Abstract
Directionality of yeast mating-type switching has been attributed to differences in chromatin structure for the left arm of chromosome III. We have mapped the structure of ∼45 kbp of the left arm of chromosome III in a and α cells in logarithmically growing cultures and in a cells during switching. Distinctive features of chromatin structure were the occurrence of DNase I-hypersensitive sites in the promoter region of nearly every gene and some replication origins and the presence of extended regions of positioned nucleosomes in ∼25% of the open reading frames. Other than the recombination enhancer, chromatin structures were identical in the two cell types. Changes in chromatin structure during switching were confined to the recombination enhancer. This unbiased analysis of an extended region of chromatin reveals that significant features of organized chromatin exist for the entire region, and these features are largely static with respect to mating type and mating-type switching. Our analysis also shows that primary chromatin structure does not cause the documented differences in recombinational frequency of the left arm of chromosome III in yeast a and α cells.
Mating-type interconversion in Saccharomyces cerevisiae is directional. HO endonuclease initiates switching by making a double-strand cut at the MAT locus on chromosome III. Repair of this break in an α cell preferentially uses silent mating-type information encoded at HMRa, while in an a cell, recombination usually takes place with sequences located at HMLα (4). The recombination enhancer (RE), located ∼30,000 bp from the left end of chromosome III and highly conserved between yeast species, controls this directionality (23, 33, 34, 36). In a cells, the RE has nuclease-hypersensitive sites indicating unusual DNA geometry and footprints suggesting protein binding sites. Some of the protein candidates which may create these footprints are the two transcription activators, Fkh1p and Fkh2p, and their associated protein, Ndd1p, which were shown to bind to the RE directly only in a cells (22). In α cells, organized chromatin abutting the binding site for Mcm1p and Matα2p replaces the active a-cell configuration (28). How these structural differences at the RE facilitate interactions between MAT and HMLα in a cells is not known. It is known, however, that extensive segments of the left arm of chromosome III are altered in their recombination properties in the two cell types. In a cells, at least 40 kbp at the left end of this chromosome is in a hyperactive state for recombination, while over 100 kbp in this region is recombinationally “cold” in α cells (35).
Alterations in chromatin structure have been invoked as a possible explanation of these differences in recombination potential (35). Chromatin structure of only two regions of chromosome III has been reported. RE structures are strikingly different in α and a cells. HML has a distinctive chromatin structure, but it is identical in the two cell types (29). Differences in the chromatin structures of the left arm of chromosome III in the two cell types and changes in chromatin structure accompanying switching are unstudied and unknown. In this study, we have mapped the chromatin structure of ∼45 kbp of the left arm of chromosome III. The analysis was carried out for both α and a cells and for the latter cell type during mating-type switching.
The single most informative probe of chromatin structure is probably DNase I (11, 18, 20, 27, 32). In general, DNase I hypersensitivity marks the sites where chromatin is more open, making the hypersensitive site synonymous with enhancer, promoter, replication origin, or other similar feature of chromatin. All of these features enhance the utility of DNase I as the primary instrument in dissection of chromatin structure of large regions of a genome.
In the process of addressing the role of chromatin in mating-type switching, we have carried out the first nonbiased study of chromatin structure of an extensive region in an organism whose genomic sequence is known. In contrast to higher organisms, where chromatin structure undergoes major changes when genes are repressed or transcribed, hypersensitive sites are present at the 5′ ends of most genes and some replication origins in the left arm of yeast chromosome III. A surprising finding is the presence of organized arrays of positioned nucleosomes on ∼25% of the genes examined. A unique change in chromatin structure occurs at the RE during mating-type interconversion in a cells, even though the structures of the remainder of the left arm of chromosome III are nearly identical in the two cell types. The chromatin structures of the entire region are the same for the two cell types except in the RE. This is in contrast to expectations of chromatin structural differences of the left arm of chromosome III as the explanation for observed cell-type-specific differences in recombinational frequency (35).
MATERIALS AND METHODS
Cell growth.
Standard yeast medium, YP, with an appropriate carbon source (2% dextrose [YPD] or 2.5% lactose [YPL]) was used. For comparison of the chromatin structures of a and α cells, FY24 α and FY23 a cells (31) were grown at 30°C to an optical density at 600 nm (OD600) of 1.0 in YPD. JKM161a (ho HMLα mata HMRa ade1-112 lys5 leu2-3 ura3-52 trp::hisG ade3::GalHO), provided by J. Haber, was used for chromatin structure analysis of switching cells. Induction of HO expression was done as described previously (30). Cells were grown to an OD600 of 0.2 in 900 ml of YPL at 30°C. They were synchronized by adding α-factor to a concentration of 10 μg/ml. When >90% of the cells were unbudded, 100 ml of 20% (wt/vol) galactose was added to induce HO expression and cutting for 30 min. After induction, cells were harvested by filtration and suspended in prewarmed YPD. At appropriate time points, NaN3 was added to 1 liter of culture to a final concentration of 0.2% and cells were harvested.
Nuclear DNA preparation and analysis.
Nuclei from 1 liter of cells of OD600 0.4 to 0.5 were prepared as described previously (15a). Nuclear DNA was cut with DNase I (Worthington) at concentrations of 0.1 to 0.8 U/ml at 37°C for 5 min. DNA from switching cells for the kinetics experiment was recovered from cells treated as described previously (30).
Southern blots.
DNA from equal cell numbers (based on weight of cell pellets) was purified, cut with BamHI, subjected to electrophoretic separation on a 1.2% agarose gel, transferred to Hybond-NX membrane (Amersham), cross-linked with UV light, and hybridized with specific probes. The probes were prepared by PCR amplification of ∼250-bp regions abutting each BamHI-cut site. The specificity of each probe was determined by running BLAST searches for probe sequences. An undigested control in the maps also serves as a measure of the specificity of each probe, because it is expected that cross-hybridization would result in the appearance of multiple bands in these samples. Probes were gel purified and random prime labeled with [α-32P]dATP. Blots were exposed to a PhosphorImager screen and analyzed with ImageQuant v.5.
RESULTS
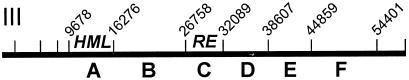
Restriction endonuclease BamHI cuts the leftmost ∼55 kbp of chromosome III into fragments that range from 6 to 11 kbp in size (Fig. 1). Since an individual indirect end label experiment can map 5 to 7 kbp of chromatin at a medium-level resolution, mapping of the entire region of interest was possible using a single set of DNase I digests repetitively separated by gel electrophoresis and/or repetitively probed on Southern blots. Since all the probes were generated by PCR amplification of genomic DNA, were about the same length, were labeled in parallel under the same conditions for random prime labeling, and were hybridized and washed identically, it is possible to make semiquantitative conclusions about relative nuclease susceptibilities from inspection of the blots. Mapping of the first 8 kbp of the chromosome was not possible due to common subtelomeric sequences between chromosome III and other chromosomes. Figure 2 presents indirect end label maps of chromatin from 9 to 54 kilo-map units (kmu) for both a and α cells at several levels of DNase I digestion. Naked genomic DNA was also cut with DNase I and mapped. The digestion of naked DNA with DNase I generated uniform smears in almost all cases, and we did not observe bands corresponding to the hypersensitive sites detected in the chromatin maps (data not shown). Features of the genome sequence are presented at the side of each experimental map and summarized at the bottom of each data set, together with vertical arrows that indicate major DNase I cutting sites. A list of the genetic features within the mapped region, obtained from the Saccharomyces Genome Database (http://www.yeastgenome.org), and the transcriptional frequency of most of the genes (7) are summarized in Table 1.
FIG. 1.
BamHI sites on the leftmost 58 kbp of yeast chromosome III. Restriction sites are indicated by the vertical tic marks and by the coordinates of the sites for those used for mapping. Locations of HML and the recombination enhancer are indicated. Letters below the line indicate the subfigure in Fig. 2 presenting the chromatin map for that region.
FIG.2.
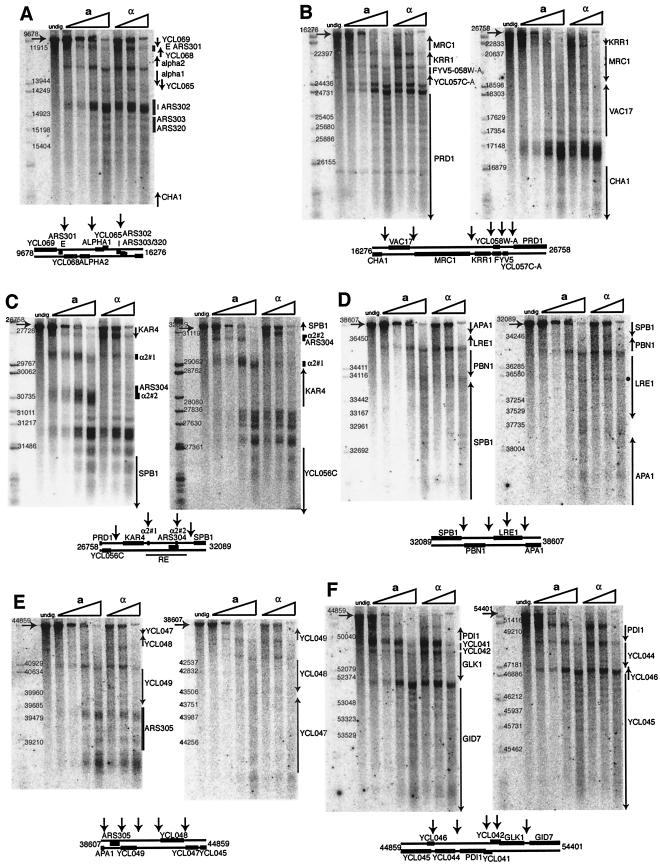
Chromatin structure of an extensive region of the left arm of chromosome III. For each panel, an undigested control is shown together with four levels of digestion of a-cell nuclei (0.1, 0.2, 0.4, and 0.8 U/ml) and three levels of digestion of α-cell nuclei (0.2, 0.4, and 0.8 U/ml). The genetic features of each region are shown to the right of each gel pattern and below, where the arrows indicate major DNase I digestion features. Numbers adjacent to size standards are coordinates for chromosome III. Each parent fragment is shown with an arrow and the coordinate for the restriction site to the left of each gel. All maps are bidirectional except for that presented in panel A, where similar subtelomeric sequences between chromosomes precluded mapping to the left of 9678. (A) Region from 9678 to 16276. Locations of two ARS sequences are shown, as are the positions of the E and I silencers. (B) Region from 16276 to 26758. (C) Region from 26758 to 32089. Locations of the two Mcm1p/Matα2p binding sites in the recombination enhancer are shown. (D) Region from 32089 to 38607. (E) Region from 38607 to 44859. The location of ARS 305 is indicated. (F) Region from 44859 to 54401.
TABLE 1.
Features and expression profiles between coordinates 9,000 and 55,000 of chromosome III
| Feature | Locus description | Expression level (no. of copies/cell) | Transcription frequency (mRNAs/h) |
|---|---|---|---|
| YCL069W | Hypothetical ORF | n/aa | n/a |
| ARS301 | ARS | ||
| YCL068C | Hypothetical ORF | 0.2 | n/a |
| YCL067C | HMLα2 | n/a | n/a |
| YCL066W | HMLα1 | n/a | n/a |
| YCL065W | Hypothetical ORF | n/a | n/a |
| ARS302 | ARS | ||
| ARS303 | ARS | ||
| ARS320 | ARS | ||
| YCL064C | CHA1 catabolic serine (threonine) dehydratase | 4.5 | 16 |
| YCL063W | VAC17 | 0.2 | 0.4 |
| YCL061C | MRC1 protein involved in replication checkpoint | 0.2 | 0.6 |
| YCL059C | KRR1 involved in cell division and spore germination | 1.9 | 5.8 |
| YCL058W-A | Identified by homology to Ashbya gossypii | n/a | n/a |
| YCL058C | FYV5 | n/a | n/a |
| YCL057C-A | Hypothetical ORF, has similarity to proteins in Schizosaccharomyces pombe, Caenorhabditis elegans, and Drosophila melanogaster | n/a | n/a |
| YCL057W | PRD1 | 0.9 | n/a |
| YCL056C | Protein of unknown function; green fluorescent protein fusion protein localizes to the cytoplasm in punctate pattern | 2.0 | 3.5 |
| YCL055W | KAR4 transcription factor involved in karyogamy | 1.3 | 3.3 |
| ARS304 | ARS | ||
| YCL054W | SPB1; putative methyltransferase | 1.8 | 3.3 |
| YCL052C | PBN1; protease B, nonderepressible form | 0.9 | 0.5 |
| YCL051W | LRE1; involved in laminarase resistance | 0.2 | n/a |
| YCL050C | APA1 | 4.3 | n/a |
| ARS305 | ARS | ||
| YCL049C | Hypothetical ORF | 0.7 | n/a |
| YCL048W | Hypothetical ORF | n/a | n/a |
| YCL047C | Hypothetical ORF | 0.8 | 1.9 |
| YCL046W | Hypothetical ORF | n/a | n/a |
| YCL045C | Hypothetical ORF | 1.6 | 1.9 |
| YCL044C | Hypothetical ORF | 0.3 | 0.2 |
| YCL043C | PDI1; protein disulfide isomerase | 6.7 | 11.6 |
| YCL042W | Hypothetical ORF | 0.4 | 0.4 |
| YCL041C | Protein required for cell viability | n/a | n/a |
| YCL040W | GLK1; glucokinase | 3.7 | 3.8 |
| YCL039W | GID7 | 0.2 | 0.2 |
n/a, for indicated ORFs, data are not present in the analysis of Holstege et al. (7).
Chromatin structure features common to both cell types.
Most of the structure map was very similar for the two cell types. Specifically, there was no indication of a generalized increase in nuclease susceptibility for the highly recombination-competent a-cell nuclei compared to the nuclei from less recombinogenic α cells. Visual comparison of the rate of disappearance of the parent restriction endonuclease DNA fragment in the two cell types during the time course of DNase I digestion (Fig. 2) revealed minor variations but no consistently enhanced rate of nuclease digestion for a versus α cells. There is one significant exception to this generalization. For a region where chromatin structure was known to differ for the two cell types, the RE (28), the parent fragment was lost at lower nuclease concentrations in a- than in α-cell nuclei (Fig. 2C), consistent with a protected chromatin domain in α cells. Because the DNase I sensitivity of the RE mirrors known chromatin structure differences, the absence of similar differential nuclease sensitivity for other regions strongly suggests that there are no global differences in chromatin structure of a and α cells for the leftmost 55 kb of chromosome III.
The first region mapped, from 9 to 16 kmu, includes the HMLα locus (Fig. 2A). The E and I silencers which flank the locus were hypersensitive to DNase I; these sites are also locations of autonomous replicating sequences (ARSs) 301 and 302, respectively. Whether the altered chromatin structure results from silencer or replication origin function is uncertain. Two additional ARSs in this region, 303 and 320, were not nuclease sensitive. As expected from the high-resolution analysis of this locus (28), the intergenic promoter region between the α1 and α2 genes was also nuclease sensitive, in spite of the transcriptional inactivity of the two genes. This is an exception to the generality that silenced gene promoters are nuclease insensitive while transcriptionally competent gene promoters are nucleosome free and nuclease sensitive. The possibility of a role for Rap1p binding in both repression and maintaining a nucleosome-free sensitive site is attractive (38). Both in the YCL065 open reading frame (ORF) and between the I silencer and the 3′ end of the CHA1 locus, there were patterns of nuclease sensitivity, a “ladder,” indicative of arrays of positioned nucleosomes.
Mapping the next region, from 16 to 27 kmu, revealed a number of features of chromatin structure which were characteristic of much of the entire ∼45-kbp segment of chromosome III (Fig. 2B). Hypersensitive sites occurred near the 5′ and/or 3′ ends of ORFs, most frequently the former. Strong hypersensitive sites were present in two intergenic promoters of divergently transcribed genes, YCL057-PRD1 and YCL063 (VAC17)-CHA1. The hypersensitivity at the CHA1 promoter is consistent with previously published data (12). Note that there are two distinct hypersensitive sites marking the 5′ end of YCL057 and PRD1. Separate hypersensitive sites for the 5′ end of some genes were possible to observe when the region between two genes was long enough to resolve in the mapping gels. Some genes, predicted to have low transcriptional activity (Table 1) (7), had positioned nucleosomes over extensive regions of the ORF. Examples in the Fig. 2B map are PRD1 and VAC17, which displayed long stretches of regular DNase I cutting sites with a periodicity of ∼160 bp. These regions, suggestive of highly organized, sequence-specific chromatin structure, were unexpected. Both genes are low-activity genes with 0.9 or 0.2 mRNA molecule per cell, respectively (7). The coexistence of a domain of positioned nucleosomes within an ORF with a nuclease-hypersensitive promoter region 5′ of the gene suggests that establishment of transcriptional competency can proceed in the absence of extensive levels of transcription. Note that all these data derive from studies of unique, genomic yeast genes, obviating population considerations that may accompany studies of multicopy minichromosomes. The caveat that structures and expression may vary in a cell-cycle-dependent fashion is apparent to us; we note that there is no indication of cell cycle-specific regulation of the cited genes (19).
The region from 27 to 33 kmu of chromosome III contains the recombination enhancer. The only cell-type-specific features of chromatin structure were located in this segment of the chromosome (Fig. 2C) and are discussed in depth below.
From 32 to 39 kmu, centromere proximal of the RE, little was distinctive in the DNase I map of chromatin structure (Fig. 2D). Nuclease-sensitive sites were present at the intergenic 3′ ends of a gene pair, SPB1-PBN1. An unusual hypersensitive site was present in the middle of the LRE1 gene, which was highlighted with a dot located to the right of the mapping data in Fig. 2D. In mapping nearly 45 kbp of chromatin, this is the only DNase I-hypersensitive site that was located internally in an ORF sequence. Diffuse ladders of cutting sites were present in the 3′ halves of the SPB1 and APA1 genes. These were not nearly as sharp, defined bands as those present to the right of the I silencer or in the PRD1 and YCL063 ORFs. The pattern in the 32- to 39-kmu region is suggestive of an imprecisely organized chromatin structure.
Many features of DNase I cutting in the 39- to 45-kmu region of chromosome III were reminiscent of inferred structures for more distal regions of the chromosome (Fig. 2E). Hypersensitive regions were present in the 5′ flanking regions of YCL048 and YCL049. Diffuse ladders of cutting sites suggested a poorly organized set of positioned nucleosomes in the ORFs for YCL047 and YCL049. Differing from features of previously described regions was the presence of a long segment with distinctive chromatin structure in a genomic sequence that lacked any identified ORF, downstream of YCL049. A DNase I-hypersensitive site 3′ of the ORF abutted three diffuse cutting sites, suggesting the presence of three nucleosomes. The region was bracketed on the opposite end by a strong DNase I-hypersensitive site at the location of ARS305. Whether the chromatin structure of this region is critical for function of this ARS or is a consequence of the activity of the replication origin remains to be determined.
The density of genes in the final chromatin region whose structure was mapped, 45 to 54 kmu of chromosome III, is very high, making it difficult to distinguish the several DNase I-hypersensitive sites in association with the 5′ or 3′ flanking sequences of a particular gene or ORF (Fig. 2F). This region did exemplify two different types of gene structure. YCL039 (GID7) and YCL045 are predicted to have low activity (0.2 and 1.6 copies of mRNA, respectively). Both of these ORFs were packaged in arrays of positioned nucleosomes. In contrast, PDI1 and GLK1 are genes of moderate activity (6.7 and 3.7 copies of mRNA, respectively). These two genes have strong DNase I-hypersensitive sites at their 5′ and 3′ flanking regions. The presence of two different types of genes in close proximity to each other indicates that long-range organization of chromatin regions with similar transcriptional regulation and chromatin structure, thought to occur in some cases in higher organisms (1, 5, 10), is not a rule in yeast.
Distinctive chromatin structures in a- or α-cell nuclei.
High-resolution analysis of micrococcal nuclease (MNase) digestion sites in the RE previously showed major differences in chromatin structure between a and α cells (28). This conclusion was reinforced by the results of the current DNase I study of the region of chromosome III from 27 to 33 kmu (Fig. 2C). The RE DNA contains two binding sites for Mcm1p and Matα2p, α2#1 and α2#2, separated by ∼1,350 bp, and a region with extensive TTTG/A repeat sequences just to the right of α2#1 (14).
To the left of α2#1, covering the KAR4 ORF, positioned nucleosomes were present in the nuclei of both cell types. In contrast, the DNase I pattern of this region indicated two domains. The ORF for KAR4 was generally nuclease resistant, with diffuse, minor cutting sites marking linker regions between nucleosomes that were not precisely positioned. The intergenic promoter region between the 5′ ends of KAR4 and YCL056 is more sensitive to DNase I in both cell types. One strong site adjacent to YCL056 probably relates to its transcription; the origin of the other sites in this region is obscure. At the other end of the RE, a strong DNase I-hypersensitive site was present in both cell types flanking the YCL054 ORF. Three diffuse cutting sites occupy the ∼200-bp space between this site and the 5′ end of the ORF.
Both the operator sites, α2#1 and α2#2, are DNase I hypersensitive in a cells. The cut sites encompass a broad region, larger than the 11-bp site that interacts with Mcm1p (16), a trans-acting factor that binds these sites in a cells. Other proteins, Fkh1p, Fkh2p, and Ndd1p, were shown to bind to the RE domains around the α2#1 operator (22). In α cells, site α2#1 was slightly sensitive to DNase I while site α2#2 was not cut preferentially at all. This is surprising, since the half-life of Matα2p is very short (6) and the protein is expected to be absent from its binding site in isolated nuclei from α cells (13). Thus, only Mcm1p should bind these DNA sequences in isolated nuclei.
Between the α2#1 operator and the hypersensitive site to the 5′ side of SPB1 in α cells, a continuous ladder of DNase I cutting sites signaled the presence of an array of positioned nucleosomes. Organized chromatin spanned the second operator without apparent interruption, at least at the level of resolution afforded by indirect end label mapping of DNase I cutting sites. The highly organized chromatin in α cells (28) may fold into a higher-order structure that precludes access of the nuclease to the second operator. The region between the two operators was relatively resistant to this nuclease and was featureless in its digestion pattern in a cells.
Chromatin structure changes during mating-type interconversion in a-cells.
We next evaluated changes in chromatin structure during mating-type interconversion in a cells. Normally, expression of HO endonuclease is confined to mother cells in the G1 phase after cell division (21). This group comprises a small proportion of a logarithmically growing yeast culture, so others have developed a system that allows for highly synchronous switching of an entire yeast culture (2); we have employed this system for our study of chromatin structure during switching.
In this system, randomly growing cultures of a-cell S. cerevisiae are treated with the peptide α-factor to arrest cells in G1. The cells used are an a-cell strain that expresses the HO endonuclease gene under galactose (GAL) control. The culture is changed to galactose medium to induce synthesis of HO endonuclease. After 30 min, cells are transferred to glucose medium, which represses expression of GAL-controlled genes. Cells then progress through a typical mating-type interconversion.
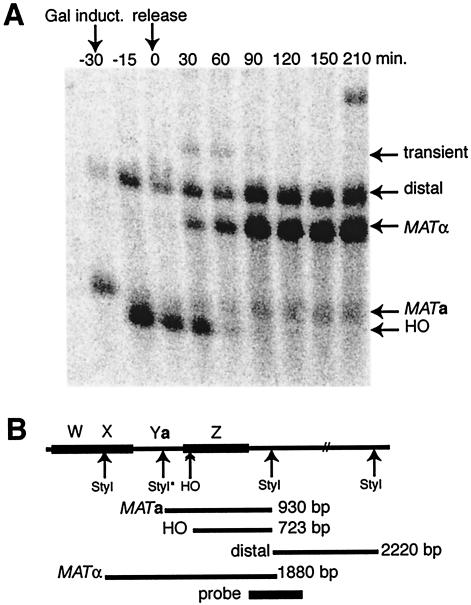
Others have devised a Southern blotting protocol that allows one to follow the time course of all aspects of switching in a cells (2). Distinctive DNA fragments mark the MATa locus, prior to or after cutting by HO endonuclease, the MATα locus, and a loading control derived from common sequences to the right of MAT (Fig. 3B). Figure 3A documents the switching kinetics in our experiments. At −30 min (end of α-factor synchronization, beginning of HO induction), cells are MATa as shown by the MATa fragment. At −15 (mostly) and 0 (completely) min, HO has cut the MAT locus. The fragments are now shorter than the MATa species. Over the next 60 to 90 min, interconversion occurs, with the bulk of the cells becoming MATα, signaled by the large MATα fragment. A small fraction of cells correct the double-strand DNA break by recombination at HMRa, leading to regeneration of the MATa fragment. An important control for experiments which require isolation of nuclei and DNase I digestion at closely spaced time points during synchronized switching is demonstration that arrest of cellular metabolism with sodium azide freezes the DNA changes during switching. Nuclei isolated from azide-treated cells arrested at the beginning or end of galactose induction of HO expression (−30 and 0 min) or 30 min into switching (30) contained DNA fragments identical to DNA isolated by a rapid, hot phenol/glass bead lysis procedure (data not shown).
FIG. 3.
Synchronized switching of mating type. (A) Time course of induction of HO endonuclease and mating-type interconversion. DNA cut with StyI was blotted with a probe whose location is indicated in panel B. As shown in panel B, this probe detects a cell-type-independent distal 2,220-bp fragment, an 1,880-bp fragment from ΜΑΤα, a 930-bp fragment from MATa, and a 723-bp fragment when MAT has been cut by the HO endonuclease. Yeast a cells, arrested at G1 by incubation with α factor, are induced to express HO in galactose (Gal induct.; −30) and simultaneously released from the cell cycle block and transferred to dextrose medium to initiate switching (release; 0 min). Additional samples were taken over the next 3.5 h, as indicated.
We next evaluated chromatin structure throughout the ∼45-kbp region shown in Fig. 2 during synchronized switching of a cells. Aliquots of cultures were taken after α-factor arrest, at the end of galactose induction of HO endonuclease expression, and at intervals during the first 60 min of switching. Sodium azide was added to arrest metabolism, and nuclei were prepared and digested with various amounts of DNase I. Purified DNA was then cut to completion with BamHI and analyzed by indirect end labeling.
The region from 9 to 16 kmu contains HMLα, the donor for recombinational repair of the double-strand break at MATa, and is therefore a strong candidate for showing changes in chromatin structure during switching. In contrast to this expectation, there were no detectable gross changes in DNase I digestion patterns in this region of chromosome III during the switching event (Fig. 4A). The minor DNase I-sensitive site at the E silencer, the major site at the I silencer, and the sites at the intergenic promoter and 3′ end of the α1 gene were unaltered from arrested cells to near completion of the mating type switch. The nucleosome ladder pattern extending rightward from the I silencer appeared to be less well defined during switching, particularly at the 20- to 40-min points (Fig. 4A). The region downstream of CHA1 appears to become more sensitive to DNase I from 30 to 40 min. This sensitivity may be caused by the replication origin function of the ARSs around that region, but this remains to be confirmed. During synchronized switching, DNase I susceptibility of the rest of the region shown in Fig. 2 was essentially identical to that in randomly growing a cells, except for the recombination enhancer (Fig. 4B).
FIG. 4.
Chromatin structure during mating-type interconversion. Nuclei from azide-arrested samples taken during synchronized mating-type switching were digested with DNase I and mapped by indirect end labeling. (A) HML region, comparable to that shown in Fig. 2A. (B) RE region.
Chromatin structure of the RE differed between a and α cells (Fig. 2C). Additional differences from the resting state in a cells were found when RE chromatin structure was examined during a synchronized switching event (Fig. 4B). Chromatin structure of the RE in the a-cell population arrested at the G1/S boundary by α-factor was similar to that in randomly growing cells. Structure was unchanged after induction of HO endonuclease expression. In contrast to these similarities, a new DNase I-sensitive site (highlighted with a dot on the right side of the mapping picture) appeared early in the switching process, increased in susceptibility, and then faded away as the switching process was completed. The site was located just to the right of the α2#2 operator, downstream from the transcription start sites for one of the three noncoding RNAs transcribed from the recombination enhancer, identified by Szeto et al. (23). At the end of switching, the RE chromatin must be remodeled from the a-cell structure, characterized by hypersensitive sites, to the α-cell array of positioned nucleosomes. Even though the majority of cells have α information at MAT after 60 min of switching (Fig. 3A), chromatin structure at the RE has features characteristic of the a-cell pattern, e.g., similar nuclease sensitivity of the two operators (Fig. 4B).
DISCUSSION
Increasingly, a role for chromatin structure is being implicated in studies of DNA function in transcription, replication, recombination, and repair. Many of the studies leading to these conclusions are inferential, using genetic or other indirect approaches. Actual studies of chromatin structure have generally been confined to areas of particular interest to the investigator; promoters, enhancers, locus control regions, and such. In contrast, we report here an unbiased examination of the DNase I susceptibility of an extensive region of yeast chromosome III. The region is of interest in its own right, since chromatin structure has been proposed as a contributory factor to directionality of mating-type interconversion (4). This inference derives from the low recombinational activity of the leftmost 100 kbp of this chromosome in α cells and the oppositely high recombinational capacity of at least 40 kbp of the same region in a cells. A corollary to the hypothesis that chromatin structure is a factor in directionality of mating-type interconversion is that there might be changes in chromatin structure during the switching event.
Chromatin structural features of the left arm of chromosome III.
The mapped region includes 30 genes or ORFs, six potential replication origins, two known silencers at HMLα, and two Matα2p/Mcm1p operators at the recombination enhancer. Thus, the ∼45-kbp chromatin segment includes most of the elements commonly found in the yeast genome and is a valid model for large-scale characterization of chromatin.
As suspected from previous studies of specific genes, DNase I-hypersensitive sites are present at the 5′ flanking regions of ORFs. The surprising finding is that a hypersensitive site is present in the promoter region of nearly every gene in the region examined. This conclusion must be tempered slightly by the possibility that some of the sites might arise from chromatin features at the 3′ end of neighboring genes. Given the parsimonious use of DNA in the yeast genome and the limited resolution of these long-range maps, we cannot distinguish between the two possibilities for most of the genes. The generality of occurrence of DNase I-sensitive sites in 5′ flanking regions favors a hypothesis that every ORF that is competent for transcription will have chromatin structure that leads to such a site. Occurrence of hypersensitive sites in the promoter region of nearly all genes, whether the gene is active at different levels or inactive, suggests that some basic chromatin structural feature exists at every promoter. The identification of this structural element, proteins involved in its creation, and how it relates to further aspects of transcriptional control will be important in understanding eukaryotic gene regulation. Some of the factors that might contribute to creation of hypersensitive sites are binding of transcription factors, changes in DNA confirmation caused by protein binding, altered or remodeled nucleosomes, and absence of nucleosomes at the promoter regions.
While hypersensitive sites are present upstream of many of the genes in our ∼45-kbp structure map, there is no striking correlation between intensities of cutting and transcription levels. Most yeast genes are transcriptionally competent; only a few are truly silenced in the fashion of larger eukaryote heterochromatic genes. This is not the case for genes that are truly silenced: examples are silent mating-type loci, a-cell-specific genes in α cells, and the RE in α cells. A clear example of DNase I sensitivity that correlates with active regions of chromatin and contrasts with chromatin that is silenced is found in the RE, where the two Matα2p/Mcm1p binding sites are DNase I sensitive in a cells but less sensitive or not sensitive, for α2#1 or α2#2, respectively, as part of an organized chromatin domain in α cells. The correlation of chromatin structure with transcription of the silent RNAs from the RE is clear, where in a cells the silent RNAs are transcribed downstream of the both operators but not in α cells (23).
In addition to transcriptional competence, other features of chromatin structure reflect DNA function—a prime example is replication origin activity. Replication origin ARS305 is hypersensitive and has chromatin structural features flanking the consensus sequence; this is known to be a functional origin in actively growing yeast (8). In contrast, ARS303 and ARS320, nearer the telomere, are not known to be active and are not DNase I sensitive (25). The remaining two ARS consensus sequences are hypersensitive, but this may relate to their being part of the two silencers, E and I, at HMLα (3). The parallel between locus control regions in higher organisms and these silencers is obvious. Domain regulation appears to involve chromatin regions where trans-acting factors interact with cis-acting elements, creating either transcriptionally competent or repressed chromatin domains. The protein-binding site is often nuclease sensitive, reflecting interactions (or their dissociation during chromatin isolation) of the trans-acting factors. The resultant chromatin domain can be nuclease sensitive, when it includes active genes, or nuclease resistant, when its constituent genes are repressed. In contrast, regulation of individual genes usually involves chromatin structure that is specific for the gene.
The most surprising feature of this unbiased investigation of a large region of chromosome III in yeast is occurrence of a number of regions where DNase I susceptibility indicates regions containing positioned nucleosomes. Cutting of linker regions occurs somewhat more frequently than core particle sequences, leading to a fuzzy DNA ladder when DNase I is used to assess regions of positioned nucleosomes (20). The contrast with an MNase digestion pattern is apparent in comparison of the data in this work (e.g., Fig. 2C) with previous MNase maps (28). Given this limitation in mapping chromatin structure by DNase I digestion, it is quite striking that at least seven sites occur in the mapped region where data strongly suggest the existence of an extended (>5) region of positioned nucleosomes. The frequency of such domains of positioned nucleosomes (∼25% of ORFs) is much higher than we anticipated. Possible mechanisms for generating such organized chromatin domains include sequence/structure-specific interactions with histones, usually thought to result from nonisotropic flexibility or bending of DNA (17), statistical positioning of nucleosomes resulting from exclusion of histone binding by sequence-specific DNA binding protein(s) (9), active organization of repressed domains by a repressor and corepressor that interact with histones (15), or nonhistone protein binding to specific DNA sequences separated by a number of base pairs that is at or near an integral multiple of the nucleosome repeat length (24), ∼160 bp for budding yeast. The distinctive observation about chromatin structure made in the present study, suggesting a much wider occurrence of positioned nucleosomes in organized chromatin domains than previously envisioned, reinforces the possible role of chromatin structure in regulation of expression of a wider cast of genes and demands more extensive high-resolution analysis of chromatin structure in studies of transcriptional regulation. Of interest, one ORF with positioned nucleosomes that overlapped an oppositely transcribed gene and did not have a promoter nuclease-sensitive site, YCL046 (Fig. 2F), was expressed at a nearly 10-fold-higher level in the Young laboratory genomewide analysis of transcriptional frequency in yeast strains depleted of nucleosomes by removing expression of histone H4 (37). None of the other genes identified as having positioned nucleosomes was dramatically up-regulated by nucleosome depletion.
Chromatin structure and mating-type interconversion (switching).
When homothallic yeasts divide, the mother cell expresses HO endonuclease. This enzyme cleaves genomic DNA at the MAT locus, forcing the cell to heal the double-strand break by a recombinational event using HMLα or HMRa as the donor locus. The donor is selected based on the sequences at MAT; nearly all a cells recombine with HMLα, while most α cells use HMRa as donor (21). Directionality results from sequences at the recombination enhancer (4). When RE is activated in a cells, ∼100 kbp of the left arm of chromosome III, in general, and HML, in particular, are highly available for recombination. In contrast, in α cells, the RE is unavailable for binding of presumed trans-acting factors and the left arm of chromosome III is recombinationally cold (35). A repressive chromatin structure encompasses the RE in α cells (28); propagation of this structure has been suggested to lead to recombinational restriction for ∼40 kbp of the left arm of chromosome III. Although differences in chromatin structure have been invoked to explain the differences in recombinational capacity for this region of chromosome III in a and α cells, no experimental analysis of chromatin for the extensive regions proposed to differ in the two cell types has been available until the present study.
The surprising result of this experimental analysis is that there are no differences in the DNase I chromatin maps between a- and α-cell nuclei other than those at the RE. Particularly revealing are the closely similar rates of decrease of the parental restriction endonuclease DNA fragment during DNase I digestion of the two cell-type nuclei, indicating similar overall chromatin structures over the extensive region analyzed (Fig. 2). Validity of this analysis is testified to by differences in parental fragment digestion at the RE, where chromatin from a cells is known to have nuclease accessibility in excess of that from yeast α-cell nuclei (28). Since general DNase I sensitivity did mirror known chromatin structure differences at the RE, the absence of similar differential nuclease sensitivity for other regions strongly supports the argument that there are no global, pervasive differences in chromatin structure of a and α cells for the leftmost 55 kb of chromosome III. Thus, if general chromatin structure plays a role in the differences in recombination capacity for the left arm of chromosome III in the two cell types, the structural elements responsible either were not preserved in isolated nuclei or were not detected by DNase I digestion. While we have recently developed a DNase I in vivo expression method for chromatin structure studies (26), the level of expression of the nuclease is suitable for analysis of single-strand nicking by primer extension only. Higher activities will be necessary to apply in vivo DNase I expression to double-strand cuts studied by indirect end label analysis. Alternatively, recombinational frequency differences may result from other, higher-order chromatin structure or unknown features of nuclear organization in the two cell types.
Even more surprising is the near lack of detectable changes in the primary chromatin structure during the switching event. Clearly, the bulk of the population of a cells undergoes recombinational repair using donor sequences at HMLα. This major alteration in DNA sequence, including recombination and replication, occurs in the absence of detectable changes in chromatin structure as mapped by DNase I digestion and indirect end label analysis of HMLα. Since we deal with structural analysis of a population of cells, it is conceivable that any individual cell goes through the switching recombination in a very short time, making detection of its individual chromatin structure changes impossible in the face of most molecules being pre- or postswitching. We do, however, detect chromatin structure changes at the recombination enhancer that are temporally correlated with the switching process. Since we can monitor chromatin changes at the RE during switching, we suggest that chromatin structure alterations at the HM loci must be localized to a small region and rapidly progressing from identical pre- to postswitching structures and therefore undetectable in our analyses of chromatin structure at selected time points. Alternatively, the recombination process may occur with the participants sequestered in a region of the nucleus inaccessible to DNase I and thereby be experimentally transparent in terms of chromatin structure changes.
The one region that does have significant alterations in chromatin structure during the switching process is the RE, the same region which exhibits distinctive chromatin structures in nuclei from the two yeast mating types. The two operators where Mcm1p can bind are hypersensitive to DNase I in a cells. During the time course of synchronized mating-type interconversion, a region of chromatin just proximal to the proximal operator becomes hypersensitive to DNase I. Nuclease sensitivity appears, peaks, and decreases over a time course consistent with the timing of mating-type switching in these a cells. This observation is consistent with transient binding of a factor(s) that activates the recombination enhancer for its role in the mating-type switching process. Some of the candidates were shown to directly bind to the RE (22). Whether these factors contribute to transcription of the sterile RNAs and whether these noncoding transcripts (23) play a role in activation of an extensive region of the left arm of chromosome III for recombination remain to be determined. We have evidence suggesting that transcription, rather than the transcript, is critical for switching directionality (S.E. and R.T.S., unpublished observations), in agreement with a prescient suggestion from James Broach (23).
Acknowledgments
This work was supported by National Institutes of Health to R.T.S. (GM 52311).
We thank Jim Haber for strains and the members of the Simpson and Workman laboratories and Joe Reese for discussions.
REFERENCES
- 1.Anguita, E., C. A. Johnson, W. G. Wood, B. M. Turner, and D. R. Higgs. 2001. Identification of a conserved erythroid specific domain of histone acetylation across the alpha-globin gene cluster. Proc. Natl. Acad. Sci. USA 98:12114-12119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Connolly, B., C. I. White, and J. E. Haber. 1988. Physical monitoring of mating type switching in Saccharomyces cerevisiae. Mol. Cell. Biol. 8:2342-2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dubey, D. D., L. R. Davis, S. A. Greenfeder, L. Y. Ong, J. Zhu, J. R. Broach, C. S. Newlon, and J. A. Huberman. 1991. Evidence suggesting that the ARS elements associated with silencers of the yeast mating-type locus HML do not function as chromosomal DNA replication origins. Mol. Cell. Biol. 11:5346-5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haber, J. E. 1998. Mating-type gene switching in Saccharomyces cerevisiae. Annu. Rev. Genet. 32:561-599. [DOI] [PubMed] [Google Scholar]
- 5.Hebbes, T. R., A. L. Clayton, A. W. Thorne, and C. Crane-Robinson. 1994. Core histone hyperacetylation comaps with generalized DNase I sensitivity in chicken beta globin chromosomal domain. EMBO J. 13:1823-1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hochstrasser, M., M. J. Ellison, V. Chau, and A. Varshavsky. 1991. The short-lived MAT alpha2 transcriptional regulator is ubiquitinated in vivo. Proc. Natl. Acad. Sci. USA 88:4606-4610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Holstege, F. C. P., E. G. Jennings, J. J. Wyrick, T. I. Lee, C. J. Hengartner, M. R. Green, T. R. Golub, E. S. Lander, and R. A. Young. 1998. Dissecting the regulatory circuitry of a eukaryotic genome. Cell 95:717-728. [DOI] [PubMed] [Google Scholar]
- 8.Huang, R. Y., and D. Kowalski. 1996. Multiple DNA elements in ARS305 determine replication origin activity in a yeast chromosome. Nucleic Acids Res. 24:816-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kornberg, R. D., and L. Stryer. 1988. Statistical distributions of nucleosomes: nonrandom locations by a stochastic mechanism. Nucleic Acids Res. 16:6677-6690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Litt, M. D., M. Simpson, F. Recillas-Targa, M. N. Prioleau, and G. Felsenfeld. 2001. Transitions in histone acetylation reveal boundaries of three separately regulated neighboring loci. EMBO J. 20:2224-2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lutter, L. C. 1989. Digestion of nucleosomes with deoxyribonucleases I and II. Methods Enzymol. 170:264-269. [DOI] [PubMed] [Google Scholar]
- 12.Moreira, J. M., and S. Holmberg. 1998. Nucleosome structure of the yeast CHA1 promoter: analysis of activation-dependent chromatin remodeling of an RNA-polymerase-II-transcribed gene in TBP and RNA pol II mutants defective in vivo in response to acidic activators. EMBO J. 17:6028-6038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Murphy, M. R., M. Shimizu, S. Y. Roth, A. M. Dranginis, and R. T. Simpson. 1993. DNA-protein interactions at the S. cerevisiae alpha2 operator in vivo. Nucleic Acids Res. 21:3295-3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oliver, S. G., Q. J. M. van der Aart, M. L. Agostini-Carbone, M. Aigle, L. Alberghina, D. Alexandraki, G. Antoine, R. Anwar, J. P. G. Ballesta, P. Benit, G. Berben, E. Bergantino, N. Biteau, P. A. Bolle, M. Bolotin-Fukuhara, A. Brown, A. J. P. Brown, J. P. Buhler, C. Carcano, G. Carignani, H. Cederberg, R. Chanet, R. Contreras, M. Crouzet, B. Daignan-Fornier, E. Defoor, M. Delgado, J. Demolder, C. Doira, E. Dubois, B. Dujon, A. Dusterhoft, D. Erdmann, M. Esteban, F. Fabre, C. Fairhead, G. Faye, H. Feldmann, W. Fiers, M. C. Francingues-Gaillard, L. Franco, L. Frontali, H. Fukuhara, L. J. Fuller, P. Galland, M. E. Gent, D. Gigot, V. Gilliquet, A. Goffeau, M. Grenson, P. Grisanti, L. A. Grivell, M. de Haan, M. Haasemann, D. Hatat, J. Hoenicka, J. Hegemann, C. J. Herbert, F. Hilger, S. Hohmann, C. P. Hollenberg, K. Huse, F. Iborra, K. J. Indge, K. Isono, C. Jacq, M. Jacquet, C. M. James, J. C. Jauniaux, Y. Jia, A. Jimenez, A. Kelly, U. Kleinhauns, P. Kreisi, G. Lanfranchi, C. Lewis, C. G. van der Linden, G. Lucchini, K. Lutzenkirchen, M. J. Maat, L. Mallet, G. Mannhaupt, E. Martegani, A. Mathieu, C. T. C. Maurer, D. McConnell, R. A. McKee, F. Messenguy, H. W. Mewes, F. Molemans, M. A. Montague, M. Muzi Falconi, L. Nevas, C. S. Newlon, D. Noone, C. Pallier, L. Panzeri, B. M. Pearson, J. Perea, P. Phillippsen, et al. 1992. The complete DNA sequence of yeast chromosome III. Nature 357:38-46. [DOI] [PubMed] [Google Scholar]
- 15.Ravindra, A., K. Weiss, and R. T. Simpson. 1999. High-resolution structural analysis of chromatin at specific loci: Saccharomyces cerevisiae silent mating-type locus HMRa. Mol. Cell. Biol. 19:7944-7950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15a.Roth, S. Y., and R. T. Simpson. 1991. Yeast minichromosomes. Methods Cell Biol. 35:289-314. [PubMed] [Google Scholar]
- 16.Sauer, R. T., D. L. Smith, and A. D. Johnson. 1988. Flexibility of the yeast alpha2 repressor enables it to occupy the ends of its operator, leaving the center free. Genes Dev. 2:807-816. [DOI] [PubMed] [Google Scholar]
- 17.Shrader, T. E., and D. M. Crothers. 1989. Artificial nucleosome positioning sequences. Proc. Natl. Acad. Sci. USA 86:7418-7422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Simpson, R. T., and D. W. Stafford. 1983. Structural features of a phased nucleosome core particle. Proc. Natl. Acad. Sci. USA 80:51-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Spellman, P. T., G. Sherlock, M. Q. Zhang, V. R. Iyer, K. Anders, M. B. Eisen, P. O. Brown, D. Botstein, and B. Futcher. 1998. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol. Biol. Cell 9:3273-3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Staynov, D. Z., and Y. G. Proykova. 1998. Quantitative analysis of DNase I digestion patterns of oligo- and polynucleosomes. J. Mol. Biol. 279:59-71. [DOI] [PubMed] [Google Scholar]
- 21.Strathern, J. N., and I. Herskowitz. 1979. Asymmetry and directionality in production of new cell types during clonal growth: the switching pattern of homothallic yeast. Cell 17:371-381. [DOI] [PubMed] [Google Scholar]
- 22.Sun, K., E. Coic, Z. Zhou, P. Durrens, and J. E. Haber. 2002. Saccharomyces forkhead protein Fkh1 regulates donor preference during mating-type switching through the recombination enhancer. Genes Dev. 16:2085-2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Szeto, L., M. K. Fafalios, H. Zhong, A. K. Vershon, and J. R. Broach. 1997. Alpha2p controls donor preference during mating type interconversion in yeast by inactivating a recombinational enhancer of chromosome III. Genes Dev. 11:1899-1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thoma, F. 1986. Protein-DNA interactions and nuclease sensitive regions determine nucleosome positions on yeast plasmid chromatin. J. Mol. Biol. 190:177-190. [DOI] [PubMed] [Google Scholar]
- 25.Vujcic, M., C. A. Miller, and D. Kowalski. 1999. Activation of silent replication origins at autonomously replicating sequence elements near the HML locus in budding yeast. Mol. Cell. Biol. 19:6098-6109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang, X., and R. T. Simpson. 2001. Chromatin structure mapping in Saccharomyces cerevisiae in vivo with DNase I. Nucleic Acids Res. 29:1943-1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weintraub, H., and M. Groudine. 1976. Chromosomal subunits in active genes have an altered conformation. Science 193:848-856. [DOI] [PubMed] [Google Scholar]
- 28.Weiss, K., and R. T. Simpson. 1997. Cell type-specific chromatin organization of the region that governs directionality of yeast mating type switching. EMBO J. 16:4352-4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weiss, K., and R. T. Simpson. 1998. High-resolution structural analysis of chromatin at specific loci: Saccharomyces cerevisiae silent mating type locus HMLα. Mol. Cell. Biol. 18:5392-5403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.White, C. I., and J. E. Haber. 1990. Intermediates of recombination during mating type switching in Saccharomyces cerevisiae. EMBO J. 9:663-673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Winston, F., C. Dollard, and S. L. Ricupero-Hovasse. 1995. Construction of a set of convenient Saccharomyces cerevisiae strains that are isogenic to S288C. Yeast 11:53-55. [DOI] [PubMed] [Google Scholar]
- 32.Wu, C., and W. Gilbert. 1981. Tissue specific exposure of chromatin structure at the 5′ terminus of the rat preproinsulin gene. Proc. Natl. Acad. Sci. USA 78:1577-1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu, C., K. Weiss, C. Yang, M. A. Harris, B. K. Tye, C. S. Newlon, R. T. Simpson, and J. E. Haber. 1998. Mcm1 regulates donor preference controlled by the recombination enhancer in Saccharomyces mating-type switching. Genes Dev. 12:1726-1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu, X., and J. E. Haber. 1996. A 700 bp cis-acting region controls mating-type dependent recombination along the entire left arm of yeast chromosome III. Cell 87:277-285. [DOI] [PubMed] [Google Scholar]
- 35.Wu, X., and J. E. Haber. 1995. MATa donor preference in yeast mating-type switching: activation of a large chromosomal region for recombination. Genes Dev. 9:1922-1932. [DOI] [PubMed] [Google Scholar]
- 36.Wu, X., J. K. Moore, and J. E. Haber. 1996. Mechanism of MATα donor preference during mating-type switching of Saccharomyces cerevisiae. Mol. Cell. Biol. 16:657-668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wyrick, J. J., F. C. Holstege, E. G. Jennings, H. C. Causton, D. Shore, M. Grunstein, E. S. Lander, and R. A. Young. 1999. Chromosomal landscape of nucleosome-dependent gene expression and silencing in yeast. Nature 402:418-421. [DOI] [PubMed] [Google Scholar]
- 38.Yu, L., and R. H. Morse. 1999. Chromatin opening and transactivator potentiation by RAP1 in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:5279-5288. [DOI] [PMC free article] [PubMed] [Google Scholar]