Abstract
The large protein kinases, ataxia-telangiectasia mutated (ATM) and ATM-Rad3-related (ATR), orchestrate DNA damage checkpoint pathways. In budding yeast, ATM and ATR homologs are encoded by TEL1 and MEC1, respectively. The Mre11 complex consists of two highly related proteins, Mre11 and Rad50, and a third protein, Xrs2 in budding yeast or Nbs1 in mammals. The Mre11 complex controls the ATM/Tel1 signaling pathway in response to double-strand break (DSB) induction. We show here that the Mre11 complex functions together with exonuclease 1 (Exo1) in activation of the Mec1 signaling pathway after DNA damage and replication block. Mec1 controls the checkpoint responses following UV irradiation as well as DSB induction. Correspondingly, the Mre11 complex and Exo1 play an overlapping role in activation of DSB- and UV-induced checkpoints. The Mre11 complex and Exo1 collaborate in producing long single-stranded DNA (ssDNA) tails at DSB ends and promote Mec1 association with the DSBs. The Ddc1-Mec3-Rad17 complex associates with sites of DNA damage and modulates the Mec1 signaling pathway. However, Ddc1 association with DSBs does not require the function of the Mre11 complex and Exo1. Mec1 controls checkpoint responses to stalled DNA replication as well. Accordingly, the Mre11 complex and Exo1 contribute to activation of the replication checkpoint pathway. Our results provide a model in which the Mre11 complex and Exo1 cooperate in generating long ssDNA tracts and thereby facilitate Mec1 association with sites of DNA damage or replication block.
The maintenance of genome stability is critical to the survival and propagation of all cellular organisms. In response to DNA damage, cells activate a complex signaling pathway to promote DNA repair and induce cell cycle arrest. When DNA replication is blocked, cells also activate signaling pathways to facilitate DNA synthesis and prevent entry into M phase. These signaling pathways are called DNA damage or replication checkpoints in eukaryotes (11). These checkpoint pathways transmit signals through evolutionarily conserved protein kinases (1, 64). Two large protein kinases, ataxia-telangiectasia mutated (ATM) and ATM-Rad3-related (ATR), are the central components of the checkpoint pathway in mammalian cells. Whereas ATM responds primarily to double-strand breaks (DSBs) of DNA, ATR functions in responses to DSB induction as well as other types of DNA insults, such as UV-induced lesions and stalled replication. These large kinases regulate the activation of two downstream protein kinases, Chk1 and Chk2.
In the budding yeast Saccharomyces cerevisiae, MEC1 and TEL1 encode ATR- and ATM-related proteins, respectively. Mec1 responds to a variety of types of DNA damage and plays a central role in DNA damage and replication checkpoint controls (29), whereas Tel1 plays only a minor role in response to DSBs (35, 39, 50). Mec1 physically interacts with Ddc2 (also called Lcd1 and Pie1), a protein related to the mammalian ATR-interacting protein ATRIP (6, 41, 47, 59). Mec1 and Ddc2 function in the form of the Mec1-Ddc2 complex, and both localize to sites of DNA damage and replication block (23, 25, 30, 32, 40, 48). Replication protein A (RPA) binds single-stranded DNA (ssDNA), which is generated during DNA repair and replication processes (61). Recent evidence supports a model in which Ddc2 recognizes DNA damage by interacting with RPA-coated ssDNA and enables the Mec1-Ddc2 complex to associate with sites of DNA damage (65). Similar to Mec1, Tel1 localizes to DSB lesions (38). However, whether the Tel1 localization requires RPA function is not known. Kinases related to mammalian Chk1 and Chk2 are called Chk1 and Rad53, respectively, in budding yeast (64). Rad53 plays a critical role in checkpoint responses to DNA damage throughout the cell cycle as well as DNA replication block (29), whereas Chk1 acts at least in part at G2/M (49). After DNA damage or replication block, Rad53 becomes phosphorylated and activated in a Mec1- and Tel1-dependent manner (39, 50, 58). Rad53 activation leads to cell cycle arrest and transcription of genes required for DNA synthesis and repair (11, 29). Chk1 is phosphorylated after DNA damage as well, and its phosphorylation requires Mec1 function (49).
Phosphorylation and activation of Rad53 are also dependent on DDC1, MEC3, RAD17, and RAD24 (29). The Ddc1, Mec3, and Rad17 proteins are homologous to the mammalian Rad9, Hus1, and Rad1, respectively, and are all structurally related to PCNA (64). Rad24, a mammalian Rad17 homolog, is structurally similar to components of replication factor C (RFC) (64). Ddc1, Mec3, and Rad17 form a PCNA-related complex (64), whereas Rad24 forms an RFC-related complex with Rfc2, Rfc3, Rfc4, and Rfc5 (17, 36). The Rad24 complex regulates the recruitment of the Ddc1 complex to sites of DNA damage (25, 32). Localization of the Ddc1 complex to sites of DNA damage is also dependent on RPA function (66). The Rad24 and Ddc1 complexes play crucial roles in the activation of the Mec1 signaling pathway (29) but appear to function in parallel with the Tel1 pathway (56).
The Mre11 complex consists of two highly conserved proteins, Mre11 and Rad50, and a third protein, either Xrs2 in budding yeast or Nbs1 in mammals (8, 20). The Mre11 complex mediates diverse functions in several aspects of DNA metabolism, including DSB repair and telomere maintenance. The Mre11 complex is implicated in two different repair processes of DSB: homologous recombination and nonhomologous end joining. In DSB processing, the Mre11 complex is primarily involved in the generation of the 3′ ssDNA tails. Exonuclease 1 (Exo1) is highly conserved among eukaryotes and possesses 5′ to 3′ double-stranded DNA (dsDNA) exonuclease activity (13, 52). In budding yeast, Exo1 acts in concert with the Mre11 complex in DNA repair and homologous recombination (34, 54). Although the Mre11 complex behaves as 5′ to 3′ exonuclease in vivo, Mre11 orthologs have 3′ to 5′ dsDNA exonuclease and ssDNA endonuclease activities in vitro (8, 20). Moreover, nuclease-deficient mre11 mutants show the wild-type level of DSB processing in mitosis (33). Thus, how nuclease activities of the Mre11 complex contribute to ssDNA accumulation remains to be determined.
In addition to DSB repair, the Mre11 complex is required for checkpoint responses after DSB induction (8). Studies of mammalian cells have established that the Mre11 complex controls the ATM signaling pathway (22, 43). Mutations in ATM lead to ataxia-telangiectasia, and cells from these patients are hypersensitive to DSB-inducing agents and defective in the checkpoint activation. Hypomorphic mutations in Mre11 and Nbs1 cause ataxia-telangiectasia-like disease and Nijmegen breakage syndrome, respectively, disorders that are phenotypically similar to ataxia-telangiectasia. ATM activation correlates with autophosphorylation on Ser 1981 (2), and this phosphorylation requires functions of the Mre11 complex (4, 57). Recent evidence suggests that the Mre11 complex modulates the substrate recognition of ATM by direct interaction (26). Furthermore, the Mre11 complex controls the accumulation of ATM at DSB lesions (24). In budding yeast, the Mre11 complex controls the Tel1 signaling pathway (9, 56). Tel1 interacts with Xrs2 in a manner dependent on the Xrs2 C terminus (38). The C-terminal truncation impairs the Tel1 localization to DSBs and diminishes Tel1-dependent signaling (38). Thus, the functions of ATM/Tel1 and the Mre11 complex in the cellular DNA damage response are conserved between mammals and budding yeast. Although the Mre11 complex is required for activation of the ATM/Tel1 signaling pathway, it does control the ATM/Tel1-independent pathway. For example, in budding yeast, the Mre11 complex is essential for cell survival after DNA damage (8, 20), but no apparent survival defect is associated with tel1Δ mutation (35, 39, 50). Furthermore, the Mre11 complex is required for the G2/M-phase DNA damage checkpoint after DSB induction (19), whereas Tel1 contributes little to the checkpoint responses (39).
In this study, we show that the Mre11 complex functions in concert with Exo1 to activate the Mec1 signaling pathway. The Mre11 complex and Exo1 collaborate in ssDNA accumulation at DSB ends and promote Mec1 association with DSBs. In contrast, the Mre11 complex and Exo1 are dispensable for Ddc1 association with DSBs. The Mre11 complex and Exo1 are not only required for checkpoint responses to DSBs but are also involved in checkpoint activation after UV irradiation. Moreover, they contribute to activation of DNA replication checkpoints as Mec1 does. We propose a model in which the Mre11 complex and Exo1 cooperate in generating long ssDNA tails and thereby facilitate Mec1 association with sites of DNA damage or stalled replication.
MATERIALS AND METHODS
Plasmids and strains.
To express the HA-tagged Chk1 protein, the DNA sequences encoding the HA epitopes were attached in frame to the C-terminus end of Chk1. The entire region of CHK1, lacking the termination codon and the 3′-untranslated region, was amplified by PCR. The CHK1-HA plasmid (YCpT-CHK1-HA) was constructed by ligation of the EcoRI-BglII-treated PCR fragment and a BamHI-SalI fragment encoding four hemagglutinin (HA) epitopes with an EcoRI-SalI-linearized YCplac22 (16). The plasmids to construct mre11Δ and exo1Δ mutations were obtained from H. Tsubouchi, H. Ogawa, and T. Ogawa (54). YCpA-GAL-HO and YCpT-RAD53-HA were previously described (38). Cells carrying the rfa1-t11 mutation (55) were obtained by transforming the pKU2-t11 plasmid (obtained from K. Umezu and R. Kolodner) after linearization by PCR. Other strain constructs were described elsewhere (25, 37, 38, 59). All the strains used in this study are isogenic to KSC1516 (37) and are listed in Table 1.
TABLE 1.
Strains used in this studya
| Strain | Genotype |
|---|---|
| KSC1560 | sml1Δ::LEU2 |
| KSC1562 | xrs2Δ::hisG sml1Δ::LEU2 |
| KSC1563 | xrs2-11::KanMX sml1Δ::LEU2 |
| KSC1661 | tel1Δ::KanMX sml1Δ::LEU2 |
| KSC1561 | mec1Δ::LEU2 sml1Δ::LEU2 |
| KSC1620 | xrs2Δ::hisG |
| KSC1933 | mre11Δ::hisG |
| KSC1846 | exo1Δ::URA3 |
| KSC1979 | xrs2Δ::hisG exo1Δ::URA3 |
| KSC1980 | mre11Δ::hisG exo1Δ::URA3 |
| KSC1982 | rfa1-t11 |
| KSC1983 | rfa1-t11 xrs2Δ::hisG |
| KSC1512 | MEC1-HA::TRP1 |
| KSC1632 | MEC1-HA::TRP1 xrs2Δ::hisG |
| KSC1633 | MEC1-HA::TRP1 exo1Δ::URA3 |
| KSC1634 | MEC1-HA::TRP1 xrs2Δ::hisG exo1Δ::URA3 |
| KSC1984 | MEC1-HA::TRP1 rfa1-t11 |
| KSC1985 | MEC1-HA::TRP1 rfa1-t11 xrs2Δ::hisG |
| KSC1637 | DDC1-HA::TRP1 |
| KSC1638 | DDC1-HA::TRP1 xrs2Δ::hisG |
| KSC1639 | DDC1-HA::TRP1 exo1Δ::URA3 |
| KSC1640 | DDC1-HA::TRP1 xrs2Δ::hisG exo1Δ::URA3 |
| KSC1986 | DDC1-HA::TRP1 rfa1-t11 |
| KSC1987 | DDC1-HA::TRP1 rfa1-t11 xrs2Δ::hisG |
All the strains are isogenic to KSC1516 (MATa-inc ADH4cs::HIS2 ade1 his2 leu2 trp1 ura3) (37).
Immunoblotting.
Immunoblotting was performed as described previously (37). To examine Rad53 phosphorylation after exposure to phleomycin or UV light, cells carrying YCpT-RAD53-HA were grown in yeast extract-peptone-dextrose (YEPD) and then arrested at G1 or G2/M after a 2-h incubation with 6 μg of α-factor/ml or 15 μg of nocodazole/ml, respectively. After arrest, cells were treated with 50 μg of phleomycin/ml in the presence of α-factor or nocodazole. Alternatively, cells were spread out on YEPD plates and immediately irradiated with a 254-nm UV lamp at 75 J/m2. After UV irradiation, cells were released into fresh YEPD medium containing α-factor or nocodazole. To examine Rad53 phosphorylation after hydroxyurea (HU) treatment, cells were arrested at G1 after incubation with 6 μg of α-factor/ml for 2 h and then released into medium containing 10-mg/ml HU. To examine the HO-induced Rad53 phosphorylation, cells carrying YCpA-GAL-HO and YCpT-RAD53-HA were grown in selective medium containing sucrose and then arrested with 15 μg of nocodazole/ml for 2 h at G2/M. To induce HO expression, galactose was added to the culture to 2% at a final concentration. Cells carrying YCpT-CHK1-HA were treated similarly to monitor the phosphorylation status of Chk1.
Phleomycin and UV synchrony experiments and cell survival assays.
Phleomycin and UV synchrony experiments were performed as previously described (39, 59). Log-phase cultures were arrested with 15-μg/ml nocodazole for 2 h to synchronize cells at G2/M. For exposure to phleomycin, cells were incubated with 50-μg/ml phleomycin for 1 h. Cells were then washed to remove both nocodazole and phleomycin and released in fresh YEPD. For exposure to UV light, cells arrested at G2/M were spread on YEPD plates and irradiated with a 254-nm UV lamp at 75 J/m2. Cells were then washed to remove nocodazole and released into fresh YEPD. At timed intervals, cells were withdrawn and stained with DAPI (4′,6′-diamidino-2-phenylindole) for microscopic examination. Cell viability after exposure to phleomycin or UV light was determined as described previously (39, 59).
Measurement of DSB processing rate.
DNA degradation at DSB ends was monitored as previously described (37, 38). To prepare strand-specific probes, DNA fragments were amplified from genomic DNA by PCR with oligonucleotide primers flanked by a T7 promoter sequence. The sequences of the oligonucleotides are available upon request. 32P-labeled RNA probes were synthesized with T7 RNA polymerase.
ChIP assay.
The chromatin immunoprecipitation (ChIP) assay was performed essentially as described previously (38). Anti-HA (12CA5) antibodies and 0.1% sodium deoxycholate were replaced by anti-HA (16B2) antibodies and 0.05% sodium dodecyl sulfate, respectively. PCR was performed in a nonsaturating condition, in which the rate of PCR amplification is proportional to the concentration of substrate and cycling.
Immunofluorescence microscopic analysis.
The culture was synchronized in the G1 phase by addition of 6-μg/ml α-factor for 2 h. Cells were then washed to remove α-factor and released into YEPD containing 10-mg/ml HU. Aliquots of cells were removed and processed for indirect immunofluorescence microscopy as described previously (59).
RESULTS
Phleomycin-induced Rad53 phosphorylation in cells carrying tel1, xrs2, or mec1 mutations.
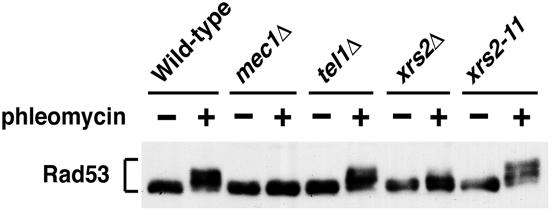
To further understand the functional link between the Mre11 complex and Tel1, we compared Rad53 phosphorylation in xrs2, tel1Δ, and mec1Δ mutants after DNA damage (Fig. 1). Phosphorylation of Rad53 can be detected as slower-migrating forms on immunoblots. Cells expressing HA-tagged Rad53 were arrested at G2/M phase with nocodazole and then incubated with the DSB-inducing agent phleomycin. After treatment with phleomycin, cells were subjected to immunoblotting analysis with anti-HA antibodies. Whereas xrs2Δ mutants exhibited a partial defect in Rad53 phosphorylation compared with wild-type cells, no phosphorylation defect was observed in tel1Δ mutants (39). The xrs2-11 mutation confers the same phenotype as tel1Δ mutation (38). Consistently, efficient phosphorylation was detected in xrs2-11 mutants. In contrast, mec1Δ mutants showed a significant phosphorylation defect (39). These results raise a possibility that the Mre11 complex may function in the same signaling pathway as Mec1 does.
FIG. 1.
Effect of mec1Δ, tel1Δ, or xrs2 mutations on phleomycin-induced Rad53 phosphorylation. Cells carrying YCpT-RAD53-HA were arrested at G2/M with nocodazole, and untreated (−) or treated (+) with phleomycin, maintaining the G2/M arrest for 60 min. Cells were then collected and subjected to immunoblotting analysis with anti-HA antibodies. Strains used are wild type (KSC1560), mec1Δ (KSC1561), tel1Δ (KSC1661), xrs2Δ (KSC1562), and xrs2-11 (KSC1563). All the strains contain an sml1Δ mutation that suppresses the lethality of mec1Δ mutants (63).
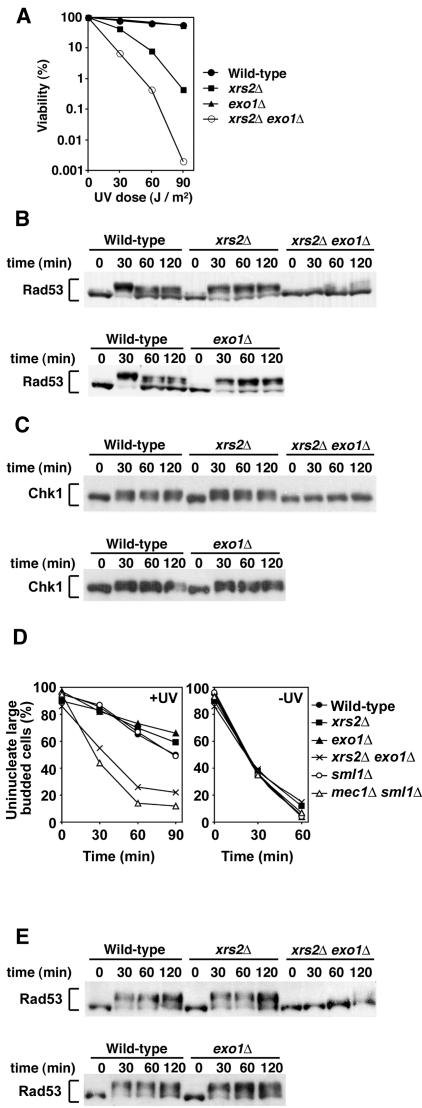
Effect of exo1Δ mutation in combination with xrs2Δ or mre11Δ mutation on cellular responses to phleomycin-induced DNA damage.
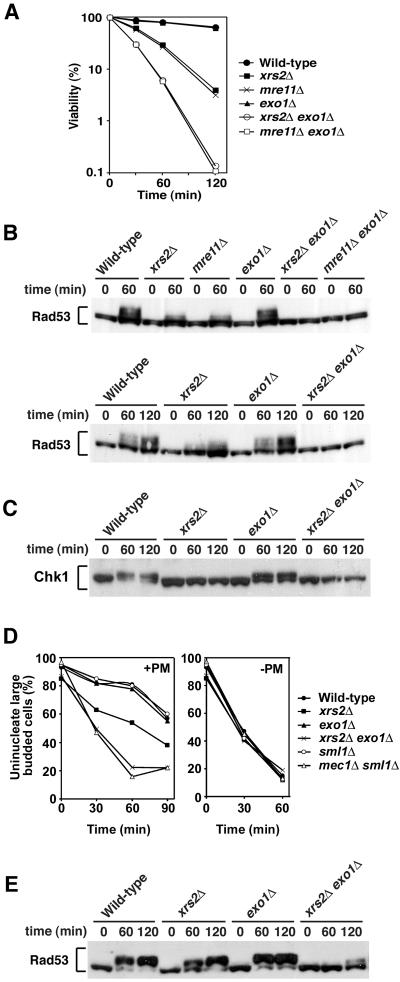
Exo1 possesses a 5′ to 3′ exonuclease activity and acts redundantly with the Mre11 complex in homologous recombination and other aspects of DNA damage metabolism (34, 54). We therefore investigated whether the Mre11 complex and Exo1 cooperate in checkpoint responses to phleomycin-induced DNA damage. We first tested the effect of exo1Δ mutation on cell survival of xrs2Δ or mre11Δ mutants after phleomycin treatment (Fig. 2A). The introduction of exo1Δ mutation exacerbated sensitivity of xrs2Δ and mre11Δ mutants to phleomycin, although the exo1Δ single mutation did not confer sensitivity. We then monitored the phosphorylation status of Rad53 in xrs2Δ exo1Δ and mre11Δ exo1Δ double mutants after phleomycin treatment at the G2/M phase (Fig. 2B). Rad53 was phosphorylated in exo1Δ mutants as efficiently as in wild-type cells. However, its phosphorylation was decreased in xrs2Δ or mre11Δ single mutants and was not detected in xrs2Δ exo1Δ or mre11Δ exo1Δ double mutants. No difference in Rad53 phosphorylation was observed in cells carrying mre11Δ or xrs2Δ mutations, consistent with the current view that Mre11, Rad50, and Xrs2 form a complex and function together (8, 20). In addition to Rad53, Chk1 controls the G2/M-phase DNA damage checkpoint pathway (49). We then monitored Chk1 phosphorylation after phleomycin treatment (Fig. 2C). Chk1 was phosphorylated in wild-type and exo1Δ mutants, whereas its phosphorylation was undetectable in xrs2Δ single or xrs2Δ exo1Δ double mutants.
FIG. 2.
Overlapping function of the Mre11 complex and Exo1 in response to phleomycin-induced DNA damage. (A) Effect of mre11Δ, xrs2Δ, and exo1Δ mutations on cell viability after exposure to phleomycin. Cells were grown in log phase and incubated with phleomycin. At the indicated times, aliquots of cells were collected and the viability was estimated. Strains used are wild type (KSC1516), mre11Δ (KSC1933), xrs2Δ (KSC1562), exo1Δ (KSC1846), mre11Δ exo1Δ (KSC1980), and xrs2Δ exo1Δ (KSC1979). (B) Effect of mre11Δ, xrs2Δ, and exo1Δ mutations on phleomycin-induced Rad53 phosphorylation at G2/M. Cells carrying YCpT-RAD53-HA were arrested with nocodazole and incubated with phleomycin maintaining the arrest. Cells were then collected at the indicated time and analyzed as in Fig. 1 to detect Rad53 phosphorylation. Strains used are the same as in panel A. (C) Effect of mre11Δ, xrs2Δ, and exo1Δ mutations on phleomycin-inducedChk1 phosphorylation at G2/M. Cells carrying YCpT-CHK1-HA were analyzed as in panel B to detect Chk1 phosphorylation. Strains used are wild type (KSC1516), xrs2Δ (KSC1562), exo1Δ (KSC1846), and xrs2Δ exo1Δ (KSC1979). (D) Cell cycle progression delay after phleomycin treatment in G2/M phase. Cells were synchronized with nocodazole at G2/M and then treated with phleomycin (+PM) or untreated (−PM). At the indicated times after release from nocodazole, the percentage of uninucleate large-budded cells was scored after DAPI staining. Strains used are wild type (KSC1516), xrs2Δ (KSC1562), exo1Δ (KSC1846), xrs2Δ exo1Δ (KSC1979), sml1Δ (KSC1560), and mec1Δ sml1Δ (KSC1561). (E) Effect of mre11Δ, xrs2Δ, and exo1Δ mutations on phleomycin-induced Rad53 phosphorylation at G1. Cells carrying YCpT-RAD53-HA were arrested with α-factor and incubated with phleomycin maintaining the arrest. Cells were then collected at the indicated time and analyzed as in panel B. Strains used are the same as in panel C.
We next examined the cell cycle arrest after exposure to phleomycin in xrs2Δ, exo1Δ, and their double mutants by monitoring mitotic division (Fig. 2D). When cell cultures were released from nocodazole arrest after phleomycin treatment, wild-type cells showed delayed nuclear division. Meanwhile, xrs2Δ single mutants proceeded through mitosis slightly faster than wild-type cells. Although exo1Δ mutation alone did not affect the process, its introduction further accelerated the mitotic progression in xrs2Δ mutants. We previously showed that mec1Δ mutants are defective in the mitotic delay after phleomycin treatment, whereas no apparent defect is associated with tel1Δ mutation (39). As a control, we also analyzed the mitotic progression in mec1Δ mutants (Fig. 2D). Similar defects were found in mec1Δ single and xrs2Δ exo1Δ double mutants.
Previous results indicated that the Mre11 complex is involved in checkpoint responses to DSB induction at G1 phase as well (19). We then monitored the phosphorylation status of Rad53 in G1-arrested xrs2Δ exo1Δ double mutants after phleomycin treatment (Fig. 2E). Again, Rad53 was phosphorylated in exo1Δ mutants as efficiently as in wild-type cells, whereas phosphorylation was partially decreased in xrs2Δ single mutants. Phosphorylation was further decreased in xrs2Δ exo1Δ double mutants. Together, these results indicate that the Mre11 complex collaborates with Exo1 in activating the Mec1-dependent checkpoint pathway after phleomycin treatment.
Effect of xrs2Δ and exo1Δ mutations on Mec1 and Ddc1 association with DSBs.
Mec1 forms a complex with Ddc2 (41, 47, 59), and the Mec1-Ddc2 complex localizes to sites of DNA damage (25, 32, 48). Rad24 forms an RFC-related complex and recruits the Ddc1-Mec3-Rad17 complex to DNA lesions (25, 32). Although the Mec1 and Ddc1 complexes are independently recruited to DNA lesions (25, 32), the activation of the Mec1 signaling pathway requires functions of the Rad24 and Ddc1 complexes (29). It is thus possible that the Mre11 complex and Exo1 control localization of the Mec1 and Ddc1 complexes to DNA lesions. To address this possibility, we examined the association of Mec1 and Ddc1 with the HO-induced DSB in xrs2Δ and exo1Δ mutants by ChIP assay. HO is a sequence-specific endonuclease that generates DSB. We used an experimental system in which cells contain a single HO cleavage site at the ADH4 locus (Fig. 3A), and HO is expressed from GAL-HO plasmid after incubation with galactose (38). The HO-induced DSB at the ADH4 locus is not efficiently repaired by homologous recombination and thereby activates the DNA damage checkpoint pathway at G2/M but not at G1 (42, 53).
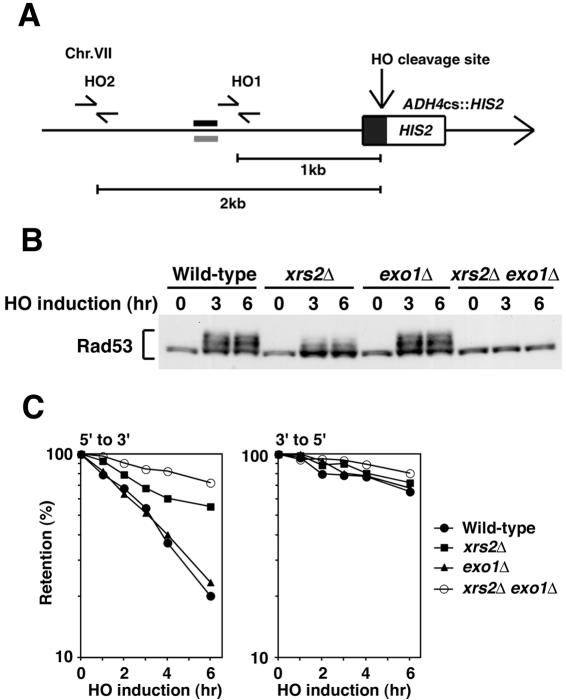
FIG. 3.
Effect of xrs2Δ and exo1Δ mutations on cellular responses to DSBs. (A) Schematic of the HO cleavage site at the ADH4 locus (ADH4cs). An HO cleavage site, marked with HIS2, was introduced at the ADH4 locus on chromosome VII. The primer pairs were designed to amplify regions 1 and 2 kb apart from the HO cleavage site. An arrow represents the telomere. The black and gray bars indicate probes to examine the rate of degradation of the DSB ends (38). (B) Rad53 phosphorylation in response to HO-induced DSBs. Cells carrying YCpT-RAD53-HA and YCpA-GAL-HO were grown in sucrose and treated with nocodazole. After arrest at G2/M, the culture was incubated with galactose to induce HO expression. Aliquots were harvested at the indicated time points and analyzed as in Fig. 1. Strains used are wild type (KSC1516), exo1Δ (KSC1846), xrs2Δ (KSC1562), and xrs2Δ exo1Δ (KSC1979). (C) Degradation of the HO-induced DSB ends. Wild-type (KSC1516), exo1Δ (KSC1846), xrs2Δ (KSC1562), and xrs2Δ exo1Δ (KSC1979) cells carrying YCpA-GAL-HO were treated as in panel B. Purified DNAs were fixed to a membrane and hybridized with RNA probes, each complementary to the 5′- to 3′- or 3′- to 5′-degrading strand.
We first monitored Rad53 phosphorylation after HO expression in xrs2Δ and exo1Δ mutants (Fig. 3B). Cells expressing Rad53-HA were transformed with the GAL-HO plasmid. Transformed cells were grown initially in sucrose to repress HO expression and then transferred to medium containing nocodazole to arrest at G2/M. After arrest, galactose was added to induce HO expression. Cells were collected at various times and subjected to immunoblotting analysis. Rad53 was phosphorylated in exo1Δ single mutants as efficiently as in wild-type cells. However, phosphorylation was partially decreased in xrs2Δ mutants and was not detectable in xrs2Δ exo1Δ double mutants. Thus, similar to phleomycin-induced lesions, HO-induced DSBs activate checkpoint responses in an Xrs2- and Exo1-dependent manner.
DSB ends are degraded primarily by 5′ to 3′ exonuclease activities, generating 3′-ended single-stranded DNA tails (60). To confirm an overlapping role of the Mre11 complex and Exo1 in DSB processing, we monitored the DNA degradation rate at DSB ends after HO expression in xrs2Δ and exo1Δ mutant cells (Fig. 3C). Cells carrying the GAL-HO plasmid were grown as above to induce HO expression. After HO expression, cells were collected at various times to prepare genomic DNA. Purified DNA was fixed to membrane filters and hybridized with strand-specific RNA probes complementary to sequences near the cleavage site at the ADH4 locus (38). The rate of 5′ to 3′ degradation of the DNA ends was partially decreased in xrs2Δ mutants as found previously (21, 38), whereas no degradation defect was observed in exo1Δ mutants. Degradation was much slower in xrs2Δ exo1Δ double mutants than in xrs2Δ single mutants, although some residual degradation was observed in the double mutants. In contrast, xrs2Δ, exo1Δ, and their double mutations had little effect on the 3′ to 5′ degradation. DSB induction by HO does not require the functions of the Mre11 complex and Exo1 (54). These results indicate that the Mre11 complex and Exo1 collaborate in the ssDNA generation at DSB ends, consistent with the previous findings that they play an overlapping role in DNA damage repair (34, 54).
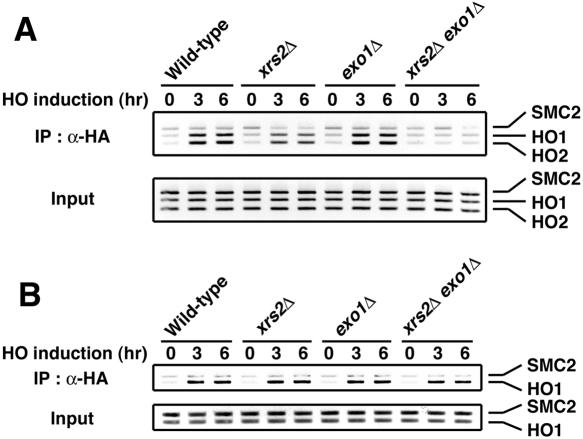
We then tested whether Mec1 associates with the HO-induced DSB in xrs2Δ and exo1Δ mutants by ChIP assay (Fig. 4A). Cells expressing HA-tagged Mec1 were transformed with the GAL-HO plasmid and grown as above to induce HO expression. Cells were collected at various times, and extracts prepared after formaldehyde cross-linking were sonicated and subjected to immunoprecipitation with anti-HA antibodies. Coprecipitated DNA was extracted and amplified by PCR using a primer set corresponding to regions near the HO restriction site or primers for the SMC2 locus containing no cleavage site. As found previously (25, 38), Mec1 associated with the HO-induced DSB in wild-type cells; PCR amplified sequences near the HO restriction site after incubation with galactose but not a control site in the SMC2 locus. Mec1 associated with the DSB efficiently in exo1Δ single mutants as well. However, the Mec1 association was partially decreased in xrs2Δ single mutants and was undetectable in xrs2Δ exo1Δ double mutants. We also examined Ddc1 association with the HO-induced DSB in those xrs2Δ and exo1Δ mutants (Fig. 4B). Cells expressing HA-tagged Ddc1 were transformed with the GAL-HO plasmid and were examined as above by ChIP assay. Similar to the Mec1 association, Ddc1 association with DSBs occurred efficiently in exo1Δ single mutants. However, unlike the Mec1 association, the Ddc1 association was neither affected by xrs2Δ single nor xrs2Δ exo1Δ double mutation. Thus, Mec1 association with DSBs correlates with the length of ssDNA at the DSB ends, whereas the generation of long ssDNA tracts is not critical for the Ddc1 association.
FIG. 4.
Effect of xrs2Δ and exo1Δ mutations on association of Mec1 and Ddc1 with the HO-induced DSB. (A) Association of Mec1 with DSBs. Cells expressing HA-tagged Mec1 were transformed with YCpA-GAL-HO plasmid. Transformed cells were grown in sucrose and treated with nocodazole. After arrest at G2/M, the culture was incubated with galactose to induce HO expression, while part of the culture was maintained in sucrose to repress HO expression. Aliquots of cells were collected at the indicated times and subjected to chromatin immunoprecipitation. PCR was carried out with the primers for the HO cleavage site at the ADH4 locus and for the control SMC2 locus (see Fig. 3A). PCR products from the respective input extracts are shown below. Strains used here are wild type (KSC1512), exo1Δ (KSC1633), xrs2Δ (KSC1632), and xrs2Δ exo1Δ (KSC1634). (B) Association of Ddc1 with the HO-induced DSB. Cells expressing Ddc1-HA were analyzed as in panel A. Strains used here are wild type (KSC1637), xrs2Δ (KSC1638), exo1Δ (KSC1639), and xrs2Δ exo1Δ (KSC1640).
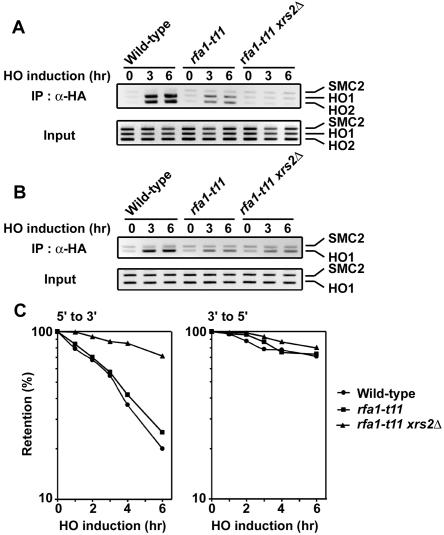
Synergetic effect of rfa1-t11 and xrs2Δ mutations on Mec1 and Ddc1 association with DSBs.
Single-stranded regions of DNA are covered with RPA that acts in most aspects of cellular DNA metabolism (61). RPA is composed of three subunits that are encoded by RFA1, RFA2, and RFA3. Recent evidence provided a model in which the RPA-coated ssDNA is an important structure to recruit the Mec1-Ddc2 and Ddc1-Mec3-Rad17 complexes to sites of DNA damage (65, 66). A mutation in RFA1, rfa1-t11, impairs association of the Mec1 and Ddc1 complexes with DNA lesions but does not affect binding of RPA to ssDNA (65). It was possible that limited ssDNA generation would further decrease association of Mec1 and Ddc1 with DSBs in rfa1-t11 mutants. We then compared association of Mec1 and Ddc1 with the HO-induced DSB in rfa1-t11 single and rfa1-t11 xrs2Δ double mutants (Fig. 5A and B). Consistent with the previous findings (65, 66), association of Mec1 and Ddc1 with DSBs was partially defective in rfa1-t11 single mutants. As expected, Mec1 association was further decreased and undetectable in rfa1-t11 xrs2Δ double mutants (Fig. 5A). However, introduction of xrs2Δ mutation did not affect Ddc1 association with DSBs in rfa1-t11 mutants (Fig. 5B).
FIG. 5.
Effect of rfa1-t11 mutation on DSB association of Mec1 and Ddc1 in xrs2Δ mutants. (A) Association of Mec1 with DSBs. Cells expressing Mec1-HA were analyzed as in Fig. 4A. Strains used here are wild type (KSC1512), rfa1-t11 (KSC1984), and rfa1-t11 xrs2Δ (KSC1985). (B) Association of Ddc1 with the HO-induced DSB. Cells expressing Ddc1-HA were analyzed as in panel A. Strains used here are wild type (KSC1637), rfa1-t11 (KSC1986), and rfa1-t11 xrs2Δ (KSC1987). (C) Degradation of the HO-induced DSB ends. Wild-type (KSC1516), rfa1-t11 (KSC1982), and rfa1-t11 xrs2Δ (KSC1983) cells carrying YCpA-GAL-HO were analyzed as in Fig. 3C.
We also monitored the DNA degradation rate at DSB ends in rfa1-t11 xrs2Δ mutant cells (Fig. 5C). Cells carrying the GAL-HO plasmid were grown as above to induce HO expression. After HO expression, cells were collected at various times to prepare genomic DNA. Purified DNA was subjected to blot hybridization analysis. The rfa1-t11 mutation alone did not affect the rate of 5′ to 3′ degradation of the DNA ends, but introduction of the rfa1-t11 mutation reduced the degradation rate in xrs2Δ mutants. No defect in the 3′ to 5′ degradation was found in rfa1-t11 single or rfa1-t11 xrs2Δ double mutants. These results are consistent with the model in which the generation of long ssDNA tracts promotes Mec1 association with DSBs but not Ddc1 association.
Effect of xrs2Δ and mre11Δ mutations on cellular responses to UV irradiation.
Exposure of cells to UV light results in checkpoint activation, and this UV-induced checkpoint activation depends on Mec1 functions. We thus investigated the effect of xrs2Δ and exo1Δ mutations on cellular responses to UV irradiation. We first examined the effect of xrs2Δ and exo1Δ mutations on viability loss after UV irradiation (Fig. 6A). Whereas xrs2Δ single mutants were sensitive to UV light, exo1Δ single mutants were as resistant as wild-type cells. However, the introduction of exo1Δ mutation increased UV sensitivity of xrs2Δ mutants. We next monitored UV-induced Rad53 phosphorylation in xrs2Δ and exo1Δ mutants (Fig. 6B). Cells expressing Rad53-HA were arrested with nocodazole at G2/M. After arrest, cells were irradiated with UV light and subjected to immunoblotting analysis. Rad53 was phosphorylated in xrs2Δ and exo1Δ single mutants within 30 min after irradiation, as found in wild-type cells. However, phosphorylation was decreased in xrs2Δ exo1Δ double mutants: no phosphorylation was observed 30 min after irradiation, although some phosphorylation was detected at later time points. We also monitored Chk1 phosphorylation after UV irradiation (Fig. 6C). Whereas xrs2Δ exo1Δ double mutation decreased UV-induced phosphorylation, no apparent phosphorylation defect was observed in either of the single mutants. We then examined the G2/M-phase cell-cycle arrest in xrs2Δ and exo1Δ mutants after UV irradiation (Fig. 6D). After UV irradiation, both xrs2Δ and exo1Δ mutants delayed mitotic progression as efficiently as wild-type cells. In contrast, xrs2Δ exo1Δ double mutants underwent mitosis faster than wild-type cells. To address the effect on activation of the G1-phase checkpoint pathway, we monitored Rad53 phosphorylation after UV irradiation in G1-arrested cells (Fig. 6E). Again, Rad53 phosphorylation was significantly decreased in xrs2Δ exo1Δ double mutants after UV irradiation, whereas no phosphorylation defect was associated with xrs2Δ or exo1Δ single mutants. Cells containing mec1 mutations failed to delay mitotic progression and became defective in Rad53 phosphorylation after exposure to UV light, whereas tel1Δ mutants did not (50, 58, 59). Thus, the Mre11 complex and Exo1 are functionally overlapped with Mec1 in UV-induced checkpoint responses.
FIG. 6.
Redundant role of Xrs2 and Exo1 in cellular responses to UV irradiation. (A) Cell viability after exposure to UV light. Viability was determined after UV irradiation at the indicated dosages. Strains used are the same as in Fig. 2C. (B) Rad53 phosphorylation after UV irradiation at G2/M. The same cells as in panel A were transformed with YCpT-RAD53-HA. Transformed cells were grown in log phaseand treated with nocodazole. After arrest at G2/M, the culture was irradiated with UV light. Cells were harvested at the indicated times and analyzed as in Fig. 1. (C) Chk1 phosphorylation after UV irradiation at G2/M. The same cells as in panel A were transformed with YCpT-CHK1-HA and analyzed as in panel B. (D) G2/M-phase cell cycle progression after exposure to UV light. Cells were grown in YEPD and arrested with nocodazole. After synchronization at G2/M, cells were irradiated with UV (+UV) or mock-treated (−UV). At the indicated times after release of cultures from nocodazole, the percentage of uninucleate large-budded cells was scored after DAPI staining. Strains used are the same as in Fig. 2D. (E) Rad53 phosphorylation after UV irradiation at G1. Cells carrying YCpT-RAD53-HA were treated with α-factor. After arrest at G1, the culture was irradiated with UV light. Cells were harvested at the indicated times and analyzed as in panel B. Strains used are the same as in panel A.
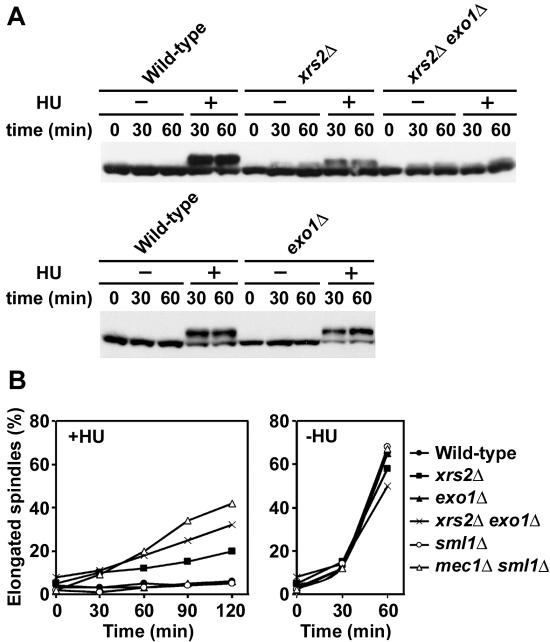
Effect of xrs2Δ and exo1Δ mutations on checkpoint responses to DNA replication block.
Mec1 controls checkpoint responses to DNA replication block as well (11). We therefore addressed whether the Mre11 complex and Exo1 control the replication checkpoint pathway. We first examined HU-induced Rad53 phosphorylation in xrs2Δ and exo1Δ mutants (Fig. 7A). Cells expressing Rad53-HA were synchronized with α-factor at G1 and released into medium containing HU. As a control, cells were also released into medium lacking HU. After release, cells were collected at various time points and subjected to immunoblotting analysis. In wild-type and exo1Δ single mutant cells, Rad53 was similarly phosphorylated in the presence of HU, whereas no phosphorylation was detected in the absence of HU. Background phosphorylation was detected in xrs2Δ single or xrs2Δ exo1Δ double mutants after release from G1 arrest. However, HU treatment increased Rad53 phosphorylation more significantly in xrs2Δ single mutants compared with xrs2Δ exo1Δ double mutants. We next monitored cell-cycle arrest in response to DNA replication block (Fig. 7B). Cells were synchronized with α-factor at G1 and released into medium containing HU. Cells were collected at various time points, and their nuclear and microtubular structures were analyzed by immunofluorescence microscopic analysis. In the presence of HU, most wild-type cells were arrested as large-budded cells with short spindles. Similar efficient arrest was observed in exo1Δ single mutants. Although both xrs2Δ single and xrs2Δ exo1Δ double mutants showed defects in blocking mitotic entry after release into HU, xrs2Δ exo1Δ double mutants were more defective than xrs2Δ single mutants. We note that xrs2Δ single and xrs2Δ exo1Δ double mutations partially delayed mitotic entry even in the absence of HU, consistent with the background Rad53 phosphorylation during S-phase progression. These results suggest that the Mre11 complex and Exo1 control activation of the replication checkpoint responses as well.
FIG. 7.
Effects of xrs2Δ and exo1Δ mutations on checkpoint responses to DNA replication block. (A) Rad53 phosphorylation after HU treatment. Cells were transformed with YCpT-RAD53-HA. Transformed cells were grown in log phase and treated with α-factor. After arrest at G1, cells were released into medium with HU (+) or without HU (−). Cells were harvested at the indicated times and analyzed as in Fig. 1. Strains used are the same as in Fig. 2C. (B) Mitotic entry after exposure to HU. Cells were grown in YEPD and arrested with α-factor at G1. Cells were then released into medium with or without HU. At the indicated times after release, the percentage of cells with elongated spindles was scored after staining with DAPI and antitubulin antibodies. Strains used are the same as in Fig. 2D.
DISCUSSION
Several studies have established that the Mre11 complex regulates activation of the Tel1 signaling pathway. In this study we demonstrate that the Mre11 complex also controls the Mec1 signaling pathway. The Mre11 complex collaborates with Exo1 in producing long 3′ ssDNA tails at DSB ends and promotes Mec1 association with the DSBs. Activation of the Mec1 signaling pathway requires the functions of the Rad24 and Ddc1 complexes. The Rad24 complex recruits the Ddc1 complex to sites of DNA damage. However, association of Ddc1 with DSBs does not require the generation of long ssDNA tracts. Mec1 is not only required for DSB-induced checkpoint responses but also involved in UV-induced checkpoint responses. Accordingly, the Mre11 complex and Exo1 play an overlapping role in activation in both DSB- and UV-induced checkpoints. The Mre11 complex and Exo1 are also involved in checkpoint responses to stalled DNA replication. Our results suggest that the Mre11 complex and Exo1 collaborate in generating long ssDNA tracts and thereby enable Mec1 to associate with DNA lesions.
Several lines of evidence have indicated that the Mre11 complex controls checkpoint responses after DSB induction (9, 19, 56). Mutations in MRE11, RAD50, or XRS2 cause a partial defect in checkpoint responses after γ-irradiation (19). Here we also show that the Mre11 complex contributes in part to checkpoint activation in response to HO-induced DSB or after phleomycin treatment. The Mre11 complex collaborates with Exo1 in DNA damage repair and homologous recombination (34, 54). Interestingly, introduction of exo1Δ mutation further impairs the checkpoint responses in mre11Δ or xrs2Δ mutants, although exo1Δ mutation alone does not. Mec1 plays a central role in activation of the checkpoint responses, whereas Tel1 contributes little to the responses (38, 39). Together, these findings indicate that the Mre11 complex and Exo1 control the Mec1 signaling pathway after DSB induction.
DSB ends are primarily degraded by 5′ to 3′ exonuclease activities, and this DSB processing is dependent on the Mre11 complex function (21, 28). We also confirmed overlapping roles of the Mre11 complex and Exo1 in the DNA degradation at DSB ends. Whereas xrs2Δ mutation delays the 5′ to 3′ DNA degradation, no apparent degradation delay is associated with exo1Δ mutation. However, introduction of the exo1Δ mutation enhanced degradation defect in xrs2Δ mutants. Little effect on the opposite 3′ to 5′ DNA degradation is observed in these single and double mutants. HO incision occurs similarly in these single and double mutants (54). Thus, the Mre11 complex and Exo1 cooperate in producing long ssDNA tracts at DSB ends. However, other proteins are likely involved in activation of the Mec1 pathway, because some residual 5′ to 3′ degradation is still observed in xrs2Δ exo1Δ double mutants (34, 54). To understand the link of the Mre11 complex and Exo1 to the Mec1 signaling pathway, we examined Mec1 association with DSBs in the xrs2Δ and exo1Δ mutants. Mec1 association with DSBs was partially decreased in xrs2Δ single mutants compared with wild-type cells, and was undetectable in xrs2Δ exo1Δ double mutants. Together, these results suggest that the Mre11 complex and Exo1 collaborate in generation of long ssDNA tails at DSB ends, thereby leading to activation of the Mec1 signaling pathway.
DSBs result from breakage of the sugar-phosphate backbone after genotoxic stress. In addition, various adductive DNA products are generated in cells that suffer the effects of genotoxic agents. UV radiation produces two major classes of DNA lesions, the cyclobutane pyrimidine dimers and 6-4 photoproducts (14). The bulky adducts on these photolesions distort the DNA helix and become substrates for nucleotide excision repair (NER) (10). NER is a multistep process that involves bipartite damage recognition, oligonucleotide excision, and gap-filling DNA synthesis (10). Removal of damaged nucleotides, followed by two DNA incisions, leaves single-stranded regions consisting of approximately 24 to 32 nucleotides (10). This ssDNA region might be involved in activation of the Mec1 signaling pathway. Although NER represents a major repair system for UV-induced photoproducts, the NER-independent mechanism contributes to repair of UV-induced lesions as well. Indeed, the Mre11 complex and Exo1 are required for UV-induced damage repair (38, 44). If the ssDNA region could not be filled immediately, endonucleases might cleave the ssDNA region, giving rise to a DSB. This DSB generation might require functions of the Mre11 complex, because Mre11 proteins possess ssDNA endonuclease activities in vitro. In any cases, once generated, DSBs would be processed by the Mre11 complex and Exo1, generating long ssDNA tracts which efficiently recruit Mec1. Alternatively, NER-independent mechanisms might convert UV-induced lesions into DSBs. If the above model were the case, NER dysfunction could also cause a checkpoint defect after UV irradiation, but would not eliminate the checkpoint response. RAD14 in budding yeast is implicated in the NER pathway (10). Consistently, rad14Δ mutations confer a defect in Rad53 phosphorylation immediately after UV irradiation (15), but its phosphorylation becomes detectable at a later time point (62).
DNA replication block after HU treatment activates checkpoint responses in a Mec1-dependent manner. Accordingly, the Mre11 complex and Exo1 are both required for phosphorylation of Rad53 after HU treatment. These results imply that the stalled replication forks are converted to lesions containing DSBs, where the Mre11 complex and Exo1 contribute to generation of long ssDNA tracts. Even in the absence of HU, xrs2Δ and xrs2Δ exo1Δ mutants exhibited a background Rad53 phosphorylation in S phase. In contrast, no apparent background phosphorylation was detected at G1 or G2/M phase (for example, see Fig. 2). Thus, the S-phase progression by itself produces DNA lesions in xrs2Δ single and xrs2Δ exo1Δ double mutants. It is possible that DSBs that arise during normal DNA replication are not repaired in xrs2Δ mutants (7). The accumulating DSB lesions might be degraded into ssDNAs by nucleases other than the Mre11 complex and Exo1, thereby activating the Mec1 signaling pathway even in the absence of HU.
Activation of the Mec1 signaling pathway also requires functions of Rad24, Ddc1, Mec3, and Rad17. Rad24 (related to human Rad17) forms an RFC-related complex with small subunits of RFC, whereas Ddc1, Mec3, and Rad17 (Rad9, Hus1, and Rad1 in humans) display sequence relatedness to PCNA and form a heterotrimer complex. As RFC loads PCNA on a primer-template junction, the Rad24 (human Rad17) complex loads the Ddc1 (human Rad9) complex on DNA in vitro (3, 12, 31, 66). Consistently, Rad24 recruits the Ddc1 complex to sites of DNA damage in vivo (25, 32). Association of the Ddc1 complex with DNA lesions also requires the function of the ssDNA-binding protein RPA. In fact, Ddc1 association with DSBs is decreased in cells carrying rfa1-t11, a mutation in RFA1 encoding the large subunit of RPA (66). Although the rfa1-t11 mutation alone does not affect DNA degradation rate at DSBs, introduction of the rfa1-t11 mutation further delayed ssDNA accumulation in xrs2Δ mutants. However, Ddc1 association with DSB in rfa1-t11 xrs2Δ double mutants was similar to that in rfa1-t11 single mutants. Ddc1 association with DSBs does not require generation of long ssDNA tracts; Ddc1 accumulates at DSBs in xrs2Δ exo1Δ mutants as efficiently as in wild-type cells. RPA thus contributes to aspects other than stabilization of long ssDNA tracts to localize the Ddc1 complex to DNA lesions. More defective DNA degradation was observed in rfa1-t11 xrs2Δ double mutants, suggesting that RPA could control the recruitment of nucleases other than the Mre11 complex. The RPA requirement was also examined in vitro using the yeast Ddc1 or human Rad9 complex, but its requirement might depend on the DNA substrate structures (3, 12, 31, 66). Circular DNA substrates with small gaps or nicks can serve as good loading templates for the yeast and human complexes in the absence of RPA (3, 31), whereas circular substrates with extended ssDNA regions require RPA for loading (66). When a linear DNA substrate is used, RPA appears to stimulate loading of the complex on the DNA and inhibit its sliding off from the substrate (12). Thus, RPA plays an important role in loading the Ddc1 complex and its homologs on the DNA substrate, but its biochemical role has not been precisely determined.
Mec1 is involved not only in DNA damage checkpoint but also telomere maintenance (45). Telomeres are shortened in tel1Δ mutants compared with wild-type cells (18). Although mec1Δ mutation by itself does not affect the telomere length (18), introduction of mec1Δ mutation can shorten telomeres in tel1Δ mutants (5). The Mre11 complex controls not only the Tel1 pathway (38, 56) but also part of the Mec1 pathway in response to DNA damage. However, the Mre11 complex and Tel1 control the same pathway in maintenance of telomere length (46). Exo1 is required for checkpoint response in mre11Δ mutants but dispensable for telomere maintenance in mre11Δ mutants (27, 34). One explanation could be that although the Mre11 complex is essential for telomere maintenance, the nuclease activity is not required for the maintenance. There is no good correlation between the length of telomere and ability to process DSB ends among mre11 mutants (27, 33). Alternatively, proteins other than the Mre11 complex and Exo1 might play a more important role in generation of ssDNA tracts at telomere ends. Although Mec1 is recruited to telomere ends (51), it is not clear whether its localization requires the function of the Mre11 complex or Exo1.
In summary, we provide evidence indicating that the Mre11 complex and Exo1 collaborate in generating long ssDNA tract at sites of DNA damage and promoting Mec1 association with the DNA damage sites. Previous studies have demonstrated that the Mre11 complex is essential for activation of the Tel1 signaling pathway. The Mre11 complex thus governs two separate Mec1 and Tel1 signaling pathways in budding yeast. In mammals, it has been well demonstrated that the Mre11 complex and ATM act in the same DNA damage checkpoint pathway. Recently, however, there is accumulating evidence supporting the theory that the Mre11 complex may control the ATR pathway during DSB metabolism (4). Similar to the Mre11 complex, Exo1 is highly conserved from yeast to humans. It is possible that the Mre11 complex and Exo1 control activation of the ATR signaling pathway through a similar mechanism.
Acknowledgments
We thank T. Wakayama for plasmid and strain construction and R. Kolodner, H. Ogawa, T. Ogawa, H. Tsubouchi, and K. Umezu for materials. We also thank C. Newlon and her lab members for helpful discussion.
This work was partially supported by the Foundation of UMDNJ.
REFERENCES
- 1.Abraham, R. T. 2001. Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes Dev. 15:2177-2196. [DOI] [PubMed] [Google Scholar]
- 2.Bakkenist, C. J., and M. B. Kastan. 2003. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature 421:499-506. [DOI] [PubMed] [Google Scholar]
- 3.Bermudez, V. P., L. A. Lindsey-Boltz, A. J. Cesare, Y. Maniwa, J. D. Griffith, J. Hurwitz, and A. Sancar. 2003. Loading of the human 9-1-1 checkpoint complex onto DNA by the checkpoint clamp loader hRad17-replication factor C complex in vitro. Proc. Natl. Acad. Sci. USA 100:1633-1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carson, C. T., R. A. Schwartz, T. H. Stracker, C. E. Lilley, D. V. Lee, and M. D. Weitzman. 2003. The Mre11 complex is required for ATM activation and the G2/M checkpoint. EMBO J. 22:6610-6620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chan, S. W., J. Chang, J. Prescott, and E. H. Blackburn. 2001. Altering telomere structure allows telomerase to act in yeast lacking ATM kinases. Curr. Biol. 11:1240-1250. [DOI] [PubMed] [Google Scholar]
- 6.Cortez, D., S. Guntuku, J. Qin, and S. J. Elledge. 2001. ATR and ATRIP: partners in checkpoint signaling. Science 294:1713-1716. [DOI] [PubMed] [Google Scholar]
- 7.Costanzo, V., K. Robertson, M. Bibikova, E. Kim, D. Grieco, M. Gottesman, D. Carroll, and J. Gautier. 2001. Mre11 protein complex prevents double-strand break accumulation during chromosomal DNA replication. Mol. Cell 8:137-147. [DOI] [PubMed] [Google Scholar]
- 8.D'Amours, D., and S. P. Jackson. 2002. The Mre11 complex: at the crossroads of dna repair and checkpoint signalling. Nat. Rev. Mol. Cell Biol. 3:317-327. [DOI] [PubMed] [Google Scholar]
- 9.D'Amours, D., and S. P. Jackson. 2001. The yeast Xrs2 complex functions in S phase checkpoint regulation. Genes Dev. 15:2238-2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de-Laat, W. L., N. G. Jaspers, and J. H. Hoeijmakers. 1999. Molecular mechanism of nucleotide excision repair. Genes Dev. 13:768-785. [DOI] [PubMed] [Google Scholar]
- 11.Elledge, S. J. 1996. Cell cycle checkpoints: preventing an identity crisis. Science 274:1664-1672. [DOI] [PubMed] [Google Scholar]
- 12.Ellison, V., and B. Stillman. 2003. Biochemical characterization of DNA damage checkpoint complexes: clamp loader and clamp complexes with specificity for 5′ recessed DNA. PLoS Biol. 1:E33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fiorentini, P., K. N. Huang, D. X. Tishkoff, R. D. Kolodner, and L. S. Symington. 1997. Exonuclease I of Saccharomyces cerevisiae functions in mitotic recombination in vivo and in vitro. Mol. Cell. Biol. 17:2764-2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Friedberg, E. C., G. C. Walker, and W. Siede. 1995. DNA repair and mutagenesis. ASM Press, Washington, D.C.
- 15.Giannattasio, M., F. Lazzaro, M. P. Longhese, P. Plevani, and M. Muzi-Falconi. 2004. Physical and functional interactions between nucleotide excision repair and DNA damage checkpoint. EMBO J. 23:429-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gietz, R. D., and A. Sugino. 1988. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene 74:527-534. [DOI] [PubMed] [Google Scholar]
- 17.Green, C. M., H. Erdjument-Bromage, P. Tempst, and N. F. Lowndes. 2000. A novel Rad24 checkpoint protein complex closely related to replication factor C. Curr. Biol. 10:39-42. [DOI] [PubMed] [Google Scholar]
- 18.Greenwell, P. W., S. L. Kronmal, S. E. Porter, J. Gassenhuber, B. Obermaier, and T. D. Petes. 1995. TEL1, a gene involved in controlling telomere length in S. cerevisiae, is homologous to the human ataxia telangiectasia gene. Cell 82:823-829. [DOI] [PubMed] [Google Scholar]
- 19.Grenon, M., C. Gilbert, and N. F. Lowndes. 2001. Checkpoint activation in response to double-strand breaks requires the Mre11/Rad50/Xrs2 complex. Nat. Cell Biol. 3:844-847. [DOI] [PubMed] [Google Scholar]
- 20.Haber, J. E. 1998. The many interfaces of Mre11. Cell 95:583-586. [DOI] [PubMed] [Google Scholar]
- 21.Ivanov, E. L., N. Sugawara, C. I. White, F. Fabre, and J. E. Haber. 1994. Mutations in XRS2 and RAD50 delay but do not prevent mating-type switching in Saccharomyces cerevisiae. Mol. Cell. Biol. 14:3414-3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kastan, M. B., and D. S. Lim. 2000. The many substrates and functions of ATM. Nat. Rev. Mol. Cell Biol. 1:179-186. [DOI] [PubMed] [Google Scholar]
- 23.Katou, Y., Y. Kanoh, M. Bando, H. Noguchi, H. Tanaka, T. Ashikari, K. Sugimoto, and K. Shirahige. 2003. S-phase checkpoint proteins Tof1 and Mrc1 form a stable replication-pausing complex. Nature 424:1073-1083. [DOI] [PubMed] [Google Scholar]
- 24.Kitagawa, R., C. J. Bakkenist, P. J. McKinnon, and M. B. Kastan. 2004. Phosphorylation of SMC1 is a critical downstream event in the ATM-NBS1-BRCA1 pathway. Genes Dev. 18:1423-1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kondo, T., T. Wakayama, T. Naiki, K. Matsumoto, and K. Sugimoto. 2001. Recruitment of Mec1 and Ddc1 checkpoint proteins to double-strand breaks through distinct mechanisms. Science 5543:867-870. [DOI] [PubMed] [Google Scholar]
- 26.Lee, J. H., and T. T. Paull. 2004. Direct activation of the ATM protein kinase by the Mre11/Rad50/Nbs1 complex. Science 304:93-96. [DOI] [PubMed] [Google Scholar]
- 27.Lee, S. E., D. A. Bressan, J. H. Petrini, and J. E. Haber. 2002. Complementation between N-terminal Saccharomyces cerevisiae mre11 alleles in DNA repair and telomere length maintenance. DNA Repair (Amsterdam) 1:27-40. [DOI] [PubMed] [Google Scholar]
- 28.Lee, S. E., J. K. Moore, A. Holmes, K. Umezu, R. D. Kolodner, and J. E. Haber. 1998. Saccharomyces Ku70, Mre11/Rad50 and RPA proteins regulate adaptation to G2/M arrest after DNA damage. Cell 94:399-409. [DOI] [PubMed] [Google Scholar]
- 29.Longhese, M. P., M. Foiani, M. Muzi-Falconi, G. Lucchini, and P. Plevani. 1998. DNA damage checkpoint in budding yeast. EMBO J. 17:5525-5528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lucca, C., F. Vanoli, C. Cotta-Ramusino, A. Pellicioli, G. Liberi, J. Haber, and M. Foiani. 2004. Checkpoint-mediated control of replisome-fork association and signalling in response to replication pausing. Oncogene 23:1206-1213. [DOI] [PubMed] [Google Scholar]
- 31.Majka, J., and P. M. Burgers. 2003. Yeast Rad17/Mec3/Ddc1: a sliding clamp for the DNA damage checkpoint. Proc. Natl. Acad. Sci. USA 100:2249-2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Melo, J. A., J. Cohen, and D. P. Toczyski. 2001. Two checkpoint complexes are independently recruited to sites of DNA damage in vivo. Genes Dev. 21:2809-2821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moreau, S., J. R. Ferguson, and L. S. Symington. 1999. The nuclease activity of Mre11 is required for meiosis but not for mating type switching, end joining, or telomere maintenance. Mol. Cell. Biol. 19:556-566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moreau, S., E. A. Morgan, and L. S. Symington. 2001. Overlapping functions of the Saccharomyces cerevisiae Mre11, Exo1 and Rad27 nucleases in DNA metabolism. Genetics 159:1423-1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morrow, D. M., D. A. Tagle, Y. Shiloh, F. S. Collins, and P. Hieter. 1995. TEL1, an S. cerevisiae homolog of the human gene mutated in ataxia telangiectasia, is functionally related to the yeast checkpoint gene, MEC1. Cell 82:831-840. [DOI] [PubMed] [Google Scholar]
- 36.Naiki, T., T. Shimomura, T. Kondo, K. Matsumoto, and K. Sugimoto. 2000. Rfc5, in cooperation with Rad24, controls DNA damage checkpoints throughout the cell cycle in Saccharomyces cerevisiae. Mol. Cell. Biol. 20:5888-5896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Naiki, T., T. Wakayama, D. Nakada, K. Matsumoto, and K. Sugimoto. 2004. Association of Rad9 with double-strand breaks through a Mec1-dependent mechanism. Mol. Cell. Biol. 24:3277-3285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nakada, D., K. Matsumoto, and K. Sugimoto. 2003. ATM-related Tel1 associates with double-strand breaks through an Xrs2-dependent mechanism. Genes Dev. 17:1957-1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nakada, D., T. Shimomura, K. Matsumoto, and K. Sugimoto. 2003. The ATM-related Tel1 protein of Saccharomyces cerevisiae controls a checkpoint response following phleomycin treatment. Nucleic Acids Res. 31:1715-1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Osborn, A. J., and S. J. Elledge. 2003. Mrc1 is a replication fork component whose phosphorylation in response to DNA replication stress activates Rad53. Genes Dev. 17:1755-1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Paciotti, V., M. Clerici, G. Lucchini, and M. P. Longhese. 2000. The checkpoint protein Ddc2, functionally related to S. pombe Rad26, interacts with Mec1 and is regulated by Mec1-dependent phosphorylation in budding yeast. Genes Dev. 14:2046-2059. [PMC free article] [PubMed] [Google Scholar]
- 42.Pellicioli, A., S. E. Lee, C. Lucca, M. Foiani, and J. E. Haber. 2001. Regulation of Saccharomyces Rad53 checkpoint kinase during adaptation from DNA damage-induced G2/M arrest. Mol. Cell 7:293-300. [DOI] [PubMed] [Google Scholar]
- 43.Petrini, J. H. 2000. The Mre11 complex and ATM: collaborating to navigate S phase. Curr. Opin. Cell Biol. 12:293-296. [DOI] [PubMed] [Google Scholar]
- 44.Qiu, J., M. X. Guan, A. M. Bailis, and B. Shen. 1998. Saccharomyces cerevisiae exonuclease-1 plays a role in UV resistance that is distinct from nucleotide excision repair. Nucleic Acids Res. 26:3077-3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ritchie, K. B., J. C. Mollory, and T. D. Petes. 1999. Interactions of TLC1 (which encodes the RNA subunit of telomerase), TEL1, and MEC1 in regulating telomere length in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 19:6065-6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ritchie, K. B., and T. D. Petes. 2000. The Mre11p/Rad50p/Xrs2p complex and the Tel1p function in a single pathway for telomere maintenance in yeast. Genetics 155:475-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rouse, J., and S. P. Jackson. 2000. LCD1: an essential gene involved in checkpoint control and regulation of the MEC1 signalling pathway in Saccharomyces cerevisiae. EMBO J. 19:5793-5800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rouse, J., and S. P. Jackson. 2002. Lcd1p recruits Mec1p to DNA lesions in vitro and in vivo. Mol. Cell 9:857-869. [DOI] [PubMed] [Google Scholar]
- 49.Sanchez, Y., J. Bachant, H. Wang, F. Hu, D. Liu, M. Tetzlaff, and S. J. Elledge. 1999. Control of the DNA damage checkpoint by chk1 and rad53 protein kinases through distinct mechanisms. Science 286:1166-1171. [DOI] [PubMed] [Google Scholar]
- 50.Sanchez, Y., B. A. Desany, W. J. Jones, Q. Liu, B. Wang, and S. J. Elledge. 1996. Regulation of RAD53 by the ATM-like kinase MEC1 and TEL1 in yeast cell cycle checkpoint pathways. Science 271:357-360. [DOI] [PubMed] [Google Scholar]
- 51.Takata, H., Y. Kanoh, N. Gunge, K. Shirahige, and A. Matsuura. 2004. Reciprocal association of the budding yeast ATM-related proteins Tel1 and Mec1 with telomeres in vivo. Mol. Cell 14:515-522. [DOI] [PubMed] [Google Scholar]
- 52.Tishkoff, D. X., N. S. Amin, C. S. Viars, K. C. Arden, and R. D. Kolodner. 1998. Identification of a human gene encoding a homologue of Saccharomyces cerevisiae EXO1, an exonuclease. Cancer Res. 58:5027-5031. [PubMed] [Google Scholar]
- 53.Toczyski, D. P., D. J. Galgoczy, and L. H. Hartwell. 1997. CDC5 and CKII control adaptation to the yeast DNA damage checkpoint. Cell 90:1097-1106. [DOI] [PubMed] [Google Scholar]
- 54.Tsubouchi, H., and H. Ogawa. 2000. Exo1 roles for repair of DNA double-strand breaks and meiotic crossing over in Saccharomyces cerevisiae. Mol. Biol. Cell 11:2221-2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Umezu, K., N. Sugawara, C. Chen, J. E. Haber, and R. D. Kolodner. 1998. Genetic analysis of yeast RPA1 reveals its multiple functions in DNA metabolism. Genetics 148:989-1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Usui, T., H. Ogawa, and J. H. Petrini. 2001. A DNA damage response pathway controlled by Tel1 and the Mre11 complex. Mol. Cell 7:1255-1266. [DOI] [PubMed] [Google Scholar]
- 57.Uziel, T., Y. Lerenthal, L. Moyal, Y. Andegeko, L. Mittelman, and Y. Shiloh. 2003. Requirement of the MRN complex for ATM activation by DNA damage. EMBO J. 22:5612-5621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vialard, J. E., C. S. Gilbert, C. M. Green, and N. F. Lowndes. 1998. The budding yeast Rad9 checkpoint protein is subjected to Mec1/Tel1-dependent hyperphosphorylation and interacts with Rad53 after DNA damage. EMBO J. 17:5679-5688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wakayama, T., T. Kondo, S. Ando, K. Matsumoto, and K. Sugimoto. 2001. Pie1, a protein interacting with Mec1, controls cell growth and checkpoint responses in Saccharomyces cerevisiae. Mol. Cell. Biol. 21:755-764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.White, C. I., and J. E. Haber. 1990. Intermediates of recombination during mating type switching in Saccharomyces cerevisiae. EMBO J. 9:663-673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wold, M. S. 1997. Replication protein A: a heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu. Rev. Biochem. 66:61-92. [DOI] [PubMed] [Google Scholar]
- 62.Zhang, H., J. Taylor, and W. Siede. 2003. Checkpoint arrest signaling in response to UV damage is independent of nucleotide excision repair in Saccharomyces cerevisiae. J. Biol. Chem. 278:9382-9387. [DOI] [PubMed] [Google Scholar]
- 63.Zhao, X., E. G. D. Muller, and R. Rothstein. 1998. A suppressor of two essential checkpoint genes identifies a novel protein that negatively affects dNTP pool. Mol. Cell 2:329-340. [DOI] [PubMed] [Google Scholar]
- 64.Zhou, B.-B. S., and S. J. Elledge. 2000. The DNA damage response: putting checkpoints in perspective. Nature 408:433-439. [DOI] [PubMed] [Google Scholar]
- 65.Zou, L., and S. J. Elledge. 2003. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 300:1542-1548. [DOI] [PubMed] [Google Scholar]
- 66.Zou, L., D. Liu, and S. J. Elledge. 2003. Replication protein A-mediated recruitment and activation of Rad17 complexes. Proc. Natl. Acad. Sci. USA 100:13827-13832. [DOI] [PMC free article] [PubMed] [Google Scholar]