Abstract
The physiological role of an orphan G protein-coupled receptor, LGR5, was investigated by targeted deletion of this seven-transmembrane protein containing a large N-terminal extracellular domain with leucine-rich repeats. LGR5 null mice exhibited 100% neonatal lethality characterized by gastrointestinal tract dilation with air and an absence of milk in the stomach. Gross and histological examination revealed fusion of the tongue to the floor of oral cavity in the mutant newborns and immunostaining of LGR5 expression in the epithelium of the tongue and in the mandible of the wild-type embryos. The observed ankyloglossia phenotype provides a model for understanding the genetic basis of this craniofacial defect in humans and an opportunity to elucidate the physiological role of the LGR5 signaling system during embryonic development.
The leucine-rich repeat-containing, G protein-coupled receptors (LGRs) designated LGR4 through LGR8 are structurally similar to receptors for gonadotropins and thyrotropin (11, 12). These receptors are characterized by a large N-terminal extracellular domain containing leucine-rich repeats followed by a seven-transmembrane region. Phylogenetic analyses showed three LGR subgroups: the glycoprotein hormone receptors; the subgroup of LGR4, LGR5, and LGR6; and a third subgroup represented by LGR7 and LGR8 recently found to be receptors for the relaxin family ligands (13, 15, 23). Evolutionary analyses suggested that these three subgroups of LGRs existed before the divergence of vertebrates and invertebrates (10). LGR5, also known as GPR49, HG38, or FEX, is a large protein consisting of 18 extracellular leucine-rich repeats together with a seven-transmembrane region (8, 12, 17). Unlike LGRs in the other two subgroups, the ligands and physiological functions for the LGR4, LGR5, and LGR6 genes are unclear.
Ankyloglossia is a rare human craniofacial defect associated with difficulties in an infant's ability to breast-feed and defective speech articulation (6, 16, 18). In patients with this disorder, the lingual frenulum, which attaches the tongue to the floor of the oral cavity, extends to the tip of the tongue, thereby preventing optimal tongue movement. Limitation of tongue movement may vary from very mild to complete fusion of the tongue to the floor of the mouth. Ankyloglossia in breast-feeding infants can cause ineffective latch, inadequate milk transfer, and maternal nipple pain, resulting in slower weight gain and untimely weaning. Some cases of ankyloglossia are associated with cleft palate (CPX; MIM 303400; OMIM database, Johns Hopkins University, Baltimore, Md.) and are inherited as an X-linked disorder caused by mutations in TBX22, a T-box transcription factor gene located in Xq21 (3).
In an attempt to elucidate the physiological roles of the subgroup of orphan LGRs consisting of LGR4, LGR5, and LGR6, we performed gene deletion experiments with LGR5 using a mouse model. Here, we report that the LGR5 null mice exhibit neonatal lethality associated with ankyloglossia characterized by fusion of the tongue to the floor of the mouth leading to the inability to nurse and neonatal mortality.
MATERIALS AND METHODS
Generation and genotyping of LGR5 null mice.
A targeting vector was constructed in which exon 18 of the LGR5 gene was replaced with a neomycin resistance gene derived from the MC1 neo vector (Fig. 1A). Lex-1 embryonic stem (ES) cells were electroporated with the targeting vector before selection of the cells expressing the targeted allele for the generation of chimeric mice. LGR5 mutant mice were obtained by mating chimeric mice with C57BL/6 mice. Mice were housed in the Stanford Research Animal Facility, and all procedures were approved by the Institutional Care and Use Committee. Intercross litters were obtained by pairing heterozygous male and female animals overnight. The day the vaginal plug was observed was considered as embryonic day 0.5 (E0.5).
FIG. 1.
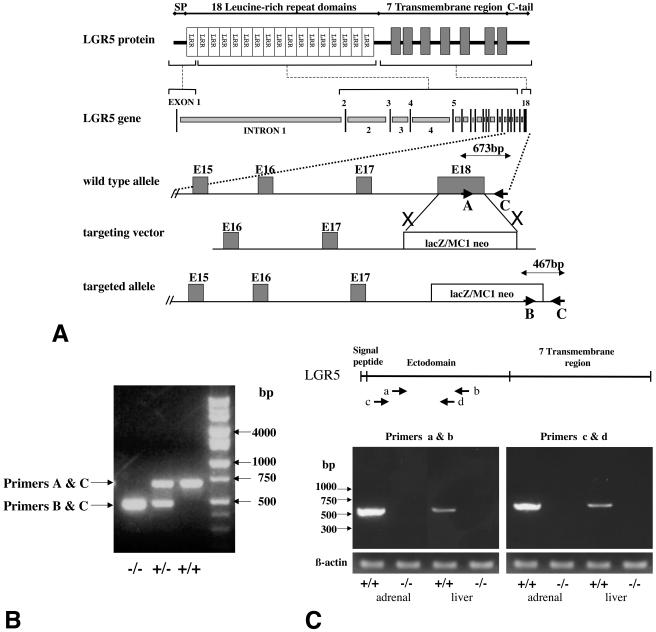
Targeted disruption of the LGR5 genes and genotyping of LGR5 null mice. (A) Schematic representation of LGR5 genomic DNA, the targeting vector, and the disrupted gene. The LGR5 gene contains 18 exons, with exon 1 encoding the signal peptide for secretion (SP) and the N-terminal leucine-rich repeat domain. Exons 2 to 17 encode the 17 leucine-rich repeat (LRR) domains, and exon 18 encodes the seven-transmembrane region plus the C-terminal tail. In the targeting vector, the transgene containing the LacZ-MC1 neo cassette replaced exon 18 of the LGR5 gene, leading to the loss of LGR5 expression in the targeted allele. (B) PCR genotyping of wild-type (+/+), heterozygous (+/−), and LGR5 null (−/−) mice. Triplex PCR was performed with genomic DNA as the template together with three primers. Primers A and C allowed the amplification of an LGR5 gene fragment (673 bp) in the wild-type allele, whereas primers B and C amplified a chimeric gene fragment (467 bp) in the mutant allele. In heterozygous animals, both PCR products were generated. (C) Lack of expression of the LGR5 ectodomain-LacZ transgene in LGR5 null mice. (Top panel) Location of two sets of primers used to amplify the ectodomain of LGR5. (Lower panel) Amplification of LGR5 transcripts (a and b primers, 524 bp; c and d primers, 540 bp) in wild type (+/+) but not LGR5 null (−/−) mice. The levels of β-actin serve as positive controls.
For genotyping, genomic DNA was extracted from tail tips of neonates with a genomic DNA extraction kit (Promega, Madison, Wis.). PCR was carried out with three primers (Fig. 1A): upstream primer A (5′-CCTCTTTGCTAAACCTCACC-3′) located in exon 18 of LGR5; the second upstream primer, B (5′-GCAGCGCATCGCCTTCTATC-3′), located at the 3′ end of the targeting vector; and the downstream primer, C (5′-ACGAGTCTTCTCACTATGGG-3′), located at the 3′ end of the targeting vector. Two PCR products were expected: a 673-kb product from wild-type alleles amplified by primers A and C and a 467-kb product from the targeted alleles amplified by primers B and C.
Estimation of LGR5 transcript levels based on real-time RT-PCR.
Total RNA was extracted from different organs by employing the RNAeasy kit (QIAGEN, Valencia, Calif.) before cDNA synthesis using Advantage reverse transcriptase (BD Bioscience, Franklin Lakes, N.J.) and the oligo(dT) primer (BD Bioscience, Franklin Lakes, N.J.). PCR was performed in the SmartCycler (Cepheid, Inc., Sunnyvale, Calif.) according to the manufacturer's protocol. To test if the transgene containing the N-terminal extracellular domain (ectodomain) of LGR5 was expressed in the LGR5 null mice, we performed additional reverse transcription-PCR (RT-PCR) by amplifying the LGR5 ectodomain region with two pairs of primers: sense primers a (5′-CCTCTGCTTCCTAGAAGAGTTAC-3′) and c (5′TCAGTATGAACAACATCAGTCAG-3′) and antisense primers b (5′-CTAGTTCCTTAAGGTTGGAGAGT-3′) and d (5′-TTGCAGTGGGGAATTCATCAAGGTTATTAT-3′). The primers for β-actin were sense primer 5′-GGACCTGACGGACTACCTCATG-3′ and antisense primer 5′-TCTTTGATGTCACGCACGATTT-3′. The following primers and probes were used for real-time PCR: LGR5 sense 5′-CTTCCGAATCGTCGATCTTC-3′ and antisense 5′-AACGATCGCTCTCAGGCTAA-3′, probe 5′-6-carboxy-fluorescein (FAM)-TCACTCTGGCAGCGCTGGAA-6-carboxy-fluorescein (FAM)-CTCTTCTACCTGGCGCTCTGCTTG-6-carboxy-tetramethyl-rhodamine (TAMRA)-3′, β-actin sense 5′-GGACCTGACGGACTACCTCATG-3′ and antisense 5′-TCTTTGATGTCACGCACGATTT-3′, and probe 5′FAM-CCTGACCGAGCGTGGCTACAGCTTC-TAMRA-3′. To determine the copy number of target transcripts, LGR5 and β-actin cDNAs were used to generate calibration curves by plotting the threshold cycle (Ct) versus the known copy number for each plasmid template. The copy numbers for target samples were determined according to the calibration curve. To correct for differences in RNA extraction, data were normalized by dividing the copy number of the target cDNA by that of β-actin.
Expression of recombinant antigenic epitopes for LGR5 and generation of LGR5 antibody.
The primary sequences encoding the ectodomain of human LGR5 cDNA were used to predict their antigenicity (14). A region corresponding to amino acids 22 to 141 of LGR5 was amplified from the plasmid containing full-length LGR5 cDNA and subcloned into the pET21a vector (Novagen, EMD Biosciences, San Diego, Calif.). Primer pairs for the selected epitope were designed as follows: sense 5′-GGCAGCTCTCCCAGGTCT-3′ and antisense 5′-GCAGAATTTGCGAAGCCTTCAA-3′. Expression of recombinant protein was induced in Escherichia coli strain BL21trxB(DE3) (Novagen, EMD Biosciences) with 1 mM isopropyl-1-thio-β-d-galactopyranoside at 37°C for 4 h. Recombinant protein with six-His tag was then purified from the inclusion body dissolved in 8 M urea by metal chelate chromatography. The protein was emulsified in Freund's adjuvant before injection into rabbits to generate polyclonal antibodies to LGR5.
Generation of the ectodomain of LGR5 in eukaryotic cells and immunoblotting analyses.
A eukaryotic cell expression plasmid encoding the entire ectodomain (amino acid residues 22 to 508) of human LGR5 was transfected into human 293T cells. Clonal cell lines stably expressing a recombinant protein encoding the ectodomain of LGR5, named 5BP, were grown in Dulbecco's modified Eagle's medium (DMEM) combined with F-12 medium (DMEM/F-12) with 10% fetal bovine serum. After the cells were confluent, media were replenished with serum-free DMEM/F-12. Three days later, the media were collected, centrifuged, and filtered through 0.22-μm-pore-diameter filters (Corning, Cambridge, Mass.). Conditioned media containing recombinant 5BP were then purified by metal chelate chromatography. To verify the specificity of the LGR5 antibody, conditioned media containing the soluble 5BP were analyzed with immunoblots. Samples were fractionated by electrophoresis in a 7.5% polyacrylamide gel. Some samples were pretreated with N-glycosidase F (New England Biolabs, Inc., Beverly, Mass.) to confirm the glycoprotein nature of 5BP. Proteins were transferred to polyvinylidene difluoride membranes (Hybond-P; Amersham Pharmacia Biotech, Piscataway, N.J.). Subsequently, immunoblotting was performed with rabbit polyclonal antibodies to LGR5 followed by incubation with a horseradish peroxidase-conjugated donkey anti-rabbit immunoglobulin G before immunofluorescent imaging with the ECL enhanced chemiluminescence Western blot system (Amersham).
Gross morphology as well as histological and immunohistochemical analyses.
Newborn pups were euthanized with CO2 and fixed in toto by immersion in Bouin's fixative for 16 h before paraffin embedding. Blocks were sectioned at a 4-μm thickness and stained with hematoxylin and eosin by standard procedures. Immunohistochemical analysis was performed with rabbit polyclonal antibodies against LGR5. Substitution for the primary antibody with rabbit preimmune serum served as the negative control. Staining was performed with the Histostain-SPAEC kit following the manufacturer's instructions (Zymed Laboratories, South San Francisco, Calif.).
Data analysis.
All experimental data are presented as the mean ± standard error. Statistical analysis was performed with Statview 4.5 (Abacus Concepts, Berkeley, Calif.) software. Data were analyzed with the Mann-Whitney U test. Significance was accepted at P < 0.05.
RESULTS
Based on recombinant DNA targeting, the LGR5 gene was disrupted in mice following the replacement of exon 18 encoding the seven-transmembrane region with a transgene containing selection markers (Fig. 1A). ES cells expressing the targeted allele were selected to derive chimeric mice for subsequent generation of LGR5 null mice. Genotyping was performed at birth by multiplex PCR analyses to amplify LGR5 gene fragments with or without the transgene (Fig. 1B). Because our β-galactosidase staining in mutant mice suggested a lack of expression of the transgene encoding the ectodomain of LGR5 fused to LacZ (data not shown), we further performed RT-PCR analyses in neonates to completely rule out the expression of LGR5 transcripts in mutant animals. Using two pairs of primers corresponding to the ectodomain of LGR5, RT-PCR analyses indicated a lack of expression of this region in LGR5 null mice as compared with the high expression in adrenal gland and liver of wild-type littermates (Fig. 1C). Comparable levels of expression of the β-actin transcripts in both mutant and wild-type animals served as positive controls.
Based on the expected Mendelian ratio of wild-type and mutant animals following mating of heterozygous mice, embryonic lethality of LGR5 null mice could be excluded (Table 1). Although no significant difference in body weight was observed at birth among different groups of animals (P > 0.05), all the LGR5 null mice died within 24 h after parturition.
TABLE 1.
Neonatal lethality of LGR5 null micea
| Parameter | Mouse group
|
Total | ||
|---|---|---|---|---|
| Wild type | Heterozygous | Homozygous | ||
| No. of pups | 32 | 74 | 35 | 141 |
| No. of neonatal deaths | 2 | 4 | 35 | 41 |
| Body wt at birth (g) | 1.45 ± 0.18 | 1.48 ± 0.17 | 1.39 ± 0.12 | |
Genotyping was performed for intercrosses of heterozygous mice at the day of birth. In a total of 141 mice from 15 litters, the ratio of wild-type, heterozygous, and LGR5 null mice was 1:2.3:1.1. Although wild-type and heterozygous mice showed limited neonatal lethality (2 of 32 = 6.3% and 4 of 74 = 5.4%, respectively), all 35 of the LGR5 null newborns died within 24 h of birth.
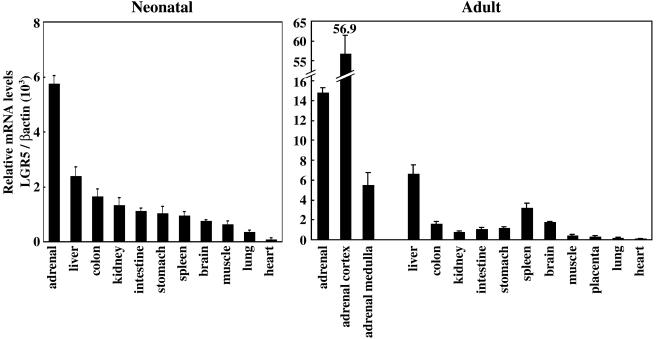
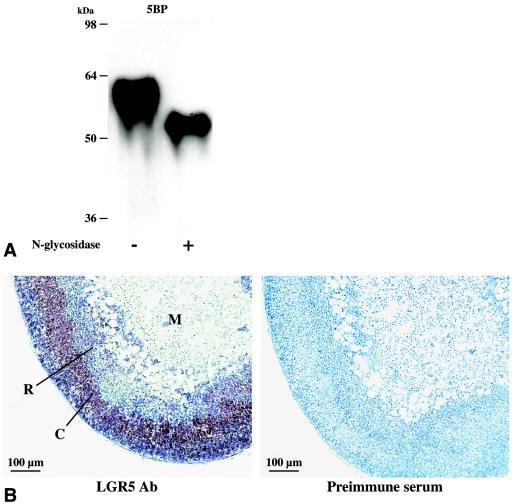
To further understand the role that deletion of LGR5 plays in the death of LGR5 null mice, the expression of LGR5 in diverse tissues was analyzed by performing real-time PCR and immunostaining in wild-type mice. As shown in Fig. 2, LGR5 transcripts were detected in multiple tissues, with adrenal glands and liver showing the highest levels in both neonatal and adult animals. In adrenal glands, the LGR5 transcript was higher in the cortex but significantly lower in the medulla. We further generated a fragment (amino acids 22 to 141) of LGR5 in prokaryotic cells and purified the epitope-tagged protein for the generation of LGR5 antibodies. As shown in Fig. 3A, immunoblotting utilizing this antibody detected the purified recombinant 5BP derived from eukaryotic cells as an N-linked glycoprotein. Using this antibody, immunohistochemical analyses indicated that LGR5 was highly expressed in the outer adrenal cortex, with minimal levels in the medulla and zona reticularis (Fig. 3B). Although the LGR5 gene was expressed in adrenal gland, no difference in the morphology of the gland was found in LGR5 null newborn mice (data not shown). Likewise, no abnormal liver histology was apparent.
FIG. 2.
Tissue expression pattern of LGR5 in mice. Real-time RT-PCR analyses were performed to estimate LGR5 transcript levels in diverse tissues from neonatal (left panel) and adult (right panel) mice. Results are expressed as the ratios between LGR5 and β-actin transcripts for normalization.
FIG. 3.
Characterization of LGR5 antibodies and immunostaining of LGR5 in adrenal gland. (A) Immunoblot analyses of affinity-purified 5BP corresponding to the ectodomain of LGR5. Some samples were pretreated with N-glycosidase to remove N-linked carbohydrate side chains. (B) Immunostaining of LGR5 in the adrenal glands of adult mice. Ab, antibody; C, cortex; M, medulla; R, zona reticularis.
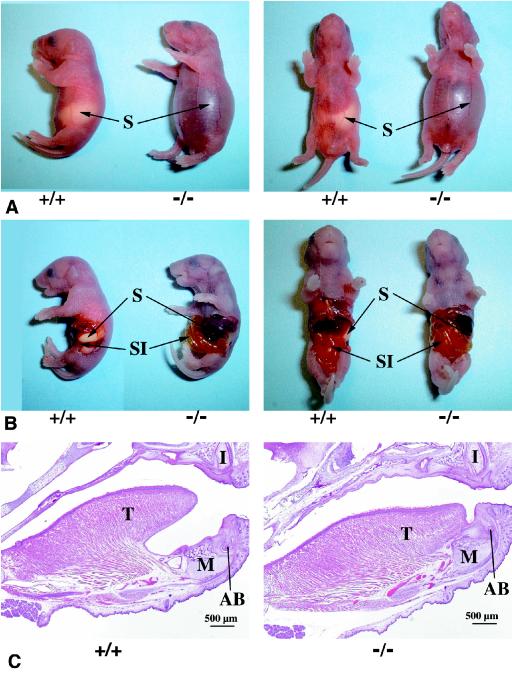
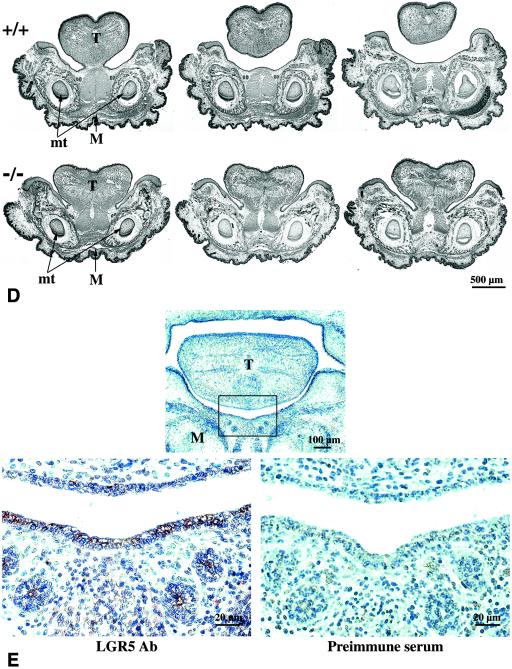
Close examination of the LGR5 null neonates revealed a gradual distension of the abdomen after parturition. As shown in Fig. 4A, wild-type animals had milk in their stomachs but the stomachs of LGR5 null mice were empty and filled with air, presumably as the result of aerophagia associated with a suckling defect. The mutant animals became gradually cyanotic with gasping respirations. By 12 h after birth, the entire gastrointestinal tract had become distended (Fig. 4B). The perinatal lethality of these animals was due partly to respiratory failure as the result of pressure against the diaphragm from the markedly distended abdomen. Because earlier studies in mice linked craniofacial defects such as cleft palate to suckling-related neonatal lethality (1, 6, 7, 21), we searched for possible anatomical abnormalities in LGR5 mutant mice. Although a cleft palate was not found, gross examination of the oral cavity revealed immobilized tongues adherent to the floor of the oral cavity. Histological sagittal sections of the head revealed that the tongue in LGR5 null mice was fused along the entire ventral surface to the floor of the oral cavity, in contrast to the expected conformation as seen in wild-type mice (Fig. 4C). In transverse serial sections of the mandible region, persistent attachment of the tongue proper of the mandible was also apparent (Fig. 4D). To determine if LGR5 proteins were expressed in this region during early development, immunostaining was performed in transverse sections from wild-type embryos at E14.5. As shown in Fig. 4E, LGR5 staining could be found in the epithelium of the tongue and the epithelium and mesenchyme of the mandible.
FIG. 4.
Dilated gastrointestinal tract and abnormal craniofacial development in LGR5 null mice. (A) The wild-type LGR5 (+/+) neonates had milk in their stomachs (S), whereas the LGR5 null (−/−) neonates had dilated stomachs without milk. (B) The entire gastrointestinal tract of LGR5 null mice was dilated without milk, whereas the wild-type mice had a normal appearance. SI, small intestine. (C) In sagittal sections of the craniofacial region, the LGR5 null mice showed fusion of the tongue (T) to the mandible (M), whereas these two regions are separated in the wild-type mice. Identification of similar structures in the upper and lower jaws indicates the sections were taken at the same level. I, developing upper incisor; AB, developing alveolar bone of lower incisor. (D) In transverse serial sections of the mandible region, the tongue of LGR5 null mice was attached to the mandible, whereas the tongue of the wild-type animals in the same region was connected only in a posterior section. Similar sections are reflected by the morphology of the molar teeth (mt), and more anterior sections are shown on the right. (E) Immunostaining of LGR5 antigen in the epithelium of the tongue and the epithelium and mesenchyme of the mandible at E14.5. Ab, antibody. The boxed area in the upper panel is enlarged in the lower panels.
DISCUSSION
The present studies in LGR5 null mice indicated that this orphan receptor is involved in craniofacial development and the mutant mice provide an animal model for the investigation of the genetic basis of ankyloglossia. All LGR5 null mice died within 24 h of birth. This phenotype is similar to that of neonatal mice with a cleft palate. Although for humans, cleft palate is not life-threatening, mice with cleft palate die within 24 h of birth, showing an abdomen dilated with air due to a suckling defect (1, 7, 21). In humans, ankyloglossia is reported to cause breast-feeding difficulties and results in slower weight gain (2, 24). Although there are no reports describing mice with ankyloglossia unaccompanied by cleft palate, the observed neonatal lethality of all LGR5 null mice suggests that ankyloglossia alone could prevent proper suckling in mice.
During embryonic development, the tongue is formed from foregut endoderm and by E13 the distal end of the tongue is freed from the floor of the mouth. Programmed cell death and resorption of the developing skeletal muscle in the ventral anterior region free the tongue, and normally a thin tissue band, the lingual frenulum, remains as the only attachment. Disturbances of this process result in an anteriorily extended and/or shortened frenulum leading to the ankyloglossia phenotype. Findings of ankyloglossia in LGR5 mutant mice together with the expression of LGR5 in the epithelium of the tongue and tissues within the mandible of wild-type animals suggest a role of the LGR5 signaling pathway in proper tongue development. Our immunohistochemical localization of LGR5 in the tongue and the mandible in wild-type mice is consistent with an earlier report based on in situ hybridization analyses (8). At E11.5, LGR5 transcripts were found in the epithelium and mesenchyme overlaying the mandibular cleft. At E13.5, the signal persisted in this region together with signals in the most lateral aspects of the tongue.
Although no cleft palate was found in LGR5 null mice, mutations in a transcriptional factor, TBX22, have been reported in families with both cleft palate and ankyloglossia (3). Of interest, in situ hybridization analyses of Tbx22 expression in mouse embryos indicated that this gene is expressed in the mesenchyme of the inferior nasal septum, the posterior palatal shelf before fusion, the base of the tongue, and lateral region of the mandible (4, 9). Although Tbx22 mutant mice have not been reported, the localization of Tbx22 in the lingual frenulum correlated with the ankyloglossia phenotype in humans. Many different ligand signaling systems have been implicated in craniofacial development. These include several transforming growth factor β (TGF-β) ligands, fibroblast growth factors (FGFs), hedgehog paralogs, wingless genes, platelet-derived growth factors, and endothelin-1 (6). Recent studies further emphasized the involvement of FGF, sonic hedgehog, and TGF-β/BMP ligand signaling systems in palate formation (19). Of interest, both FGF receptor 2b (FGFR2b) and FGF10 null mice exhibited cleft palate and a partial ankylosis of the tongue associated with defective epithelialization between the floor of the mouth and the tongue (22). The present observations of the ankyloglossia phenotype in LGR5 null mice underlie important roles of G protein-coupled receptors during tongue formation and suggest possible interactions of diverse ligand signaling systems during craniofacial development. The present study provides the first mouse model to understand the human ankyloglossia phenotype without associated cleft palate.
In addition to expression in the craniofacial region, LGR5 transcripts have been found in other tissues. Northern blot analysis showed that LGR5 is expressed in skeletal muscle, placenta, spinal cord, and various regions of the brain (12, 17). In situ hybridization analyses further demonstrated the expression of LGR5 in adrenal medulla, male and female gonads, and the olfactory bulb of adult mice (8). The present LGR5 expression analyses, in general, confirmed these earlier reports. However, we detected both LGR5 transcripts and antigens mainly in adrenal cortex, inconsistent with an earlier in situ hybridization analysis showing high LGR5 transcripts in adrenal medulla (8). The basis for these discrepancies is unclear.
In addition to its role in craniofacial formation during embryonic development, LGR5 may also play important roles in adult life. A recent study demonstrated the overexpression of LGR5 in human hepatocellular carcinomas with β-catenin mutations, suggesting that LGR5 may be involved in tumorigenesis (25). Future identification of the cognate ligand for LGR5 and the elucidation of the signaling pathway for this G protein-coupled receptor could provide a better understanding of the molecular mechanisms of tongue development and the physiological roles of this subgroup of LGRs conserved between vertebrates and invertebrates (5, 20).
Acknowledgments
This study was supported by National Institutes of Health grant HD23273 to A.J.W.H.
We gratefully acknowledge Pauline L. Chu for histology processing and C. Spencer for editorial assistance. We thank Lexicon Genetics, Inc. (The Woodlands, Tex.), for providing the mutant animals and the personnel of the Research Animal Facility (Stanford University School of Medicine) for care and monitoring of animals.
REFERENCES
- 1.Asada, H., Y. Kawamura, K. Maruyama, H. Kume, R. G. Ding, N. Kanbara, H. Kuzume, M. Sanbo, T. Yagi, and K. Obata. 1997. Cleft palate and decreased brain gamma-aminobutyric acid in mice lacking the 67-kDa isoform of glutamic acid decarboxylase. Proc. Natl. Acad. Sci. USA 94:6496-6499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berg, K. L. 1990. Tongue-tie (ankyloglossia) and breastfeeding: a review. J. Hum. Lact. 6:109-112. [DOI] [PubMed] [Google Scholar]
- 3.Braybrook, C., K. Doudney, A. C. Marcano, A. Arnason, A. Bjornsson, M. A. Patton, P. J. Goodfellow, G. E. Moore, and P. Stanier. 2001. The T-box transcription factor gene TBX22 is mutated in X-linked cleft palate and ankyloglossia. Nat. Genet. 29:179-183. [DOI] [PubMed] [Google Scholar]
- 4.Bush, J. O., Y. Lan, K. M. Maltby, and R. Jiang. 2002. Isolation and developmental expression analysis of Tbx22, the mouse homolog of the human X-linked cleft palate gene. Dev. Dyn. 225:322-326. [DOI] [PubMed] [Google Scholar]
- 5.Eriksen, K. K., F. Hauser, M. Schiott, K. M. Pedersen, L. Sondergaard, and C. J. Grimmelikhuijzen. 2000. Molecular cloning, genomic organization, developmental regulation, and a knock-out mutant of a novel leu-rich repeats-containing G protein-coupled receptor (DLGR-2) from Drosophila melanogaster. Genome Res. 10:924-938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Francis-West, P., R. Ladher, A. Barlow, and A. Graveson. 1998. Signalling interactions during facial development. Mech. Dev. 75:3-28. [DOI] [PubMed] [Google Scholar]
- 7.Halford, M. M., J. Armes, M. Buchert, V. Meskenaite, D. Grail, M. L. Hibbs, A. F. Wilks, P. G. Farlie, D. F. Newgreen, C. M. Hovens, and S. A. Stacker. 2000. Ryk-deficient mice exhibit craniofacial defects associated with perturbed Eph receptor crosstalk. Nat. Genet. 25:414-418. [DOI] [PubMed] [Google Scholar]
- 8.Hermey, G., A. Methner, H. C. Schaller, and I. Hermans-Borgmeyer. 1999. Identification of a novel seven-transmembrane receptor with homology to glycoprotein receptors and its expression in the adult and developing mouse. Biochem. Biophys. Res. Commun. 254:273-279. [DOI] [PubMed] [Google Scholar]
- 9.Herr, A., D. Meunier, I. Muller, A. Rump, R. Fundele, H. H. Ropers, and U. A. Nuber. 2003. Expression of mouse Tbx22 supports its role in palatogenesis and glossogenesis. Dev. Dyn. 226:579-586. [DOI] [PubMed] [Google Scholar]
- 10.Hsu, S. Y. 2003. New insights into the evolution of the relaxin-LGR signaling system. Trends Endocrinol. Metab. 14:303-309. [DOI] [PubMed] [Google Scholar]
- 11.Hsu, S. Y., M. Kudo, T. Chen, K. Nakabayashi, A. Bhalla, P. J. van der Spek, M. van Duin, and A. J. Hsueh. 2000. The three subfamilies of leucine-rich repeat-containing G protein-coupled receptors (LGR): identification of LGR6 and LGR7 and the signaling mechanism for LGR7. Mol. Endocrinol. 14:1257-1271. [DOI] [PubMed] [Google Scholar]
- 12.Hsu, S. Y., S. G. Liang, and A. J. Hsueh. 1998. Characterization of two LGR genes homologous to gonadotropin and thyrotropin receptors with extracellular leucine-rich repeats and a G protein-coupled, seven-transmembrane region. Mol. Endocrinol. 12:1830-1845. [DOI] [PubMed] [Google Scholar]
- 13.Hsu, S. Y., K. Nakabayashi, S. Nishi, J. Kumagai, M. Kudo, O. D. Sherwood, and A. J. Hsueh. 2002. Activation of orphan receptors by the hormone relaxin. Science 295:671-674. [DOI] [PubMed] [Google Scholar]
- 14.Jameson, B. A., and H. Wolf. 1988. The antigenic index: a novel algorithm for predicting antigenic determinants. Comput. Appl. Biosci. 4:181-186. [DOI] [PubMed] [Google Scholar]
- 15.Kumagai, J., S. Y. Hsu, H. Matsumi, J. S. Roh, P. Fu, J. D. Wade, R. A. Bathgate, and A. J. Hsueh. 2002. INSL3/Leydig insulin-like peptide activates the LGR8 receptor important in testis descent. J. Biol. Chem. 277:31283-31286. [DOI] [PubMed] [Google Scholar]
- 16.Lalakea, M. L., and A. H. Messner. 2003. Ankyloglossia: does it matter? Pediatr. Clin. N. Am. 50:381-397. [DOI] [PubMed] [Google Scholar]
- 17.McDonald, T., R. Wang, W. Bailey, G. Xie, F. Chen, C. T. Caskey, and Q. Liu. 1998. Identification and cloning of an orphan G protein-coupled receptor of the glycoprotein hormone receptor subfamily. Biochem. Biophys. Res. Commun. 247:266-270. [DOI] [PubMed] [Google Scholar]
- 18.Moore, G. E. 1995. Molecular genetic approaches to the study of human craniofacial dysmorphologies. Int. Rev. Cytol. 158:215-277. [DOI] [PubMed] [Google Scholar]
- 19.Murray, J. C., and B. C. Schutte. 2004. Cleft palate: players, pathways, and pursuits. J. Clin. Investig. 113:1676-1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nishi, S., S. Y. Hsu, K. Zell, and A. J. Hsueh. 2000. Characterization of two fly LGR (leucine-rich repeat-containing, G protein-coupled receptor) proteins homologous to vertebrate glycoprotein hormone receptors: constitutive activation of wild-type fly LGR1 but not LGR2 in transfected mammalian cells. Endocrinology 141:4081-4090. [DOI] [PubMed] [Google Scholar]
- 21.Proetzel, G., S. A. Pawlowski, M. V. Wiles, M. Yin, G. P. Boivin, P. N. Howles, J. Ding, M. W. Ferguson, and T. Doetschman. 1995. Transforming growth factor-beta 3 is required for secondary palate fusion. Nat. Genet. 11:409-414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rice, R., B. Spencer-Dene, E. C. Connor, A. Gritli-Linde, A. P. McMahon, C. Dickson, I. Thesleff, and D. P. Rice. 2004. Disruption of Fgf10/Fgfr2b-coordinated epithelial-mesenchymal interactions causes cleft palate. J. Clin. Investig. 113:1692-1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sudo, S., J. Kumagai, S. Nishi, S. Layfield, T. Ferraro, R. A. Bathgate, and A. J. Hsueh. 2003. H3 relaxin is a specific ligand for LGR7 and activates the receptor by interacting with both the ectodomain and the exoloop 2. J. Biol. Chem. 278:7855-7862. [DOI] [PubMed] [Google Scholar]
- 24.Wilton, J. M. 1990. Sore nipples and slow weight gain related to a short frenulum. J. Hum. Lact. 6:122-123. [DOI] [PubMed] [Google Scholar]
- 25.Yamamoto, Y., M. Sakamoto, G. Fujii, H. Tsuiji, K. Kenetaka, M. Asaka, and S. Hirohashi. 2003. Overexpression of orphan G-protein-coupled receptor, Gpr49, in human hepatocellular carcinomas with beta-catenin mutations. Hepatology 37:528-533. [DOI] [PubMed] [Google Scholar]