Abstract
Carbonic anhydrase III is a cytosolic protein which is particularly abundant in skeletal muscle, adipocytes, and liver. The specific activity of this isozyme is quite low, suggesting that its physiological function is not that of hydrating carbon dioxide. To understand the cellular roles of carbonic anhydrase III, we inactivated the Car3 gene. Mice lacking carbonic anhydrase III were viable and fertile and had normal life spans. Carbonic anhydrase III has also been implicated in the response to oxidative stress. We found that mice lacking the protein had the same response to a hyperoxic challenge as did their wild-type siblings. No anatomic alterations were noted in the mice lacking carbonic anhydrase III. They had normal amounts and distribution of fat, despite the fact that carbonic anhydrase III constitutes about 30% of the soluble protein in adipocytes. We conclude that carbonic anhydrase III is dispensable for mice living under standard laboratory husbandry conditions.
The mammalian carbonic anhydrases reversibly hydrate carbon dioxide, thus generating both bicarbonate and hydrogen ions for maintenance of pH homeostasis (30, 34). At least 15 different mammalian proteins with carbonic anhydrase structure are known, 11 of which are catalytically active (16). The existence of multiple isozymes underscores the importance of the reaction in a variety of physiologic functions including acid-base balance, respiration, urinary acidification, and bone resorption (5). The isozymes vary in developmental expression, tissue distribution, and subcellular location. Carbonic anhydrase isozyme III (Car3) has several characteristics which distinguish it from the other isozymes, especially its low specific activity, which is only ∼3% of that of Car2 (13, 14). Car3 had been thought to also possess intrinsic tyrosine phosphatase activity, but that was subsequently found to be due to a contaminating phosphatase (12).
The enzyme is remarkably abundant in skeletal muscle (3) and adipocytes (31), constituting up to 8 and 25% of the soluble fraction of these tissues. Car3 expression is negligible in preadipocytes, becoming substantial upon differentiation (21), but the mechanism of differentiation-dependent Car3 expression is not understood. Despite the notable abundance of Car3 in fat and muscle, its function is unknown, although it has been implicated in fatty acid metabolism (22). Car3 could facilitate rapid conversion of glycolytic intermediates to oxaloacetate and citrate and stimulate their incorporation into fatty acids. However, adipocyte Car3 expression in obese mice is lower than that in lean mice (21, 32). Exposure of differentiated mouse adipocytes to insulin decreased Car3 expression by 90% while expression of Car2 was unchanged (20).
Car3 has two reactive sulfhydryl groups which can conjugate to glutathione through a disulfide link, a process termed S-glutathionylation (4, 19). Car3 is rapidly glutathionylated in vivo and in vitro when cells are exposed to oxidative stresses, and it is one of the most carbonylated proteins in rodent liver. These observations have led to the suggestion that the enzyme plays a role in the cellular response to oxidative stresses, including reperfusion injury and aging (2, 35). Expression of Car3 in cells lacking the protein protects them from hydrogen peroxide-induced apoptosis, while expression of the closely related Car2 does not (24).
We utilized targeted gene deletion to create a mouse lacking Car3, for the purpose of elucidating the normal function of Car3. We now report studies which characterize this knockout mouse.
MATERIALS AND METHODS
Targeted disruption of Car3.
Genomic DNA including Car3 was isolated from a mouse 129SvJ genomic library (Stratagene, La Jolla, Calif.), and the genomic organization of Car3 was determined by restriction digestions and sequencing as described previously (11). The Car3 gene targeting construct (Fig. 1A) was generated by subcloning a 4.0-kb NotI/XbaI genomic fragment containing the 5′ end of the locus and a 6.7-kb SpeI/NheI fragment containing the 3′ end in a pPNT backbone (36). The resulting vector replaces the 4.2-kb XbaI/SpeI fragment containing exons 3, 4, and 5 of Car3 with a PGKneo gene.
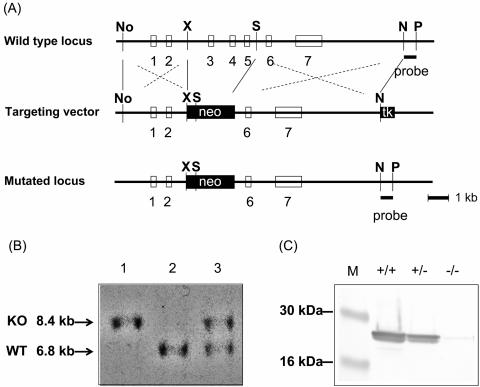
FIG. 1.
Targeted disruption of Car3. (A) Gene deletion strategy. Exons are depicted as open boxes. Filled boxes are selectable markers containing the genes for neomycin resistance (neo) and thymidine kinase (tk). Exons 3, 4, and 5 were replaced with a neomycin resistance gene. Abbreviations for the restriction sites: No, NotI; X, XbaI; S, SpeI: N, NheI; P, PmeI. (B) Southern blot analysis of genomic DNA. Genomic DNA was prepared from littermates obtained by mating heterozygotes. SpeI/PmeI-cleaved genomic DNA from a wild-type mouse (WT) and a mutant mouse (KO) was hybridized to the probe, distinguishing the wild-type allele (6.8 kb) from the targeted allele (8.4 kb). (C) Western blot analysis of liver from 6-month-old male mice. Each lane contained 20 μg of total protein. Car3 was reduced in the heterozygote and absent in the homozygote. M denotes molecular mass standards.
The targeting vector was linearized with NotI and transfected into TC1 embryonic stem cells by electroporation as described previously (7). One hundred eighteen colonies resistant to Geneticin were analyzed for homologous recombination by Southern blotting and PCR. Embryonic stem cell genomic DNA was isolated by proteinase K digestion (100 μg/ml) in 100 mM Tris-5 mM EDTA-200 mM sodium chloride-0.2% sodium dodecyl sulfate (SDS), pH 8.0. Five micrograms of SpeI/PmeI-digested genomic DNA was separated on an 0.6% agarose gel. After transfer to a Hybond-N+ nylon membrane (Amersham), hybridization was performed with the flanking sequence probe, an 0.5-kb NheI/PmeI fragment, representing the region 3′ of the genomic sequence (Fig. 1A). For genotyping by PCR, primers M29F and M11R were generated as described previously (11). Eight clones were selected and verified by PCR and extensive restriction analysis with the use of flanking probes to demonstrate that the locus was targeted correctly. Two clones were injected into C57BL/6J blastocysts and implanted into (CBA × C57BL/6J)F1 foster mothers. Male chimeras from two clones selected by agouti color were mated to C57BL/6 females to confirm germ line transmission. Agouti offspring were tested for the targeted Car3 allele by Southern blotting or PCR of tail DNA. Chimeric males were bred to wild-type 129SvEv females to generate mice heterozygous for the Car3 deletion in a pure 129SvEv background.
Anatomical examinations.
Detailed gross and histological examinations were performed on two male and two female knockout mice, plus one wild-type mouse of each sex. All of the mice were well hydrated and well muscled and contained ample body fat. The heart, liver, and kidneys of each mouse were weighed. Representative samples of the following organs or tissues were obtained for histopathology: brain, liver, spleen, pancreas, kidneys, urinary bladder, adrenal glands, heart, lung, thyroid gland, salivary gland, submandibular lymph node, thymus, eyes, hind limb skeletal muscle, tongue, stomach, small intestine, cecum, colon, skin, bone marrow, brown adipose tissue, testis, epididymis, ovary, and uterus. Tissues were fixed in 10% neutral buffered zinc formalin (Z-fix; Anatech, Battle Creek, Mich.) and processed for routine histologic examination at 6 μm. Tissues were stained with hematoxylin and eosin.
For electron microscopic studies, samples of liver, brown fat, white fat, and skeletal muscle (gastrocnemius and soleus) from Car3+/+ and Car3−/− mice were fixed in 2.5% glutaraldehyde-0.1 M cacodylate buffer and postfixed in 1% osmium tetroxide. Tissues were dehydrated through alcohol and propylene oxide and then embedded in Eponate 12 resin (Ted Pella, Redding, Calif.). Thin sections were prepared with a Leica Ultracut UCT ultramicrotome and stained with uranyl acetate and lead citrate. Sections were examined under a Philips 201 electron microscope.
Kaplan-Meier survival analysis was calculated with the WinSTAT plug-in for Microsoft Excel (Robert Finch Software, Calumet City, Ill.).
Biochemical and physiological analyses.
Fatty acid analysis of whole-tissue extracts was performed in the Laboratory of Membrane Biochemistry and Biophysics, National Institute on Alcohol Abuse and Alcoholism, by gas chromatography-mass spectrometry as described previously (23). Tissue carbonyl content was determined both spectrophotometrically and by Western blotting of SDS-polyacrylamide gels (18) Carbonic anhydrase activity was determined as described previously (12).
Western blots were performed with antisera to recombinant Car3 (12). Tissues were homogenized in Nonidet P-40 lysis buffer (1% Nonidet P-40, 25 mM Tris-HCl [pH 7.5], 140 mM NaCl, 1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, 1 μg of aprotinin/ml, 1 μg of leupeptin/ml, 1 μg of pepstatin/ml, and 1 mM sodium vanadate). Proteins in the lysates were separated by electrophoresis on SDS-10% polyacrylamide gels and then transferred to nitrocellulose membranes (Millipore, Bedford, Mass.). After being blocked with 5% nonfat milk in phosphate-buffered saline (150 mM NaCl, 8 mM Na2HPO4, 2 mM KH2PO4, 3 mM KCl, pH 7.4) containing 0.1% Tween 20, the filters were incubated with Car3 antibody, followed by incubation with anti-rabbit immunoglobulin G coupled to horseradish peroxidase (Kirkegaard & Perry Laboratories, Gaithersburg, Md.).
Contractile parameters were determined for isolated whole soleus muscle (9) and in fiber bundles of soleus muscle as described previously (37). Briefly, muscles or fiber bundles were incubated in modified Krebs-Henseleit solution (120 mM NaCl, 4 mM KCl, 1 mM KH2PO4, 1.2 mM MgSO4, 25 mM NaHCO3, 1 mM CaCl2, and 5.5 mM glucose); equilibrated with 95% O2-CO2, pH 7.4, at 25°C; and directly stimulated under isometric conditions at optimal length with supramaximal voltage. Single-twitch and tetanic contractions (50 Hz, 1.5 s) were analyzed for tension and contraction times. The fatigue protocol for fiber bundles elicited one 1.2-s tetanic contraction at 15 Hz every 5 s for 45 min. At the end of the fatigue protocol, myosin heavy chain content was analyzed (15). A hyperoxia challenge (8) was administered by placing the caged mice, with food and water, inside a large AtmosBag (Sigma-Aldrich) previously flushed with oxygen. After the bag was taped closed, oxygen was bled into the bag at a constant flow to maintain the oxygen concentration at ≥99%, monitored by a MiniOx I oxygen analyzer (Catalyst Research, Pittsburgh, Pa.). Animals were observed at least every 1 to 2 h; moribund animals were removed and killed.
Exercise tolerance was assessed on a motor-driven treadmill (Columbus Instruments, Columbus, Ohio). Three 12-month-old male mice of each genotype were familiarized with the treadmill the day before formal testing by running them twice at 0.2 m/min for 5 min. The next day they were then tested at a treadmill speed of 0.3 m/min, running until they failed to move despite an electrical stimulus. Each mouse was tested five different times, and the results were averaged to give the endurance time for the mouse.
Microarray analyses. (i) RNA and array preparation.
RNA was prepared from three male and three female wild-type and knockout mice by extraction with phenol-guanidine isothiocyanate (5) (Trizol reagent; Invitrogen, Carlsbad, Calif.). The aqueous phase was brought to 70% ethanol, applied to an RNeasy Midi spin column, and processed according to the directions of the manufacturer (Qiagen, Valencia, Calif.). Brain, kidney, liver, soleus (a type I or slow-twitch muscle), and gastrocnemius (a type II or fast-twitch muscle) were separately extracted from each of the 12 mice, but because of the small yield from the soleus, samples were combined from the three animals within each sex-genotype group. Six arrays (three typical and three dye-flipped) were hybridized for each sex and tissue type (four for soleus). Evaluation and analysis of the large volume of data generated by this number of arrays required additional techniques, described below.
Microarrays were prepared in the Microarray Research Facility of the National Institute of Allergy and Infectious Diseases, printed as described by the Brown laboratory (http://cmgm.stanford.edu/pbrown/protocols/index.html). The 13,443 oligonucleotide probes were 70-mers (Mouse Genome Oligo Set Version 1.0; Qiagen); the corresponding genes are listed at http://oligos.qiagen.com/arrays/omad.php and include carbonic anhydrases 1, 2, 3, 4, 5a, 5b, 6, 7, 13, 14, and 15 and carbonic anhydrase-like 1. Total RNA (20 μg) was labeled with Cy3 or Cy5 with the Fairplay Microarray labeling kit (Stratagene). Hybridizations were performed with a procedure adapted from the work of Hegde and colleagues (10). The microarrays were prehybridized for 1 h at 42°C in a buffer of 750 mM sodium chloride-750 mM sodium citrate-1% bovine serum albumin-0.1% SDS at pH 7.0. The slides were washed twice with water and once with isopropanol and then dried by centrifugation at 1,000 × g for 3 min (Beckman Allegra 6 centrifuge; Fullerton, Calif.). The labeled targets were brought to 30 ml in 10 mM Tris-1 mM EDTA, pH 8.0, to which was added 4 μg of yeast tRNA (Sigma, St. Louis, Mo.), 1 μg of poly(dA)40-60 (Amersham Pharmacia, Piscataway, N.J.), and 10 μg of human Cot-1 DNA (Invitrogen). The targets were denatured by being heated to 98°C for 2 min and then mixed with 33 μl of hybridization buffer (1.5 M sodium chloride, 1.5 M sodium citrate, 0.2% SDS, pH 7.0). Hybridization was carried out in a sealed, humidified chamber submerged in a water bath at 42°C for 14 to 18 h.
(ii) Measurement of change in expression.
Fluorescence scanning and image analysis were performed using Axon Gene Pix Pro software (Axon Instruments, Union City, Calif.). Signal values for each channel were taken as the difference between the fluorescence pixel mean and the background pixel median. In cases where the pixel mean for a spot did not exceed the calculated background mean by more than twice the background pixel standard deviation, the raw (non-background-subtracted) fluorescence value was used. Expression change was calculated for each spot as log (base 2) of the ratio of signal in the two channels. A value of 2 is a fourfold increase in expression relative to the reference sample. A value of −2 conveys a decrease in expression to one-fourth of that in the reference sample.
(iii) Normalization and evaluation.
For each array, overall signal in both channels was balanced by nonlinear regression with the Loess normalization. Proc Loess (SAS Institute, Cary, N.C.) was used with the following parameters to fit expression change versus the average signal for both channels: smooth = 0.7, degree = 1, and iterations = 10. This method seeks to balance the two fluorescence channels by setting the average expression change to zero at every level of average signal. Utilizing the assumption that the overwhelming majority of spots will have expression change equal to zero, the method seeks to correct for bias due to differences in dye performance and other systematic inequalities between the two fluorescence channels, whether or not the magnitude of the inequality is dependent on the magnitude of the fluorescence signal.
Each set of matching spots (that is, the same gene over all arrays) was also checked for dye bias by testing for a difference in expression change when the Cy3-Cy5 dye used on the reference sample differed (“dye flip” trials). By design, this is a comparison of three arrays to another three arrays. In some cases fewer than three qualifying values are available per group.
Expression change was detected by testing against the null hypothesis that there was no difference of the sample from the reference sample by using a one-sample t test with Proc Univariate (SAS). A “volcano plot” convention was used to display the results (38).
RESULTS
Disruption of the Car3 gene by homologous recombination.
The gene targeting strategy was successful, as demonstrated by Southern, Northern, and Western blotting (Fig. 1). We genotyped 870 pups born from heterozygous matings to assess whether the expected number of knockout and heterozygote pups was affected. The ratio of Car3+/+ to Car3+/− to Car3−/− was 23:51:26, as expected for unbiased Mendelian inheritance. The ratios were similar for males and females. No differences in fertility or in litter size were noted for either male or female mice.
Growth and life span.
We determined the postnatal growth patterns for the mice as an integrated assessment of biological health and also because Car3 is particularly rich in adipocytes. Sex-specific growth curves of the wild-type, heterozygous, and knockout animals were superimposable through 80 days of life, when measurements were stopped. We recorded age at death of animals allowed to reach their life span. No effect on life span was observed (Fig. 2).
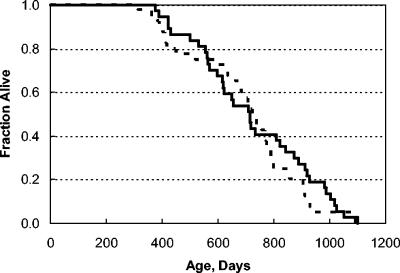
FIG. 2.
Kaplan-Meier survival analysis. No difference in survival of male and female mice was detected, so they were combined for the analysis shown here. We monitored a cohort of 36 Car+/+ (broken line) and 40 Car−/− (solid line) mice. There was no difference between the groups (P > 0.4).
Histological and biochemical examinations.
Detailed gross and histological examinations were also performed as described in Materials and Methods. No difference in organ weights was noted, and knockout mice had a normal distribution and amount of fat. Light and electron microscopic examination of adipocytes, liver, and both slow- and fast-twitch muscle revealed no differences among the genotypes. No significant lesions were found in any of the mice, except for mild multifocal, unilateral testicular mineralization in one of the Car3-deficient mice. The distribution of fatty acids in serum, muscle, liver, and fat was analyzed by gas chromatography-mass spectrometry for two male wild-type and two knockout mice, but it was not changed by deletion of Car3 (data not shown). Myosin heavy chain I is found in slow-twitch muscles, which are also rich in Car3. We determined its content in the soleus muscle from each of six Car3+/+ and Car3−/− animals and found no significant difference (45% ± 8% versus 45% ± 11%). Myosin heavy chain I was absent, and myosin heavy chain IId comprised 100 and 92% of myosin heavy chain from extensor digitorum longus muscle of Car3+/+ and Car3−/− mice, respectively.
Effect of stress.
As mentioned above, Car3 is abundant in muscle, especially slow-twitch muscles such as the soleus. Habitus and gait were normal to our observation. We then attempted to uncover a possible role for the enzyme by testing the animals' endurance with a treadmill test. As shown in Table 1, knockout mice performed as well as did their wild-type littermates.
TABLE 1.
Treadmill endurance testa
| Genotype | Mean running time (min) | SD |
|---|---|---|
| Car3+/+ | 8.9 | 1.0 |
| Car3+/− | 8.8 | 0.6 |
| Car3−/− | 8.2 | 1.5 |
The groups did not differ, as assessed with the unpaired t test (P > 0.4).
Car3 may have a function in defense against oxidative stress, as the protein is known to have one of the greatest extents of oxidative modification (2). We therefore tested the response of the mice to exposure to ∼100% oxygen. Adult rodents typically die with pulmonary edema after 3 to 4 days of exposure, but other organs including brain and liver demonstrate oxidative damage within the first day of exposure (33). No difference was detected in time of survival between the wild-type and Car3-knockout animals (Table 2).
TABLE 2.
Hyperoxia challengea
| Genotype | Mean survival (h) | SD |
|---|---|---|
| Car3+/+ | 66.1 | 6.7 |
| Car3+/− | 74.2 | 6.0 |
| Car3−/− | 67.9 | 5.4 |
Survival of the wild type and homozygous knockouts did not differ, as assessed with the unpaired t test (P > 0.5). The heterozygotes did survive slightly longer than the wild type did (P = 0.02) or the homozygous knockouts (P = 0.04) did.
If Car3 has a role in oxidative defense, then mice lacking the protein may carry an increased burden of oxidatively damaged molecules. The carbonyl content of cellular proteins provides an integrated assessment of this cellular burden (17). We quantitated the total carbonyl content in male and female mice with the three genotypes Car3+/+, Car3+/−, and Car3−/−. No significant difference was found among the three groups in brain, fat, heart, kidney, liver, lung, muscle, ovary, spleen, testes, or uterus. We also used the Western blot method for carbonyl (18). While semiquantitative, the method can detect changes in specific protein bands which could be missed when determining the total content. While no differences were found among the three genotypes in any tissue, the methodology lacks the sensitivity to detect substantive changes in oxidation of low-abundance proteins.
Contractile properties of isolated muscle.
The contractile properties of whole soleus muscle, rich in Car3, were compared between wild-type and Car3-knockout animals (Table 3). The Car3−/− muscles had shorter half-relaxation times for both single and tetanic twitches. Car3−/− solei had a minor reduction of tetanic force when normalized to their cross-sectional area. Soleus fiber bundles from Car3−/− mice also exhibited shorter relaxation times for both single-twitch and tetanic contractions (data not shown). Cote and colleagues reported that inhibition of Car3 by millimolar concentrations of methazolamide rendered mouse soleus muscle more resistant to fatigue (6). We therefore tested the fatigability of soleus muscle from our mice. Fatigue was induced in both whole soleus and isolated fiber bundles (Fig. 3); there was no difference in fatigability between Car3+/+ and Car3−/− animals.
TABLE 3.
Contractile parameters of whole soleus musclea
| Parameter | Car3+/+ | Car3−/− | P |
|---|---|---|---|
| No. of muscles studied | 8 | 7 | |
| Area (cm2) | 0.005 ± 0.001 | 0.005 ± 0.001 | Not significant |
| Weight (mg) | 7.1 ± 1.0 | 7.1 ± 1.5 | Not significant |
| Twitch tension (N/cm2) | 2.7 ± 0.6 | 2.2 ± 0.5 | Not significant |
| Time to peak (ms) | 40.5 ± 5.0 | 37.1 ± 3.4 | Not significant |
| Half-relaxation time (ms) | 50 ± 9 | 40 ± 3 | P < 0.05 |
| Tetanic tension (N/cm2) | 18.2 ± 2.0 | 14.5 ± 2.7 | P < 0.05 |
| Half-relaxation time (ms) | 97 ± 12 | 78 ± 8 | P < 0.05 |
| Twitch tension/tetanic tension ratio | 0.15 ± 0.03 | 0.15 ± 0.01 | Not significant |
Results are given as the means ± standard deviations. Probability was calculated with the unpaired t test. The animals were 26 ± 10 weeks of age. Muscles were immersed in Krebs-Henseleit bicarbonate solution equilibrated with 95% O2-5% CO2 at 25°C.
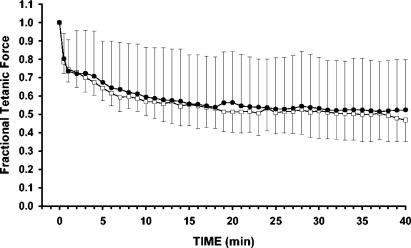
FIG. 3.
Fatigability for soleus fiber bundles from Car3−/− (□) and Car3+/+ (•) mice. The results are shown as the fraction of force obtained before starting the fatigue protocol. Under the conditions described in Table 1, muscle fibers were subjected to a series of tetanic contractions elicited every 5 s for 40 min, with a stimulation frequency of 15 Hz. The means and standard deviations are plotted.
Microarray analyses.
We considered that hypotheses for the functional role of Car3 might be deduced from comparison of the gene expression patterns of wild-type and knockout animals by microarray analysis. To improve the robustness of the analysis, we performed the microarray analyses on multiple tissues from both male and female mice, as detailed in Materials and Methods. When data from all analyses were combined and analyzed as described in Materials and Methods, only a single change was detected and that was the expected loss of expression of Car3 in the knockout animals. These microarray analyses were performed on mRNA obtained from animals raised under standard husbandry conditions. It remains to be determined whether exposure to specific stresses would lead to differential responses.
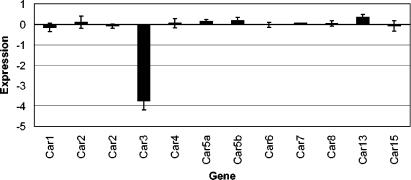
The microarray included probes for 10 other mouse carbonic anhydrase isozymes (Fig. 4). A statistically significant increase in expression of Car13 was detected, although the increase was only ∼30% ± 10%. Whether the Car13 protein and activity are changed and what the physiological impact is remain to be investigated, particularly as the first characterization of Car13 has just been published (16).
FIG. 4.
Relative expression of carbonic anhydrase genes. The bars give the expression in the knockout Car−/− animals relative to that in the wild-type Car+/+ mice, measured by microarray. The expression value is the average of the log (base 2) ratio of Car−/− to Car+/+ as described in Materials and Methods. Each mean was calculated from arrays run on 17 liver samples (6 male, 11 female). Error bars show the standard errors of the means. Student's t test revealed statistically significant changes for Car3 (P < 10−6) and for Car13 (P < 0.02), although the actual increase in Car13 was only about 30% ± 10%.
DISCUSSION
In 1972 Noltmann and colleagues purified “basic muscle protein” from rabbit muscle, which constituted 1 to 2% of the total protein extracted with their protocol (1). After identifying it as Car3, they commented, “It was inconceivable at the time that a major component of the solubilizable protein of skeletal muscle had not yet been identified” (25). More than 30 years later the function of that protein remains an enigma. Mice lacking Car3 develop normally, are fertile, and have a normal life span under current husbandry conditions.
Lack of Car3 in the slow muscle soleus does not affect the fiber type composition, as assessed by analysis of myosin heavy chain I. Important contraction parameters such as time-to-peak and single-twitch force are unaltered, suggesting that the major muscular functions, the calcium release mechanism, and the contractile apparatus are not affected. It is not clear why the half-relaxation times of single twitches and of tetani are reduced. The observation is difficult to interpret because these relaxation times are affected by a multitude of cellular parameters, including pH, phosphate concentration, ADP level, and others. We conclude that, although minor disturbances may have occurred in the muscle cell, its major properties appear unchanged.
Given the large number of carbonic anhydrases, it may be that other isozymes are sufficient for normal cellular function when Car3 is lacking. While possible, this is somewhat unattractive as an explanation because the specific activity of Car3 as a carbonic anhydrase is so low, being ∼3% of that of Car2. Furthermore, loss of Car2 has a profound phenotype in humans, causing Guibaud-Vainsel syndrome or marble brain disease, a syndrome which includes osteopetrosis, renal tubular acidosis, and cerebral calcification (29).
Alternatively, it may be that Car3 is required for an effective response to specific stimuli or stresses which mice do not encounter in the laboratory animal facility. This consideration led us to compare wild-type and Car3-deficient mice on the exercise treadmill and upon exposure to hyperoxia. These stresses did not reveal differences between the genotypes. We suspect that there will be stresses to which the knockout animals respond poorly, although we have not yet identified them.
As implied above when we noted its low specific activity, Car3 may not actually function as a carbonic acid anhydrase. Identification of a new catalytic activity acquired by Car3 would likely provide insight into the protein's physiological role. Although entirely speculative, an intriguing possibility is that Car3 has evolved into a percarbonic acid anhydrase. While carbonic anhydrase mediates the reaction of carbon dioxide with water, H2O + CO2 ⇆ H2CO3, a percarbonic anhydrase would mediate the reaction of carbon dioxide with hydrogen peroxide, H2O2 + CO2 ⇆ H2CO4. Richardson and colleagues have studied the chemistry of this reaction and shown that, under their solvent conditions, percarbonic acid is a powerful oxidizing agent which could readily oxidize methionine residues of proteins to methionine sulfoxide (26, 27).
Whether acting as a percarbonic anhydrase or as another enzyme, Car3 expression decreased the level of reactive oxygen species when expressed in NIH 3T3 cells and protected them against cytotoxic concentrations of hydrogen peroxide (24). Thus, Car3 may function as a member of the defense system against oxidative stresses, a function which could explain the large fraction of Car3 which is itself oxidatively modified in vivo (2, 28).
Acknowledgments
We thank Norman Salem and James Loewke of the Laboratory of Membrane Biochemistry and Biophysics, National Institute on Alcohol Abuse and Alcoholism, for performing the fatty acid analyses. We are grateful to Michael Wilson of the Research Technology Branch, National Institute of Allergy and Infectious Diseases, for guiding the microarray analyses. We thank Hans-Peter Kubis of the Zentrum Physiologie at the Medizinische Hochschule Hannover for the electrophoretic analyses of muscle myosin.
REFERENCES
- 1.Blackburn, M. N., J. M. Chirgwin, G. T. James, T. D. Kempe, T. Parsons, A. M. Register, K. D. Schnackerz, and E. A. Noltmann. 1972. Pseudoisoenzymes of rabbit muscle phosphoglucose isomerase. J. Biol. Chem. 247:1170-1179. [PubMed] [Google Scholar]
- 2.Cabiscol, E., and R. L. Levine. 1995. Carbonic anhydrase III. Oxidative modification in vivo and loss of phosphatase activity during aging. J. Biol. Chem. 270:14742-14747. [DOI] [PubMed] [Google Scholar]
- 3.Carter, N. D. 1991. Hormonal and neuronal control of carbonic anhydrase III gene expression in skeletal muscle, p. 247-256. In S. J. Dodgson, R. E. Tashian, G. Gross, and N. D. Carter (ed.), The carbonic anhydrases: cellular physiology and molecular genetics. Plenum Publishing Corp., New York, N.Y.
- 4.Chai, Y. C., C.-H. Jung, C.-K. Lii, S. S. Ashraf, S. Hendrich, B. Wolf, H. Sies, and J. A. Thomas. 1991. Identification of an abundant S-thiolated rat liver protein as carbonic anhydrase III. Characterization of S-thiolation and dethiolation reactions. Arch. Biochem. Biophys. 284:270-278. [DOI] [PubMed] [Google Scholar]
- 5.Chomczynski, P., and N. Sacchi. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162:156-159. [DOI] [PubMed] [Google Scholar]
- 6.Cote, C. H., N. Jomphe, A. Odeimat, and P. Fremont. 1994. Carbonic anhydrase in mouse skeletal muscle and its influence on contractility. Biochem. Cell Biol. 72:244-249. [DOI] [PubMed] [Google Scholar]
- 7.Deng, C., A. Wynshaw-Boris, F. Zhou, A. Kuo, and P. Leder. 1996. Fibroblast growth factor receptor 3 is a negative regulator of bone growth. Cell 84:911-921. [DOI] [PubMed] [Google Scholar]
- 8.Frank, L., J. R. Bucher, and R. J. Roberts. 1978. Oxygen toxicity in neonatal and adult animals of various species. J. Appl. Physiol. 45:699-704. [DOI] [PubMed] [Google Scholar]
- 9.Geers, C., and G. Gros. 1990. Effects of carbonic anhydrase inhibitors on contraction, intracellular pH and energy-rich phosphates of rat skeletal muscle. J. Physiol. 423:279-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hegde, P., R. Qi, K. Abernathy, C. Gay, S. Dharap, R. Gaspard, J. E. Hughes, E. Snesrud, N. Lee, and J. Quackenbush. 2000. A concise guide to cDNA microarray analysis. BioTechniques 29:548-556. [DOI] [PubMed] [Google Scholar]
- 11.Kim, G., T. Lee, A. Wynshaw-Boris, and R. L. Levine. 2001. Nucleotide sequence and structure of the mouse carbonic anhydrase III gene. Gene 265:37-44. [DOI] [PubMed] [Google Scholar]
- 12.Kim, G., J. Selengut, and R. L. Levine. 2000. Carbonic anhydrase III: the phosphatase activity is extrinsic. Arch. Biochem. Biophys. 377:334-340. [DOI] [PubMed] [Google Scholar]
- 13.Koester, M. K., L. M. Pullan, and E. A. Noltmann. 1981. The p-nitrophenyl phosphatase activity of muscle carbonic anhydrase. Arch. Biochem. Biophys. 211:632-642. [DOI] [PubMed] [Google Scholar]
- 14.Koester, M. K., A. M. Register, and E. A. Noltmann. 1977. Basic muscle protein, a third genetic locus isoenzyme of carbonic anhydrase? Biochem. Biophys. Res. Commun. 76:196-204. [DOI] [PubMed] [Google Scholar]
- 15.Kubis, H. P., and G. Gros. 1997. A rapid electrophoretic method for separating rabbit skeletal muscle myosin heavy chains at high resolution. Electrophoresis 18:64-66. [DOI] [PubMed] [Google Scholar]
- 16.Lehtonen, J., B. Shen, M. Vihinen, A. Casini, A. Scozzafava, C. T. Supuran, A. K. Parkkila, J. Saarnio, A. J. Kivela, A. Waheed, W. S. Sly, and S. Parkkila. 2004. Characterization of CA XIII, a novel member of the carbonic anhydrase isozyme family. J. Biol. Chem. 279:2719-2727. [DOI] [PubMed] [Google Scholar]
- 17.Levine, R. L. 2002. Carbonyl modified proteins in cellular regulation, aging, and disease. Free Radic. Biol. Med. 32:790-796. [DOI] [PubMed] [Google Scholar]
- 18.Levine, R. L., N. Wehr, J. A. Williams, E. R. Stadtman, and E. Shacter. 2000. Determination of carbonyl groups in oxidized proteins. Methods Mol. Biol. 99:15-24. [DOI] [PubMed] [Google Scholar]
- 19.Lii, C. K., Y. C. Chai, W. Zhao, J. A. Thomas, and S. Hendrich. 1994. S-thiolation and irreversible oxidation of sulfhydryls on carbonic anhydrase III during oxidative stress: a method for studying protein modification in intact cells and tissues. Arch. Biochem. Biophys. 308:231-239. [DOI] [PubMed] [Google Scholar]
- 20.Lynch, C. J., W. A. Brennan, Jr., T. G. Vary, N. D. Carter, and S. J. Dodgson. 1993. Carbonic anhydrase III in obese Zucker rats. Am. J. Physiol. 264:E621-E630. [DOI] [PubMed] [Google Scholar]
- 21.Lynch, C. J., K. M. McCall, M. L. Billingsley, L. M. Bohlen, S. P. Hreniuk, L. F. Martin, L. A. Witters, and S. J. Vannucci. 1992. Pyruvate carboxylase in genetic obesity. Am. J. Physiol. 262:E608-E618. [DOI] [PubMed] [Google Scholar]
- 22.Lyons, G. E., M. E. Buckingham, S. Tweedie, and Y. H. Edwards. 1991. Carbonic anhydrase III, an early mesodermal marker, is expressed in embryonic mouse skeletal muscle and notochord. Development 111:233-244. [DOI] [PubMed] [Google Scholar]
- 23.Moriguchi, T., J. Loewke, M. Garrison, J. N. Catalan, and N. Salem, Jr. 2001. Reversal of docosahexaenoic acid deficiency in the rat brain, retina, liver, and serum. J. Lipid Res. 42:419-427. [PubMed] [Google Scholar]
- 24.Raisanen, S. R., P. Lehenkari, M. Tasanen, P. Rahkila, P. L. Harkonen, and H. K. Vaananen. 1999. Carbonic anhydrase III protects cells from hydrogen peroxide-induced apoptosis. FASEB J. 13:513-522. [DOI] [PubMed] [Google Scholar]
- 25.Register, A. M., M. K. Koester, and E. A. Noltmann. 1978. Discovery of carbonic anhydrase in rabbit skeletal muscle and evidence for its identity with “basic muscle protein. ” J. Biol. Chem. 253:4143-4152. [PubMed] [Google Scholar]
- 26.Richardson, D. E., C. A. Regino, H. Yao, and J. V. Johnson. 2003. Methionine oxidation by peroxymonocarbonate, a reactive oxygen species formed from CO2/bicarbonate and hydrogen peroxide. Free Radic. Biol. Med. 35:1538-1550. [DOI] [PubMed] [Google Scholar]
- 27.Richardson, D. E., H. R. Yao, K. M. Frank, and D. A. Bennett. 2000. Equilibria, kinetics, and mechanism in the bicarbonate activation of hydrogen peroxide: oxidation of sulfides by peroxymonocarbonate. J. Am. Chem. Soc. 122:1729-1739. [Google Scholar]
- 28.Rokutan, K., J. A. Thomas, and H. Sies. 1989. Specific S-thiolation of a 30 KDa cytosolic protein from rat liver under oxidative stress. Eur. J. Biochem. 179:233-239. [DOI] [PubMed] [Google Scholar]
- 29.Sly, W. S., D. Hewett-Emmett, S. J. Dodgson, Y. S. Yu, and R. E. Tashian. 1983. Carbonic anhydrase II deficiency identified as the primary defect in the autosomal recessive syndrome of osteopetrosis with renal tubular acidosis and cerebral calcification. Proc. Natl. Acad. Sci. USA 80:2752-2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sly, W. S., and P. Y. Hu. 1995. Human carbonic anhydrases and carbonic anhydrase deficiencies. Annu. Rev. Biochem. 64:375-401. [DOI] [PubMed] [Google Scholar]
- 31.Spicer, S. S., Z. H. Ge, R. E. Tashian, D. J. Hazen-Martin, and B. Schulte. 1990. Comparative distribution of carbonic anhydrase isoenzymes III and II in rodent tissues. Am. J. Anat. 187:55-64. [DOI] [PubMed] [Google Scholar]
- 32.Stanton, L. W., P. A. Ponte, R. T. Coleman, and M. A. Snyder. 1991. Expression of CA III in rodent models of obesity. Mol. Endocrinol. 5:860-866. [DOI] [PubMed] [Google Scholar]
- 33.Starke-Reed, P. E., and C. N. Oliver. 1989. Protein oxidation and proteolysis during aging and oxidative stress. Arch. Biochem. Biophys. 275:559-567. [DOI] [PubMed] [Google Scholar]
- 34.Tashian, R. E. 1989. The carbonic anhydrases: widening perspectives on their evolution, expression and function. Bioessays 10:186-192. [DOI] [PubMed] [Google Scholar]
- 35.Thomas, J. A., B. Poland, and R. Honzatko. 1995. Protein sulfhydryls and their role in the antioxidant function of protein S-thiolation. Arch. Biochem. Biophys. 319:1-9. [DOI] [PubMed] [Google Scholar]
- 36.Tybulewicz, V. L., C. E. Crawford, P. K. Jackson, R. T. Bronson, and R. C. Mulligan. 1991. Neonatal lethality and lymphopenia in mice with a homozygous disruption of the c-abl proto-oncogene. Cell 65:1153-1163. [DOI] [PubMed] [Google Scholar]
- 37.Wetzel, P., T. Liebner, and G. Gros. 1990. Carbonic anhydrase inhibition and calcium transients in soleus fibers. FEBS Lett. 267:66-70. [DOI] [PubMed] [Google Scholar]
- 38.Wolfinger, R. D., G. Gibson, E. D. Wolfinger, L. Bennett, H. Hamadeh, P. Bushel, C. Afshari, and R. S. Paules. 2001. Assessing gene significance from cDNA microarray expression data via mixed models. J. Comput. Biol. 8:625-637. [DOI] [PubMed] [Google Scholar]