Abstract
Transcriptional activity of the TATA-binding protein (TBP) is controlled by a variety of proteins. The BTAF1 protein (formerly known as TAFII170/TAF-172 and the human ortholog of Saccharomyces cerevisiae Mot1p) and the NC2 complex composed of NC2α (DRAP1) and NC2β (Dr1) are able to bind to TBP directly and regulate RNA polymerase II transcription both positively and negatively. Here, we present evidence that the NC2α subunit interacts with BTAF1. In contrast, the NC2β subunit is not able to associate with BTAF1 and seems to interfere with the BTAF1-TBP interaction. Addition of NC2α or the NC2 complex can stimulate the ability of BTAF1 to interact with TBP. This function is dependent on the presence of ATP in cell extracts but does not involve the ATPase activity of BTAF1 nor phosphorylation of NC2α. Together, our results constitute the first evidence of the physical cooperation between BTAF1 and NC2α in TBP regulation and provide a framework to understand transcription functions of NC2α and NC2β in vivo.
Initiation of gene transcription by eukaryotic RNA polymerase II (pol II) is tightly controlled by a multitude of regulatory factors. The concerted action of these factors results in the formation of the pol II preinitiation complex (32, 33). Recent studies have deepened our knowledge of the regulation of the steps leading to this. It is also becoming clear that the mode and sequence of the recruitment of the basal transcription factors vary among promoters (7). The preinitiation complex consists of several basal transcription factors, including TATA-binding protein (TBP), which plays a central role in the assembly process. This is underscored by the fact that transcription of the majority of cellular genes in vivo requires TBP (11). In human cells several factors were shown to bind directly to and regulate the activity of TBP in pol II transcription. The best-studied factors are TBP-associated factors (TAFs), which together with TBP form the TFIID complex (25, 40). Others include the BTAF1 protein (TAFII170/TAF-172) and the NC2 (Dr1-DRAP1) complex.
BTAF1 and its Saccharomyces cerevisiae ortholog Mot1p form stable complexes with TBP in cell extracts (37, 38, 43, 44; for a review, see reference 35). The observation that a large proportion of TBP is complexed with BTAF1 as the B-TFIID complex gives rise to the notion that BTAF1 is an important regulator of TBP function (44). Indeed, in vitro studies show that the B-TFIID complex is able to bind promoter DNA and support transcription (18, 34, 44). On the other hand, BTAF1 and Mot1p proteins contain dATPase activity, which is involved in the dissociation of TBP from DNA in an ATP-dependent stroke. This activity can explain their negative effect on transcription (5, 34, 36). In accordance with a dual role of Mot1p in transcription, mRNA expression profiling and mutational analyses indicate that Mot1p affects transcription both positively and negatively (1, 6, 10, 12, 15, 26, 27, 39). The positive role of Mot1p is strengthened by observations that it is present at the sites of certain promoters upon their activation and is indispensable for TBP binding and transcription at these promoters (1, 10, 15). To reconcile the opposite effects of BTAF1 and Mot1p on transcription, it has been hypothesized that these proteins can liberate TBP from nonpromoter sites in an ATP-dependent reaction and deliver it onto the promoter DNA (15, 30). Despite recent advances in our knowledge of BTAF1 and Mot1p function (35) several questions concerning the regulation of BTAF1 and Mot1p remain unexplored. In particular the identity and mechanism of protein factors that regulate BTAF1 and Mot1p function are poorly understood.
The NC2 (Dr1-DRAP1) complex represents another regulator of TBP function. NC2 consists of two subunits, NC2α (DRAP1) and NC2β (Dr1), which form a stable complex via histone fold domains at their amino-terminal ends (17, 22, 29). NC2 was identified in in vitro experiments as an inhibitor of pol II transcription (28). The NC2 complex interacts with TBP bound to TATA box DNA and blocks the association of the basal transcription factors TFIIA and TFIIB, resulting in nonproductive TBP-TATA complexes (17, 29). In addition to its inhibitory role on TATA box-containing promoters, NC2 was also isolated as a factor required for efficient transcription from downstream promoter element-containing promoters in Drosophila melanogaster extracts (46). As in the case of Mot1p, in vivo studies with yeast indicate that NC2 plays both negative and positive roles in transcription (4, 26, 39). Specifically, Bur6p (yeast NC2α) was shown by mRNA expression profiling to stimulate expression of a subset of yeast genes. Promoter occupancy of yeast NC2α correlated with promoter activity, suggesting a positive role in transcription (4, 14).
Genetic analysis of yeast suggested separate roles of NC2α and NC2β in gene expression (23). A recent study by Collart and colleagues indicated that NC2α can be isolated without NC2β from yeast cell extracts and that this is dependent on growth conditions (8). In addition, NC2α binding correlated with active promoters, whereas binding of the NC2 complex correlated with repressed promoters (8). Genetic experiments with mice established a crucial role for the NC2α subunit during gastrulation (20). This led to the finding that NC2α, but not NC2β, can bind to the FoxH1 transcription factor and can inhibit its binding to DNA (20). In addition, distinct tissue distributions of NC2α and NC2β mRNAs have been reported (29).
The observation that BTAF1 (Mot1p) and NC2 regulate similar steps in transcription raises the possibility of a functional interaction. Firstly, biochemical analyses indicate that they both have negative and positive roles in transcription (5, 17, 29, 44, 46). Secondly, Mot1p and NC2α were isolated in the same genetic screen of yeast (24, 39). Thirdly, Mot1p and NC2α are required for expression of certain yeast promoters, such as promoters of GAL1 and GAL10 and the TATA-less promoters of HIS3 and HIS4 (26, 39). Finally, Mot1p and NC2α regulate an overlapping set of genes in yeast as revealed by mRNA expression profiling (1, 10, 14).
Here we present evidence that BTAF1 and B-TFIID can interact directly with isolated human NC2α. The NC2β subunit of the NC2 complex does not interact with BTAF1 but seems to be able to disrupt B-TFIID. Moreover, NC2α can stimulate association of BTAF1 with TBP in an ATP-dependent reaction. Our results provide a molecular basis for previous observations of functional links between BTAF1 (Mot1p) and NC2α (Bur6p). They also suggest a novel mode of TBP regulation by a previously unknown mechanism involving ATP hydrolysis.
MATERIALS AND METHODS
Plasmids.
Plasmids encoding LexA fusions of BTAF1 fragments used in a yeast two-hybrid screen were described previously (36). pJG-NC2α and pJG-NC2β were obtained by inserting NC2α and NC2β coding sequences flanked by EcoRI sites into EcoRI-digested pJG4-5. All NC2α cDNA mutants and deletions in the pJG4-5 vector were created by standard PCR-based techniques. pET-hisNC2α, pET-hisNC2β, and pDr1 were described previously (17, 19). pACYC-NC2α was created by inserting the NdeI-BamHI fragment from pET-hisNC2α into NdeI-BamHI-digested pACYC-11b. pACYC-NC2β was created by inserting the NdeI-BamHI fragment from pDr1 into NdeI-BamHI-digested pACYC-11b. pACYC-hisNC2β was created by inserting the NcoI (blunted)-BamHI fragment of pET-hisNC2β into XhoI (blunted)-BamHI-digested pACYC-11b. All described pET-hisNC2α mutants and deletions were created by standard PCR-based techniques. pGEX-NC2α was created by inserting the NdeI-BamHI fragment from pET-hisNC2α into pGEX-2T with the modified polylinker. pGEX-NC2β was created by inserting the blunted NheI-BamHI fragment from pDr1 into SmaI-digested pGEX-2T. pGST-BTAF1(505-891) was created by inserting the appropriate BTAF1 cDNA fragment into pGEX-2T with the modified polylinker. pGST-ΔTFIIS was described previously (36). Plasmid pTM3 containing cDNA coding for BTAF1 mutant K1297A was constructed using PCR-based mutagenesis and used to construct a BTAF1 K1297A-expressing vaccinia virus as described previously (34).
Protein expression and purification.
Escherichia coli BL21(DE3) strains expressing an appropriate protein or pair of proteins were cultured on a 2-liter scale and lysed by lysozyme treatment as described previously (36). To obtain NC2(hisα/β), NC2(α/hisβ), NC2(GSTα/hisβ), and NC2(hisα/GSTβ) complexes, bacteria were transformed with the appropriate pair of pET- and pACYC-based plasmids and grown in the presence of 50 μl of ampicillin/ml and 30 μg of chloramphenicol/ml. All hexahistidine-tagged proteins or protein complexes were purified on a 5-ml Ni-nitrilotriacetic acid (NiNTA)-agarose column (QIAGEN) as the first step. After lysate loading, the column was washed with NiNTA wash buffer (50 mM Tris-HCl [pH 8.0], 300 mM KCl, 20% glycerol, 0.1 mM EDTA, 20 mM imidazole, 1 mM phenylmethylsulfonyl fluoride [PMSF], 0.5 mM β-mercaptoethanol). Proteins were eluted with NiNTA wash buffer containing 300 mM imidazole. Proteins or protein complexes containing the glutathione S-transferase (GST) tag were purified on a 1.5-ml glutathione-agarose column (Amersham Biosciences). After loading, the column was washed with GST wash buffer (50 mM Tris-HCl [pH 8.0], 300 mM KCl, 20% glycerol, 1 mM EDTA, 1 mM PMSF, 1 mM dithiothreitol [DTT]) and eluted with GST wash buffer containing 10 mM reduced glutathione (Sigma).
After the NiNTA agarose step, NC2(hisα/β) and NC2(α/hisβ) protein complexes were purified further on a MonoS HR 5/5 column (Amersham Biosciences). NiNTA column fractions were dialyzed against buffer A (20 mM HEPES-KOH [pH 7.9], 20% glycerol, 0.5 mM EDTA, 0.5 mM PMSF, 1 mM DTT) containing 100 mM KCl and loaded onto the column. Bound proteins were eluted using a linear gradient of 100 to 600 mM KCl in buffer A. Human recombinant hisTBP was purified on NiNTA-agarose as described previously (42), adjusted to buffer A containing 100 mM KCl, and loaded onto a 5-ml heparin column (Amersham Biosciences). Bound TBP was eluted with a linear gradient of 100 to 500 mM KCl in buffer A. All purified proteins were dialyzed against buffer A containing 100 mM KCl and stored at −80°C.
Native B-TFIID was purified from HeLa cells as described previously (34). Expression and purification of recombinant B-TFIID in HeLa cells using the T7 polymerase-driven vaccinia virus-based system was described previously (34). Expression and purification of recombinant hisBTAF1 in HeLa cells using the endogenous promoter-driven vaccinia virus-based system was described previously (45). HeLa or RK13 cell lysates containing overexpressed wild-type or K1297A BTAF1 proteins were obtained by coinfection of cells with vaccinia viruses expressing T7 polymerase and the appropriate BTAF1 protein (34). Cells were lysed in ELB buffer (50 mM HEPES-KOH [pH 7.9], 150 mM KCl, 0.01% Triton X-100, 20% glycerol, 0.5 mM EDTA, 0.5 mM PMSF, 1 μg of aprotinin/ml, 1 μg of leupeptin/ml, 1 μg of pepstatin/ml).
Protein-protein interaction assays.
A yeast two-hybrid screen of a human peripheral blood leukocyte cDNA library (Origene) was performed using LexA-BTAF1(505-1133) as bait according to standard procedures (47). The strength of the interactions was determined by quantification of β-galactosidase activity in yeast lysates as described previously (48). For the GST pull-down assay (see Fig. 2B), approximately 1 μg of GST-tagged proteins or protein complexes was bound to 10 μl of glutathione-agarose beads for 1 h at 4°C in binding buffer (50 mM Tris-HCl [pH 8.0], 100 mM KCl, 20% glycerol, 0.5 mM EDTA, 0.5 mM PMSF, 1 mM DTT, 0.01% Triton X-100). The beads were washed three times with binding buffer, and BTAF1-enriched lysate or purified hisTBP was subsequently added in 350 μl of binding buffer and incubated for 2 h at 4°C. The beads were washed three times in binding buffer, and bound BTAF1 or hisTBP proteins were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis followed by Western blotting and detected by using specific antibodies. Another GST pull-down assay (see Fig. 2A) was performed as described above using bacterial lysates of GST fusion proteins and purified NC2 proteins. In additional experiments (see Fig. 4, 5, and 6), equal amounts of GST-TBP or GST-ΔTFIIS from bacterial lysates were preincubated with glutathione-agarose beads as described above. After washing, BTAF1-containing lysates or partially purified hisBTAF1 and NC2 proteins were added to the reaction mixture as indicated in the figure legends and processed as described above. Additional reagents were added along with BTAF1 and NC2 proteins as indicated in the figure legends. A hexahistidine pull-down assay (see Fig. 2C) was performed as described (see Fig. 2B) except that 10 μl of NiNTA agarose beads was used, and the binding buffer lacked EDTA and contained 0.5 mM β-mercaptoethanol instead of DTT.
FIG. 2.
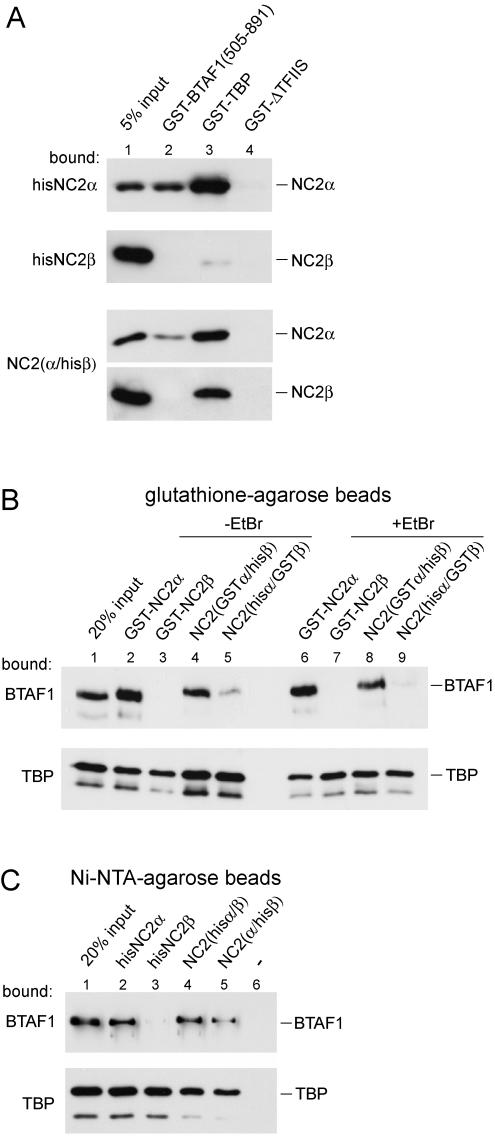
Direct interaction of BTAF1 with NC2α but not with NC2β. (A) The BTAF1 505-891 fragment interacts directly with NC2α. GST pull-down was performed as described in Materials and Methods by using bacterial lysates containing equal amounts of GST-BTAF1 (505-891) (lane 2), GST-TBP (lane 3), and GST-ΔTFIIS (lane 4). Purified hisNC2α, hisNC2β, or NC2(α/hisβ) was added as indicated. (B) NC2 proteins were expressed and purified to near homogeneity (unpublished data). GST-NC2α, GST-NC2β, NC2(GSTα/hisβ), and NC2(hisα/GSTβ) were tested for their ability to bind BTAF1 and hisTBP. The lysate of RK13 cells overexpressing BTAF1 (3 μl; top panel) or purified hisTBP (20 ng; bottom panel) were incubated with glutathione-agarose beads coated with different NC2 proteins (as indicated above the lanes) in the absence (lanes 2 to 5) or presence(lanes 6 to 9) of 20 μg of ethidium bromide (EtBr)/ml as described in Materials and Methods. This concentration of ethidium bromide was shown to severely diminish TBP-DNA and B-TFIID-DNA interaction in EMSA (data not shown). Bound proteins were visualized by immunoblotting using TBP- or BTAF1-specific antibodies. (C) Hexahistidine pull-down was performed as described for the GST pull-down shown in panel B except that NiNTA-agarose beads, purified hisNC2α (lane 2), hisNC2β (lane 3), NC2(hisα/β) (lane 4), or NC2(α/hisβ) (lane 5) was used. GST-TBP was used as the source of TBP in the bottom panel.
FIG. 4.
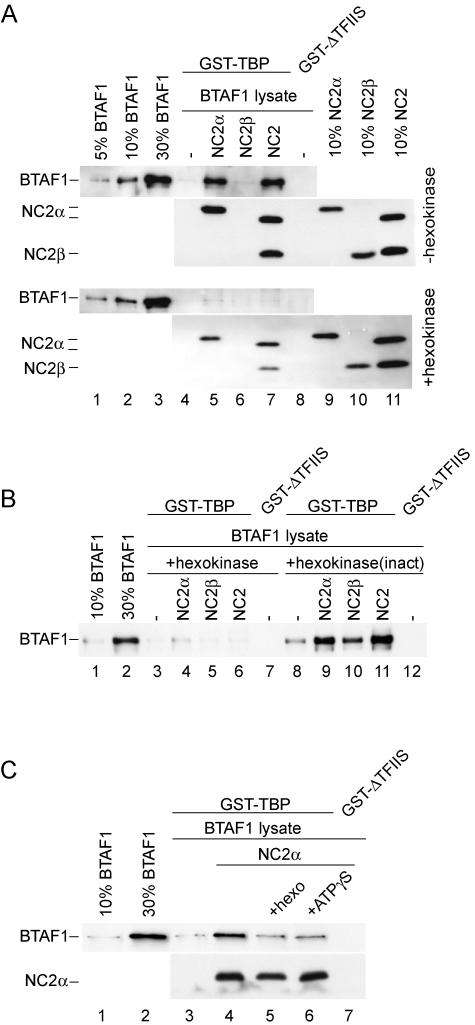
NC2α stimulates the interaction of BTAF1 with TBP in an ATP-dependent manner. (A) RK13 cell lysate (1 μl) containing overexpressed BTAF1 was incubated with glutathione-agarose beads coated with GST-TBP (lanes 4 to 7) or GST-ΔTFIIS (lane 8) in the absence (lanes 4 and 8) or presence of 6.5 pmol of hisNC2α (lane 5), hisNC2β (lane 6), and NC2(α/hisβ) (lane 7) as described in Materials and Methods. Lanes 4 to 8 in the bottom panel contained 10 U of hexokinase and 10 mM glucose. (B) Hexokinase activity is necessary to abolish NC2α-mediated binding of BTAF1 and TBP. RK13 cell lysate (1 μl) containing overexpressed BTAF1 was incubated with glutathione-agarose beads coated with GST-TBP (lanes 3 to 6 and 8 to 11) or GST-ΔTFIIS (lanes 7 and 12) in the absence (lanes 3, 7, 8, and 12) or presence of 6.5 pmol of hisNC2α (lanes 4 and 9), hisNC2β (lanes 5 and 10), and NC2(α/hisβ) (lanes 6 and 11). Lanes 3 to 7 contained 10 U of hexokinase and 10 mM glucose. Lanes 8 to 12 contained 10 U of hexokinase inactivated by boiling for 3 min [hexokinase(inact)] and 10 mM glucose. (C) Excess ATP analogue inhibits NC2α-stimulated BTAF1-TBP interaction. RK13 cell lysate (1 μl) containing overexpressed BTAF1 was incubated with glutathione-agarose beads coated with GST-TBP (lanes 3 to 6) or GST-ΔTFIIS (lane 7) in the absence (lanes 3 and 7) or presence of 6.5 pmol of hisNC2α (lanes 4 to 6). Lane 5 contained 4 U of hexokinase (hexo) and 5 mM glucose. Lane 6 contained 2 mM ATPγS. All reaction mixtures additionally contained 5 mM magnesium chloride.
FIG. 5.
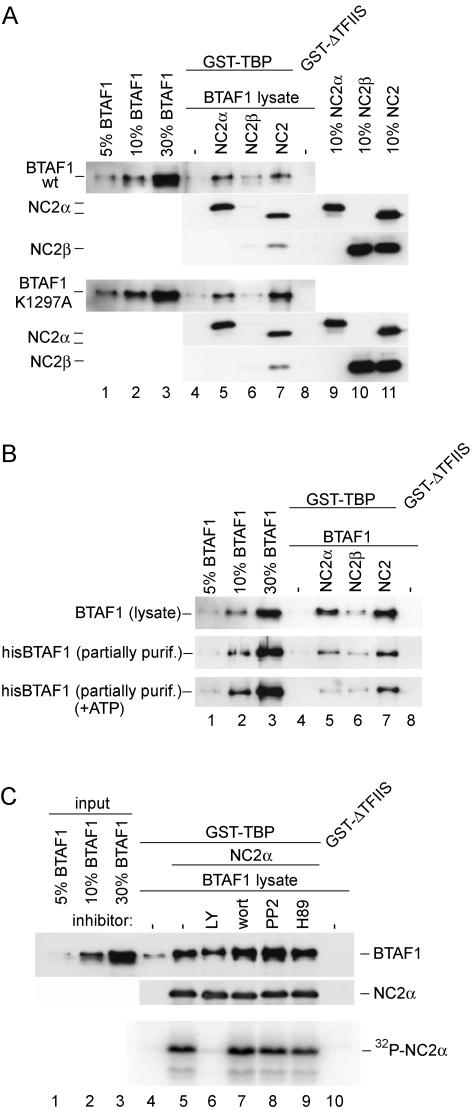
ATP hydrolysis by BTAF1 or NC2α phosphorylation is not required for its binding to TBP in the presence of NC2α. (A) The ATPase-deficient mutant of BTAF1 binds to TBP in the presence of NC2α. Reactions were performed as described for Fig. 4A. BTAF1-containing HeLa cell lysates were diluted with the lysate of cells infected with T7 polymerase-expressing virus to obtain equal concentrations of wild-type and mutant BTAF1. (B) ATP does not increase BTAF1-TBP interaction in the presence of NC2α. Reactions were performed as described for Fig. 4A, except that the reaction mixtures shown in the top panel received 0.5 μl of BTAF1-enriched RK13 cell lysate, and those shown in the middle and bottom panels received 0.5 μl of partially purified (purif.) hisBTAF1. Reaction mixtures shown in the bottom panel also contained 1 mM ATP, 20 U of creatine phos-phokinase, and 10 mM creatine phosphate. (C) NC2α phosphorylation is not required for BTAF1-TBP interaction. The reaction was performed as described for panel A, except that 10 times more GST-TBP, hisNC2α, and BTAF1-containing lysate was added. Twenty microcuries of [32P]γATP and 5 mM MgCl2 were included in all the reactions. Lane 6 included 50 μM LY194002 (LY), lane 7 contained 10 μM wortmannin (wort), lane 8 contained 50 μM PP2, and lane 9 contained 50 μM H89. The top panel represents the immunoblotting experiment, whereas the bottom panel represents the autoradiogram.
FIG. 6.
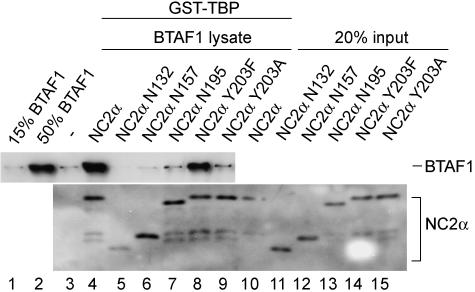
The BTAF1-NC2α interaction is necessary for stimulation of BTAF1 binding to TBP. BTAF1-enriched RK13 cell lysate (1 μl) and GST-TBP-coated beads were incubated in the absence (lane 3) and in the presence (lanes 4 to 9) of 100 pmol of the indicated hisNC2α mutant proteins as described in Materials and Methods. hisNC2α proteins were detected using anti-His antibodies in a Western blotting experiment.
DNA binding activity assay.
The electrophoretic mobility shift assay (EMSA) was performed essentially as described previously (34). Radiolabeled AdMLP probe (−53 to −12), 5 ng of hTBP, 3 to 5 ng of recombinant B-TFIID, and the NC2 amounts indicated in the figure legends were used. For the supershift experiments 500 ng of anti-NC2α monoclonal antibody or 1 μg of affinity-purified polyclonal antibody PF299 directed against BTAF1 was used (34).
RESULTS
Isolation of NC2α as a BTAF1 interactor by yeast two-hybrid screening.
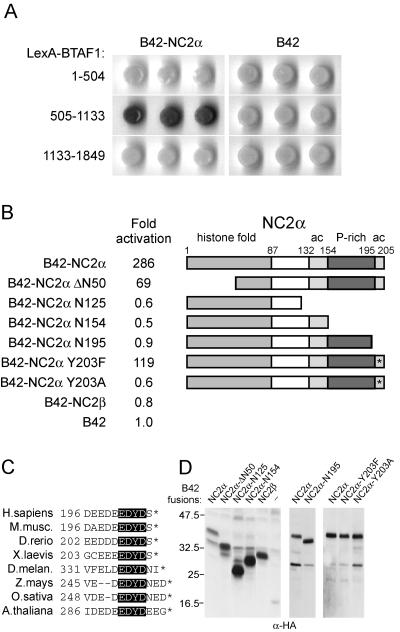
In order to identify interactors of BTAF1 we performed yeast two-hybrid screens with different parts of the BTAF1 protein of a human cDNA library. Using a LexA fusion of the middle part of human BTAF1 (residues 505 to 1133) as the bait (36), we identified a cDNA clone encoding NC2α. The isolated cDNA coded for a truncated form of NC2α and lacked the majority of the histone fold (NC2α ΔN50). Further analysis showed that full-length NC2α can also interact with the BTAF1 construct and that the interaction is fourfold more efficient than that of NC2α ΔN50 (Fig. 1B). We found that NC2α interacted only with the central BTAF1 fragment of residues 505-1133 and not with the 1-504 or 1133-1849 fragment (Fig. 1A). The interaction was specific to the NC2α subunit of the NC2 complex, since NC2β was unable to interact with BTAF1 (Fig. 1B). We tested further amino-terminal truncations of NC2α. Proteins lacking the first 74, 83, 100, and 120 amino acids interacted, but very weakly, with the BTAF1 505-1133 fragment (data not shown). Several other NC2α constructs lacking various domains were also tested in this way. Deletion of the extreme carboxyl-terminal acidic domain (residues 196 to 205) of NC2α resulted in the complete loss of interaction with BTAF1 (Fig. 1B). This acidic domain of NC2α is highly conserved and in the human protein contains eight acidic residues and an invariant tyrosine (Fig. 1C). Therefore, we tested whether tyrosine 203 within this motif is involved in BTAF1 binding by creating NC2α constructs with mutations to phenylalanine or alanine at this position. Strikingly, NC2α Y203F retained its ability to interact, whereas NC2α Y203A was completely deficient in the interaction with the BTAF1 505-1133 fragment (Fig. 1B). This shows that the aromatic ring, but not the hydroxyl group of Y203, is essential for the interaction with BTAF1. We verified the expression of the NC2α constructs and found that the mutants were expressed to levels equal to or higher than those of the wild-type protein (Fig. 1D).
FIG. 1.
NC2α interacts with BTAF1 in the yeast two-hybrid system. (A) S. cerevisiae strain EGY48 was transformed with plasmids expressing LexA fusions of the human BTAF1 fragments indicated, a B42 fusion with full-length NC2α, and pSH18-34 LacZ reporter plasmid. To score for the interacting proteins, the indicated yeast strains were grown on plates containing 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) as a β-galactosidase substrate. (B) Yeast strain EGY48 was transformed with a plasmid expressing a LexA fusion of human BTAF1 (505-1133), pSH18-34 plasmid, and B42 fusions of full-length NC2α, NC2β, or the indicated NC2α mutants. β-Galactosidase activity in the cell lysates was expressed as the induction relative to the activity in lysates of yeast containing the empty B42 vector control, pSH18-34, and the LexA-BTAF1 (505-1133) plasmid. The standard deviation was less than 30% in each case. To the right are schematic representations of the NC2α fragments for each B42 construct. The different domains are indicated as follows: ac, acidic domain; P-rich, proline-rich domain. (C) The carboxyl-terminal acidic domain of NC2α is highly conserved in most metazoans. Alignment of NC2α from various organisms was performed using the Clustal W program (41). The invariant EDYD motif is highlighted. H. sapiens, Homo sapiens; M. musc., Mus musculus; D. rerio, Danio rerio; X. laevis, Xenopus laevis; D. melan., D. melanogaster; Z. mays, Zea mays; O. sativa, Oryza sativa; A. thaliana, Arabidopsis thaliana. (D) Expression of B42 fusion proteins in yeast lysates. Equal amounts of yeast cells were lysed, and levels of B42-NC2 subunit expression were visualized by immunoblotting with anti-HA antibodies. To the left of the panel the molecular weights of comigrated marker proteins are indicated.
In conclusion, we have identified the NC2α subunit of NC2 as an interaction partner for BTAF1 and mapped the BTAF1-interacting region. The conserved acidic region at the carboxyl terminus of NC2α is indispensable for BTAF1 interaction, but other domains, including the histone fold, also contribute to BTAF1 binding.
BTAF1 and NC2α interact in vitro.
To confirm the interaction between BTAF1 and NC2α we tested their ability to interact in a GST pull-down assay. For this, we expressed both NC2 subunits as hexahistidine-tagged proteins in bacteria and purified them to near homogeneity (unpublished data). In addition, we obtained recombinant NC2 complex by coexpression of NC2α and hexahistidine-tagged NC2β (unpublished data). Gel filtration analysis of the NC2 complex indicated the absence of free NC2 subunits in these preparations (data not shown). Amino acids 505 to 891 of BTAF1 were expressed as a GST fusion protein, which was able to bind recombinant NC2α (Fig. 2A). GST-TBP was also able to interact with NC2α. Consistent with the yeast two-hybrid results, the GST-BTAF1 protein fragment did not interact with NC2β, whereas GST-TBP retained small amounts of NC2β (Fig. 2A). The TFIIS transcription elongation factor (GST-ΔTFIIS) was used as a negative control and did not retain detectable amounts of NC2α or NC2β. Testing of the NC2 complex in this assay revealed only weak binding of NC2α and not NC2β to GST-BTAF1 (505-891). In contrast, both subunits were efficiently binding to GST-TBP.
As the next step, GST fusions of NC2α, NC2β, or the NC2 complex were analyzed for their ability to bind full-length BTAF1 (Fig. 2B). To isolate NC2 complexes with the proper subunit stoichiometry, NC2α and NC2β were coexpressed as GST or hexahistidine fusion proteins in bacteria. These recombinant NC2 complexes were isolated by NiNTA affinity chromatography for the hexahistidine protein followed by glutathione affinity chromatography for the other subunit, which was expressed as a GST fusion protein. By this strategy we ensured that the recombinant subunits were only present in a complexed form (unpublished data). Subsequent gel filtration analysis confirmed the absence of free NC2 subunits or aggregates in these preparations (data not shown). As expected from the yeast two-hybrid data, recombinant full-length BTAF1 overexpressed in the RK13 cell lysate was able to bind to GST-NC2α but not to GST-NC2β or the control proteins GST-ΔTFIIS and GST (Fig. 2B, top panel, lanes 2 and 3, and data not shown). Importantly, the interaction of BTAF1 and NC2α did not depend on DNA, since the addition of ethidium bromide did not affect binding of BTAF1 (Fig. 2B, top panel, compare lanes 2 and 6). The lack of interaction between NC2β and BTAF1 was specific for this pair of proteins. The TBP control experiment revealed that both GST-NC2α and GST-NC2β, but not GST-ΔTFIIS or GST, retained TBP efficiently (Fig. 2B, bottom panel, lanes 2, 3, 6, and 7, and data not shown).
As the next step we investigated the ability of BTAF1 to bind to the NC2 complex. Strikingly, BTAF1 interaction with NC2 depended on the specific tags fused to the NC2 subunits. BTAF1 was specifically retained on the glutathione beads coated with NC2 via NC2α(GSTα/hisβ) but not via NC2β(hisα/GSTβ) (Fig. 2B, top panel, lanes 4, 5, 8, and 9). This is surprising since both preparations of the NC2 complex were purified to remove noncomplexed subunits. Also, both preparations of NC2 were equally active in binding TBP (Fig. 2B, bottom panel, lanes 4, 5, 8, and 9). To corroborate these results and ensure that the effect seen was not due to a specific preparation of the NC2 complex or the affinity tag used for purification, we repeated the experiment using NiNTA-agarose beads. As expected, recombinant hisNC2α, but not hisNC2β, was able to interact with BTAF1 (Fig. 2C, top panel, lanes 2 and 3). Similar to the results illustrated in Fig. 2B, the NC2 complex preparations immobilized via the NC2α subunit were more efficient in binding BTAF1 (Fig. 2C, top panel, lanes 4 and 5). As shown previously, TBP was able to interact with all NC2 proteins tested (Fig. 2C, bottom panel). It is possible that activities present in the cell lysates modify and/or disrupt NC2 complexes (see Fig. 5A; data not shown). However, it is important to note that we obtained identical results with partially purified BTAF1 preparations (data not shown). In summary, the in vitro binding experiments show that BTAF1 is able to directly interact with the NC2α but not the NC2β subunit of the NC2 complex.
NC2α but not NC2β or NC2 complex can bind to B-TFIID-DNA complexes.
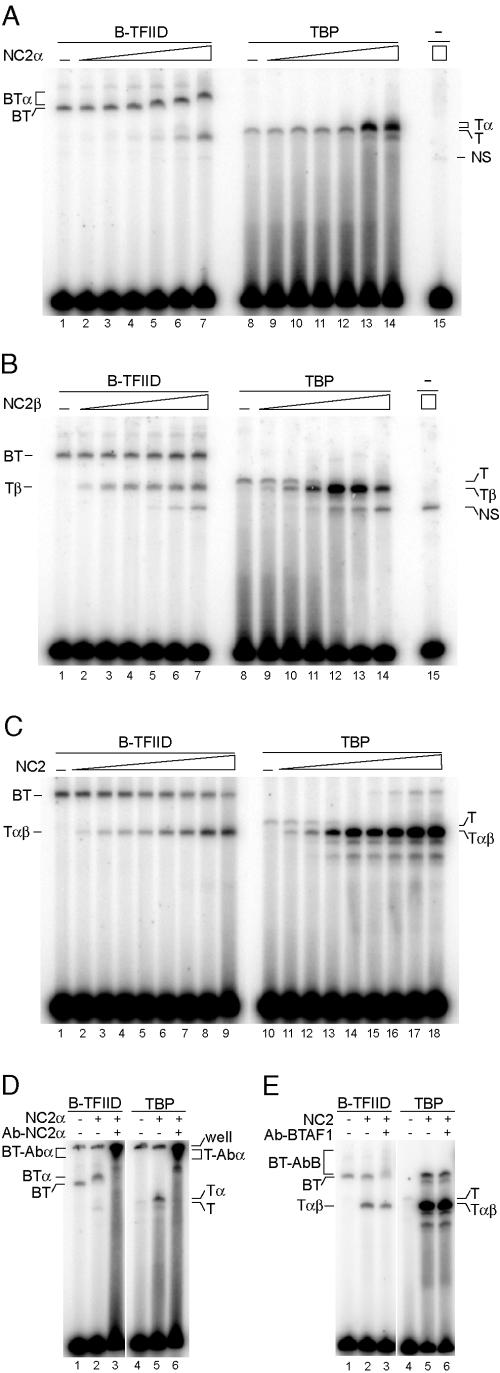
Since both BTAF1 and NC2 can form ternary complexes with TBP and TATA box DNA (5, 16, 29, 34), we determined the effect of the NC2 subunits on the interaction between B-TFIID or TBP and TATA box-containing promoter DNA. We utilized the EMSA for this purpose. Both B-TFIID and TBP are able to bind stably to an adenovirus major late promoter TATA box under our assay conditions (34; unpublished data). Addition of recombinant hisNC2α protein resulted in formation of a slower-migrating complex when B-TFIID or TBP was bound to the TATA box (Fig. 3A). The presence of NC2α in these complexes was confirmed by addition of recombinant hisNC2β (unpublished data) and by supershift experiments. Addition of anti-NC2α antibody, but not a control antibody, resulted in a supershift only when NC2α was present in the reactions (Fig. 3D and data not shown). Addition of recombinant hisNC2β to the B-TFIID-DNA complex did not result in formation of a slower-migrating complex. Instead, a specific, faster-migrating complex was formed at high concentrations of NC2β (Fig. 3B, lanes 1 to 7). In contrast, NC2β formed an abundant NC2β-TBP-DNA complex at 100-fold-lower concentrations, and this complex migrated faster than the TBP-DNA complex (Fig. 3B, lanes 8 to 14). The reason for this increased mobility is unclear. Strikingly, the complex formed upon the addition of NC2β to B-TFIID migrated at the same position as the NC2β-TBP-DNA complex (Fig. 3B, compare lanes 1 to 7 with lanes 8 to 14). The B-TFIID preparation used in these experiments was highly purified and lacked any noncomplexed TBP (34). These experiments indicate that NC2β is not able to bind B-TFIID and suggests that at higher concentrations NC2β may capture TBP transiently released from B-TFIID and form a ternary NC2β-TBP-DNA complex (see below).
FIG.3.
Analysis of the interaction between NC2 proteins and DNA-bound B-TFIID or TBP. (A) NC2α binds to both B-TFIID and TBP on DNA. EMSA was performed as described in Materials and Methods with 3 to 5 ng of B-TFIID (lanes 1 to 7) or 5 ng of TBP (lanes 8 to 14). Lanes 2 to 7 and 9 to 14 received increasing amounts of hisNC2α (0.135, 0.45, 1.35, 4.5, 13.5, and 45 pmol). Lane 15 received 45 pmol of hisNC2α. (B) NC2β can dissociate B-TFIID-DNA and stimulate formation of a TBP-NC2β-DNA complex. EMSA was performed as described for panel A. Lanes 2 to 7 and 9 to 14 received increasing amounts of hisNC2β (0.153, 0.51, 1.53, 5.1, 15.3, and 51 pmol). Lane 15 received 51 pmol of hisNC2β. (C) NC2 can dissociate B-TFIID-DNA complex. EMSA was performed with 3 to 5 ng of B-TFIID (lanes 1 to 9) or with 5 ng of TBP (lanes 10 to 18). Lanes 2 to 9 and 11 to 18 received increasing amounts of NC2(hisα/β) (0.72, 2.4, 7.2, 24, and 72 fmol, and 0.24, 0.72, and 2.4 pmol). (D) Analysis of the B-TFIID-NC2α-DNA complex by specific antibodies. EMSA was performed as described for panel A with B-TFIID (lanes 1 to 3) or TBP (lanes 4 to 6). Lanes 2, 3, 5, and 6 received 4.5 pmol of hisNC2α. Lanes 3 and 6 received 500 ng of anti-NC2α antibody. (E) BTAF1 is absent from the TBP-NC2-DNA complex. EMSA was performed as described for panel D. Lanes 2, 3, 5, and 6 received 0.24 pmol of NC2(hisα/β). Lanes 3 and 6 received 1 μg of anti-BTAF1 antibody PF299. Abbreviations: T, TBP; BT, B-TFIID; α, NC2α; β, NC2β; αβ, NC2 complex; Abα, anti-NC2α antibody; AbB, anti-BTAF1 antibody; NS, nonspecific interaction.
Next, we tested the NC2 complex for its ability to interact with DNA-bound B-TFIID. As in the case of NC2β, addition of NC2 to B-TFIID resulted in the formation of a complex migrating at the same position as the NC2-TBP-DNA ternary complex (Fig. 3C, compare lanes 1 to 9 with lanes 10 to 18). We analyzed the resulting protein-DNA complexes using specific antibodies. Higher-migrating B-TFIID was supershifted with anti-BTAF1 antibodies, whereas the faster-migrating complex formed upon addition of NC2 to B-TFIID was not affected. This excludes the presence of BTAF1 in the faster-migrating complex (Fig. 3E, compare lanes 2 and 3). The use of NC2 complex preparations, in which one of the subunits was fused to GST, resulted in slower-migrating complexes, confirming the presence of both NC2 subunits in these complexes (unpublished data). Moreover, endogenous HeLa-derived NC2 was able to dissociate B-TFIID-DNA complex with similar efficiency, excluding specific effects of affinity tags used for NC2 purification (unpublished data). It is important to note that the concentrations of NC2β or NC2 required to form a complex with TBP and DNA were 100-fold higher when B-TFIID was used than with TBP (Fig. 3B, compare lanes 7 and 10, and C, compare lanes 6 and 11). This difference suggests that TBP in the B-TFIID complex is shielded from the binding by NC2β or NC2. In accordance with this we notice that the addition of NC2β or NC2 to B-TFIID does not coincide with the significant reduction of the B-TFIID-DNA complex formation. This observation is in line with the fact that most of the B-TFIID is not complexed with DNA under EMSA conditions. In addition, free BTAF1 released from B-TFIID by NC2(β) may compete with NC2(β) for TBP.
In conclusion, the EMSA analysis indicates that NC2α, but not NC2β or the NC2 complex, can form a quaternary complex with B-TFIID and DNA. Higher concentrations of NC2β or NC2 seem to disrupt a B-TFIID-DNA complex, and formation of a NC2(β)-TBP-DNA complex is observed. This indicates that on DNA, NC2(β) and NC2 can compete with BTAF1 to bind TBP.
NC2α can stimulate BTAF1 binding to TBP in an ATP-dependent manner.
Our analysis indicates that NC2α is able to bind to TBP or BTAF1 in the absence of NC2β (Fig. 2B and C and 3A). Although a significant fraction of BTAF1 can efficiently form a heterodimer with TBP in cells (45; L. A. Pereira and H. T. M. Timmers, unpublished observations), the interaction between isolated forms of TBP and BTAF1 is relatively inefficient in vitro (5). Therefore, we tested whether NC2 or its NC2α subunit plays a role in the association of BTAF1 with TBP. We used GST-TBP to investigate its interaction with BTAF1 in the absence or the presence of the NC2 subunits. As expected, TBP-coated beads retained only low amounts of full-length BTAF1 (Fig. 4A, top panel, lane 4). In contrast, addition of NC2α or the NC2 complex to the binding reaction mixture resulted in a dramatic increase in BTAF1-TBP interaction. NC2β had no effect on the BTAF1-TBP interaction (Fig. 4A, top panel, lanes 4 to 8). Analysis of the amounts of NC2 subunits retained on the GST-TBP beads indicated that both NC2α and NC2 complexes, but not isolated NC2β, were bound efficiently (Fig. 4A, top panel, lanes 5 to 7). We noted, however, that the ratio of NC2α to NC2β retained in the presence of the BTAF1 lysate is increased (Fig. 4A, compare lane 7 and 11). Therefore, we cannot exclude that released NC2α rather than the NC2 complex is responsible for the observed stimulation of BTAF1 binding to GST-TBP.
BTAF1 possesses an ATPase activity that modulates its binding to TBP (5). Also, NC2α can be phosphorylated both in vivo and in vitro (17, 29). Since we were using a cell lysate as the source of BTAF1, we tested whether ATP was required for NC2α stimulation of BTAF1 binding to TBP. To this end hexokinase and glucose were added to deplete ATP from the cell lysates (21). Addition of hexokinase to the GST pull-down reaction mixtures containing NC2α and BTAF1 resulted in a decrease in NC2α- and NC2-mediated stimulation of BTAF1 binding to TBP (Fig. 4A, lanes 5 and 7, compare top and bottom panels). The levels of NC2α and NC2 binding to the TBP were slightly reduced by the hexokinase treatment (Fig. 4A, compare top and bottom panels), but this cannot account for the hexokinase effect on BTAF1 binding. Hexokinase activity was crucial for this effect, because the addition of heat-inactivated hexokinase did not decrease BTAF1-TBP binding (Fig. 4B, lanes 4 to 9). Interestingly, addition of active hexokinase to reaction mixtures lacking NC2α also diminished the basal level of BTAF1 binding to TBP (Fig. 4A, compare lanes 4 and 5, and B, compare lanes 3 and 8). This could suggest that cell lysates contain levels of endogenous NC2α or the NC2 complex sufficient to promote BTAF1 binding to TBP but not detectable by our Western blot analysis (Fig. 4A, lane 4, and data not shown). Addition of an excess amount of the ATP analogue ATPγS to the GST pull-down reaction mixtures had an effect similar to that of the addition of hexokinase (Fig. 4C, compare lanes 3, 4, 5, and 6), which indicates that the stimulation depends on ATP hydrolysis. Collectively, these results indicate that addition of both NC2α and the NC2 complex can stimulate the BTAF1-TBP interactions and that ATP hydrolysis is involved in this stimulation.
ATP hydrolysis by BTAF1 and NC2α phosphorylation do not influence NC2α-enhanced BTAF1 binding to TBP.
An explanation for the ATP dependence could be a requirement for ATP hydrolysis by BTAF1. We used a BTAF1 mutant which is defective in ATP hydrolysis to investigate this possibility. Mutation K1297A in human BTAF1 is located in a conserved loop of the ATPase domain and corresponds to the K1303A mutation in yeast Mot1p. This mutation prevents Mot1p from binding and hydrolyzing ATP (2, 3). Wild-type or mutant BTAF1 was equally capable of binding TBP in the presence of NC2α (Fig. 5A, compare lanes 4 and 5). This suggests that an ATP-dependent activity exists in the crude lysates, which is required for NC2α stimulation of BTAF1-TBP interaction. To investigate this further, we used in the GST pull-down assay a partially purified preparation of hexahistidine-tagged BTAF1, which displays high ATPase activity (45). As evident from Fig. 5B, hisBTAF1 interaction with TBP in the presence of NC2α was significantly reduced compared with BTAF1 derived from a cell lysate (Fig. 5B, lane 5, compare top and middle panels). However, the enhancing activity of NC2α was not diminished completely. Nevertheless, addition of ATP to the hisBTAF1-containing reaction mixtures did not restore binding to TBP to levels similar to that of BTAF1 from a cell lysate (Fig. 5B, lane 5).
We investigated whether NC2α phosphorylation occurs under our assay conditions and could be required for efficient BTAF1-TBP interaction. Phosphorylation of GST-TBP bound NC2α, but not of TBP or BTAF1, could be detected (Fig. 5C, bottom panel, lane 5, and data not shown). We used a panel of kinase inhibitors to abolish NC2α phosphorylation. LY294002, an inhibitor of casein kinase II and phosphatidylinositol 3-kinase, was able to completely inhibit NC2α phosphorylation (Fig. 5C, bottom panel, lane 6). In contrast, wortmannin (inhibitor of phosphatidylinositol 3-kinase), H89 (protein kinase A inhibitor), and PP2 (Src family tyrosine kinase inhibitor) did not reduce NC2α phosphorylation (Fig. 5C, bottom panel, lanes 7, 8, and 9). Interestingly, lack of NC2α phosphorylation did not affect stimulation of the BTAF1-TBP interaction (Fig. 5C, top panel, compare lanes 5 and 6).
Together, the results shown in Fig. 5 indicate that the ATPase activity of BTAF1 does not play a role in NC2α-stimulated binding between BTAF1 and TBP. Also, inhibition of NC2α phosphorylation does not prevent NC2α-mediated stimulation of BTAF1 binding to TBP.
The carboxyl-terminal acidic region and the conserved tyrosine are required for NC2α stimulation of the BTAF1-TBP interaction.
To test whether the interaction between NC2α and BTAF1 is directly responsible for the increase in BTAF1-TBP interaction we used NC2α mutants identical or similar to that described for the yeast two-hybrid assay (Fig. 1B). The addition of NC2α mutant proteins defective for interaction with BTAF1 to the GST pull-down reaction mixtures containing GST-TBP and BTAF1 did not result in increased BTAF1 retention (Fig. 6). In contrast, the mutant NC2α Y203F protein, which retained its ability to bind BTAF1 in the yeast two-hybrid assay, also enhanced the binding of BTAF1 to TBP similarly to the wild-type NC2α (Fig. 6, compare lanes 4 and 8). This strong correlation between the abilities of NC2α to bind BTAF1 and to enhance its interaction with TBP indicates that NC2α binding to BTAF1 is crucial for this function. We note that the truncation of the carboxyl-terminal acidic domain alone (NC2α N195) or together with the proline-rich domain (NC2α N157) of NC2α did not result in the decrease of its binding to TBP (Fig. 6, lanes 4, 6, and 7). Deletion of both acidic domains and the proline-rich domain (NC2α N132) resulted in somewhat weaker interaction with TBP (Fig. 6, lane 5).
Taken together our experiments indicate that the NC2α-dependent stimulation of the BTAF1-TBP interaction is not dictated by its ability to interact with TBP. Instead, NC2α stimulation correlates with the ability to interact with BTAF1.
DISCUSSION
The TBP plays a crucial role in eukaryotic gene transcription and is subjected to control at many steps (25, 35, 40). Here, we report a novel interaction between two TBP regulators, BTAF1 and NC2α. This interaction was first detected in yeast two-hybrid assays. Subsequent DNA binding analyses showed that NC2α can form a quaternary complex with BTAF1, TBP, and DNA. NC2β does not interact with BTAF1 and seems to disrupt BTAF1-TBP interactions by competing for TBP binding. Surprisingly, we find that NC2α can stimulate the interaction of TBP with BTAF1 in an ATP-dependent manner. As indicated by NC2α mutant analysis, the stimulation correlates closely with the ability of the NC2α subunit to bind to the BTAF1 protein.
Mapping of the NC2α interaction with BTAF1 and TBP.
The yeast two-hybrid assay was used to map the BTAF1-NC2α interaction. We found that the central region of BTAF1 was responsible for the interaction (Fig. 1A). In contrast to this, BTAF1 interaction required several parts of human NC2α protein. A carboxyl-terminal aromatic residue (Y203) embedded in a stretch of acidic amino acids seems to play a crucial role in the BTAF1 interaction. Although integrity of the histone fold domain is not essential, it clearly contributes to the strength of the interaction (Fig. 1B). In contrast, NC2α interaction with NC2β requires an intact histone fold (17, 22, 29). This supports our conclusion that NC2β is not involved in the interaction with BTAF1. We also observe the interaction of isolated NC2α with TBP (Fig. 2 and 3A). Previous studies by EMSA analysis did not reveal this interaction, which could be due to the specific assay conditions and the amounts of NC2 subunits used (17, 29). We show that the second conserved acidic domain and the proline-rich domain outside of the histone fold of NC2α are dispensable for TBP binding. The deletion of both acidic domains and the proline-rich domain reduces the interaction only slightly (Fig. 6). The crystal structure of NC2-TBP-DNA indicates that the histone fold of NC2α is largely responsible for its interaction with TBP, but it does not exclude other contact points (22). In contrast to NC2α, in vitro interaction of free NC2β with TBP was described previously (19).
Comparison of yeast mRNA expression profiles as determined by DNA microarrays reveals an overlap between Mot1p- and Bur6p (yeast NC2α)-dependent genes (1, 4, 10, 14). This suggests that these proteins cooperate in transcription of a subset of yeast genes. Although the BTAF1-NC2α interaction would support this, it is unclear whether the yeast orthologs behave similarly. First, the central region of BTAF1 is its least conserved part (35). Secondly, the acidic motif present at the extreme carboxyl terminus of metazoan NC2α is absent from its yeast counterparts (Fig. 1C). In this respect it is important to note that Auble and colleagues have reported formation of a yeast NC2-Mot1p-TBP-DNA complex in EMSA (9). This differs from our findings that human NC2 does not bind to a BTAF1-TBP-DNA complex. Rather, we observe dissociation into an NC2-TBP-DNA complex (Fig. 3).
Functional consequences of BTAF1-NC2α interaction.
Several lines of evidence support the formation of NC2α-BTAF1-TBP-DNA complexes. Firstly, we detect this complex by EMSA and confirm its composition by use of specific antibodies (Fig. 3A and D). Secondly, pull-down assays confirm the formation of NC2α-BTAF1 and NC2α-BTAF1-TBP complexes (Fig. 2B and C and data not shown). Thirdly, the NC2α surfaces required for BTAF1 and TBP binding are different, and the NC2α-BTAF1 interaction is required for efficient BTAF1 binding to TBP (Fig. 1 and 6). Additionally, TBP interacts with the amino-terminal third (36), whereas NC2α interacts with the middle part of BTAF1 (Fig. 1A). However, we also note that mammalian cell extracts do not contain a significant pool of free NC2α (S. Gilfillan and M. Meisterernst, unpublished observations), which implies that NC2α-BTAF1-TBP-DNA complex formation in cells would require liberation of NC2α from the NC2 complex.
In contrast to NC2α, NC2β was unable to bind to BTAF1 or B-TFIID in our assays (Fig. 1, 2, and 3B). At high concentrations NC2β seems to disrupt B-TFIID-DNA complexes, which could result from capturing transiently released TBP. In the case of the NC2 complex we observed the same (Fig. 3C). It is interesting that the results of GST and hexahistidine pull-down assays depend on which subunit is tagged for immobilization (Fig. 2). BTAF1 binding is reduced when immobilization occurs via the NC2β subunit. Two independent events may account for this result. Firstly, it is possible that BTAF1 competes with NC2β for binding to NC2α. Indeed, we observe partial disruption of the NC2 complex in GST pull-down assays (Fig. 4A and 5A and data not shown). Alternatively, factors present in cell lysates might destabilize the NC2 complex, possibly as the result of phosphorylation. Both NC2 subunits are known to be phosphorylated both in vivo and in vitro, but the modification sites and kinases involved have not been characterized in full detail (8, 17, 19, 29). In this respect, we show that casein kinase II is, most likely, involved in the phosphorylation of NC2α in our experimental setup (Fig. 5C). Interestingly, casein kinase II was implicated in the destabilization of TBP-independent NC2 complex binding to DNA (17). However, our kinase inhibitor experiment (Fig. 5C) argues against involvement of casein kinase II in stimulation of BTAF1-TBP interaction.
Overall, our results suggest that TBP surfaces utilized by NC2β and BTAF1 overlap. Similarly, NC2β and BTAF1 are likely to use overlapping surfaces of NC2α. Since the histone fold of NC2α is sufficient for the interaction with NC2β (17, 29), we hypothesize that BTAF1 makes contacts with the histone fold of NC2α, and this could compete with binding of NC2β.
We have tested several hypotheses in regard to the function of the interaction of BTAF1 and NC2α. Similarly to Mot1p-TBP complex (18), B-TFIID exhibits relaxed specificity of binding to non-TATA sequences (data not shown). Interaction with NC2α does not influence this property of B-TFIID (data not shown). Also, the B-TFIID complex of BTAF1 and TBP is able to support transcription by pol II reconstituted with highly purified transcription factors in vitro (34, 44). Assays employing purified TBP showed that NC2α does not affect basal transcription from the adenovirus major late promoter in vitro (17, 29, 49). NC2α also does not affect transcription reaction mixtures reconstituted with B-TFIID (data not shown). In contrast, the NC2 complex is capable of inhibiting TBP- and B-TFIID-containing transcription reactions with similar efficiency (data not shown). Thus, in our present assays isolated NC2α has no effect on B-TFIID-driven transcription. However, the NC2α- and ATP-dependent activity stimulating association of BTAF1 and TBP may be involved in this, and this activity is likely to be missing from our transcription reactions utilizing highly purified factors.
NC2α and ATP regulate BTAF1 function.
The interaction between BTAF1 and TBP in vitro was shown to be relatively inefficient (5). On the other hand, B-TFIID is one of the main TBP-containing complexes in higher eukaryotic cells (44). We present data which can explain this. NC2α is able to drastically increase the efficiency of BTAF1 binding to TBP in vitro but only when ATP is present during this process (Fig. 4). We also show that the interaction between NC2α and BTAF1 is required for the efficient binding of BTAF1 to TBP (Fig. 6).
There are several scenarios that could account for the ATP involvement in NC2α-stimulated binding of BTAF1 to TBP. ATP hydrolysis by BTAF1 could be required for its interaction with the NC2α subunit and TBP. In this hypothesis NC2α could serve as a modulator of the ATPase activity of BTAF1. However, NC2α does not seem to influence the ATPase activity of B-TFIID and does not modulate the ability of B-TFIID to dissociate from DNA in the presence of ATP (data not shown). Moreover, the K1297A mutant of BTAF1, which is incapable of ATP binding and hydrolysis, is still able to bind TBP in the presence of NC2α (Fig. 5A). Also, addition of ATP to purified BTAF1 does not enhance its binding to NC2α-TBP (Fig. 5B). Collectively these data suggest that the ATP hydrolysis by BTAF1 itself is not responsible for the ATP requirement in the formation of the NC2α-BTAF1-TBP complex.
ATP-dependent stimulation most likely requires an activity present in the cell lysates. Several factors capable of hydrolyzing ATP are known to interact directly or indirectly with TBP. Those include the TAF1 and TFIIH factors in basal transcription machinery or coactivator complexes, such as Mediator (7, 13). The levels of exogenously provided TBP, NC2α, and BTAF1 in the GST pull-down assays greatly exceed their endogenous levels (data not shown). Therefore, we consider it unlikely that the hypothetical factor forms a stoichiometric complex with NC2α-BTAF1-TBP. The present data suggest that the factor(s) acts in a catalytic manner, which may point to kinase or chaperone activities. The kinase inhibitor experiment (Fig. 5C) failed to identify the relevant kinase activity.
Dynamics of TBP association with DNA and consequences for pol II transcription regulation.
Our results bear important implications for mechanisms by which NC2 components dynamically interact not only with BTAF1 but also with TBP. The finding of the BTAF1-NC2α interaction increases the multitude of interactions between these factors. It is important to note that HeLa cell extracts contain free pools of BTAF1 and NC2β but not of TBP or NC2α (5; L. A. Pereira, H. T. M. Timmers, S. Gilfillan, and M. Meisterernst, unpublished observations). We propose that transiently released NC2α could serve as a signal to mark promoter-bound TBP for the binding by BTAF1. This could facilitate BTAF1 interaction with TBP and promoter DNA. On the other hand, replacement of BTAF1 by NC2β in a BTAF1-NC2α-TBP-promoter complex would provide an effective means by which to shut down transcription. In this setup, association of NC2α with TBP and BTAF1 at the moment of delivery would give an opportunity to link two transcriptional events, activation and repression, by one factor. Although mechanistically different, similar dual roles were described for the proteasome involvement in both transcription activation and repression of active complexes (31). On the other hand, repression of the TBP-DNA complex by NC2 could be relieved by BTAF1. First, BTAF1 could contact NC2α and subsequently disrupt the NC2-TBP interaction. This is consistent with recent observations regarding yeast, which implicated NC2 binding to TBP-DNA in transcription activation (4).
It is important to stress that current experiments do not reflect the complexity of the transcription process in vivo. Therefore, it is difficult to speculate what other factors may influence the outcome of the NC2-BTAF1 interplay. Investigation of this regulatory network would require development of an in vitro transcription system, which depends both on NC2 and on BTAF1. Interestingly, the NC2 complex was implicated in transcription of downstream promoter element-containing promoters in Drosophila extracts. This raises the possibility that BTAF1 is also involved in this. Moreover, Drosophila NC2α contains regions responsible for BTAF1 binding (Fig. 1C). Additional experiments will be required to test this hypothesis.
In summary, our studies provide evidence for a direct interaction between NC2α and BTAF1. Our results give insight into the regulation of BTAF1 activity and underscore the importance of the general transcriptional regulators in the dynamics of TBP function. Our findings also provide a basis for further studies on the regulation of TBP in pol II transcription.
Acknowledgments
We thank D. Reinberg for pDr1 and X. Zhao for advice on EMSA. We acknowledge François Kavelaars and Richard van Schaik for excellent technical assistance, members of our laboratory for discussions, and Pim Pijnappel and Florence Mousson for critical reading of the manuscript.
This work was supported by The Netherlands Organization for Scientific Research NWO-MW Pionier grant 900-98-142, Human Sciences Frontier Program Organization (HSFPO) grant RG0196/1998, and European Union Improving Human Potential grant RTN2-2001-00026.
REFERENCES
- 1.Andrau, J.-C., C. J. C. van Oevelen, H. A. A. M. van Teeffelen, P. A. Weil, F. C. P. Holstege, and H. T. M. Timmers. 2002. Mot1 is essential for TBP recruitment to selected promoters during in vivo gene activation. EMBO J. 21:5173-5183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Auble, D. T., K. E. Hansen, C. G. F. Mueller, W. S. Lane, J. Thorner, and S. Hahn. 1994. Mot1, a global repressor of RNA polymerase II transcription, inhibits TBP binding to DNA by an ATP-dependent mechanism. Genes Dev. 8:1920-1934. [DOI] [PubMed] [Google Scholar]
- 3.Auble, D. T., D. Wang, K. W. Post, and S. Hahn. 1997. Molecular analysis of the SNF2/SWI2 protein family member MOT1, an ATP-driven enzyme that dissociates TATA-binding protein from DNA. Mol. Cell. Biol. 17:4842-4851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cang, Y., and G. Prelich. 2002. Direct stimulation of transcription by negative cofactor 2 (NC2) through TATA-binding protein (TBP). Proc. Natl. Acad. Sci. USA 99:12727-12732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chicca, J. J., II, D. T. Auble, and B. F. Pugh. 1998. Cloning and biochemical characterization of TAF-172, a human homolog of yeast Mot1. Mol. Cell. Biol. 18:1701-1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Collart, M. A. 1996. The NOT, SPT3, and MOT1 genes functionally interact to regulate transcription at core promoters. Mol. Cell. Biol. 16:6668-6676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cosma, P. A. 2002. Ordered recruitment: gene-specific mechanism of transcription activation. Mol. Cell 10:227-236. [DOI] [PubMed] [Google Scholar]
- 8.Creton, S., J. Q. Svejstrup, and M. A. Collart. 2002. The NC2 α and β subunits play different roles in vivo. Genes Dev. 16:3265-3276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Darst, R., A. Dasgupta, C. Zhu, J.-Y. Hsu, A. Vroom, T. Muldrow, and D. T. Auble. 2003. Mot1 regulates the DNA binding activity of free TBP in an ATP-dependent manner. J. Biol. Chem. 278:13216-13226. [DOI] [PubMed] [Google Scholar]
- 10.Dasgupta, A., R. P. Darst, K. J. Martin, C. A. Afshari, and D. T. Auble. 2002. Mot1 activates and represses transcription by direct, ATPase-dependent mechanisms. Proc. Natl. Acad. Sci. USA 99:2666-2671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davidson, I. 2003. The genetics of TATA-binding protein (TBP) and TBP-related factors. Trends Biochem. Sci. 28:391-398. [DOI] [PubMed] [Google Scholar]
- 12.Davis, J. L., R. Kunisawa, and J. Thorner. 1992. A presumptive helicase (MOT1 gene product) affects gene expression and is required for viability in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 12:1879-1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dikstein, R., S. Ruppert, and S. Tjian. 1996. TAFII250 is a bipartite protein kinase that phosphorylates the basal transcription factor RAP74. Cell 84:781-790. [DOI] [PubMed] [Google Scholar]
- 14.Geisberg, J. V., F. C. P. Holstege, R. A. Young, and K. Struhl. 2001. Yeast NC2 associates with the RNA polymerase II preinitiation complex and selectively affects transcription in vivo. Mol. Cell. Biol. 21:2736-2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Geisberg, J. V., Z. Moqtaderi, L. Kuras, and K. Struhl. 2002. Mot1 associates with transcriptionally active promoters and inhibits association of NC2 in Saccharomyces cerevisiae. Mol. Cell. Biol. 22:8122-8134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goppelt, A., and M. Meisterernst. 1996. Characterization of the basal inhibitor of class II transcription NC2 from Saccharomyces cerevisiae. Nucleic Acids Res. 24:4450-4455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goppelt, A., G. Stelzer, F. Lottspeich, and M. Meisterernst. 1996. A mechanism for repression of class II gene transcription through specific binding of NC2 to TBP-promoter complexes via heterodimeric histone fold domains. EMBO J. 15:3105-3116. [PMC free article] [PubMed] [Google Scholar]
- 18.Gumbs, O., A. M. Campbell, and P. A. Weil. 2003. High-affinity DNA binding by a Mot1p-TBP complex: implications for TAF-independent transcription. EMBO J. 22:3131-3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Inostroza, J. A., F. H. Mermelstein, I. Ha, W. S. Lane, and D. Reinberg. 1992. Dr1, a TATA-binding protein-associated phosphoprotein and inhibitor of class II gene transcription. Cell 70:477-489. [DOI] [PubMed] [Google Scholar]
- 20.Iratni, R., Y. T. Yan, C. Chen, J. Ding, Y. Zhang, S. M. Price, D. Reinberg, and M. M. Shen. 2002. Inhibition of excess nodal signaling during mouse gastrulation by the transcriptional corepressor DRAP1. Science 298:1996-1999. [DOI] [PubMed] [Google Scholar]
- 21.Jiang, Y., S. T. Smale, and J. D. Gralla. 1993. A common ATP requirement for open complex formation and transcription at promoters containing initiator or TATA elements. J. Biol. Chem. 268:6535-6540. [PubMed] [Google Scholar]
- 22.Kamada, K., F. Shu, H. Chen, S. Malik, G. Stelzer, R. G. Roeder, M. Meisterernst, and S. K. Burley. 2001. Crystal structure of negative cofactor 2 recognizing the TBP-DNA transcription complex. Cell 106:71-81. [DOI] [PubMed] [Google Scholar]
- 23.Kim, S., K. Cabane, M. Hampsey, and D. Reinberg. 2000. Genetic analysis of the Ydr1-Bur6 repressor complex reveals an intricate balance among transcriptional regulatory proteins in yeast. Mol. Cell. Biol. 20:2455-2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee, T. I., J. J. Wyrick, S. S. Koh, E. G. Jennings, E. L. Gadbois, and R. A. Young. 1998. Interplay of positive and negative regulators in transcription initiation by RNA polymerase II holoenzyme. Mol. Cell. Biol. 18:4455-4462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee, T. I., and R. A. Young. 1998. Regulation of gene expression by TBP-associated proteins. Genes Dev. 12:1398-1408. [DOI] [PubMed] [Google Scholar]
- 26.Lemaire, M., J. Xie, M. Meisterernst, and M. A. Collart. 2000. The NC2 repressor is dispensable in yeast mutated for the Sin4p component of the holoenzyme and plays roles similar to Mot1p in vivo. Mol. Microbiol. 36:163-173. [DOI] [PubMed] [Google Scholar]
- 27.Madison, J. M., and F. Winston. 1997. Evidence that Spt3 functionally interacts with Mot1, TFIIA, and TATA-binding protein to confer promoter-specific transcriptional control in Saccharomyces cerevisiae. Mol. Cell. Biol. 17:287-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meisterernst, M., and R. G. Roeder. 1991. Family of proteins that interact with TFIID and regulate promoter activity. Cell 67:557-567. [DOI] [PubMed] [Google Scholar]
- 29.Mermelstein, F., K. Yeung, J. Cao, J. A. Inostrosa, H. Erdjument-Bromage, K. Eagelson, D. Landsman, P. Levitt, P. Tempts, and D. Reinberg. 1996. Requirement of a corepressor for Dr1-mediated repression of transcription. Genes Dev. 10:1033-1048. [DOI] [PubMed] [Google Scholar]
- 30.Muldrow, T. A., A. M. Campbell, P. A. Weil, and D. T. Auble. 1999. MOT1 can activate basal transcription in vitro by regulating the distribution of TATA binding protein between promoter and nonpromoter sites. Mol. Cell. Biol. 19:2835-2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Muratani, M., and W. P. Tansey. 2003. How the ubiquitin-proteasome system controls transcription. Nat. Rev. Mol. Cell Biol. 4:192-201. [DOI] [PubMed] [Google Scholar]
- 32.Narlikar, G. J., H. Y. Fan, and R. E. Kingston. 2002. Cooperation between complexes that regulate chromatin structure and transcription. Cell 108:475-487. [DOI] [PubMed] [Google Scholar]
- 33.Orphanides, G., and D. Reinberg. 2002. A unified theory of gene expression. Cell 108:439-451. [DOI] [PubMed] [Google Scholar]
- 34.Pereira, L. A., M. P. Klejman, C. Ruhlmann, F. Kavelaars, M. Oulad-Abdelghani, H. T. M. Timmers, and P. Schultz. 2004. Molecular architecture of the basal transcription factor B-TFIID. J. Biol. Chem. 279:21802-21807. [DOI] [PubMed] [Google Scholar]
- 35.Pereira, L. A., M. P. Klejman, and H. T. M. Timmers. 2003. Roles for BTAF1 and Mot1p in dynamics of TATA-binding protein and regulation of RNA polymerase II transcription. Gene 315:1-13. [DOI] [PubMed] [Google Scholar]
- 36.Pereira, L. A., J. A. van der Knaap, V. van den Boom, F. A. van den Heuvel, and H. T. M. Timmers. 2001. TAFII170 interacts with the concave surface of TATA-binding protein to inhibit its DNA binding activity. Mol. Cell. Biol. 21:7523-7534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Poon, D., A. M. Campbell, Y. Bai, and P. A. Weil. 1994. Yeast TAF170 is encoded by MOT1 and exists in a TATA box-binding protein (TBP)-TBP-associated factor complex distinct from transcription factor IID. J. Biol. Chem. 269:23135-23140. [PubMed] [Google Scholar]
- 38.Poon, D., and P. A. Weil. 1993. Immunopurification of yeast TATA-binding protein and associated factors. J. Biol. Chem. 268:15325-15328. [PubMed] [Google Scholar]
- 39.Prelich, G. 1997. Saccharomyces cerevisiae BUR6 encodes a DRAP1/NC2α homolog that has both positive and negative roles in transcription in vivo. Mol. Cell. Biol. 17:2057-2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pugh, B. F. 2000. Control of gene expression through regulation of the TATA-binding protein. Gene 255:1-14. [DOI] [PubMed] [Google Scholar]
- 41.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Timmers, H. T. M. 1994. Transcription initiation by RNA polymerase II does not require hydrolysis of the β-γ phosphoanhydride bond of ATP. EMBO J. 13:391-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Timmers, H. T. M., R. E. Meyers, and P. A. Sharp. 1992. Composition of transcription factor B-TFIID. Proc. Natl. Acad. Sci. USA 89:8140-8144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Timmers, H. T. M., and P. A. Sharp. 1991. The mammalian TFIID protein is present in two functionally distinct complexes. Genes Dev. 5:1946-1956. [DOI] [PubMed] [Google Scholar]
- 45.van der Knaap, J. A., J. W. Borst, R. Gentz, P. C. van der Vliet, and H. T. M. Timmers. 1997. Cloning of the cDNA for the TAFII170 subunit of transcription factor B-TFIID reveals homology to global transcription regulators in yeast and Drosophila. Proc. Natl. Acad. Sci. USA 94:11827-11832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Willy, P. J., R. Kobayashi, and J. T. Kadonaga. 2000. A basal transcription factor that activates or represses transcription. Science 290:982-985. [DOI] [PubMed] [Google Scholar]
- 47.Wingfield, P. T. 2001. Identification of protein interactions. In P. T. Wingfield (ed.), Current protocols in protein science, vol. 3. John Wiley & Sons, Inc., New York, N.Y.
- 48.Winkler, G. S., T. K. Albert, C. Dominguez, Y. I. Legtenberg, R. Boelens, and H. T. M. Timmers. 2004. An altered-specificity ubiquitin-conjugating enzyme/ubiquitin-protein ligase pair. J. Mol. Biol. 337:157-165. [DOI] [PubMed] [Google Scholar]
- 49.Yeung, K., S. Kim, and D. Reinberg. 1997. Functional dissection of a human Dr1-DRAP1 repressor complex. Mol. Cell. Biol. 17:36-45. [DOI] [PMC free article] [PubMed] [Google Scholar]