Abstract
When telomerase is inactivated in Saccharomyces cerevisiae, telomeric DNA shortens with every cell division, and cells stop dividing after ∼100 generations. Survivors that form in these senescent populations and resume growing have variably amplified arrays of subtelomeric Y′ elements. We marked a chromosomal Y′ element with the his3AI retrotransposition indicator gene and found that Y′HIS3 cDNA was incorporated into the genome at ∼10- to 1,000-fold-higher frequencies in survivors compared to telomerase-positive strains. Y′HIS3 cDNA mobility was significantly reduced if assayed at 30°C, a nonpermissive temperature for Ty1 retrotransposition, or in the absence of Tec1p, a transcription factor for Ty1. Microarray analysis revealed that Y′ RNA is preferentially associated with Ty1 virus-like particles (VLPs). Genomic copies of Y′HIS3 cDNA typically have downstream oligo(A) tracts, followed by a complete Ty1 long terminal repeat and TYA1 or TYB1 sequences. These data are consistent with the use of Ty1 cDNA to prime reverse transcription of polyadenylated Y′ RNA within Ty1 VLPs. Unmarked Y′-oligo(A)-Ty1 cDNA was also detected in survivors, reaching copy numbers of ∼10−2 per genome. We propose that Y′-oligo(A)-Ty1 cDNA recombines with Y′ elements at eroding telomeres in survivors and may play a role in telomere maintenance in the absence of telomerase.
Telomeres in most eukaryotes studied consist of short tandem DNA repeat sequences that are bound by specific proteins to form protective caps. Maintenance of telomeric DNA repeats is required for genome stability and is accomplished by the enzyme telomerase. Loss or inactivation of a component of telomerase leads to progressive shortening of telomeric DNA, which increases the likelihood that individual chromosomes will lose their protective caps (3, 32). When these caps are lost, telomeric DNA is subject to DNA repair processes that can result in chromosome fusions and rearrangements (3). The loss of telomeric DNA and subsequent chromosome instability lead to apoptosis and/or cellular senescence in mammalian cells and yeasts (18, 30). In different species of yeasts, a fraction of the senescing cells recover and begin to grow well again. These “survivors” have telomeres with altered structures that are maintained by telomere recombination or have circularized chromosomes (29, 31, 33).
In Saccharomyces cerevisiae, chromosomes terminate with ∼350 bp of TG1-3/C1-3A repeats, and immediately centromere-proximal to these repeats are zero to four copies of a complex subtelomeric repeat known as Y′ (48). Two types of alternative telomere structures in S. cerevisiae telomerase-negative survivors are formed by homologous recombination: type I, with amplified arrays of subtelomeric Y′ elements separated by short stretches of telomeric repeats and type II, with long heterogeneous tracts of TG1-3 telomeric repeats (29, 42). Y′ elements are present in two varieties, Y′-L and Y′-S, which are ∼6.7 and ∼5.2 kb in length, respectively (7, 47). Both varieties contain an open reading frame (ORF) with sequence motifs characteristic of helicases (28, 47). This putative helicase activity has been confirmed in an in vitro assay using Y′-Help-1p overexpressed and purified from insect cells (47), but its function in vivo is not known. Y′ transcription has been reported to increase substantially in telomerase-negative survivors (47). This increase in transcription is consistent with a model in which arrays of Y′ elements in survivors are maintained through a process involving reverse transcription of Y′ transcripts and incorporation of the resulting cDNA onto the ends of chromosomes. However, Y′ elements lack sequences characteristic of retrotransposons and are not predicted to encode any proteins with reverse transcriptase (RT) activity (28, 47).
The possibility that Y′ cDNA might be produced and used to extend telomeric DNA was previously tested. Strains containing single Y′ elements marked with a retrotransposition indicator gene, his3AI, were constructed, and a genetic assay was used to examine the frequency of Y′HIS3 cDNA formation in telomerase-negative survivors (42). Such cDNA-based events were very rarely observed, and the authors of that study concluded that these events did not contribute significantly to telomere maintenance in survivors. The only known RT genes in the S. cerevisiae genome are those encoded by Ty long-terminal-repeat (LTR) retrotransposons. Formation of Y′ cDNA would likely depend upon Ty elements and Ty1 in particular, since Ty1 is capable of generating functional HIS3 pseudogenes from a GALhis3AI reporter construct (12) and duplications of a portion of the URA2 gene (36). Retrotransposition of Ty1 occurs very infrequently at 30°C compared to 20 to 25°C (24, 34), so the extremely low levels of incorporation of Y′ cDNA detected previously (42) could have resulted from growing assay cultures at 30°C.
The Ty1 LTR retrotransposon is activated in telomerase-negative yeast strains (37, 38). Ty1 elements produce a terminally redundant RNA that is translated into Gag (TyA1) and Gag-Pol (TyA1-TyB1) proteins (43). Ty1 proteins form cytoplasmic virus-like particles (VLPs), in which Ty1 RNA is reverse transcribed into a linear cDNA that is subsequently integrated into the genome (43). Ty1 cDNA levels and retrotransposition frequencies progressively increase during senescence of telomerase-negative yeast, reaching a peak just before the formation of survivors. After survivors form, Ty1 cDNA levels and retrotransposition frequencies often remain above wild-type levels (38). Activation of Ty1 cDNA synthesis is mediated by a DNA-damage-signaling pathway that is activated by telomere erosion (16, 22, 38).
We investigated the formation of Y′ cDNA in telomerase-negative survivors and hypothesized that Ty1 might mediate this process. Human L1 retrotransposons have been shown to be capable of generating pseudogenes of cellular transcripts and to mediate retrotransposition of Alu elements (13, 17), demonstrating that similar processes occur in other organisms. In the absence of specific knowledge about the molecular details of this process, we refer to the reverse transcription of Y′ RNA to form cDNA that is incorporated into the genome as Y′ cDNA mobility. We report the detection of Y′ cDNA mobility in telomerase-negative survivors at levels ca. 10- to 1,000-fold above those obtained with telomerase-positive strains, when grown in conditions permissive for Ty1 retrotransposition. Genetic and molecular analyses of cDNA-based events indicate that Ty1 is involved in Y′ cDNA mobility and that synthesis of Y′ cDNA is primed by using Ty1 cDNA or cDNA intermediates. The potential implications and consequences of this process for the biology of Y′, telomeres, and survivors are discussed.
MATERIALS AND METHODS
Yeast strains, media, and plasmids.
Yeast strains were grown with standard media prepared as described previously (35). Strains were derivatives of BY4741 and BY4742 (5), JC574 (MATa ade2-101 his3Δ200 lys2-801 ura3-52), or YM608 (MATα ade2-101 his3Δ200 lys2-801 trp1Δ901 ura3-52; a gift of Mark Johnston, Washington University School of Medicine, St. Louis, Mo.). To generate strains JC3840 and JC3841 (MATα his3Δ1 leu2Δ0 lys2Δ0 tlc1::LEU2 ura3Δ0 Y′his3AI[Δ1], pRS317TLC1), a tlc1::LEU2 derivative of BY4742 (JC3290) was constructed by one-step transplacement with pBLUE61::LEU2 (40). This strain was transformed with plasmid pRS317TLC1, a LYS2-marked, CEN-based plasmid containing TLC1 (42), to generate JC3292. A subtelomeric Y′ element was then marked with his3AI[Δ1], which has the artificial intron (AI) placed at a position in HIS3 that is deleted in his3Δ1 to prevent the formation of His+ prototrophs by DNA recombination between alleles (37). To accomplish this, the his3AI-URA3-his3AI insert in plasmid pSL300his3AI-URA3-his3AI (42) was amplified with primers YHIS3-1 and YHIS3-2, which introduced 48 bp of Y′ sequence on either side of the insert. The resulting PCR product was transformed into JC3292, Ura+ transformants were selected, followed by selection for Ura- cells with 5-fluoroorotic acid, to obtain a strain (JC3638) with a Y′his3AI element. To replace his3AI with his3AI[Δ1], plasmid pBJC720 containing a ClaI fragment of his3AI[Δ1] from plasmid pBJC573 (6) cloned into the ClaI site in vector pRS406 (39) was linearized with NheI and transformed into JC3638, and Ura+ transformants were selected. Ura- cells were selected by using 5-fluoorotic acid, and those in which his3AI[Δ1] had replaced his3AI were identified by PCR. The his3AI[Δ1] gene is inserted between positions 6251 and 6252 in the Y′ sequence TEL15R-YP (403 bp from the 3′ end of Y′) (14). Strain JC3287 is an est2Δ1::URA3 derivative of YM608 generated by one-step transplacement with plasmid pVL363 (26). JC3723 is a derivative of JC574 containing a Y′his3AI element, and was generated by PCR mediated two-step gene transplacement with primers YHIS3-1 and YHIS3-2 and plasmid pSL300his3AI-URA3-his3AI, as described above.
Generation and transformation of survivors.
Telomerase-negative survivors of strains JC3840 and JC3841 were generated by screening for loss of pRS317TLC1 and serial subculture. One set of survivors (denoted by C) was generated by subculture at 30°C, and the other (denoted by D) by was generated by subculture at 23°C. Cells were grown for 3 days at 30°C or for 5 days at 23°C for each subculture. Cells were patched onto yeast extract-peptone-dextrose (YPD) plates, grown overnight, and then streaked onto YPD plates (subculture 1 or SC1). Lys− segregants were serially restreaked onto YPD plates until SC5 or SC6. Individual survivor colonies, identified by faster growth after recovery from senescence, were patched onto YPD plates, grown at 30°C overnight, and then stored at 4°C.
To generate survivors in a different strain background, strains JC3287 and JC3723 were crossed, and spores with the genotypes est2Δ1::URA3, Y′his3AI, or EST2 Y′his3AI were selected. Spores of the genotype est2Δ1::URA3 were serially restreaked four times and subcultured for 7 days at 23°C. A single survivor colony from each SC5 streak was patched onto YPD, grown overnight at 30°C, and then stored at 4°C.
tec1::KanMX4 derivatives of individual survivors were generated by transformation with a tecl::KanMX4 PCR product (45), followed by selection on YPD plates containing 200 μg of G418/ml. Transformants were single-colony purified, patched onto YPD plates, grown overnight at 30°C, and then stored at 4°C. As a control, TEC1 survivors were restreaked onto YPD plates, and single colonies were patched onto YPD plates, grown overnight at 30°C, and stored at 4°C.
Y′ cDNA mobility assay.
The frequency of His+ prototroph formation in strains with Y′his3AI or Y′his3AI[Δ1] was used as a measure of Y′ cDNA mobility. Cells of survivors and telomerase-positive strains were diluted to an optical density at 600 nm of 0.005 in YPD broth and grown at 20°C for 3 days. Triplicate cultures for each strain were grown in parallel. An aliquot of 0.005 μl to 0.01 μl was plated onto YPD agar to determine the cell density of each culture. Portions (3 to 6 ml) of each culture were divided among SC-His plates, which were incubated at 30°C for 7 days. His+ frequency was calculated as the number of His+ colonies divided by the total number of cells plated. The results for the triplicate cultures were added to obtain a total His+ frequency. The Mann-Whitney rank sum test was used to compare median frequency values from different data sets for significant differences.
Southern analysis.
Genomic DNA was prepared from 6 to 10 ml of saturated cultures by glass bead disruption and phenol extraction (1). Then, 12 μg of DNA was digested and separated on 0.7 to 1% agarose gels. The DNA samples were transferred to Hybond-N membranes (Amersham), which were hybridized in NorthernMax buffer (Ambion) at 42 to 50°C overnight with 32P-labeled DNA or RNA probes. DNA probes were prepared by random primer labeling with PCR products of Y′ or TEL1 generated by using the primer pairs YPEND-UP and YPEND-DN or TEL1F and TEL1R, respectively (Table 1). YPEND-UP and YPEND-DN generate a 726-bp product from the 3′ portion of the Y′ coding region. Telomeric DNA probe was prepared by random primer labeling of an EcoRI-SalI fragment of plasmid pCA75#5 (a gift from Virginia Zakian, Princeton University, Princeton, N.J.) containing 71 bp of TG1-3/C1-3A repeat sequences. Labeled HIS3 or TYB1 sense strand transcripts were produced by in vitro transcription with plasmid pGEM-HIS3 (10) or pGEM-TYB (6). Blots were washed four or five times at a maximum stringency of 0.2 or 0.1× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.1% sodium dodecyl sulfate at 65°C. Banding patterns were visualized by autoradiography. A Storm 860 phosphorimager and ImageQuant software were used for quantification. The Southern blot assay used to measure Ty1 cDNA has been described previously (6).
TABLE 1.
List of primers used
| Primer | Sequence (5′-3′) |
|---|---|
| 3654 | CGTCTATGTGTAAGTCACC |
| ACT1PF | CTCCTACGTTGGTGATGAAGCTC |
| ACT1PR | ACCTGGGAACATGGTGGTACCA |
| AX020 | CTATTACATTATGGGTGGTATGTTGG |
| FP | TCTTTGCCTTCGTTTATCTTGCCTG |
| H3HOPA2 | TCTCCTACTTTCTCCCTTTGCAAACC |
| H3HOPS | CGCAGTTGTCGAACTTGGTTTGCAAAG |
| HIS33 | GTGGAGGGAACATCGTTGGTAC |
| HIS35 | AGCAGGCAAGATAAACGAAGGCAAAG |
| HIS3BEG | TAGTCAGGGAAGTCATAACAC |
| HIS3END | CATACTTACTGACATTCATAGG |
| HIS3HOP | TGATACATGCTCTGGCCAAG |
| HIS3HOP2 | ACCAGCCGGAATGCTTGGCCAG |
| NRP-2 | GATCGTCGACGCATTTTCTTGAAAGCTTTGCAGAG |
| TEL1COMP | CGGATTTCTGACGATATGGACCTAGGATTACAATTGTGCTCA |
| TEL1F | CGGATTTCTGACGATATGGAC |
| TEL1R | ACCAACGTACTGAAGGTATCC |
| TYAOUT2 | TCTCTGGAACAGCTGATGAAG |
| TYBOUT2 | GTGATGACAAAACCTCTTCCG |
| YBKWDS1 | TACTGCCGTGCAACAAACAC |
| YCOMP | AAGTATGGAGCAACTTGCGTGAATCTGGACTGCAGGGTCCGCAGT |
| YEXPAND | TTACTTCCTTGGTGCAATGTCCCAC |
| YHELP1 | GTATGGAGCAACTTGCGTGAATC |
| YHIS3-1 | TATTACTATGTAGAGATTCGAGCAGAGAAGTTGGAGAGTGAAGGAAATACTAGTGCTGCAGCTTTAAATATCG |
| YHIS3-2 | CAATATACTATACTACACAATACATAATCACTGACTTTCGTAACAACATCTAGATGGTCCTCTAGTACTCTC |
| YPEND-DN | CAACTTCTCTGCTCGAATCTCTAC |
| YPEND-UP | GAGAGATCTACTCTCAGATACAGAG |
| YR1 | TGCTTTCAGTCACTAATACATAACAC |
Northern analysis.
Total RNA was prepared by hot acidic phenol extraction (1) from 2 to 3 ml of saturated cultures. The final RNA samples were then precipitated with equal volumes of 4 M LiCl at 4°C for >15 min. Samples were spun at 4°C for 15 min, washed with 70% ethanol, and dissolved in Milli-Q water. RNA samples were denatured with glyoxal, separated on 1% agarose gels, and transferred to Hybond-N membranes (Amersham) in 5× SSC-0.01 N NaOH. Blots were hybridized in NorthernMax buffer (Ambion) overnight at 42 to 50°C for DNA probes or 68°C for RNA probes. 32P-labeled DNA and RNA probes were prepared as for Southern blotting. Templates for DNA probes were PCR products for Y′, as for Southern blotting analyses, or ACT1 generated with primers ACT1PF and ACT1PR (see Table 1). The HIS3 probe was prepared as for the Southern blots. Blots were washed four or five times at a maximum stringency of 0.2 or 0.1× SSC-0.1% sodium dodecyl sulfate at 65 or 68°C. Bands were visualized and quantified as for Southern blots.
PCR, inverse PCR, cloning, and sequencing.
PCR analyses of His+ events were performed with the following primers: 3654, H3HOPA2, H3HOPS, HIS33, HIS35, HIS3BEG, HIS3END, HIS3HOP, HIS3HOP2, YBKWDS1, YEXPAND, YHELP1, YR1, TYAOUT2, and TYBOUT2 (see Table 1). PCR was performed with up to 1 μg of genomic DNA and MasterAmp Taq (Epicentre). For inverse PCR, ca. 1 to 3 μg of genomic DNA was restriction digested with XhoI, ligated in a total volume of 50 μl overnight, and PCR amplified with MasterAmp Taq, and primers NRP-2 and FP (see Table 1). Unmarked Y′-Ty1 cDNA was amplified with up to 1 μg of genomic DNA, primer YHELP1, and either primer TYAOUT2 or TYBOUT2. Products of interest were cloned with the TOPO TA cloning kit (Invitrogen) and sequenced, or PCR products were sequenced directly.
Ty1 VLP preparation and microarray analysis.
Ty1 VLPs were prepared according to the standard protocol (15) from two independent galactose-induced cultures of yeast strain JB224 (ura3-167 his3Δ200), which contains the inducible Ty1 expression plasmid pJef724 (4). RNA was isolated from Ty1 VLPs exactly as described for retroviral RNA (19). Total cytoplasmic RNA was extracted from an aliquot of the same culture by disruption of cells in RNA lysis buffer, followed by phenol extraction (1). All RNA samples were digested with DNase (Promega) and purified by using an RNeasy minikit (Qiagen). RNA concentration and integrity was determined spectrophotometrically and by agarose gel electrophoresis. Microarray analysis was performed by using the Affymetrix GeneChip Yeast Genome S98 Array (Affymetrix) according to the protocols described by manufacturer in the Johns Hopkins Medicine Microarray Core Facility. Genes were annotated for analysis by using the Saccharomyces Genome Database (14).
Quantification of unmarked Y′-Ty1 cDNA by competitive PCR.
A cloned Y′-Ty1 cDNA junction containing Y′ sequences extending 17 bp downstream of the Y′ polyadenylation signal sequence, followed by a 22-bp oligo(A) tract, and the 3′ Ty1 LTR was amplified with primers YCOMP and TYBOUT2 to generate a Y′-Ty1 cDNA competitor. Primer YCOMP contains the sequence of primer YHELP1 at its 5′ end and sequence from a region 142 bp further downstream in Y′ at its 3′ end, so amplification with this primer introduced a 141-bp internal deletion of Y′ sequences into the competitor molecule. A TEL1 competitor was generated by amplification of JC3840 genomic DNA with primers TEL1COMP and TEL1R. Amplification with TEL1COMP introduced a 51-bp deletion into the competitor. Constant amounts of genomic DNA from JC3840, JC3841, JC3723:X1 EST2 spores (JC3833 and JC3834), six JC3840/1 type II survivors (4C, 10C, 26C, 10D, 12D, and 24D), four JC3840/1 type I survivors (17C, 24C, 17D, and 30D), and four JC3723:X1 type I survivors (1CA, 4AA, 5BA, and 9BA) were amplified separately with primers YHELP1 and AX020 to detect Y′-Ty1 cDNA or primers TEL1F and TEL1R to detect the single-copy gene TEL1 in the presence of 0.1 to 60 fg or 0.2 to 2 pg of competitor DNA, respectively. PCR conditions consisted of 95°C for 4 min, followed by 35 cycles (Y′-Ty1 cDNA) or 24 cycles (TEL1) of 94°C for 1 min, 60°C or 62°C for 1 min, and 72°C for 1 min, and finally by 72°C for 10 min using FailSafe Premix I and FailSafe Enzyme Mix (Epicentre). The quantities of endogenous Y′-Ty1 cDNA and TEL1 templates were determined by the quantities of competitor DNA required to generate endogenous and competitor PCR products of equal intensities when products were analyzed by agarose gel electrophoresis. Two independent DNA samples for each strain were analyzed, and the results for each strain were averaged. The copy number per genome of Y′-Ty1 cDNA was calculated by dividing the quantity of Y′-Ty1 cDNA by the quantity of TEL1 template and then dividing that ratio by 2.1 to correct for the size difference between the Y′-Ty1 cDNA and TEL1 competitors (895 and 423 bp, respectively).
RESULTS
Y′ cDNA mobility is elevated in telomerase-negative survivors.
To examine the effect of deleting telomerase-encoding genes on Y′ cDNA mobility, a genetic assay system was established by using the his3AI retrotransposition indicator gene (9, 42). The his3AI gene, which is a HIS3 gene rendered nonfunctional by an insertion of an intron that is in the opposite transcriptional orientation of HIS3, was inserted into the 3′ end of Y′, just downstream of the Y′HELP1 coding region. A functional copy of HIS3 is created only when the AI is spliced out of Y′his3AI transcripts, the Y′ HIS3 RNA is reverse transcribed, and the resulting cDNA is incorporated into the genome (see Fig. 1). This system allows Y′ cDNA mobility to be measured as the frequency of His+ prototroph formation in his3Δ strains with a copy of Y′his3AI (his3AI is used to refer to both his3AI and his3AI[Δ1] [see Materials and Methods]).
FIG. 1.
Use of his3AI to detect Y′ cDNA mobility through a genetic assay. The his3AI indicator gene was inserted into the 3′ end of Y′, downstream of the coding region, in two different strain backgrounds lacking a functional copy of HIS3. His+ prototroph formation can occur when Y′his3AI is transcribed, spliced to produce Y′HIS3 RNA, and reverse transcribed into cDNA and the cDNA is incorporated into the genome.
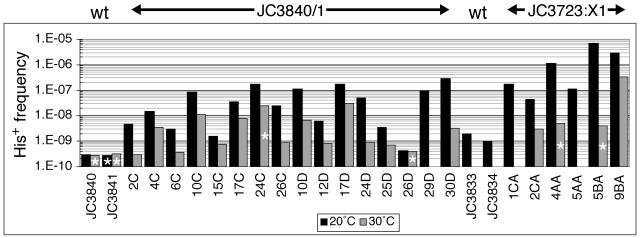
Telomerase-negative survivors were generated by serially subculturing est2Δ1::URA3, Y′his3AI spores from a cross between strains JC3723 and JC3287 (herein referred to as JC3723:X1) and tlc1::LEU2, Y′his3AI colonies (strains JC3840 and JC3841, herein referred to as JC3840/1), following segregation of a plasmid carrying TLC1 (see Materials and Methods). Individual survivor colonies and telomerase-positive control strains were grown at 20°C and assayed for the frequency of His+ prototroph formation. Totals of 16 JC3840/1 and 6 JC3723:X1 survivors were examined (Fig. 2). His+ frequencies were clearly elevated in both JC3840/1 and JC3723:X1 survivor strains compared to the telomerase-positive strains JC3840 and JC3841 and JC3723:X1 EST2 spores (JC3833 and JC3834), respectively, but the frequencies varied substantially for different survivors. The ranges for JC3840/1 or JC3723:X1 survivors were from 0.42 × 10−9 to 290 × 10−9 or from 45 × 10−9 to 7,200 × 10−9, respectively. In contrast, His+ frequencies for JC3840 and JC3841 were 0.3 × 10−9 and < 0.24 × 10−9, whereas those for two JC3723:X1 EST2 spores were 2 × 10−9 and 1 × 10−9. Similar His+ frequency values were obtained from several independent assays of these control strains (data not shown). The fold increase in His+ frequency for JC3840/1 survivors was 110, and for JC3723:X1 survivors it was 420, as determined by comparing the median frequency for each set of survivors (30 × 10−9 or 635 × 10−9, respectively) to the median value for the congenic telomerase-positive strains (0.27 × 10−9 or 1.5 × 10−9, respectively).
FIG. 2.
Y′ cDNA mobility is elevated in JC3840/1 and JC3723:X1 survivors compared to telomerase-positive control strains and is decreased when assayed at 30°C compared to 20°C. The frequency of His+ prototroph formation due to mobilization of Y′his3AI is shown for JC3840/1 survivors (2C to 30D) and the congenic telomerase-positive strains JC3840 and JC3841, as well as for JC3723:X1 survivors (1CA to 9BA) and the congenic telomerase-positive strains JC3833 and JC3834. Cultures for these assays were grown either at 20°C (black bars) or 30°C (gray bars). Bars marked with stars represent cultures with no His+ prototrophs, so the corresponding frequency is the maximum possible frequency determined as if one His+ colony had formed. JC3840/1 survivors 17C, 24C, 17D, and 30D and all JC3723:X1 survivors were type I, and the 12 remaining JC3840/1 survivors were type II.
Ty1 activity is required for high levels of Y′ cDNA mobility in telomerase-negative survivors.
Having found that the frequency of Y′ cDNA mobility was greatly elevated in many survivors, we next tested whether Ty1 was involved in this process. Our first approach was to determine His+ frequencies for 15 of the 16 JC3840/1 survivors and 4 of the 6 JC3723:X1 survivors when assayed at 30°C, as opposed to 20°C. If Ty1 RT mediates Y′ cDNA mobility, it was expected that the His+ frequency for each survivor would be reduced when assayed at 30°C (24, 34). For every survivor analyzed, the His+ frequency was reduced at 30°C (Fig. 2, gray bars) versus 20°C (Fig. 2, black bars), and in four instances no His+ events were observed at 30°C (bars with stars, Fig. 2).
The fold reduction in the His+ frequency varied among individual survivors. When median values were compared, the reductions in His+ frequency at 30°C were 28-fold for JC3840/1 survivors and 440-fold for JC3723:X1 survivors, and these reductions were statistically significant (0.005 > P > 0.001 and P = 0.025, respectively). These values include the four maximum possible His+ frequencies (bars with stars in Fig. 2) and therefore represent minimum estimates of the reduction for each set of survivors. Northern analysis of Y′ RNA and Y′his3AI RNA levels for a subset of survivors revealed that the reductions in His+ frequencies were not accompanied by equivalent reductions of Y′ transcript levels at 30°C (data not shown).
A second approach used to test for a role of Ty1 in Y′ cDNA mobility was to measure the His+ frequency in survivors lacking Tec1p, a transcription factor required for full Ty1 expression (23). The tecl::KanMX allele was introduced into a subset of JC3840/1 and JC3723:X1 survivors. Because variation in the level of Y′his3AI mobility arises simply as a result of subculturing survivors (for examples, see Fig. 3, black bars) and subculturing is necessary to generate transformants, one or two tecl::KanMX transformants of each survivor were assayed in parallel with two colonies from restreaks of the corresponding TEC1 survivors. In all eight JC3840/1 survivors and both JC3723:X1 survivors tested the His+ frequency was lower in the absence of TEC1 (Fig. 3). For 11 of the 19 teclΔ transformants examined, no His+ colonies were obtained (Fig. 3, bars with stars). The median His+ frequencies (including the maximum possible frequencies for these 11 transformants) were reduced 31-fold and 480-fold for JC3840/1 and JC3723:X1 survivors, respectively, in the absence of Tec1p (P < 0.001 or P = 0.025, respectively). Again, these are minimum estimates of the reductions, since in many cases no His+ events were observed. Northern analysis of Y′ and Y′his3AI RNA levels for a subset of strains demonstrated that there was no consistent reduction of Y′ transcript levels in tec1Δ strains compared to TEC1 controls (data not shown). Southern analysis of total cellular DNA in the same subset of strains confirmed that tec1Δ strains had lower levels of Ty1 cDNA, but the magnitude of the decrease varied among different strains (2.9- to 18-fold [data not shown]). The observations that Y′ cDNA mobility is reduced at 30°C and in the absence of Tec1p suggest a role for Ty1 expression in this process.
FIG. 3.
Y′ cDNA mobility is decreased in teclΔ survivors. The frequency of His+ prototroph formation is shown for the telomerase-positive strains JC3840 and JC3841, for JC3840/1 survivors (2C to 30D), and for JC3723:X1 survivors (4AA and 5BA). Assays were done on restreaked survivors (TEC1, black bars) or corresponding tec1::KanMX derivatives (gray bars). Multiple bars for a given survivor and genotype indicate that multiple restreaked colonies or transformants were analyzed. As for Fig. 2, bars marked with stars indicate cultures with no His+ colonies and represent maximum possible frequencies.
Type I survivors tend to have higher levels of Y′ cDNA mobility than type II survivors.
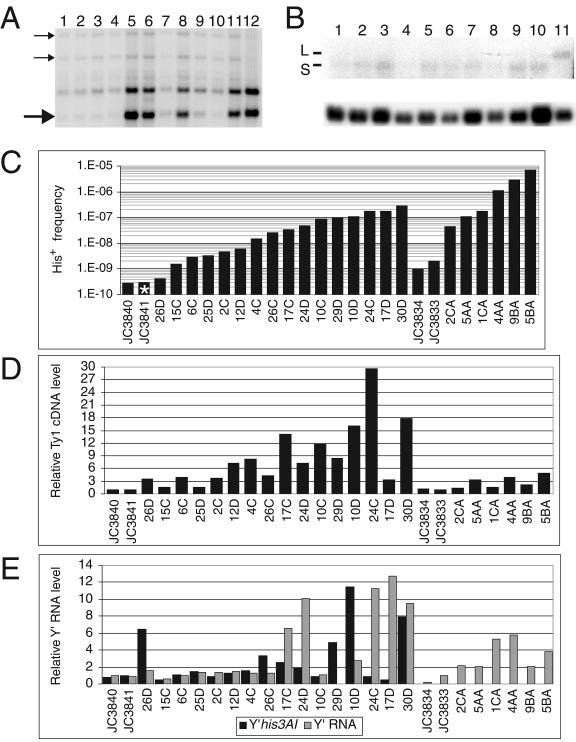
The involvement of Ty1 expression in Y′ cDNA mobility suggested that Y′ RNA may be incorporated into Ty1 VLPs and reverse transcribed by Ty1 RT to generate Y′ cDNA. Therefore, we tested whether the levels of Ty1 RT activity and Y′his3AI transcripts correlated with the frequency of Y′his3AI cDNA mobility. The level of Ty1 cDNA is an indirect but useful indicator of Ty1 RT activity, since Ty1 cDNA is not appreciably degraded in wild-type or telomerase-negative strains (38). Levels of unincorporated Ty1 cDNA were measured relative to representative single-copy genomic Ty1 elements by Southern blot analysis (6) (Fig. 4A). Y′ and Y′his3AI RNAs in all 22 survivors were quantified by Northern analysis and then compared to the level of ACT1 mRNA (Fig. 4B). Figure 4 compares the His+ frequencies of all of the survivors at 20°C from Fig. 2 to the relative levels of Ty1 cDNA and Y′ and Y′his3AI RNA (Fig. 4C through E, respectively). Survivors are arranged in ascending order based on their His+ frequency (Fig. 4C) and are present in this same order in panels D and E.
FIG. 4.
Variation in His+ frequency for different survivors is not strongly correlated with levels of Ty1 cDNA or Y′his3AI RNA. (A) Representative Southern blot of DNA samples from strains JC3840 (lanes 1 and 2), JC3841 (lanes 3 and 4), or the set D JC3840/1 survivors (lanes 5 to 12 correspond to survivors 10D to 30D). Small arrows indicate bands representing genomic Ty1 elements used to quantify the Ty1 cDNA band (large arrow). (B) A Northern blot of total RNA from JC3840 (lanes 1 and 2), JC3841 (lane 3), and set C JC3840/1 survivors (2C to 26C) probed for Y′his3AI RNA (upper panel) and reprobed for ACT1 mRNA (lower panel). L and S indicate marked Y′-L and Y′-S transcripts, respectively. his3AI was present in a Y′-S for all strains except 26C, in which case it was in a Y′-L (data not shown). Signals for Y′ and Y′his3AI RNA were normalized to ACT1 mRNA, and total Y′ transcripts levels were determined by combining the signals for Y′-L and Y′-S in each sample. (C) The His+ frequencies reported in Fig. 2 for strains assayed at 20°C are shown again with survivors in each set (JC3840/1 and JC3723:X1) grouped in order of ascending frequency values. (D) The relative levels of Ty1 cDNA determined by a Southern blot assay (see panel A) for strains grouped in the same order as in panel C. (E) The relative levels of total Y′ and Y′his3AI RNA determined by Northern blots (see panel B) for strains grouped in the same order as in panel C.
No strong correlations are evident when the data in Fig. 4C to 4E are compared. Ty1 cDNA levels varied substantially in JC3840/1 survivors. The cDNA levels were less variable in JC3723:X1 survivors and were only 1.5- to 4.8-fold higher than in congenic telomerase-positive strains. However, no clear trend between cDNA levels and His+ frequency was evident in either strain background. Only 6 of 12 JC3840/1 survivors had a >2-fold increase in Y′his3AI RNA compared to control strains, and there was no evidence for a correlation between Y′his3AI RNA levels and His+ frequency. Y′his3AI transcripts could not be detected in JC3723:X1 survivors, so it was not possible to test for a correlation between Y′his3AI RNA and His+ frequency in these survivors. In contrast to what was described in a previous report with a different yeast strain (47), Y′ RNA was not substantially elevated in most survivors (Fig. 4E). Only 5 of 12 JC3840/1 survivors and 3 of 6 JC3723:X1 survivors had a >3-fold increase in Y′ RNA.
To determine whether there is a relationship between Y′ cDNA mobility and the type of telomere structure in telomerase-negative survivors, Southern blot assays with a TG1-3/C1-3A probe were used to determine survivor type. Type I survivors have long arrays of Y′ elements at their telomeres and yield a pattern of telomeric restriction fragments that corresponds to unit length Y′ fragments and short terminal fragments. Type II survivors have heterogeneously long tracts of TG1-3/C1-3A repeats and yield many telomeric restriction fragments that are heterogeneous in length up to ∼12 kb (29, 42). Four JC3840/1 survivors were type I, and the remaining twelve were type II, whereas all six JC3723:X1 survivors were type I (Table 2).
TABLE 2.
Type I survivors tend to have higher frequencies of His+ prototroph formation
| Genotype (n) | Survivor type | His+ frequency (10−9)a
|
Y′ his3A1 DNAd
|
Y′ DNAbd
|
|||
|---|---|---|---|---|---|---|---|
| Range | Median | Range | Median | Range | Median | ||
| JC3840/1 (4) | I | 35-290 | 170 | 0.96-22 | 1.8 | 15-84 | 26 |
| JC3840/1 (12) | II | 0.42-110 | 11 | 0.33-6.3 | 1.0 | 0.27-10 | 1.5 |
| JC3723:X1 (6) | I | 45-7,200 | 635 | NDc-16 | 1.6 | 11-32 | 26 |
His+ frequencies reported in Fig. 2 for 20°C assays.
Combined signal for Y′-L and Y′-S.
ND, not detected. Y′his3A1 was no longer detected in one JC3723:X1 survivor after it was assayed for His+ prototroph formation and DNA was extracted.
Copy number relative to that for telomerase-positive strains.
For JC3840/1 survivors, the median His+ frequency was significantly higher in type I survivors than in type II survivors (P = 0.005). Relatively high His+ frequencies were also observed in the JC3723:X1 type I survivors. One type I JC3840/1 survivor (17D) had relatively low Ty1 cDNA levels compared to other JC3840/1 survivors (Fig. 4D), indicating that type I survivors did not have consistently higher levels of Ty1 cDNA. In addition, type I survivors did not have uniformly higher levels of Y′his3AI RNA compared to type II survivors (Fig. 4C). These observations are consistent with the difference in His+ frequency for type I and type II survivors being the result of a variable(s) other than Ty1 cDNA and Y′his3AI RNA levels. Y′ and Y′his3AI copy number were determined relative to the TEL1 gene or the his3Δ1 allele in JC3840/1 survivors by Southern blotting (data not shown). Since type I survivors contain amplified Y′ sequences, there is a correlation between increased Y′ copy number and higher His+ frequencies. However, there is no apparent relationship between Y′his3AI copy number and the frequency of His+ colonies formed (Table 2). In summary, the only significant correlation identified with high levels of Y′ cDNA mobility is the formation of type I survivors.
Y′ RNA is enriched in Ty1 VLPs.
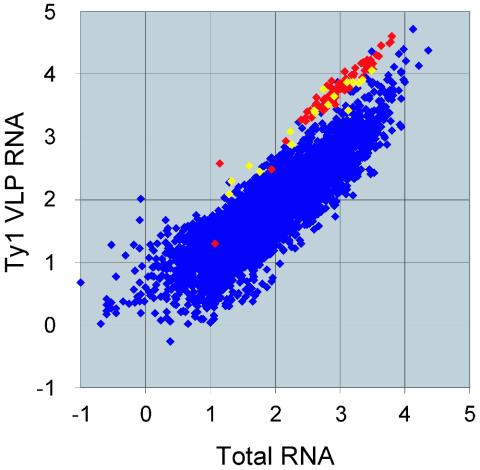
To directly test the model that Y′ RNA is reverse transcribed within Ty1 VLPs, we sought to determine whether Y′ RNA is enriched in Ty1 VLPs. Transposition was induced in two independent yeast cultures of a telomerase-positive strain harboring a galactose-inducible Ty1 element on a plasmid, and VLP fractions were prepared. VLP RNA and total RNA, prepared from aliquots of the same transposition-induced cultures, were extracted and analyzed by using the Affymetrix YG-S98 microarray (Affymetrix). A clear enrichment of all represented Y′ RNA species is observed within Ty1 VLPs in comparison to the yeast total cytoplasmic RNA population (Fig. 5). This is highlighted by the distribution of Y′-specific hybridization signals along the top edge of the data set. Analysis of signal intensities suggested that Y′ RNA was overrepresented within the Ty1 VLP RNA pool, with a mean increase (± the standard error) of (5.7 ± 0.5)-fold (range, 1.9 to 10.2). For comparison, signals corresponding to Ty1 probes indicated enrichment of these sequences by (6.2 ± 0.3)-fold (range, 1.7 to 26.8) within Ty1 VLPs. Since the strength of the signals obtained in this analysis indicated that Y′ was relatively well expressed, it is not likely that the enrichment of Y′ RNA in VLPs is an artifact (46). This finding provides good supportive evidence for the proposed model for Y′ cDNA formation.
FIG. 5.
Y′ RNA is enriched in Ty1 VLP fractions. The results of hybridization analyses by using Affymetrix microarrays are displayed as a scatter plot, with each point representing a particular yeast ORF as defined by the Saccharomyces Genome Database. Points are plotted as the log10 of the mean hybridization signal intensity, derived from two independent replicate experiments. For clarity, signals corresponding to Y′ ORFs are yellow, whereas those corresponding to Ty1 are red.
Ty1 sequences are joined to the 3′ end of Y′HIS3 cDNA.
We sought to characterize the structures of the cDNA sequences that conferred His+ phenotypes to further understand the mechanism of Y′ cDNA mobility. Sequencing of inverse PCR products obtained by using DNA from six independent His+ strains identified an oligo(A) tract and Ty1 LTR sequences adjacent to HIS3 cDNA (for examples, see Fig. 6, structures II to IV). This prompted us to screen genomic DNA from 72 independent His+ strains obtained from both JC3840/1 and JC3723:X1 survivors for Y′ or Ty1 sequences adjacent to HIS3 cDNA. The PCR was performed with primers that crossed the AI splice junction of the HIS3 cDNA in combination with primers to Y′ or Ty1 coding sequences to selectively amplify sequences adjacent to HIS3 as opposed to his3AI. Representative structures determined by PCR and by sequencing 28 of the 53 HIS3-Ty1 junction PCR products that were obtained (see below) are shown in Fig. 6. Similar structures were identified in His+ derivatives of both type I and type II survivors.
FIG. 6.
Examples of Y′HIS3 cDNA structures. Seven structures (I to VII) determined by PCR and DNA sequencing of a subset of PCR products are depicted. Y′ sequences, HIS3 sequences, and Ty1 sequences are represented by white, black, and gray boxes, respectively. A black ellipse represents TG1-3 DNA repeats and An represents an oligo(A) tract. Boxed arrowheads represent LTRs, and the direction of the arrowhead indicates the orientation. The labels TYA and TYB correspond to the indicated Ty1 ORF. Above the drawings for structures II to VII are one or two representative sequence junctions. Lowercase letters represent HIS3 or Y′ sequences, and capital letters represent the oligo(A) tract and Ty1 sequences. Subscripts are used to indicate long stretches of the same base, and the oligo(A) tract is distinguished by parentheses. Numbers above a sequence indicate the corresponding junction position in HIS3 (relative to the HIS3 ORF) or Y ′ (for TEL15R-YP) sequences, whereas numbers below indicate the junction position for Ty1-H3 (GenBank accession M18706). Boldface letters correspond to Ty1 primer-binding site or polypurine tract sequences found at the junction for structure III or IV, respectively. Sequences that are underlined represent regions of microhomology between the Y′HIS3 RNA poly(A) tail (structures V and VI) or HIS3 sequences (structure VII) and Ty1 sequences. The seven structures represent 36 independent His+ events (27 from type I and 9 from type II survivors). The two columns at the left side of the drawings indicate the fraction of these total events represented by each structure. The drawing is approximately to scale.
Of 72 independent HIS3 cDNA events analyzed, 71 (99%) contained upstream Y′ sequences (at least ∼600 bp) adjacent to HIS3 (Y′ sequences to the left of HIS3 in Fig. 6). This result is consistent with the need to transcribe the Y′his3AI element from the Y′ promoter in order for the AI to be present in the orientation necessary for subsequent splicing (Fig. 1). Downstream of HIS3 (sequences to the right in Fig. 6), sequences from the 3′ end of Y′, followed by sequences from the 5′ end of a second Y′ were detected in 10 cases (14%, see Fig. 6, structure I), and Ty1 sequences were detected in 53 cases (74%, for examples, see Fig. 6, structures II to VII). In the nine remaining cases (13%), neither a second Y′ nor Ty1 sequences were detected. These results are consistent with the involvement of Ty1 in the generation of Y′ cDNA and with Y′HIS3 cDNA being incorporated into Y′ arrays in some cases.
PCR products corresponding to structure I in Fig. 6 could indicate that Y′HIS3 cDNA is adjacent to a downstream Y′ element in a Y′ array. However, Y′ elements can also be present as extrachromosomal circles (21), and a PCR product corresponding to structure I could be obtained if Y′HIS3 cDNA was present as a circular molecule. In the nine cases for which a downstream Y′ element or Ty1 sequences were not identified, Y′HIS3 cDNA could be the terminal Y′ in a subtelomeric Y′ array. Alternatively, Y′HIS3 could be adjacent to Y′ or Ty1 sequences that were not amplified efficiently with the primers we used or to other sequences in the genome. Additional PCR analyses indicated that Y′his3AI was still present in 71 of the 72 His+ strains, and his3Δ1 was still present in all 23 JC3840/1 His+ strains, a finding consistent with Y′HIS3 cDNA being incorporated at new genomic sites, rather than frequently recombining with existing his3 alleles.
The Ty1 sequences identified adjacent to Y′HIS3 in 53 cases were present in the same transcriptional orientation as Y′HIS3 in 13 instances (for examples, see Fig. 6, structures IV and VI) or the opposite orientation in 34 instances (structures II, III, V, and VII). Ty1 sequences were present at the last six junctions in both orientations, suggesting that the Ty1 sequences were rearranged, but none of these HIS3-Ty1 junctions were sequenced. Sequence analysis of 28 HIS3-Ty1 junction PCR products determined that the junctions between Y′HIS3 and Ty1 sequences occurred at oligo(A) tracts in 27 of 28 cases. The oligo(A) tract was present either near A-rich sequences in the HIS3 promoter or ∼15 bp downstream of a Y′ polyadenylation site, AAGAAA (28), located 50 bp downstream of his3AI in Y′ (Fig. 6, structures II to VI). This finding is consistent with reverse transcription beginning at the poly(A) tail of Y′HIS3 RNA. In the one cDNA lacking an oligo(A) tract, the junction occurred between sites in the HIS3 promoter and adjacent to the Ty1 primer-binding site containing 9/11 bp of microhomology (Fig. 6, structure VII).
The junction between the oligo(A) tract of Y′HIS3 and Ty1 occurred at the 5′ or 3′ most nucleotide of the Ty1 LTR in 18 of the 28 sequenced junctions (structures II, III, and IV, Fig. 6). For one of these 18 junctions, the 3′ end of the LTR was preceded by 3 bp of the Ty1 primer-binding site (structure III), and for another of these 18 junctions, 7 bp of the Ty1 polypurine tract preceded the 5′ LTR (structure IV). Complete LTRs are only present in Ty1 cDNA, and not in Ty1 RNA, so it is likely that Ty1 cDNA served as a primer for reverse transcription of polyadenylated Y′HIS3 RNA in the formation of these events. Seven junctions occurred in TYA1 or TYB1 with the Ty1 sequences in the opposite orientation (structure V) or same orientation (structure VI) as Y′HIS3. Ty1 sequences at these junctions were circularly permuted. In two of five (structure V) and in two of two cases (structure VI) one to three A's were present in the Ty1 sequences at the junction with the oligo(A) sequence, indicating that template switching may have occurred with very limited microhomology. Altogether, structures II to VII represent 26 of the 28 sequenced junctions.
Unmarked Y′-oligo(A)-LTR-TYB1 cDNA is relatively abundant in telomerase-negative survivors.
In order to demonstrate that the junction between Y′HIS3 cDNA and Ty1 sequences was not an artifact of the His+ selection, we screened genomic DNA from 10 JC3840/1 and all 6 JC3723:X1 survivors for unmarked Y′-Ty1 cDNA junctions by PCR with a primer to the 3′ end of the Y′ coding region and a second primer to unique sequences in either TYA1 or TYB1 (near the 5′ or 3′ ends of Ty1, respectively). A discrete product of the size predicted for a junction occurring between Y′ sequence just downstream of the Y′ poly(A) signal sequence and the complete 3′ end of Ty1 was generated with the TYB1 primer from all 16 samples, as well as some faint smaller products (Fig. 7A and B). Reactions with the TYA1 primer generated a product of the size expected for a junction between Y′ sequences just downstream of the poly(A) signal sequence and the 5′ end of Ty1 from only 1 of the 16 samples, in addition to producing some smaller products in all of the reactions (data not shown). These primer pairs generated extremely faint products or no products with wild-type DNA samples (Fig. 7B and data not shown). Twelve of fifteen Y′-oligo(A)-LTR-TYB1 PCR products from eight different survivors that were cloned and sequenced contained Y′ sequences extending 12 to 17 bp beyond the Y′ poly(A) signal sequence (28), followed by an oligo(A) tract of 13 to 34 bp, and the 3′ end of the Ty1 LTR (Fig. 7A). The shorter PCR products seen in all reactions appear to represent Y′-Ty1 junctions that occur at different sites of microhomology between Y′ and Ty1 within the coding region of Y′, as determined by sequencing cloned products.
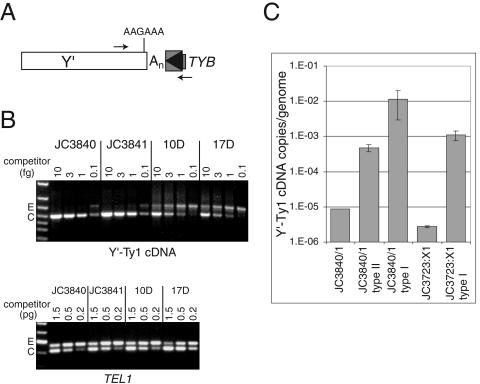
FIG. 7.
Y′-Ty1 cDNA is relatively abundant in telomerase-negative survivors. (A) A schematic representation of the most common Y′-Ty1 cDNA junction in survivors as identified by sequencing cloned PCR products. The junction depicted occurs at an oligo(A) tract (An) ∼15 bp downstream of a polyadenylation signal sequence (AAGAAA) at the 3′ end of Y′ and includes Ty1 sequences in the opposite transcriptional orientation of Y′ that begin at the exact 3′ end of the Ty1 LTR (boxed arrowhead). Small arrows above and below the junction represent primers used in competitive PCRs. (B) Representative results of quantitative PCR assays. A constant amount of genomic DNA from wild-type strains (JC3840 and JC3841) and telomerase-negative survivors (10D and 17D) was amplified with Y′ and Ty1 primers (see panel A) in the presence of various amounts of Y′-Ty1 competitor DNA (upper image) or with TEL1 primers in the presence of various amounts of TEL1 competitor DNA (lower image) (row E, endogenous product; row C, competitor product). (C) Average number of Y′-Ty1 cDNA junctions per genome determined for two wild-type strains for each strain background (JC3840/1 and JC3723:X1), six JC3840/1 type II survivors, four JC3840/1 type I survivors, and four JC3723:X1 type I survivors.
To determine whether Y′-Ty1 cDNA is abundant enough to play a role in extending eroding telomeres in survivors, we quantified the copy number per genome of Y′-Ty1 cDNA (Y′-3′ LTR junctions) by using a competitive PCR assay. The amount of a Y′-Ty1 cDNA competitor needed to produce endogenous Y′-Ty1 cDNA products and competitor products of equal intensities was determined (Fig. 7B). Various amounts of an internally deleted Y′-Ty1 competitor (made by PCR from a cloned Y′-oligo[A]-LTR-TYB1 cDNA junction) were added to PCRs with DNA from JC3840 and JC3841, two JC3723:X1 EST2 strains, six JC3840/1 type II survivors, four JC3840/1 type I survivors, and four JC3723:X1 type I survivors. The quantity of endogenous template for the single-copy gene TEL1 was also determined by using an internally deleted TEL1 competitor (Fig. 7B). A ratio of the values obtained for Y′-Ty1 cDNA and TEL1 for each sample was then used to estimate the copy number of Y′-Ty1 cDNA per genome (Fig. 7C).
The numbers of Y′-Ty1 cDNA molecules in survivors were increased 54-, 1,300-, and 400-fold over wild-type values for JC3840/1 type II, JC3840/1 type I, and JC3723:X1 type I survivors, respectively (Fig. 7C). These fold increases are minimum estimates, since even the lowest levels of competitor used did not produce endogenous products of intensities equal to competitor products in most reactions with wild-type DNA samples. The substantial increase in JC3840/1 type I survivors versus JC3840/1 type II survivors is consistent with the higher levels Y′his3AI cDNA mobility in type I survivors of this background. It is not yet known whether these junctions represent free cDNA intermediates, chromosomal sequences, or a mixture of both. The presence of Y′-Ty1 cDNA at 0.01 to 0.001 copies per genome in type I survivors is consistent with the possibility that Y′ cDNA mobility could have a role in the extension of chromosome ends in telomerase-negative survivors.
DISCUSSION
We have demonstrated that Y′ cDNA mobility is significantly elevated, more than 1,000-fold in some cases, in telomerase-negative survivors compared to telomerase-positive strains. Moreover, Ty1 plays a role in Y′ cDNA mobility, based on the following three observations: first, Y′his3AI cDNA mobility is reduced at 30°C compared to 20°C and in tec1Δ survivors; second, Y′ RNA is enriched in Ty1 VLP fractions; and third, Ty1 sequences are frequently adjacent to Y′ and Y′HIS3 cDNA. Preferential incorporation of Y′ RNA into Ty1 VLPs, even in telomerase-positive strains, suggests that Y′ may take advantage of Ty1's retrotransposition machinery in a manner that could potentially be important for the persistence of Y′ elements. This is reminiscent of the dependence of Alu sequences on the human L1 retrotransposition machinery for mobility (13). It has previously been reported that Ty1 mediates formation of pseudogenes (12, 36), but the current study demonstrates that Ty1 mobilizes an endogenous transcript at frequencies high enough to directly detect the cDNA.
A previous attempt to monitor Y′ cDNA mobility detected much lower frequencies of mobility than those reported here (42), despite the use of the same his3AI marker. However, Y′ cDNA mobility was assayed in type II survivors that were grown at 30°C in the previous study. We have shown here that type II survivors tend to have lower frequencies of Y′ cDNA mobility than type I survivors (Table 2). Moreover, growth at 30°C inhibits the frequency with which Y′ cDNA-based events are obtained (Fig. 2). We believe that these two factors alone account for the majority of the variation between our results and those previously reported (42). However, strain differences may account for a minor component of the variation.
Considerable variation was seen in the His+ frequency in different survivors (Fig. 2). Variations in Ty1 cDNA levels and in Y′his3AI RNA levels were not correlated with this variation in mobility (Fig. 4). The lack of correlation between Y′his3AI RNA levels and mobility fits with the observation that Y′ RNA is enriched in Ty1 VLPs in telomerase-positive strains, even though cDNA mobility is barely detectable in such strains. These results suggest that the increased mobility in survivors compared to telomerase-positive strains may result from an increase in the efficiency of a step(s) after Y′ transcription and incorporation into VLPs, such as Y′ cDNA formation or incorporation of the cDNA into the genome. The latter possibility is intriguing, since telomeres in telomerase-negative strains are known to be highly recombinogenic (20, 29, 42). Also, higher frequencies of mobility were observed in type I survivors, which have telomeres with long arrays of Y′ elements and short TG1-3 tracts, versus type II survivors, which have heterogeneously long TG1-3 tracts. If Y′ cDNA is incorporated at telomeres, then perhaps this process is facilitated by the structure of telomeric DNA, chromatin, or specific recombination proteins at type I telomeres. It could be that formation of a suitable recipient for recombination with Y′ cDNA is a rate-limiting step for mobility, and type I telomeres might be more suitable recipients. However, we have detected Y′ cDNA mobility in survivors derived from a rad51Δ strain (data not shown), indicating that Rad51p, which is required for formation of type I survivors (8, 25, 41), is not absolutely required for Y′ cDNA mobility. These data cannot be used to rule out incorporation of Y′ cDNA by recombination, however, since Rad51p is not required for recombination between cDNA and chromosomal sequences (11).
Priming of Y′ cDNA synthesis by Ty1 cDNA.
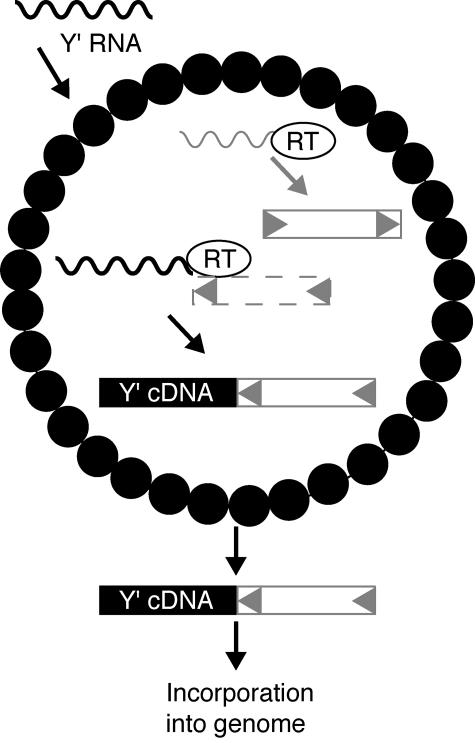
Retrovirus-like RTs are thought to require a primer to generate cDNA (2, 44). Our data demonstrate that Ty1 nucleic acids are used to prime Y′ cDNA synthesis since Y′ RNA is incorporated into Ty1 VLPs and Ty1 sequences were often identified adjacent to Y′HIS3 cDNA. Sequences of 27 of 28 HIS3-Ty1 junctions contained oligo(A) tracts, and sequences of 16 of 21 unmarked Y′-Ty1 junctions contained oligo(A) tracts (data not shown), indicating that priming often occurs at the poly(A) tail of Y′ RNA. Many junctions also contained sequences from the very 5′ or 3′ end of the Ty1 LTR, and these sequences are only present on Ty1 cDNA, not Ty1 RNA. We propose that Ty1 cDNA is frequently used to prime the synthesis of Y′ cDNA in VLPs and that the resulting chimeric cDNA molecules then leave VLPs and enter the nucleus, where they are incorporated into the genome (Fig. 8). Template switching during or at the completion of minus-strand synthesis could account for junctions in which Y′ and Ty1 sequences are in the same transcriptional orientation, whereas switching during or at the completion of plus-strand synthesis could account for junctions in which Y′ and Ty1 sequences are in opposite orientations. We cannot rule out the possibility that Ty1 RNA may prime Y′ cDNA synthesis in some instances, however. It is unclear whether second-strand synthesis might occur in VLPs or at the site of incorporation into the genome. Some Y′HIS3 cDNAs were adjacent to the 5′ end of a second Y′ element rather than Ty1 sequences (Fig. 6, structure I). In such instances, Ty1 cDNA may still prime reverse transcription of Y′ RNA, but gene conversion of Y′ elements in telomeric arrays by Y′HIS3 cDNA would result in the loss of oligo(A)-Ty1 cDNA sequences.
FIG. 8.
Model for Y′ cDNA formation. Y′ RNA (wavy black line) is incorporated into Ty1 VLPs (bounded by small black circles). Ty1 RT, which normally reverse transcribes Ty1 RNA (wavy gray line) into Ty1 cDNA (box with gray arrowheads), uses Ty1 cDNA intermediates to prime reverse transcription of Y′ RNA to generate Y′ cDNA (black box). Dashed lines for the associated Ty1 cDNA indicate uncertainty about the exact cDNA molecules used as primers. The resulting hybrid molecules containing Y′ and Ty1 cDNA sequences leave VLPs and enter the genome.
The sequences of junctions with oligo(A) tracts indicate that Ty1 RT can switch from making Ty1 cDNA to making Y′ cDNA at sites where there is very little homology (no more than one to three nucleotides) or no homology. This same sort of template switching onto a poly(A) tail with little or no homology was observed with GAL1his3AI-derived HIS3 cDNAs (12). In the case of one Y′HIS3-Ty1 structure (structure VII in Fig. 6) and some unmarked Y′-Ty1 junctions (data not shown), junctions did not involve oligo(A) tracts, and microhomology was present between HIS3 and Ty1 or Y′ and Ty1 sequences at the junction. In the cases of GAL1his3AI-derived HIS3 cDNA and duplications of a portion of URA2, microhomology-dependent template switching between the 5′ end of his3AI RNA and Ty1 was observed, and the HIS3 or URA2 sequences were flanked on both sides by Ty1 sequences (12, 36). The sequences upstream of the Y′HIS3 cDNA are still unknown, but no Ty1 sequences have been detected there by PCR, and we have thus far been unable to identify unique DNA sequences adjacent to Y′HIS3-Ty1 cDNA by using inverse PCR (data not shown).
What are the implications of Y′ cDNA mobility in survivors?
The elevation of Y′ cDNA mobility in survivors raises the question of whether this process has any role in telomere maintenance in survivors. Y′ cDNA mobility could potentially be involved in maintaining Y′ arrays in type I survivors, if Y′-Ty1 cDNA recombines with Y′ elements at chromosome ends. Depending on whether recombination occurred with a full-length or partially eroded Y′ recipient, such recombination events could add ca. 6 to 12 kb to telomeres if full-length Ty1 elements were joined to Y′ sequences. At least ∼2 kb of Ty1 sequences have been found associated with Y′HIS3 in several His+ strains (data not shown). Each S. cerevisiae telomere is estimated to lose 3 bp of telomeric DNA per cell division in the absence of telomerase (40). To compensate for this terminal loss by mobilization of Y′-Ty1 cDNA, the frequency of Y′ cDNA mobility would have to be ∼1.6 × 10−2 events/generation (assuming additions of 6 kb and considering that DNA is lost from all 32 telomeres). The copy number of Y′-Ty1 cDNA in JC3840/1 type I survivors was (1.1 ± 0.83) × 10−2 per genome (Fig. 7C), a finding consistent with the possibility that Y′ cDNA mobility is involved in maintenance of type I telomeres.
Quantification of unmarked Y′-Ty1 cDNA suggests that the frequencies of Y′his3AI cDNA mobility reported may substantially underrepresent the frequency of endogenous Y′ cDNA mobility. The presence of his3AI and the inefficiency of splicing reduce the detection of Ty1 transposition events by ∼80-fold (9, 10), so it is possible that Y′his3AI mobility is similarly reduced compared to the cDNA mobility of unmarked Y′ elements. The Y′ copy number of our type I survivors was over 20-fold higher than in congenic telomerase-positive strains, on average, suggesting that they may have ca. 200 to 600 copies of Y′ (27). These considerations suggest that the frequency of unmarked Y′ cDNA mobility could be ≥16,000-fold higher than the reported frequency of Y′his3AI mobility in type I survivors.
Y′ cDNA mobility could also reflect a more general process that might increase genetic variability during senescence and survivor formation. It is not yet known whether the mobilization of other cellular transcripts is also increased in yeast survivors. Since prior work is consistent with Ty1 having a more general role in mobilizing cellular transcripts (12), then the increased activity of Ty1 during senescence and survivor formation (38) might lead to the formation of various pseudogenes. These duplications could provide genetic variability during a time of significant stress (senescence) and potentially allow the selection of particular variants to occur. Although elevated mobilization of cDNA could also be detrimental (by insertional mutagenesis, for example), from a population standpoint it may be more advantageous to provide the potential for a fraction of cells to survive, when otherwise most or all cells might die. The observation that only a fraction of yeast cells recover from senescence induced by the absence of telomerase activity (29) is consistent with rare events or rare variations contributing to recovery.
Acknowledgments
We thank V. Lundblad and V. Zakian for kindly providing plasmids. We also thank C. Coros, K. Derbyshire, and D. Scholes for critically reviewing the manuscript. We thank Francisco Martinez Murillo of the Johns Hopkins Medicine Microarray Facility for assistance and helpful discussions. We thank the Wadsworth Center Molecular Genetics Core for DNA sequencing and oligonucleotide synthesis.
This study was supported in part by grants GM36481 (J.D.B.) and GM52072 (M.J.C.) from the National Institutes of Health.
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1993. Current protocols in molecular biology, vol. 2. John Wiley & Sons, Inc., New York, N.Y.
- 2.Bibillo, A., and T. H. Eickbush. 2002. The reverse transcriptase of the R2 non-LTR retrotransposon: continuous synthesis of cDNA on non-continuous RNA templates. J. Mol. Biol. 316:459-473. [DOI] [PubMed] [Google Scholar]
- 3.Blackburn, E. H. 2001. Switching and signaling at the telomere. Cell 106:661-673. [DOI] [PubMed] [Google Scholar]
- 4.Boeke, J. D., D. J. Garfinkel, C. A. Styles, and G. R. Fink. 1985. Ty elements transpose through an RNA intermediate. Cell 40:491-500. [DOI] [PubMed] [Google Scholar]
- 5.Brachmann, C. B., A. Davies, G. J. Cost, E. Caputo, J. Li, P. Hieter, and J. D. Boeke. 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14:115-132. [DOI] [PubMed] [Google Scholar]
- 6.Bryk, M., M. Banerjee, D. Conte, and M. J. Curcio. 2001. The Sgs1 helicase of Saccharomyces cerevisiae inhibits retrotransposition of Ty1 multimeric arrays. Mol. Cell. Biol. 21:5374-5388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chan, C. S., and B. K. Tye. 1983. Organization of DNA sequences and replication origins at yeast telomeres. Cell 33:563-573. [DOI] [PubMed] [Google Scholar]
- 8.Chen, Q., A. Ijpma, and C. W. Greider. 2001. Two survivor pathways that allow growth in the absence of telomerase are generated by distinct telomere recombination events. Mol. Cell. Biol. 21:1819-1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Curcio, M. J., and D. J. Garfinkel. 1991. Single-step selection for Ty1 element retrotransposition. Proc. Natl. Acad. Sci. USA 88:936-940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Curcio, M. J., A. M. Hedge, J. D. Boeke, and D. J. Garfinkel. 1990. Ty RNA levels determine the spectrum of retrotransposition events that activate gene expression in Saccharomyces cerevisiae. Mol. Gen. Genet. 220:213-221. [DOI] [PubMed] [Google Scholar]
- 11.Derr, L. K. 1998. The involvement of cellular recombination and repair genes in RNA-mediated recombination in Saccharomyces cerevisiae. Genetics 148:937-945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Derr, L. K., J. N. Strathern, and D. J. Garfinkel. 1991. RNA-mediated recombination in S. cerevisiae. Cell 67:355-364. [DOI] [PubMed] [Google Scholar]
- 13.Dewannieux, M., C. Esnault, and T. Heidmann. 2003. LINE-mediated retrotransposition of marked Alu sequences. Nat. Genet. 35:41-48. [DOI] [PubMed] [Google Scholar]
- 14.Dolinski, K., R. Balakrishnan, K. R. Christie, M. C. Costanzo, S. S. Dwight, S. R. Engel, D. G. Fisk, J. E. Hirschman, E. L. Hong, L. Issel-Tarver, A. Sethuraman, C. L. Theesfeld, G. Binkley, C. Lane, M. Schroeder, S. Dong, S. Weng, R. Andrada, D. Botstein, and J. M. Cherry. Saccharomyces Genome Database. [Online.] http://www.yeastgenome.org/.
- 15.Eichinger, D. J., and J. D. Boeke. 1988. The DNA intermediate in yeast Ty1 element transposition copurifies with virus-like particles: cell-free Ty1 transposition. Cell 54:955-966. [DOI] [PubMed] [Google Scholar]
- 16.Enomoto, S., L. Glowczewski, and J. Berman. 2002. MEC3, MEC1, and DDC2 are essential components of a telomere checkpoint pathway required for cell cycle arrest during senescence in Saccharomyces cerevisiae. Mol. Biol. Cell 13:2626-2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Esnault, C., J. Maestre, and T. Heidmann. 2000. Human LINE retrotransposons generate processed pseudogenes. Nat. Genet. 24:363-367. [DOI] [PubMed] [Google Scholar]
- 18.Ferreira, M. G., K. M. Miller, and J. P. Cooper. 2004. Indecent exposure: when telomeres become uncapped. Mol. Cell 13:7-18. [DOI] [PubMed] [Google Scholar]
- 19.Fu, W., and A. Rein. 1993. Maturation of dimeric viral RNA of Moloney murine leukemia virus. J. Virol. 67:5443-5449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hackett, J. A., D. M. Feldser, and C. W. Greider. 2001. Telomere dysfunction increases mutation rate and genomic instability. Cell 106:275-286. [DOI] [PubMed] [Google Scholar]
- 21.Horowitz, H., and J. E. Haber. 1985. Identification of autonomously replicating circular subtelomeric Y′ elements in Saccharomyces cerevisiae. Mol. Cell. Biol. 5:2369-2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ijpma, A. S., and C. W. Greider. 2003. Short telomeres induce a DNA damage response in Saccharomyces cerevisiae. Mol. Biol. Cell 14:987-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Laloux, I., E. Dubois, M. Dewerchin, and E. Jacobs. 1990. TEC1, a gene involved in the activation of Ty1 and Ty1-mediated gene expression in Saccharomyces cerevisiae: cloning and molecular analysis. Mol. Cell. Biol. 10:3541-3550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lawler, J. F. J., D. P. Haeusser, A. Dull, J. D. Boeke, and J. B. Keeney. 2002. Ty1 defect in proteolysis at high temperature. J. Virol. 76:4233-4240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Le, S., J. K. Moore, J. E. Haber, and C. W. Greider. 1999. RAD50 and RAD51 define two pathways that collaborate to maintain telomeres in the absence of telomerase. Genetics 152:143-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lingner, J., T. R. Cech, T. R. Hughes, and V. Lundblad. 1997. Three ever shorter telomere (EST) genes are dispensable for in vitro yeast telomerase activity. Proc. Natl. Acad. Sci. USA 94:11190-11195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Louis, E. J. 1995. The chromosome ends of Saccharomyces cerevisiae. Yeast 11:1553-1573. [DOI] [PubMed] [Google Scholar]
- 28.Louis, E. J., and J. E. Haber. 1992. The structure and evolution of subtelomeric Y′ repeats in Saccharomyces cerevisiae. Genetics 131:559-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lundblad, V., and E. H. Blackburn. 1993. An alternative pathway for yeast telomere maintenance rescues est1− senescence. Cell 73:347-360. [DOI] [PubMed] [Google Scholar]
- 30.Lundblad, V., and J. W. Szostak. 1989. A mutant with a defect in telomere elongation leads to senescence in yeast. Cell 57:633-643. [DOI] [PubMed] [Google Scholar]
- 31.McEachern, M. J., and E. H. Blackburn. 1996. Cap-prevented recombination between terminal telomeric repeat arrays (telomere CPR) maintains telomeres in Kluyveromyces lactis lacking telomerase. Genes Dev. 10:1822-1834. [DOI] [PubMed] [Google Scholar]
- 32.McEachern, M. J., A. Krauskopf, and E. H. Blackburn. 2000. Telomeres and their control. Annu. Rev. Genet. 34:331-358. [DOI] [PubMed] [Google Scholar]
- 33.Nakamura, T. M., J. P. Cooper, and T. R. Cech. 1998. Two modes of survival of fission yeast without telomerase. Science 282:493-496. [DOI] [PubMed] [Google Scholar]
- 34.Paquin, C. E., and V. M. Williamson. 1984. Temperature effects on the rate of Ty transposition. Science 226:53-55. [DOI] [PubMed] [Google Scholar]
- 35.Rose, A., and J. Broach. 1990. Propagation and expression of cloned genes in yeast: 2-micron circle-based vectors. Methods Enzymol. 185:234-279. [DOI] [PubMed] [Google Scholar]
- 36.Schacherer, J., Y. Tourrette, J. L. Souciet, S. Potier, and J. De Montigny. 2004. Recovery of a function involving gene duplication by retroposition in Saccharomyces cerevisiae. Genome Res. 14:1291-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Scholes, D. T., M. Banerjee, B. Bowen, and M. J. Curcio. 2001. Multiple regulators of Ty1 transposition in Saccharomyces cerevisiae have conserved roles in genome maintenance. Genetics 159:1449-1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Scholes, D. T., A. E. Kenny, E. R. Gamache, Z. Mou, and M. J. Curcio. 2003. Activation of a LTR-retrotransposon by telomere erosion. Proc. Natl. Acad. Sci. USA 100:15736-15741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Singer, M. S., and D. E. Gottschling. 1994. TLC1: template RNA component of Saccharomyces cerevisiae telomerase. Science 266:404-409. [DOI] [PubMed] [Google Scholar]
- 41.Teng, S. C., J. Chang, B. McCowan, and V. A. Zakian. 2000. Telomerase-independent lengthening of yeast telomeres occurs by an abrupt Rad50p-dependent, Rif-inhibited recombinational process. Mol. Cell. Biol. 6:947-952. [DOI] [PubMed] [Google Scholar]
- 42.Teng, S. C., and V. A. Zakian. 1999. Telomere-telomere recombination is an efficient bypass pathway for telomere maintenance in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:8083-8093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Voytas, D. F., and J. D. Boeke. 2002. Ty1 and Ty5 of Saccharomyces cerevisiae, p. 631-662. In N. Craig, R. Craigie, M. Gellert, and A. Lambowitz (ed.), Mobile DNA II. American Society for Microbiology, Washington, D.C.
- 44.Wilhelm, M., and F. X. Wilhelm. 2001. Reverse transcription of retroviruses and LTR retrotransposons. Cell Mol. Life Sci. 58:1246-1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Winzeler, E. A., D. D. Shoemaker, A. Astromoff, H. Liang, and K. Anderson. 1999. Functional characterization of the Saccharomyces cerevisiae genome by gene deletion and parallel analysis. Science 285:901-906. [DOI] [PubMed] [Google Scholar]
- 46.Wodicka, L., H. Dong, M. Mittmann, M. H. Ho, and D. J. Lockhart. 1997. Genome-wide expression monitoring in Saccharomyces cerevisiae. Nat. Biotechnol. 15:1359-1367. [DOI] [PubMed] [Google Scholar]
- 47.Yamada, M., N. Hayatsu, A. Matsuura, and F. Ishikawa. 1998. Y′-Help1, a DNA helicase encoded by the yeast subtelomeric Y′ element, is induced in survivors defective for telomerase. J. Biol. Chem. 273:33360-33366. [DOI] [PubMed] [Google Scholar]
- 48.Zakian, V. A. 1996. Structure, function, and replication of Saccharomyces cerevisiae telomeres. Annu. Rev. Genet. 30:141-172. [DOI] [PubMed] [Google Scholar]