 283:156-166. [DOI] [PubMed] [Google Scholar]
- 17.Kaufman, R. J. 2002. Orchestrating the unfolded protein response in health and disease. J. Clin. Investig. 110:1389-1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lassus, P., X. Opitz-Araya, and Y. Lazebnik. 2002. Requirement for caspase-2 in stress-induced apoptosis before mitochondrial permeabilization. Science 297:1352-1354. [DOI] [PubMed] [Google Scholar]
- 19.Lee, A., N. N. Iwakoshi, and L. H. Glimcher. 2003. XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Mol. Cell. Biol. 23:7448-7459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li, P., D. Nijhawan, I. Budihardjo, S. Srinivasula, M. Ahmad, E. Alnemri, and X. Wang. 1996. Cytochrome c and dATP-dependent formation of Apaf-1/caspase-9 complex initiates an apoptotic protease cascade. Cell 91:479-489. [DOI] [PubMed] [Google Scholar]
- 21.Lin, Y., A. Devin, Y. Rodriguez, and Z. G. Liu. 1999. Cleavage of the death domain kinase RIP by caspase-8 prompts TNF-induced apoptosis. Genes Dev. 13:2514-2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin, A., and M. Karin. 2003. NF-kappaB in cancer: a marked target. Semin. Cancer Biol. 13:107-114. [DOI] [PubMed] [Google Scholar]
- 23.Liu, Z. G., H. Hsu, D. V. Goeddel, and M. Karin. 1996. Dissection of TNF receptor 1 effector functions: JNK activation is not linked to apoptosis while NF-κB activation prevents cell death. Cell 87:565-576. [DOI] [PubMed] [Google Scholar]
- 24.Ma, M. H., H. H. Yang, K. Parker, S. Manyak, J. M. Friedman, C. Altamirano, Z. Q. Wu, M. J. Borad, M. Frantzen, E. Roussos, J. Neeser, A. Mikail, J. Adams, N. Sjak-Shie, R. A. Vescio, and J. R. Berenson. 2003. The proteasome inhibitor PS-341 markedly enhances sensitivity of multiple myeloma tumor cells to chemotherapeutic agents. Clin. Cancer Res. 9:1136-1144. [PubMed] [Google Scholar]
- 25.Madrid, L. V., C.-Y. Wang, D. C. Guttridge, A. J. Schottelius, A. S. Baldwin, and M. W. Mayo. 2000. Akt suppresses apoptosis by stimulating the transactivation potential of the RelA/p65 subunit of NF-κB. Mol. Cell. Biol. 20:1626-1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maniatis, T. 1999. A ubiquitin ligase complex essential for the NF-κB, Wnt/Wingless, and Hedgehog signaling pathways. Genes Dev. 13:505-510. [DOI] [PubMed] [Google Scholar]
- 27.Mayo, M. W., and A. S. Baldwin. 2000. The transcription factor NF-κB: control of oncogenesis and cancer therapy resistance. Biochim. Biophys. Acta 1470:M55-M62. [DOI] [PubMed] [Google Scholar]
- 28.McCullough, K. D., J. L. Martindale, L. Klotz, T. Aw, and N. J. Holbrook. 2001. Gadd153 sensitizes cells to endoplasmic reticulum stress by down-regulating Bcl2 and perturbing the cellular redox state. Mol. Cell. Biol. 21:1249-1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mitsiades, N., C. S. Mitsiades, V. Poulaki, D. Chauhan, G. Fanourakis, X. Gu, C. Bailey, M. Joseph, T. A. Libermann, S. P. Treon, et al. 2002. Molecular sequelae of proteasome inhibition in human multiple myeloma cells. Proc. Natl. Acad. Sci. USA 99:14374-14379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nakagawa, T., H. Zhu, N. Morishima, E. Li, J. Xu, B. Yanker, and J. Yuan. 2000. Caspase-12 mediates endoplasmic-reticulum-specific apoptosis and cytotoxicity by amyloid-beta. Nature 403:98-103. [DOI] [PubMed] [Google Scholar]
- 31.Patel, V., C. Leethanakul, and J. S. Gutkind. 2001. New approaches to the understanding of the molecular basis of oral cancer. Crit. Rev. Oral Biol. Med. 12:55-63. [DOI] [PubMed] [Google Scholar]
- 32.Ron, D. 2002. Translational control in the endoplasmic reticulum stress response. J. Clin. Investig. 110:1383-1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Russo, S. M., J. E. Tepper, A. S. Baldwin, Jr., R. Liu, J. Adams, P. Elliott, and J. C. Cusack. 2001. Enhancement of radiosensitivity by proteasome inhibition: implications for a role of NF-κB. Int. J. Radiat. Oncol. Biol. Phys. 50:183-193. [DOI] [PubMed] [Google Scholar]
- 34.Ryter, S. W., and R. M. Tyrrell. 2000. The heme synthesis and degradation pathways: role in oxidant sensitivity. Free Rad. Biol. Med. 28:289-309. [DOI] [PubMed] [Google Scholar]
- 35.Scheuner, D., B. Song, E. McEwen, C. Liu, R. Laybutt, P. Gillespie, T. Saunders, S. Bonner-Weir, and R. J. Kaufman. 2001. Translational control is required for the unfolded protein response and in vivo glucose homeostasis. Mol. Cell 7:1165-1176. [DOI] [PubMed] [Google Scholar]
- 36.Shintani, S., M. Mihara, Y. Ueyama, T. Matsumura, and D. T. Wong. 2001. Cyclin D1 overexpression associates with radiosensitivity in oral squamous cell carcinoma. Int. J. Cancer 96:159-165. [DOI] [PubMed] [Google Scholar]
- 37.Sunwoo, J. B., Z. Chen, G. Dong, N. Yeh, C. Crowl Bancroft, E. Sausville, J. Adams, P. Elliott, and C. Van Waes. 2001. Novel proteasome inhibitor PS-341 inhibits activation of nuclear factor-kappa B, cell survival, tumor growth, and angiogenesis in squamous cell carcinoma. Clin. Cancer Res. 7:1419-1428. [PubMed] [Google Scholar]
- 38.Tergaonkar, V., M. Pando, O. Vafa, G. Wahl, and I. Verma. 2002. p53 stabilization is decreased upon NFκB activation: a role for NFkappaB in acquisition of resistance to chemotherapy. Cancer Cell 1:493-503. [DOI] [PubMed] [Google Scholar]
- 39.Van Antwerp, D. J., S. J. Martin, T. Kafri, D. R. Green, and I. M. Verma. 1996. Suppression of TNF-alpha-induced apoptosis by NF-κB. Science 274:787-789. [DOI] [PubMed] [Google Scholar]
- 40.Wang, C.-Y., M. Mayo, and A. S. Baldwin. 1996. TNF- and cancer therapy-induced apoptosis potentiation by inhibition of NF-κB. Science 274:784-787. [DOI] [PubMed] [Google Scholar]
- 41.Wang, C.-Y., M. W. Mayo, R. C. Korneluk, D. V. Goeddel, and A. S. Baldwin. 1998. NF-κB antiapoptosis: induction of TRAF1 and TRAF2 and c-IAP1 and c-IAP2 to suppress caspase-8 activation. Science 281:1680-1683. [DOI] [PubMed] [Google Scholar]
- 42.Wang, C.-Y., D. C. Guttridge, M. W. Mayo, and A. S. Baldwin. 1999. NF-κB expression of the Bcl-2 homologue A1/Bfl-1 to preferentially suppress chemotherapy-induced apoptosis. Mol. Cell. Biol. 19:5923-5929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang, C.-Y., J. Cusack, R. Liu, and A. S. Baldwin. 1999. Control of inducible chemoresistance: enhanced anti-tumor therapy via increased apoptosis through inhibition of NF-κB. Nat. Med. 5:412-417. [DOI] [PubMed] [Google Scholar]
- 44.Yamada, J., S. Yoshimura, H. Yamakawa, M. Sawada, M. Nakagawa, S. Hara Y. Kaku, T. Iwama, T. Naganawa, Y. Banno, S. Nakashima, and N. Sakai. 2003. Cell permeable ROS scavengers, Tiron and Tempol, rescue PC12 cell death caused by pyrogallol or hypoxia/reoxygenation. Neurosci. Res. 45:1-8. [DOI] [PubMed] [Google Scholar]
- 45.Yamaguchi, H., K. Bhalla, and H. G. Wang. 2003. Bax plays a pivotal role in thapsigargin-induced apoptosis of human colon cancer HCT116 cells by controlling Smac/Diablo and Omi/HtrA2 release from mitochondria. Cancer Res. 63:1483-1489. [PubMed] [Google Scholar]
- 46.Yoshida, H., T. Matsui, A. Yamamoto, T. Okada, and K. Mori. 2001. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell 107:881-891. [DOI] [PubMed] [Google Scholar]
- 47.Zeng, Q., S. Chen, Z. You, F. Yang, T. E. Carey, D. Saims, and C.-Y. Wang. 2002. Hepatocyte growth factor inhibits anoikis in head and neck squamous cell carcinoma cells by activation of ERK and Akt signaling independent of NFκB. J. Biol. Chem. 277:25203-25208. [DOI] [PubMed] [Google Scholar]
- 48.Zong, W. X., C. Li, G. Hatzivassiliou, T. Lindsten, Q. C. Yu, J. Yuan, and C. B. Thompson. 2003. Bax and Bak can localize to the endoplasmic reticulum to initiate apoptosis. J. Cell Biol. 162:59-69. [DOI] [PMC free article] [PubMed] [Google Scholar]