Abstract
DNA recombination required for mating type (mat1) switching in Schizosaccharomyces pombe is initiated by mat1 imprinting. The imprinting event is regulated by mat1 cis-acting elements and by several trans-acting factors, including swi1 (for switch), swi3, swi7, and sap1. swi1 and swi3 were previously shown to function in dictating unidirectional mat1 DNA replication by controlling replication fork movement around the mat1 region and, second, by pausing fork progression around the imprint site. With biochemical studies, we investigated whether the trans-acting factors function indirectly or directly by binding to the mat1 cis-acting sequences. First, we report the identification and DNA sequence of the swi3 gene. swi3 is not essential for viability, and, like the other factors, it exerts a stimulatory effect on imprinting. Second, we showed that only Swi1p and Swi3p interact to form a multiprotein complex and that complex formation did not require their binding to a DNA region defined by the smt-0 mutation. Third, we found that the Swi1p-Swi3p complex physically binds to a region around the imprint site where pausing of replication occurs. Fourth, the protein complex also interacted with the mat1-proximal polar terminator of replication (RTS1). These results suggest that the stimulatory effect of swi1 and swi3 on switching and imprinting occurs through interaction of the Swi1p-Swi3p complex with the mat1 regions.
The fission yeast Schizosaccharomyces pombe is a haploid eukaryotic microorganism whose cells exhibit two different mating types, called P (plus) and M (minus) (18, 33). The mating type is determined by the genetic information in the mat1 locus, which contains either the mat1M or the mat1P allele, each of which encodes two transcripts essential for controlling cell type (24). A copy of the mating type genetic information is also present at the mat2P and mat3M“donor” loci that are located centromere-distal to mat1 and are transcriptionally silent. The mating type switches when a copy of either the mat2P or mat3M DNA transposes and substitutes for mat1 by DNA recombination, where the transposed genetic information is expressed (6, 12, 26).
Switching occurs spontaneously at a high frequency in “homothallic” strains (h90, i.e., homothallic, 90% sporulation) during mitotic growth, and it follows an interesting nonrandom pattern within a cell lineage (17, 28, 34). Division of an unswitchable parental cell produces two daughter cells, one of which is programmed to be switching competent and the other of which is not. After cell division of a switching-competent cell, a switched and a switching-competent daughter cell are generated. On the other hand, a switching-noncompetent cell undergoes a cell division to produce a switching-competent and a noncompetent cell. Therefore, this switching pattern in the cell lineage results in only one cell switching among four granddaughter cells after two consecutive asymmetrical cell divisions of an unswitchable cell. These rules of so-called one-in-four and consecutive switching are observed with cells of either the P or M type, where most (80 to 90%) asymmetric cell divisions and chains of consecutive switching occur generation after generation in the progeny.
Cells engineered to contain an inverted duplication of mat1 change the pattern, so that two cousin cells among four granddaughter cells switch (28). Thus, according to the strand segregation model (27, 28), this differential mating type switching by asymmetric cell division is initiated by a novel imprint that marks one specific strand of DNA at the mat1 locus, resulting in a switching-competent cell. The chemical or physical nature of the imprint is not yet known, although it is considered to be either a modification of a DNA base(s) or a nick. The imprint is generated at a specific sequence(s) in only the specific strand of double-stranded DNA, which is known to be sensitive to heat as well as to alkali and RNase T2 treatment (2, 13, 36, 42). The imprinting site is thought to be converted to a transient double-strand break during DNA replication, so the resulting double-strand break is likely to initiate recombination for mating type switching (6, 7, 16). Double-strand break sites at the mat1M and mat1P loci were mapped by genomic sequencing (36).
The mating type switching mechanism is exquisitely coupled with the process of DNA replication. mat1 is replicated unidirectionally, and strand-specific imprinting in the mat1 locus is dependent on the direction of DNA replication because the imprint is installed only when the specific strand is replicated by the lagging-strand replication complex (12, 13, 14). The regulation of direction of DNA replication at mat1 is mediated through a replication termination site (RTS1), which is located centromere-proximal to mat1 and consists of two cis-acting sequences (10, 11, 14). According to this unidirectional replication model, a replication fork moving from the centromere-proximal origin stalls at RTS1, and therefore only the fork moving in the opposite direction from a mat1-distal replication origin(s) replicates mat1. During this unidirectional replication at mat1, an imprint is installed when a lagging-strand fragment is synthesized in the upper strand (13). As a result, only one of two chromatids acquires an imprint from a round of DNA replication in the unswitchable cell, and DNA replication in the switching-competent cell results in mat1 switching of the chromatid inheriting the imprinted strand.
Several trans- and cis-acting elements are required for the imprinting process in Schizosaccharomyces pombe. In earlier research, genetic screening for mutations affecting both mating type switching and the level of double-strand break implicated swi1, swi3, and swi7 in the switching process (16, 22). Double-strand break has also been shown to efficiently initiate meiotic mat1 gene conversion (30). With this meiotic assay, it was discovered that swi1-, swi3-, and swi7-encoded factors perform the imprinting function (29). Mutations of these three genes show a similar effect and result in a reduced rate of mating type switching. swi1 has been cloned and shows significant similarity to the clock gene family in amino acid sequence (9, 14). Interestingly, it has an additional function that is not connected to the mating type switching process (37). Both swi1 and swi3 promote imprinting in two independent ways; one way is to block replication at the RTS1 site, and the other is to mediate pausing of a replication fork around the imprint site (14). The swi7 gene encodes the DNA polymerase α catalytic subunit (39). DNA polymerase α contains the primase activity that initiates synthesis of Okazaki fragments by priming RNA during general DNA replication. Apparently, in S. pombe, DNA polymerase α acquired an additional role for efficient mating type switching. The precise role of swi7 in imprinting remains unknown, but it does not seem to be related to the pausing of a replication fork around the imprint site (14). The swi3 gene has not been characterized thus far.
Deletion analysis of sequences around the imprint site has implicated cis-acting sequences in the imprinting process. The mat1 cis-acting mutation smt-0 abolishes mating type switching completely and has been shown to contain a 263-bp deletion starting from the middle of the H1 homology to a flanking region located distal to mat1 (40). Analysis of the deleted region revealed at least two different cis-acting elements, SAS1 and SAS2, both of which are required to produce an efficient level of double-strand break, and therefore, of mating type switching. These elements additively contribute to the level of mating type switching. The SAS1 element interacts with Sap1 protein, another trans-acting factor required for mating type switching (4). It is not yet known how Sap1p bound to SAS1 promotes imprinting. sap1 is essential for cell viability and is perhaps involved in chromosome organization for their proper segregation in mitosis (15).
Our aim in this study was to characterize the imprinting process by the biochemical approach. Here, we report cloning of the swi3 gene with its DNA sequence and molecular characterization. We biochemically investigated the physical interaction between the trans-acting factors involved in mating type switching after tagging them with epitopes in order to analyze whether these proteins act in the same or different steps of the imprinting pathway. We show that Swi1p interacts with Swi3p to form a complex; however, this complex does not contain Swi7p or Sap1p. Also, Swi7p does not interact with Sap1p. We also examined the binding of Swi1p and Swi3p to the mat1 chromosomal region by chromatin immunoprecipitation experiments. We found that Swi1p and Swi3p are recruited to the imprinting site and to the RTS1 site located centromere-proximal to mat1 as well.
MATERIALS AND METHODS
Yeast strains, media, and genetic techniques.
The S. pombe strains used in this study are listed in Table 1. Standard genetic techniques for strain construction and medium preparation were used as described previously (35). Yeast transformation was performed as described previously (20).
TABLE 1.
S. pombe strains used in this study
| Strain | Genotype | Source or reference(s) |
|---|---|---|
| SP492 | h90 ade6-M216 swi7 leu1-32 | 39 |
| SP976 | h90 ade6-M210 ura4-D18 leu1-32 | 13, 14 |
| SP982 | h90 his2 ade6-M216 | 14 |
| SP903 | Msmt-0 leu1-32 ade6-M216 | This study |
| E146 | h90 his2 ade6-M210 swi3-146 | 22 |
| E157 | h90 his2 ade6-M210 swi3-157 | 22 |
| SG381 | h90 ade6-M210 swi3-1 | 22 |
| SP1125 | Msmt-0 mat2p::ura4+ura4-D18 ade6-M210 | This study |
| SP73 | h90 swi3-146 leul-32 | This study |
| BSP4 | h90 ade6-M210 ura4-D18 leu1-32 swi1-myc-kanMX6 | This study |
| BSP9 | h90 ade6-M210 ura4-D18 leu1-32 swi1-GST-kanMX6 | This study |
| BSP16 | h90 his2 ade6-M216 swi3-myc-kanMX6 | This study |
| BSP33 | h90 his2 ade6-M216 swi1-GST-kanMX6 swi3-myc-kanMX6 | This study |
| BSP34 | h90 his2 ade6-M216 sap1-myc-kanMX6 | This study |
| BSP50 | Msmt-0 mat2p::ura4+ade6-M210 swi1-GST-kanMX6 swi3-myc-kanMX6 | This study |
| BSP60 | h90 ade6-M216 ura4-D18 swi1-GST-kanMX6 sap1-myc-kanMX6 | This study |
| BSP61 | h90 his2 ade6-M216 Δswi3::kanMX6 | This study |
Iodine staining.
Individual colonies were grown on sporulation medium (PMA) at 25°C. After 3 to 4 days of incubation, the PMA plate was exposed to iodine vapors. Intensity of staining reflects efficiency of mating type switching, as switching and mating produce asci that synthesize an iodine-staining starch compound during meiosis (35).
Cloning and sequencing of the swi3 gene.
Strain SP73 carrying the swi3-146 mutation was used to clone swi3 by complementation, which is exactly analogous to the procedure used to clone the swi7 gene (39). The S. pombe cDNA library, kindly provided by P. Nurse, was used to clone a swi3 cDNA copy. The DNA sequence of the swi3 gene was determined by the dideoxy method with the Taq DyeDeoxy terminator cycle sequencing kit (Applied Biosystems, Foster City, Calif.).
Construction of the swi3 deletion mutant.
An Δswi3::kanMx6 allele was constructed by PCR-based gene disruption as described previously (5). The pFA6a-kanMX6 plasmid digested with NotI was subjected to PCR with an Advantage HF Taq DNA polymerase (Clontech) with OGBL45 (89-mer) and OGBL46 (90-mer) primers. OGBL45, a forward primer, includes a stretch of sequences corresponding to the 5′ end of swi3 and to BamHI and PacI sites in the multicloning sites of plasmid pFA6a (5). OGBL46, a reverse primer, includes sequences corresponding to the 3′ end of swi3 and to the EcoRI and PmeI sites of plasmid pFA6a. The resulting DNA product from the PCR contains a 1.6-kb kanMX6 marker that is flanked by sequences just to the 5′ and 3′ ends of the swi3 open reading frame (ORF). The PCR product was introduced into strain SP982, and G418-resistant transformants were selected. Strain BSP61 carrying Δswi3::kanMx6 was chosen from the G418-resistant transformants, and its deletion was established by PCR and genomic Southern blot analysis.
Construction of epitope-tagged swi1, swi3, and sap1.
The swi1, swi3, and sap1 genes were tagged with 13 copies of the human c-myc epitope or the gene for glutathione S-transferase (GST) at the 3′ end of each endogenous locus with the procedure described previously (5). To tag the swi1 gene with 13Myc or GST, plasmids pFA6a-13Myc-kanMX6 and pFA6a-GST-kanMX6 digested with NotI were used as templates and subjected to PCR with Vent DNA polymerase (New England Biolabs) with the OGBL9 (95-mer) and OGBL1O (96-mer) primers. The OGBL9 sequence includes swi1 sequences corresponding to C-terminal codons of the gene ending just upstream of the stop codon and the BamHI and PacI sites of the multicloning site of plasmid pFA6a. The OGBL10 sequence includes swi1 sequences corresponding to the 3′ untranslated region of the gene ending just downstream of the stop codon and the EcoRI and PmeI sites of plasmid pFA6a. DNA products from the PCR were introduced into SP976, and G418-resistant transformants were selected as described previously. Strains BSP4, carrying swi1-myc-kanMx6, and BSP9, carrying swi1-GST-kanMx6, were chosen from the G418-resistant transformants after confirming their tags by PCR, genomic Southern blot, and Western blot analyses.
To tag the swi3 or sap1 gene with 13 copies of the c-myc epitope, plasmid pFA6a-13Myc-kanMX6 was subjected to PCR with the OGBL11 (95-mer) and OGBL12 (87-mer) primers or the OGBL22 (95-mer) and OGBL23 (92-mer) primers, respectively. The OGBL11 and OGBL22 primers contain swi3 and sap1 sequences, respectively, corresponding to C-terminal codons of each gene ending just upstream of the stop codons and another sequence of the vector portion, as in OGBL9. OGBL12 and OGBL23 contain swi3 and sap1 sequences, respectively, corresponding to the 3′ untranslated region of each gene ending just downstream of the stop codons and another sequence of the vector portion, as in OGBL10. DNA products from each PCR were introduced into SP982, and strains BSP16, carrying swi3-myc-kanMx6, and BSP34, carrying sap1-myc-kanMx6, were constructed. Double-tagged strains BSP33 (h90 swi1-GST-kanMx6 swi3-myc-kanMx6), BSP50 (Msmt-0 swi1-GST-kanMx6 swi3-myc-kanMx6), and BSP60 (h90 swi1-GST-kanMx6 sap1-myc-kanMx6) were constructed by genetic crosses between strains BSP9 and BSP16, BSP33 and SP1125, and BSP9 and BSP34, respectively. After construction of the tagged strains, the efficiency of mating type switching in each single- and double-tagged strain was tested by an iodine staining procedure and confirmed to be compatible with that of the untagged parental strains.
Protein analysis.
Total protein extracts were prepared from cells grown in YEA at 25°C to mid-log phase with a lysis buffer containing 5 mM EDTA, 250 mM NaCl, 0.1% (vol/vol) Nonidet P-40, 50 mM Tris-HCl (pH 7.4), 0.1 mM phenylmethylsulfonyl fluoride, and 1 μg/ml each of the protease inhibitors pepstatin, leupeptin, aprotinin, antipain, and chymostatin. Western analysis was performed as described previously (32). Immunodetection was performed by the enhanced chemiluminescence (ECL) technique described by the supplier (Amersham). Protein molecular weight standards were obtained from Amersham. An anti-c-myc mouse monoclonal antibody and an anti-GST rabbit polyclonal antibody were purchased from Roche Applied Science and Upstate, respectively. A chicken antibody against S. pombe DNA polymerase α (Swi7p) was kindly provided by T. S.-F. Wang (Stanford University).
Coimmunoprecipitation.
Coimmunoprecipitation was performed as described previously (25). Total cell extracts were preabsorbed with 50 μl of protein A- or protein G-agarose (Roche Applied Science) on a roller drum for several hours at 4°C. After centrifugation, precleared supernatant was incubated with antibody for several hours, and then 50 μl of protein A- or G-agarose was added and further incubated overnight at 4°C. After centrifugation, the supernatant was gently pulled off, and the immunoprecipitated beads were washed twice with 1 ml of Tris buffer [50 mM Tris-HCl (pH 7.4), 5 mM EDTA, 0.2% (vol/vol) Nonidet P-40, 0.1% (vol/vol) sodium deoxycholate, 0.1 mM phenylmethylsulfonyl fluoride, protease inhibitors] containing 0.2 M NaCl. Bound complexes were further washed three times with 1 ml of Tris buffer containing 0.45 M NaCl. After extensive washes, a coimmunoprecipitated antibody beads pellet was resuspended in Laemmli buffer (31) and boiled for 2 min. The recovered supernatant was separated by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE), transferred to an Immobilon P membrane, and then subjected to Western blot analysis.
Chromatin immunoprecipitation.
Chromatin immunoprecipitation was performed as described previously (19), with some modifications. Cells were grown to mid-exponential phase in YEA at 25°C and further incubated by gentle shaking for 30 min in culture medium containing a final concentration of 3% (vol/vol) formaldehyde at room temperature. The culture was transferred to an ice water bath and further fixed for 15 min. Then the culture was mixed with glycine and incubated for 5 min at 25°C. After fixed cells were washed three times with extraction buffer, total cell extracts were prepared by vortexing with glass beads, sonicating for 30 s to fragment chromatin (3 pulses of 10 s with a 30-s pause between each pulse), and then immunoprecipitated according to the procedure described above. The beads pellet obtained from immunoprecipitation was incubated with TES buffer [50 mM Tris-HCl (pH 8.0), 10 mM EDTA, 1% SDS] at 65°C overnight, and the supernatant was recovered by centrifugation. DNA was recovered by extracting the supernatant with phenol-chloroform and precipitating with ethanol.
DNA was subjected to PCR analysis (30 cycles at 94°C, 30 s; 52°C, 45 s; and 72°C, 1 min, followed by extension at 72°C, 7 min). PCR products were radiolabeled by incorporating [α-32p]dCTP (Amersham Pharmacia) and separated on 6% native polyacrylamide gels. The band signals were quantitated by phosphorimaging analysis with a Typhoon 8600 phosphorimager (Molecular Dynamics, Sunnyvale, Calif.) and ImageQuant 1.2 software. The primer sets used for MPS, RTS1, lys11, and trp5 are listed in Table 2. We included analysis of untagged swi1 and swi3 as controls in a chromatin immunoprecipitation assay.
TABLE 2.
Oligonucleotides used in this study
| Primer | Sequence (5′ to 3′) |
|---|---|
| MPS forward | CCAATTATGCTGTTCGTGTCATTC |
| MPS reverse | GGAGAAAGAGGATGGTTGATGGAGTG |
| RTS1 forward | CGGAGATGTAACGAAGCTA |
| RTS1 reverse | CTTGCAATAGAATAATCCTAC |
| lys11 forward | GCTGATCCCTCCAAGTTTGTAGAC |
| lys11 reverse | GACAATATCTTCCGACTCAGTAGC |
| trp5 forward | GTTCCAGAAGCATTAACCCAGTG |
| trp5 reverse | GACAATCTTCTGCACCCATGTAG |
| OGBL9 | AAGCGAATCGACGATTTAGAAATGGAAGGAAAATTACAAGAAATTGAACAACTCAGCGAGAATTCTTCATCGGATCGGATCCCCGGGTTAATTAA |
| OGBL10 | CATTTTACTAACAAAAATTTCAATGAACAAGCTACCCATTATTAAACTTATGATTTGAATCGAAGATATTTTATTTGAATTCGAGCTCGTTTAAAC |
| OGBL11 | ACGAGCGGAGATCCATATGTTCAAGATACAGCTGATGATGCTTTTGTCGCCCAAGACAACGATACCCAATTAGAACGGATCCCCGGGTTAATTAA |
| OGBL12 | AATGATCATGATTACTAATATTGATAAACAAAGCTGCAACAACATCTCAATCATTCATTAATGGTGAGAATTCGAGCTCGTTTAAAC |
| OGBL22 | GCCAACATGGGCGCTAATATGAATGCCATTCGTGGCGGTCTCCATTCTAGCATCTCTCCTAATCTTGGTGACCATCGGATCCCCGGGTTAATTAA |
| OGBL23 | GAGAACGAAATTCTCGAAGAAAAGAGGTTGTGTGTTACACAGGTAGGAGGAGGATGGGCGGAAAAGTTGGGAGAATTCGAGCTCGTTTAAAC |
| OGBL45 | CACCATAAGAAATGTCTACAGCTGCTTCTGATTCAGGTGTTGAAAAACTCGTCGAGGAAAACAAAAGGGCGGATCCCCGGGTTAATTAA |
| OGBL46 | AAGCTGCAACAACATCTCAATCATTCATTAATGGTGACTATTCTAATTGGGTATCGTTGTCTTGGGCGACGAATTCGAGCTCGTTTAAAC |
Nucleotide sequence accession number.
The GenBank accession number for the nucleotide sequence of the swi3 gene is AY623652.
RESULTS
Cloning and characterization of the swi3 gene.
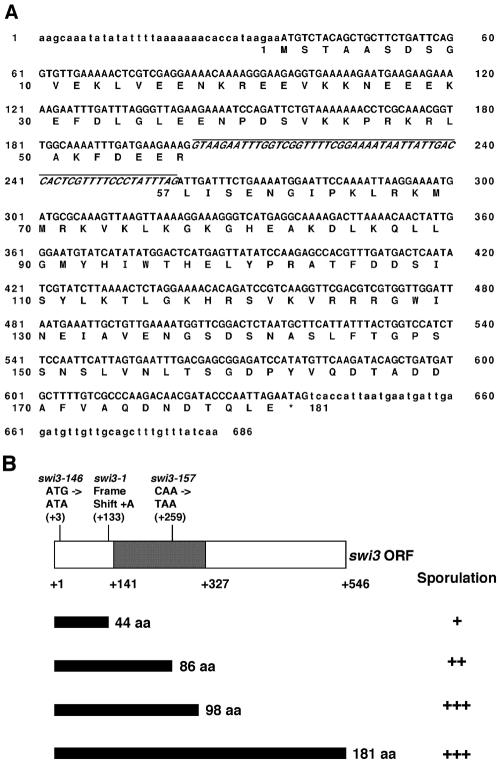
The swi3 gene was cloned by screening S. pombe genomic and cDNA libraries for plasmids that complemented the recessive swi3-146 mutation in strain SP73. The pRC14 plasmid carrying genomic DNA complementing the swi3 mutation was rescued from a transformant by transferring it into Escherichia coli and selecting for ampicillin resistance. This genomic clone was used as a probe to screen a cDNA library, resulting in the isolation of a plasmid containing a 0.6-kb yeast cDNA insert that also complemented the swi3-146 mutation. DNA sequences of the inserts from genomic and cDNA clones were determined (Fig. 1A). A single open reading frame (ORF) of 546 bp was found in the cDNA clone, which encodes a predicted protein of 181 amino acids with a calculated molecular mass of 20.6 kDa.
FIG. 1.
DNA sequence of the swi3 gene and mapping of mutations in three mutant alleles. (A) Nucleotide sequence of the swi3 gene. The exons and 5′- and 3′-flanking sequences were determined from a cDNA clone, and the intron sequence was determined from a genomic DNA clone. The exons and the intron are indicated in capital letters, and the flanking regions are indicated in lowercase letters. The deduced amino acid (aa) sequence is shown below the nucleotide sequence. The intron sequence is underlined and indicated in italics. The asterisk indicates the stop codon. (B) The nucleotide changes of three different mutations are indicated, with their positions shown in parentheses. The TIPIN-homologous region is indicated by darker shading. Predicted translated products from the mutant and wild-type ORFs are diagrammed as black bars. The effect of the alleles on sporulation is indicated by the level of sporulation in comparison with that of the wild-type control. The genomic DNA in the pRC14 plasmid, which encodes the N-terminal 98 codons of the swi3 ORF, fully complements the sporulation defect of all three mutant alleles.
Comparison of the DNA sequences between the two clones showed that the swi3 gene contained one intron of 58 bp. As a result of a search by BLAST in the databases of the NCBI, the swi3 ORF showed high similarity in amino acid sequence with mammalian and mouse timeless-interacting protein (Tipin) (2e-09 and 8e-08, respectively). The swi3-146, swi3-157, and swi3-1 mutations, causing defects in mating type switching (22), were identified by direct sequencing of PCR-amplified mutant alleles from strains E146, E157, and SG381, respectively. The swi3-146, swi3-157, and swi3-1 mutations changed the third [G to A (M1I)] and the 259th nucleotides [C to T(ochre)] and inserted A after the 132nd nucleotide (frame shift) of the wild-type swi3 ORF, respectively (Fig. 1B). swi3-157 is predicted to produce an N-terminally truncated protein with 86 amino acids. We observed that the swi3-157 mutant was partially functional in sporulation and that the defect was somewhat less than that caused by the swi3-1 mutation. Interestingly, we found that the N-terminal 98 amino acids of the swi3 ORF provided a wild-type level of switching and sporulation (Fig. 1B).
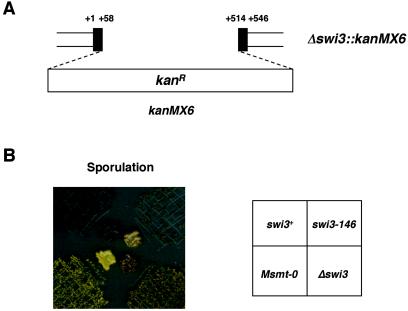
To examine the phenotype of complete-loss-of-function mutation, the swi3 gene was disrupted in strain SP982 by a PCR-based gene targeting procedure with a plasmid containing a selectable marker, kanMX6 (Fig. 2A and B). The swi3 deletion mutant was viable, with no significant reduction of growth in YEA medium. An effect of Δswi3::kanMX6 on sporulation was analyzed by iodine vapor staining of sporulated colonies as well as by microscopic observations. The Δswi3::kanMX6 allele did not abolish sporulation completely but greatly reduced it, resulting in a mottled staining pattern. Unlike the smt-0 mutant, in which mat1 switching was totally abolished, the extent of staining of the Δswi3::kanMX6 was similar to that of the swi3-146 mutant (Fig. 2B).
FIG. 2.
Δswi3 mutation reduces mating type switching. (A) The swi3 gene was disrupted by inserting a 1.6-kb kanMX6 cassette between nucleotides +58 and +514 of the ORF; +1 and +546 denote the first and last nucleotides of the swi3 ORF, respectively. (B) Comparison of the iodine-staining phenotype of homothallic wild-type swi3 (SP982), the point mutant swi3-146 (E146), the Δswi3::kanMX6 deletion mutant (BSP61), and a nonswitching Msmt-0 mutant (SP903). Efficiency of mating type switching was monitored by iodine staining of colonies sporulated on PMA medium.
Swi1p interacts with Swi3p in vivo.
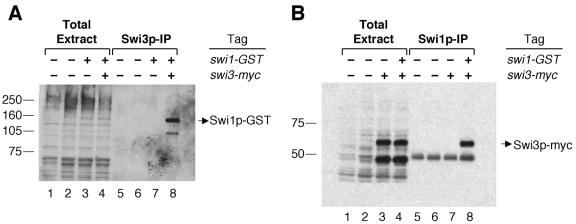
The swi1 and swi3 genes are required for mating type switching in S. pombe (16). To investigate whether the two gene products physically interact into one complex in vivo, an immunoprecipitation experiment was performed. Strain BSP33 carrying the swi1-GST and swi3-myc tags was constructed by crossing BSP9 (swi1-GST) with BSP16 (swi3-myc). As detected by Western blot analysis, the total protein extract prepared from BSP33 contained the tagged Swi1p-GST and Swi3p-myc (Fig. 3A and B, lane 4). In the first experiment, Swi3p-myc from the total protein extract was immunoprecipitated with anti-c-myc antibody, and then the immunoprecipitate was analyzed for Swi1p-GST. The result showed that a band corresponding to Swi1p-GST was present in the Swi3p-myc precipitate (Fig. 3A, lane 8). As a negative control, strain BSP9 carrying swi1-GST and untagged wild-type swi3 was likewise subjected to the immunoprecipitation experiment at the same time, and, as expected, Swi1p-GST was not detected in the precipitate (Fig. 3A, lane 7).
FIG. 3.
Swi1p interacts with Swi3p. (A) Detection of Swi1p-GST in the immunoprecipitate (IP) of Swi3p-myc. About 25 μg of total protein extracts loaded in each lane from strains SP976 (swi1 swi3, lane 1), SP982 (swi1 swi3, lane 2), BSP9 (swi1-GST swi3, lane 3), and BSP33 (swi1-GST swi3-myc, lane 4) were fractionated by SDS-8% PAGE. The protein G-agarose bead pellet from each immunoprecipitation was added with SDS sample buffer and boiled. After spinning down the beads, the same volume of supernatant of SP976 (lane 5), SP982 (lane 6), BSP9 (lane 7), and BSP33 (lane 8) was loaded and fractionated on the same gel. The proteins were transferred to an Immobilon P membrane and incubated with anti-GST antibody. We note that the second band below Swi1p-GST in lane 8 was not consistently seen in other experiments and could be a degradation product of the fusion protein. Sizes are shown to the left (in kilodaltons). (B) Detection of Swi3p-myc in the immunoprecipitate of Swi1p-GST. About 25 μg of total protein extracts from strains SP976 (lane 1), SP982 (lane 2), BSP16 (swi1 swi3-myc, lane 3), and BSP33 (lane 4) were fractionated by SDS-8% PAGE; immunoprecipitated complexes from each strain were recovered from the protein A-agarose bead pellet, loaded in lanes 5, 6, 7, and 8, respectively, and analyzed as in panel A but with anti-c-myc antibody. Swi3p-myc was detected by Western analysis as two different bands in the total extract lanes, but it was consistently observed that only the upper band was coimmunoprecipitated with Swi1p-GST. Another band below Swi3p-myc in the Swi1p-IP lanes reflects an unrelated cross-reaction of the antibody with immunoglobulin heavy chains from the polyclonal anti-GST antibody. Sizes are shown to the left (in kilodaltons).
The interaction between the two proteins was confirmed by a second experiment to test whether Swi3p-myc would also coprecipitate with Swi1p-GST in the same protein complex when a reversed immunoprecipitation was performed by pulling down Swi1p-GST in the total protein extract. The data showed that Swi3p-myc was present in the immunoprecipitate of Swi1p-GST (Fig. 3B, lane 8), demonstrating that Swi3p-myc coprecipitated with Swi1p-GST. In a negative control with strain BSP16 carrying untagged swi1 and swi3-myc, Swi3p-myc was not detected in the precipitate with anti-GST antibody (Fig. 3B, lane 7). Both sets of results indicate that the interaction between Swi1p and Swi3p is specific and that the two proteins are present in the same complex in vivo.
Swi1p-Swi3p complex, Swi7p, and Sap1p exist as separate entities in vivo.
swi1, swi3, and swi7 act in a similar manner, as they show a stimulatory effect for efficient switching by regulating mat1 imprinting. Furthermore, swi1 and swi3 perform imprinting during DNA replication (14). Given that swi7 encodes a catalytic subunit of DNA polymerase α that is required for general DNA replication, like the functions of swi1 and swi3, it probably regulates imprinting during DNA replication. Thus, it was determined whether the swi7 gene acts on the same step of the mating type switching pathway where the swi1 and swi3 genes function. Sap1p, which binds to SAS1 located near the imprint site, is another trans-acting factor required for the imprinting pathway. As with the leaky effect observed with other swi mutations, SAS1 deletion does not abolish mating type switching completely (4). Thus, Sap1p also seems to have a stimulatory effect on mating type switching. Therefore, we investigated biochemically whether Sap1p and Swi7p interact with each other and whether there is any interaction between these proteins and the Swi1p-Swi3p complex in vivo.
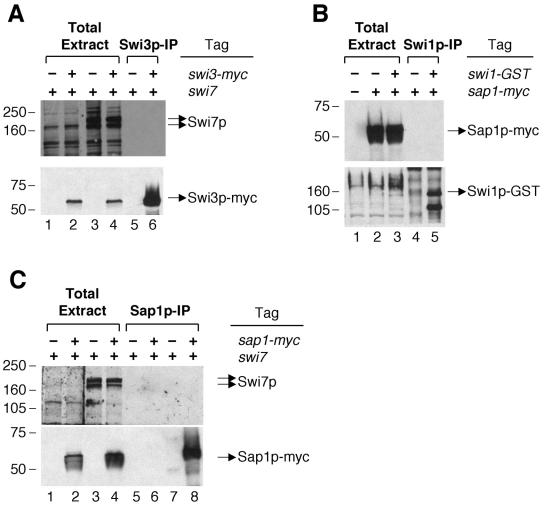
Cell lysate from BSP33 (swi1-GST swi3-myc) was incubated with anti-c-myc antibody to precipitate Swi3p-myc. Then it was determined whether Swi7p is coimmunoprecipitated with Swi3p-myc. The data showed that Swi7p did not coprecipitate with Swi3p-myc (Fig. 4A, lane 6), which was similarly shown in a negative control immunoprecipitation with untagged Swi3p (Fig. 4A, lane 5). Additionally, we performed immunoprecipitation of Swi1p-GST and found that Swi7p did not coprecipitate with it (data not shown). Next, we analyzed the interaction between Swi1p-GST and Sap1p-myc with a strain carrying swi1-GST and sap1-myc. The data showed that Sap1p-myc was not present in the immunoprecipitated complexes with Swi1p-GST (Fig. 4B, lane 5). Likewise, we also observed no interaction between Swi7p and Sap1-myc (Fig. 4C, lane 8). Taken together, these results demonstrate that the Swi1p-Swi3p complex does not interact with Swi7p and also not with Sap1p in vivo and that Swi7p does not interact with Sap1p.
FIG. 4.
Separate forms of Swi1p-Swi3p, Swi7p, and Sap1p in vivo. (A) Analysis of Swi7p in the Swi3p-myc immunoprecipitate (IP). About 25 μg of total protein extracts from BSP9 (swi1-GST swi3, lanes 1 and 3) and BSP33 (swi1-GST swi3-myc, lanes 2 and 4) were loaded in each lane. Immunoprecipitates with anti-c-myc antibody from BSP9 (lane 5) and BSP33 (lane 6) were loaded on the same gel. After blotting the gel onto an Immobilon P membrane, the proteins in lanes 1 and 2 were incubated with preimmune IgY as a negative control, and the proteins in lanes 3 to 6 were incubated with anti-polymerase α (Swi7p) antibody (upper panel). The lower panel is a control for the Swi3p-myc immunoprecipitation. After stripping off previous primary and secondary antibodies, the same blot was reprobed with anti-c-myc antibody to show the presence of immunoprecipitated Swi3p-myc in specific lanes (lower panel). (B) Analysis of Sap1p-myc in the Swi1p-GST immunoprecipitate. Total protein extracts from SP982 (swi1 sap1), BSP34 (swi1 sap1-myc), and BSP60 (swi1-GST sap1-myc) were loaded in lanes 1, 2, and 3, respectively, and immunoprecipitates of Swi1p-GST from BSP34 and BSP60 were loaded in lanes 4 and 5, respectively. The Western blot was probed with anti-c-myc antibody (upper panel) and then reprobed with anti-GST antibody later (lower panel) as described for panel A. (C) Analysis of Swi7p in the Sap1p-myc immunoprecipitate. Total protein extracts from SP982 and BSP34 were loaded in lanes 1 and 3 and lanes 2 and 4, respectively, and immunoprecipitates of Sap1p-myc from SP982 and BSP34 were loaded in lanes 7 and 8, respectively. Lanes 5 and 6 contained mock precipitates with SP982 and BSP34 lysates, respectively. In Western analysis, lanes 1 and 2 were incubated with preimmune IgY, and lanes 3 to 8 were incubated with anti-polymerase α (Swi7p) antibody (upper panel). The blot was reprobed with anti-c-myc antibody as a control for Sap1p-myc immunoprecipitation (lower panel). We note that Swi7p was detected as multiple bands by Western analysis, which was also observed in other studies (1, 41).
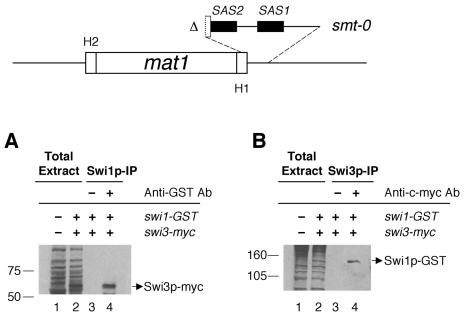
Analysis of Swi1p-Swi3p complex formation in the smt-0 mutant.
Mating type switching is totally abolished in the smt-0 mutant, as it lacks imprinting at the mat1 locus; thus, the deleted sequences in the mutant (Fig. 5) are indispensable for imprinting. It is well known that in cellular functions such as transcription, the formation of functional multiprotein complexes depends on the cis-acting elements to which they are binding. We therefore asked whether Swi1p-Swi3p complex formation depends on the mat1 cis-acting sequences required for imprinting. To determine this, we analyzed both tagged proteins in immunoprecipitation experiments with an Msmt-0 strain containing both the swi1-GST and swi3-myc tags. The results showed that both Swi1p-GST and Swi3p-Myc were present in both immunocomplexes, one with anti-GST and the other with anti-c-myc antibody (Fig. 5A and B). Thus, the Swi1p-Swi3p complex exists in vivo in the smt-0 background. This indicates that the interaction between Swi1p and Swi3p does not depend on the cis-acting sequences defined by the smt-0 mutation.
FIG. 5.
Interaction of Swi1p with Swi3p in the smt-0 mutant. A schematic representation of the 263-bp deletion of the smt-0 mutation is shown at the top. The deletion includes both SAS1 and SAS2 sequences, shown as black boxes. Immunoprecipitation (IP) was performed as described for Fig. 3 and 4. (A) Total protein extracts from SP982 (swi1 swi3) and BSP50 (swi1-GST swi3-myc) were loaded in lanes 1 and 2, respectively. With the total protein extract of BSP50, a mock precipitate of Swi1p-GST with protein A-agarose beads (lane 3) and the immunoprecipitate of the protein with anti-GST antibody (lane 4) were analyzed for Swi3p-myc by Western analysis. (B) Total protein extracts from SP982 and BSP50 were loaded in lanes 1 and 2, respectively. A mock precipitate of Swi3p-myc with protein G-agarose beads and the immunoprecipitate of Swi3p-myc with anti-c-myc antibody were loaded in lanes 3 and 4, respectively. The gel was subjected to Western analysis to detect Swi1p-GST.
Swi1p and Swi3p are bound to the mat1 locus.
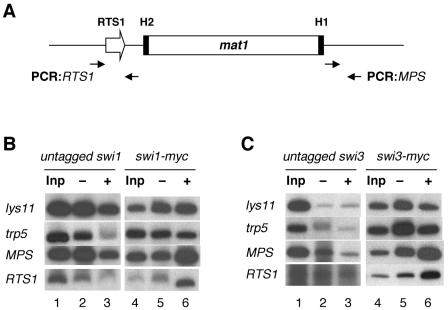
swi1 and swi3 facilitate imprinting by promoting unidirectional DNA replication of mat1. Both genes function to mediate termination of DNA replication at the RTS1 site located upstream of the imprint site; additionally, they function to pause replication fork movement near the imprint site (14). To examine whether Swi1p and Swi3p directly localize and bind to these regions to perform their roles, we performed chromatin immunoprecipitation experiments with strains containing swi1-myc or swi3-myc. These strains are derivatives of untagged strains SP976 and SP982, which were used as negative controls for the DNA binding experiments. Either c-myc epitope-tagged Swi1p or Swi3p was precipitated with or without anti-c-myc antibody in separate experiments, and enrichment of a region around the imprinting site (MPS) and of the RTS1 region in the immunoprecipitates was analyzed. To standardize the chromatin immunoprecipitation assay, parts of the coding regions of the lys11 and trp5 genes, which are unrelated to swi1 and swi3, were also analyzed in precipitates.
The results showed that these regions of lys11 and trp5 were not enriched in the precipitates, as expected (Fig. 6B and C, lanes 5 and 6). In contrast, both the MPS and RTS1 regions were enriched in Swi1p-myc (Fig. 6B, lanes 5 and 6) and Swi3p-myc (Fig. 6C, lanes 5 and 6) precipitates. We found about fivefold enrichment of these regions after normalizing to the signals from the lys11 and trp5 region controls. Compared to this result, chromatin immunoprecipitation of untagged Swi1p or Swi3p did not result in enrichment of these regions (Fig. 6B and C, lanes 2 and 3), indicating that the enrichment of the MPS and RTS1 regions was specific and due to the presence of tagged Swi1p or Swi3p. Taken together, the results of these experiments suggest that both Swi1p and Swi3p bind directly to the mat1 locus as well as to the RTS1 site.
FIG. 6.
Localization of Swi1p and Swi3p to the mat1 locus. (A) A schematic diagram of mat1 and its MPS and RTS1 regions amplified by PCR is shown. Primer sets for PCR (see Materials and Methods) are represented by filled-in arrows and were used to amplify 234-bp and 473-bp fragments of MPS and RTS1, respectively. (B) Chromatin immunoprecipitation analysis. Related SP976 (untagged swi1) and BSP4 (swi1-myc) strains were used to determine the localization of Swi1p-myc to the MPS and RTS1 regions. (C) Chromatin immunoprecipitation analysis of related strains SP982 (untagged swi3) and BSP16 (swi3-myc) was used to determine the localization of Swi3p-myc to the MPS and RTS1 regions. DNA recovered from each chromatin precipitate was analyzed by hot PCR with the indicated primer sets (see panel A) and deoxynucleoside triphosphates, including [α-32p]dCTP. Inp (input) denotes DNA extracted from the total cell extract that was subjected to immunoprecipitation experiments. It was loaded after manyfold dilution; −, mock precipitation without antibody; +, precipitation with anti-c-myc antibody. The lys11 and trp5 primer sets (see Materials and Methods) were used to amplify fragments of 345 bp for lys11 and 370 bp for trp5 for nonspecific DNA binding controls in both panels B and C.
DISCUSSION
S. pombe switches mating type in a single cell by a unique cellular system mechanistically different in fundamental ways from those evolved in other switching organisms. Several previous studies showed that mating type switching depends on a unique strand-specific imprint installed during mat1 DNA replication in one cell cycle. Replication of the imprinted chromosome in the next cell division results in a switch, following a one-in-four switching pattern in cell pedigrees. Molecular studies to define an imprinting process revealed that it is regulated by direction of DNA replication (13, 14). It is not known, however, how imprinting occurs during DNA replication or what its physical identity is. The mating type switching mutations swi1, swi3, and swi7 cause defects in the imprinting-dependent switching pathway, providing us with genetic and biochemical tools to define the process of imprinting in molecular terms.
The swi1 and swi7 genes were cloned previously (38, 39). Interestingly, swi7 encodes a catalytic subunit of DNA polymerase α, which was a discovery initially suggesting a possible involvement of DNA replication in the switching pathway. In this study, we isolated and characterized the swi3 gene, encoding a novel protein, and we applied a biochemical approach to find out if interactions occur between trans-acting factors. This is a direct way to identify whether these factors act in the same or different steps of the imprinting pathway. By coimmunoprecipitation experiments, we found that Swi1p and Swi3p interact and exist in a complex in vivo. Our results biochemically prove our previous proposition that the functions of Swi1p and Swi3p are closely related, since these proteins affect unidirectional mat1 DNA replication, which is necessary for imprinting (14).
The swi1 gene encodes a protein that is a homologue of the timeless protein and Saccharomyces cerevisiae Tof1p (14). The timeless gene, dtim, was originally identified in Drosophila melanogaster as a clock gene which controls circadian rhythms. Its identification was followed by discovery of the mouse (mTim), human (hTim), and Caenorhabditis elegans (Tim-1) homologues. Unlike dtim, its homologues do not have a clock function. Interestingly, searching for Swi3p in the NCBI databases identified novel human and mouse genes encoding a timeless-interacting protein (TIPIN) that shows significant homology to Swi3p. TIPIN was recently shown to make a complex with the timeless protein in vivo (21), suggesting that the Swi1p-Swi3p complex may be a TIM/TIPIN homologue. The function of mTIM along with mTIPIN in the mouse is unrelated to the clock mechanism, but they are necessary for cellular growth and differentiation and chromosome cohesion (8, 21). It is not yet known how the timeless complex performs its function, but both subunits play complementary roles; for example, mTIM promotes the nuclear localization of mTIPIN, and mTIPIN disrupts the ability of mTIM to form homomultimeric complexes (21). Therefore, it will be interesting to investigate whether Swi1p and Swi3p also display functional interactions similar to those of TIM and TIPIN.
Previously, the function of swi1 in mat1 switching was investigated by employing a partially deleted swi1 mutation that left the timeless conserved domain intact. This mutation, like other swi1 point mutations producing the intact timeless domain, exhibited a leaky phenotype for mat1 switching as it retained residual switching activity. To further define the function of this gene, we analyzed a swi1-disrupted mutant deleted for the timeless domain, spanning 266 codons at the N terminus of the ORF. The swi1-Δ266 null mutant was viable and exhibited a leaky phenotype for mating type switching similar to that of other swi1 mutants (unpublished data). Likewise, we found that a swi3 deletion mutant also grew normally, indicating that the swi3 gene is also not required for cell viability. This is unlike the other two mating type switching genes, swi7 and sap1, both of which are essential for cell viability (3, 39). It was also observed that mating type switching was not totally abolished in the swi3 null mutant, a phenotype similarly observed with other swi3 point mutants. Together, these results suggest that the Swi1p-Swi3p complex in S. pombe, a mouse TIM/TIPIN homologue, is not essential for viability but facilitates mating type switching. As Δswi1 and Δswi3 mutants switch mating type at a reduced rate, we deduce that Swi1p and Swi3p are not direct components of the imprinting activity.
Immunoprecipitation assays demonstrated that the Swi1p-Swi3p complex does not associate with either Swi7p or Sap1p. In a previous study with two-dimensional gel analysis, the effects of the swi1 and the swi3 mutations on the movement of DNA replication forks at mat1 were found to be different from those of the swi7 and smt-0 mutations (14). The results from our biochemical experiment support the idea that the Swi1p-Swi3p complex performs its function in a step of the imprinting pathway where Swi7p or Sap1p does not participate. swi7 and sap1 have some common characteristics: for example, they are essential genes, and, unlike swi1 and swi3, their roles in imprinting do not seem to be related to the pausing of replication forks at mat1. Therefore, we investigated to determine whether these two factors exist in a complex to act on a step different from the one involving the Swi1p-Swi3p complex. We observed by a coimmunoprecipitation experiment that Swi7p and Sap1p did not associate in a complex. This indicates that Swi7p and Sap1p probably act in different steps of the imprinting pathway.
We addressed whether formation of the Swi1p-Swi3p complex requires binding of these proteins to the cis-acting sequences essential for imprinting. For this purpose, we repeated a coimmunoprecipitation experiment in the smt-0 background, where SAS1, a binding site for Sap1p, and SAS2 are deleted, resulting in complete abolition of mating type switching. Interestingly, we still observed an interaction between the two proteins in the smt-0 mutant. This result indicates that their interaction does not depend on the presence of the cis-acting sequences. One possibility is that Swi1p and Swi3p have roles similar to those of TIM and TIPIN in assembling into an active complexed form before they interact with mat1. Alternatively, each protein may bind to a region outside SAS1 and SAS2, where they initially associate before the complex functions in mat1 switching.
In a previous study, it was shown that swi1 and swi3 control the polarity of mat1 replication in two ways, one by replication fork pausing at the mat1 locus and another by preventing the fork moving in the opposite direction from passing through a termination site. Interestingly, the dual functions can be separated in Swi1p, as the replication termination effect at RTS1 is disrupted while the pausing effect at the nearby imprint site is still intact in the swi1-rtf3 mutant (14). In this paper, we demonstrate by chromatin immunoprecipitation experiments that both Swi1p and Swi3p bind to sequences near the imprint site as well as at RTS1. Recently, the interaction of Swi1p around these regions was also observed, similar to our results in an independent investigation (23). Here, we investigated the interaction of both Swi1p and Swi3p with these regions because these proteins were identified as a multiprotein complex. We found that both of these proteins were binding near the imprint site as well as at the RTS1 region. Therefore, we suggest that the Swi1p-Swi3p complex facilitates imprinting by performing at least two functions, both of which require interaction of the complex with these regions. Two more genes, rtf1 and rtf2, were reported to affect replication termination through RTS1 (10). Therefore, at least four different trans-acting factors act at RTS1 for replication termination, which may be achieved through interactions between these factors and by their binding to this region.
Further definition of the roles of the Swi1p-Swi3p complex as well as other trans-acting factors is needed to better understand how imprinting is catalyzed and to define the connection between the functions of this complex and those of other trans-acting factors, such as swi7 and sap1. The imprint was characterized in previous studies as either an alkali-labile and RNase-sensitive modification(s) or a single-strand break (2, 13, 23, 42). Therefore, the chemical nature of the imprint in vivo needs to be further defined to understand how the key process of imprinting occurs. Most importantly, a factor for making an imprint still remains to be identified. The biochemical interactions between trans-acting factors and their binding to the imprinting region and the RTS1 region shown here would form a basis for future studies to further define the mechanism of imprinting that is well defined genetically but poorly understood biochemically.
Acknowledgments
We thank P. Young and D. Beach for an S. pombe genomic library, P. Nurse for a cDNA library, J.-Q. Wu and J. Bahler for plasmids containing the KanMX6 modules, and H. Gutz and H. Schmidt for yeast mutant strains. We also thank T. S.-F. Wang for polyclonal chicken IgY antibody against S. pombe DNA polymerase α.
This research was sponsored by the Center for Cancer Research of the National Cancer Institute, Department of Health and Human Services.
The contents of this publication do not necessarily reflect the views or policies of the Department of Health and Human Services, nor does the mention of trade names, commercial products, or organizations imply endorsement from the United States Government.
REFERENCES
- 1.Ahmed, S., S. Saini, S. Arora, and J. Singh. 2001. Chromodomain protein Swi6-mediated role of DNA polymerase alpha in establishment of silencing in fission yeast. J. Biol. Chem. 276:47814-47821. [DOI] [PubMed] [Google Scholar]
- 2.Arcangioli, B. 1998. A site- and strand-specific DNA break confers asymmetric switching potential in fission yeast. EMBO J. 17:4503-4510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arcangioli, B., T. D. Copeland, and A. J. S. Klar. 1994. Sap1, a protein that binds to sequences required for mating-type switching, is essential for viability in Schizosaccharomyces pombe. Mol. Cell. Biol. 14:2058-2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arcangioli, B., and A. J. S. Klar. 1991. A novel switch-activating site (SAS1) and its cognate binding factor (Sap1) required for efficient mat1 switching in Schizosaccharomyces pombe. EMBO J. 10:3025-3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bahler, J., J.-Q. Wu, M. S. Longtine, N. G. Shah, A. McKenzie III, A. B. Steever, A. Wach, P. Philippsen, and J. R. Pringle. 1998. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14:943-951. [DOI] [PubMed] [Google Scholar]
- 6.Beach, D. H. 1983. Cell type switching by DNA transposition in fission yeast. Nature 305:682-687. [Google Scholar]
- 7.Beach, D. H., and A. J. S. Klar. 1984. Rearrangements of the transposable mating-type cassettes of fission yeast. EMBO J. 3:603-610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chan, R. C., A. Chan, M. Jeon, T. F. Wu, D. Pasqualone, A. E. Rougvie, and J. Meyer. 2003. Chromosome cohesion is regulated by a clock gene paralogue TIM-1. Nature 423:1002-1009. [DOI] [PubMed] [Google Scholar]
- 9.Clayton, J. D., C. P. Kyriacou, and S. M. Reppert. 2001. Keeping time with the human genome. Nature 409:829-831. [DOI] [PubMed] [Google Scholar]
- 10.Codlin, S., and J. Z. Dalgaard. 2003. Complex mechanism of site-specific DNA replication termination in fission yeast. EMBO J. 22:3431-3440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dalgaard, J. Z., and A. J. S. Klar. 2001. A DNA replication-arrest site RTS1 regulates imprinting by determining the direction of replication at mat1 in S. pombe. Genes Dev. 15:2060-2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dalgaard, J. Z., and A. J. S. Klar. 2001. Does S. pombe exploit the intrinsic asymmetry of DNA synthesis to imprint daughter cells for mating-type switching? Trends Genet. 17:153-157. [DOI] [PubMed] [Google Scholar]
- 13.Dalgaard, J. Z., and A. J. S. Klar. 1999. Orientation of DNA replication establishes mating-type switching pattern in S. pombe. Nature 400:181-184. [DOI] [PubMed] [Google Scholar]
- 14.Dalgaard, J. Z., and A. J. S. Klar. 2000. swi1 and swi3 perform imprinting, pausing, and termination of DNA replication in S. pombe. Cell 102:745-751. [DOI] [PubMed] [Google Scholar]
- 15.de Lahondes, R., V. Ribes, and B. Arcangioli. 2003. Fission yeast Sap1 protein is essential for chromosome stability. Eukaryot. Cell 2:910-921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Egel, R., D. H. Beach, and A. J. S. Klar. 1984. Genes required for initiation and resolution steps of mating-type switching in fission yeast. Proc. Natl. Acad. Sci. USA 81:3481-3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Egel, R., and B. Eie. 1987. Cell lineage asymmetry in Schizosaccharomyces pombe: Unilateral transmission of a high frequency state of mating-type switching in diploid pedigrees. Curr. Genet. 12:429-433. [Google Scholar]
- 18.Egel, R., J. Kohli, P. Thuriaux, and K. Wolf. 1980. Genetics of the fission yeast Schizosaccharomyces pombe. Annu. Rev. Genet. 14:77-108. [DOI] [PubMed] [Google Scholar]
- 19.Ekwall, K., and J. F. Partridge. 1999. Fission yeast chromosome analysis: fluorescence in-situ hybridization (FISH) and chromatin immunoprecipitation (CHIP), p. 47-57. In W. A. Bickmore (ed.), Chromosome structural analysis: a practical approach. Oxford University Press, Oxford, United Kingdom.
- 20.Gietz, D., A. S. Jean, R. A. Woods, and R. H. Schiestl. 1992. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 20:1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gotter, A. L. 2003. Tipin, a novel timeless-interacting protein, is developmentally coexpressed with Timeless and disrupts its self-association. J. Mol. Biol. 331:167-176. [DOI] [PubMed] [Google Scholar]
- 22.Gutz, H., and H. Schmidt. 1985. Switching genes in Schizosaccharomyces pombe. Curr. Genet. 9:325-331. [DOI] [PubMed] [Google Scholar]
- 23.Kaykov, A., A. M. Holmes, and B. Arcangioli. 2004. Formation, maintenance and consequences of the imprint at the mating-type locus in fission yeast. EMBO J. 23:930-938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kelly, M., J. Burke, A. J. S. Klar, and D. Beach. 1988. Four mating-type genes control sexual differentiation in the fission yeast. EMBO J. 7:1537-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Keys, D. A., B.-S. Lee, J. A. Dodd, T. T. Nguyen, L. Vu, E. Fantino, L. M. Burson, Y. Nogi, and M. Nomura. 1996. Multiprotein transcription factor UAF interacts with the upstream element of the yeast RNA polymerase I promoter and forms a stable preinitiation complex. Genes Dev. 10:887-903. [DOI] [PubMed] [Google Scholar]
- 26.Klar, A. J. S. 2001. Differentiated parental DNA chain causes stem cell pattern of cell-type switching in Schizosaccharomyces pombe, p 17-36. In D. R. Marshak, R. L. Gardner, and D. Gottlieb (ed.), Stem cell biology. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 27.Klar, A. J. S. 1987. Differentiated parental DNA strands confer developmental asymmetry on daughter cells in fission yeast. Nature 326:466-470. [DOI] [PubMed] [Google Scholar]
- 28.Klar, A. J. S. 1990. The developmental fate of fission yeast cells is determined by the pattern of inheritance of parental and grandparental DNA strands. EMBO J. 9:1407-1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Klar, A. J. S., and M. J. Bonaduce. 1993. The mechanism of fission yeast mating-type interconversions: evidence for two types of epigenetically inherited chromosomal imprinted events. Cold Spring Harb. Symp. Quant. Biol. 58:457-465. [DOI] [PubMed] [Google Scholar]
- 30.Klar, A. J. S., and L. M. Miglio. 1986. Initiation of meiotic recombination by double-strand DNA breaks in S. pombe. Cell 46:725-731. [DOI] [PubMed] [Google Scholar]
- 31.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 32.Lee, B.-S., and M. R. Culbertson. 1995. Identification of an additional gene required for eukaryotic nonsense mRNA turnover. Proc. Natl. Acad. Sci. USA 92:10354-10358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leupold, U. 1950. Die vererbung von homothallie and heterothallie bei Schizosaccharomyces pombe. CR. Trav. Lab. Carlsberg Ser. Physiol. 24:381-480. [Google Scholar]
- 34.Miyata, H., and M. Miyata. 1981. Mode of conjugation in homothallic cells of Schizosaccharomyces pombe. J. Gen. Appl. Microbiol. 27:365-371. [Google Scholar]
- 35.Moreno, S. A., A. J. S. Klar, and P. Nurse. 1991. Molecular genetics analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 267:795-823. [DOI] [PubMed] [Google Scholar]
- 36.Nielsen, O., and R. Egel. 1989. Mapping the double-strand breaks at the mating-type locus in fission yeast by genomic sequencing. EMBO J. 8:269-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Noguchi, E., C. Noguchi, L.-L. Du, and P. Russell. 2003. Swi1 prevents replication fork collapse and controls checkpoint kinase Cds1. Mol. Cell. Biol. 23:7861-7874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schmidt, H. 1987. Strains of Schizosaccharomyces pombe with a disrupted swi1 gene still show some mating-type switching. Mol. Gen. Genet. 210:485-489. [DOI] [PubMed] [Google Scholar]
- 39.Singh, J., and A. J. S. Klar. 1993. DNA polymerase-α is essential for mating- type switching in fission yeast. Nature 361:271-273. [DOI] [PubMed] [Google Scholar]
- 40.Styrkarsdottir, U., R. Egel, and O. Nielsen. 1993. The smt-0 mutation which abolishes mating-type switching in fission yeast is a deletion. Curr. Genet. 23:184-186. [DOI] [PubMed] [Google Scholar]
- 41.Tan, S., and T. S.-F. Wang. 2000. Analysis of fission yeast primase defines the checkpoint response to aberrant S-phase initiation. Mol. Cell. Biol. 20:7853-7866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vengrova, S., and J. Z. Dalgaard. 2004. RNase-sensitive DNA modification(s) initiates S. pombe mating-type switching. Genes Dev. 18:794-804. [DOI] [PMC free article] [PubMed] [Google Scholar]